Abstract
Long non-coding RNAs (lncRNAs) are novel regulators for post-transcriptional gene expression, and altered lncRNAs function and expression are associated with tumorigenesis and cancer progression, although the biological functions of most lncRNAs in various cancer types and their underlying regulatory interactions have remained largely elusive. Our previous study identified microRNA (miR)-181a as a regulator of Kruppel-like factor 6 (KLF6). In the present study, a bioinformatical analysis was performed to identify the novel lncRNA CR749391 as a potential regulator of miR-181a that contains four putative binding sites. Subsequent in vitro experiments in gastric cancer (GC) cells demonstrated that CR749391 interacted with miR-181a to regulate KLF6 expression. First, a direct binding interaction was confirmed using luciferase reporter and RNA immunoprecipitation and pull-down assays. In addition, CR749391 was observed to be downregulated in GC compared with that of normal gastric cell lines. A functional study also revealed that CR749391 depletion in normal gastric epithelial cells promoted cell viability, migration and invasion, and conferred resistance to apoptosis, whereas ectopic CR749391 overexpression had the opposite effect in GC cells and inhibited in vivo tumor growth. In addition, CR749391 was observed to be downregulated in GC compared with that of normal gastric tissues, which was associated with KLF6 but inversely associated with miR-181a levels. Overall, the CR749391/miR-181a regulatory interaction and association between CR749391 and KLF6 may enhance the current understanding of GC pathogenesis, although CR749391 association with GC prognosis needs further study. The current study could provide a novel approach for lncRNA-mediated targeted GC therapy.
Keywords: long non-coding RNA, Kruppel-like factor 6, microRNA sponge, gastric cancer
Introduction
Gastric cancer (GC) ranks as the fifth most commonly diagnosed malignancy in the world and the third leading cause of cancer-associated death. GC is the most common type of malignant gastrointestinal tumor in East Asia, Eastern Europe, and certain parts of South America (1–4); patients are frequently diagnosed at an advanced stage following by excessive proliferation and metastasis. The prognosis for GC patients remains unsatisfactory, in spite of marked progress in chemoradiotherapy and surgical techniques (5,6). Numerous oncogenes and tumor suppressor genes have vital roles in GC carcinogenesis; however, to date, there are no useful biomarkers identified and improved the comprehensive management of GC patients (6). Therefore, the discovery of novel regulatory molecules in the genesis, progression and underlying molecular mechanisms of GC are of interest for both basic science and clinical treatment.
The dysregulation of various non-coding RNAs has been detected during GC development (7). Long non-coding RNAs (lncRNAs) are now regarded as important regulators of carcinogenesis and have attracted vast attention due to their roles in the pathogenesis of various cancer types. LncRNAs are transcripts with lengths ranging from 200,000 to 100,000 nucleotides and lack any detectable open reading frame. Thus far, lncRNAs have been reported to modulate gene expression by regulating gene transcription, post-transcriptional mRNA processing, or epigenetic modifications (8,9).
A hypothesis of competing endogenous RNAs (ceRNAs) has been proposed to describe the regulatory mechanism of lncRNAs (10,11). The hypothesis assumes that lncRNAs act as miRNA sponges, which compete with mRNAs to bind to miRNAs and reduce the inhibitory effect of miRNA on the expression of mRNA, thereby enhancing the expression of mRNA. Indeed, this assumption has been supported by numerous studies (12–14). For instance, lncRNA HOTAIR modulates the proliferation, migration, and invasion of GC cells and may be regarded as a ceRNA by sponging miR-331-3p (1). Furthermore, several studies have indicated that lncRNA HOTAIR also acts as a sponge for miR-152 and miR-126 (15–17). Similarly, it has been reported that lncRNA maternally expressed 3 (MEG3) causes an upregulation of B-cell lymphoma (Bcl)-2 via its ceRNA activity on miR-181a, which regulates the expression of genes controlling cell proliferation, migration, invasion, and apoptosis in GC (18). Despite the abundant results, the function of lncRNAs in tumorigenesis and tumor progression remains to be explored.
Krüppel-like factor 6 (KLF6), belonging to the family of Krüppel-like transcription factors, has been reported to act as a tumor suppressor in various types of human cancer, including GC (19–21). A previous study from our group showed that KLF6 is a target of miR-181a and plays a vital role in the regulation of GC progression (22). In the present study, a bioinformatics analysis was performed and identified lncRNA CR749391, which is localized at the downstream of the KLF6 gene in the pathway, while KLF6 contains four putative miR-181a binding sites. Therefore, it was hypothesized that lncRNA CR749391 could act as a sponge for miR-181a to modulate KLF6 expression in GC. To test this hypothesis, various in vitro experiments and afunctional analysis were performed to reveal that lncRNA CR749391 was able to regulate cell proliferation, migration, invasion, and apoptosis and also inhibited tumor growth in a nude mouse model. Most importantly, an inverse association between CR749391 and miR-181a and a positive association with KLF6 were observed in GC tissues.
Materials and methods
Cell culture and tissue preparation
GC BGC-823, SGC-7901, and MGC-803 cell lines, normal gastric epithelial GES-1 cells, and 293 cells were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The cells were cultured with Dulbecco's modified Eagle's medium with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) at37°C in a humidified atmosphere with 5% CO2. Furthermore, 11 fresh GC samples and the adjacent non-cancerous tissues (sampled 10 cm away from the cancer and harboring no tumor cells as confirmed by histology) were collected by either surgical resection or endoscopy biopsy. The present study was approved by the Medical Ethics Committee of Guangzhou Medical University (Guangzhou, China) and all tissues were obtained with informed consent from each patient before being enrolled into this study.
Vector construction and luciferase activity assay
To estimate the potential targets of CR749391, a bioinformatics analysis was performed using an online tool, Targetscan version 7.2 (http://www.targetscan.org) and found that miR-181a could be one of the targets. The full length of CR749391 were then amplified from human complementary (c)DNA using polymerase chain reaction (PCR) and subcloned into the pcDNA3.1 mammalian expression vector (Invitrogen; Thermo Fisher Scientific, Inc.). The primers, which were designed by Primer Premier v5.0 (PREMIER Biosoft International), were as follows: The forward primer was linked with BamHI restriction enzyme site (underlined), 5-CGCGGATCCGAATGTTATTTAATGGTGCCAAT-3′, while the reverse with XbaI restriction enzyme site (underlined), 5′-GCTCTAGATGAGTGTTCATTGCAGTTTT-3′. To construct luciferase reporter vectors, the 3′-untranslated region (UTR) of KLF6 (+156-+3,543 bp), a fragment of the KLF6 3′-UTR, was amplified using PCR and subcloned into the psiCHECK2 luciferase vector (Ambion; Thermo Fisher Scientific, Inc.). These KLF6 PCR primers were as follows: Forward with XhoI restriction enzyme site (underlined), 5′-CCGCTCGAGGGGAGCAGAGAGGTGGATCCT-3′ and reverse with NotI restriction enzyme site (underlined), 5′-TAGAGCGGCCGCTTGGCAGTGATGTCATCTTTTATTT-3′. The siRNAs that target CR749391 (si-CR749391), miR-181a, siR-NC, and miR-NC were designed and synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China). All positive clones were obtained by transformation in E. coli DH5α cells grown on LB plates with 50 µg/ml ampicillin.
These amplified pcDNA3.1 plasmids (Invitrogen; Thermo Fisher Scientific, Inc.) were verified by a restriction enzyme digestion and DNA sequencing before use. For the luciferase reporter assay, luciferase activity was determined using a dual luciferase assay kit (cat. no. E1910; Promega, Madison, WI, USA) according to the kit's instructions. In brief, HEK-293 cells were cultured at 37°C overnight under 5% CO2 and then transiently transfected with siCR749391 and miR-181 mimics or negative control (NC) plasmids for 48 h using Lipofectamine 2000 (Invitrogen). The total cellular protein was then extracted and assayed using the Dual-Luciferase reporter assay system (Promega Corporation). After that, all firefly luciferase activities were normalized to that of Renilla luciferase activity accordingly.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was isolated from cells or tissues using RNAiso Plus (Takara, Dalian, China) and quantified using a NanoDrop 2000c Spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific, Inc.). The first-strand cDNA was synthesized using the Revert Aid First Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.). After mixing the RNA samples with the kit reagents, the samples were incubated at 25°C for 5 min, 42°C for 60 min, and 70°C for 5 min. qPCR amplification was performed in an Applied Biosystems® Step-One plus real-time PCR System (Thermo Fisher Scientific, Inc.) using SYBR green-I Master PCR mix (Thermo Fisher Scientific, Inc.) with gene-specific primers; GAPDH was used for normalization. PCR primers used were as follows: Homo sapiens-miR-181a forward, 5′-ACACTCCAGCTGGGAACATTCAACGCTGTCGG-3′ and reverse, 5′-CTCAACTGGTGTCGTGGA-3′; GAPDH forward, 5′-TCATGAAGTGTGACGTGGACATC-3′ and reverse, 5′-CAGGAGGAGCAATGATCTTGATCT-3′; lncRNA CR749391 forward, 5′-TGAGTGCAGTGCCTGCAGAG-3′ and reverse, 5′-TCAGGTTGGCTTGGGCACATC-3′; and KLF6 forward, 5′-CAAGGGAAATGGCGATGCCT-3′ and reverse, 5′-CTTTTCTCCTGTGTGCGTCC-3′.qPCR was performed at 95°C for 10 min, followed by 40 cycles of 95°C for 10 sec and 60°C for 1 min, and the relative miRNA and mRNA expression in GC cells and tissues were calculated using the 2−∆∆Cq method according previous studies (23).
RNA binding protein immunoprecipitation assay
RNA immunoprecipitation (RIP) was performed using the EZMagna RIP kit (cat. no. 17-701; EMD Millipore, Billerica, MA, USA) following the manufacturer's protocol. In particular, BGC-823 cells at 80–90% confluency were trypsinized and then lysed in a complete RIP lysis buffer (100 mM KCl, 5 mM MgCl2, 10 mM HEPES pH 7.0, and 0.5% NP40) supplemented with DTT and RNase Inhibitor (both Promega Corporation) on ice for 5 min. Lysates (100 µl) were then incubated at 4°C overnight with 100 µl RIP buffer containing magnetic beads conjugated with human anti-argonaute 2, RISC catalytic component (Ago2) antibodies (cat. no. ab32381; Abcam, Cambridge, UK), or normal mouse immunoglobulin G (IgG; cat. no. A0216; Beyotime Institute of Biotechnology, Haimen, China) as a negative control, at 4°Covernight. Next, the samples were incubated with a Proteinase K solution (cat. no. ST533; Beyotime Institute of Biotechnology) on a shaker to remove proteins at 58°C for 30 min, and the immunoprecipitated RNA was then isolated using RNAi so Plus. The RNA concentration was measured using a NanoDrop and the RNA quality was assessed with a bioanalyser (Agilent Technologies, Santa Clara, CA, USA). The resulting RNA samples were subjected to RT-qPCR analysis to demonstrate the presence of the binding targets using respective primers as above.
RNA-RNA pull-down assay
Bio-16-UTP from the T7 High Yield Transcription Kit (cat. no. K0441; Thermo Fisher Scientific, Inc.) was added to the 3′-end of miR-181a using T7 reverse transcriptase according to the manufacturer's protocol. The labeledmiR-181a was then incubated with streptavidin magnetic beads (New England Biolabs, Ipswich, MA, USA) for 2 h at 4°C. Then, RNA-tagged streptavidin magnetic beads were collected after centrifugation at 18,000 × g for 10 min at room temperature and further incubated with total RNA at 4°C for 2 h. These RNA-RNA complexes then eluted using TE buffer and purified by ethanol for RT-qPCR analysis of CR749391 and miR-181a levels.
Western blot
Total cell lysates were prepared using radioimmunoprecioitation assay buffer (150 mM NaCl, 1 mM EDTA, 50 mM Tris-HCl pH 7.4, 1% Triton X-100, 0.5% deoxycholic acid, and 0.1% SDS), and the protein concentration was measured with the bicinchoninic acid method. Equal amounts of protein samples (50 µg) were separated by SDS-PAGE on 10% gels and transferred onto polyvinylidene difluoride membranes (EMD Millipore). The membranes were then blocked with 5% non-fat milk in PBS at room temperature for 2 h and then incubated with anti-KLF6 (cat. no. sc-7158; 1:500 dilution; Santa Cruz Biotechnology, Santa Cruz, CA, USA) or anti-GAPDH (cat. no. 5174; 1:5,000 dilution; Cell Signaling Technology) antibodies at 4°C overnight, followed by incubation with secondary antibodies (cat. no. RB230194; 1:2,000 dilution; Thermo Fisher Scientific, Inc.) for 1 h at the room temperature. The membranes were then incubated with reagents from the Pierce®SuperSignal chemiluminescent detection module (Thermo Fisher Scientific, Inc.) and exposed to X-ray film to obtain images.
Cell growth assay
BGC-823 or GES-1 cells were seeded into 6-well plates at 1 day prior to transfection. At 24 h after transfection, the cells were trypsinized and reseeded into 96-well plates at a density of 1×104 cells/well in DMEM with 10% FBS. At specific time-points (24, 48 and 72 h), the cells in designated wells were examined using a growth assay with the MTS kit (cell Titer 96AQ; Promega Corporation) following the manufacturer's protocol, and the absorption was measured at 490 nm.
Flow cytometric apoptosis assay
To detect apoptotic cells, Fluorescein (FITC) Annexin V Apoptosis Detection Kit I (cat. no. 556547; BD Biosciences, San Jose, CA, USA) was used according to the manufacturer's protocol. Briefly, at 24 h after transfection, the BGC-823 or GES-1cells were collected and washed with PBS twice. The cells (1-5×105 per sample) were resuspended in 100 µl of the binding buffer. The cells were stained with 5 µl FITC Annexin V and 5 µl propidium iodide for 15 min at 25°C in the dark. Subsequently, 400 µl 1X Binding Buffer were added to each tube and analyzed by flow cytometry within 1 h.
Cell migration and invasion assays
The cell migration assay was performed using 96-well Transwell plates (pore size, 8 µm; EMD Millipore). After transfection, the cells were resuspended in serum-free medium and 1×105 cells in 100 µl medium were added to the upper chamber, while the lower chamber was filled with media containing 10% FBS as a chemoattractant. The cells were incubated for 12 h at 37°C, and, subsequently, the cells on the upper surface of the membrane were removed using cotton swabs. Finally, the cells that had transgressed through the membrane to the lower surface were fixed in ice-cold methanol for 10 min and stained with 0.5% crystal violet solution for 10 min. The number of migrated cells on the lower surface of the membrane was counted under a microscope in five fields (magnification, ×100). For the invasion assay, the plates were initially coated with Matrigel® (BD Biosciences) diluted in serum-free medium and the subsequent procedures were identical to those for the migration assay.
Nude mouse GC cell xenograft assay
Female athymic BALB/c mice (body weight, 18–22 g; age, 4–6 weeks old) were purchased from the Model Animal Research Center of Guangzhou Medical University (Guangzhou, China). The mice were raised under specific pathogen-free conditions with 12-h day and night cycles at 22±2°C with a relative humidity of 40–60% and free access to water and food ad libitum. All animal procedures were performed according to the protocols approved by the Institutional Animal Care and Use Committee at the Guangzhou Medical University (Guangzhou, China). To establish the xenograft models, 5×106 BGC-823 cells, transfected with a CR749391 or a pcDNA3.1 vector were collected and suspended in 0.1 ml PBS, and then injected subcutaneously into the right flank of BALB/c nude mice (5 mice per group). The subcutaneous tumors were allowed to grow, and the tumor volumes were measured every three days with a caliper. After 28 days, the mice were sacrificed. The tumor volumes were calculated by using the following equation: V (mm3)=AxB2/2, where A is the largest diameter and B is the perpendicular diameter. The primary tumors were excised, and the tumor tissues were used to perform RT-qPCR analysis of the CR749391 levels.
Statistical analysis
Statistical analyses were performed using SPSS 20.0 software (IBM Corp., Armonk, NY, USA). Continuous variables were expressed as the mean ± standard deviation. One-way analysis of variance with Bonferroni's post-hoc test was performed to assess differences between multiple groups. The paired tumor and adjacent normal tissues were compared using anunpaired t-test, while the correlation between levels of the different target genes was calculated using Pearson's correlation coefficient test. A two-tailed P<0.05 was considered to be statistically significant.
Results
LncRNA CR749391 interacts with miR-181a to regulate KLF6 expression
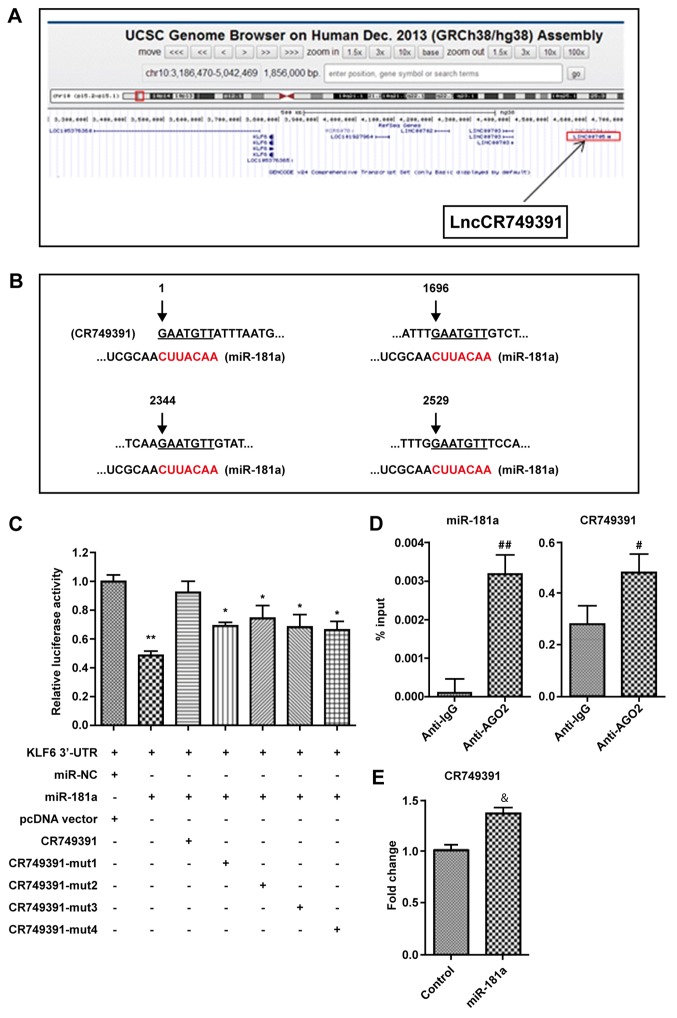
A previous study by our group reported that miR-181a modulates the behavior of GC cells by targeting KLF6 (22). By searching the University of California Santa Cruz database (http://genome.ucsc.edu/), lncRNA CR749391 was predicted to be located downstream of KLF6 and had four putative binding sites for miR-181a (Fig. 1A and B). It was thus hypothesized that lncRNA CR749391 may regulate KLF6 expression through miR-181a. To experimentally verify this, luciferase assays were performed to evaluate whether CR749391 directly binds to miR-181a to affect the luciferase activity of a reporter plasmid driven by a sequence from the 3′-UTR of KLF6. As presented in Fig. 1C, transfection of miR-181a mimics significantly decreased the luciferase activity of the KLF6 reporter plasmid, which, in turn, was recovered by co-transfection of CR749391 with miR-181a. Furthermore, the luciferase activity was partially restored by introducing a mutation in each binding site of miR-181a (Fig. 1C). To further confirm the interaction between CR749391 and miR-181a, RIP and pull-down experiments were performed. CR749391 and miRNA-181a were enriched in Ago-2 containing miRNAs relative to control IgG immunoprecipitation (Fig. 1D). Furthermore, pull-down analysis indicated that CR749391 was enriched in the miR-181a fraction compared with the control group (Fig. 1E).
Figure 1.
LncRNA CR749391 directly interacts with miR-181a. (A) CR749391 was identified as a novel lncRNA located upstream of KLF6. (B) The 4 putative binding sites of miR-181a in the CR749391 transcript sequence. (C) Luciferase reporter plasmid containing the sequence from the 3′-untranslated region of KLF6 was co-transfected with miR-181a and CR749391-coding plasmids into 293T cells. Luciferase activity was determined at 48 hours after transfection using the dual luciferase assay and the results are presented in the histogram as the relative luciferase activity normalized to Renilla activity. *P<0.05, **P<0.01 vs. the control group. (D) RNA immunoprecipitation with mouse monoclonal anti-Ago2, pre-immune IgG or 10% input from BGC 823-cell extracts. RNA levels of miR-181a and CR749391 in immunoprecipitates were determined by reverse transcription-quantitative polymerase chain reaction. #P<0.05, ##P<0.01 vs. Anti-IgG. (E) Pull-down analysis of CR749391 levels. &P<0.05 vs. Control. Values are expressed as the mean ± standard deviation (n=3). lncRNA, long non-coding RNA; miR, microRNA; chr, chromosome; mut, mutated; UTR, untranslated region; KLF, Kruppel-like factor; NC, negative control; IgG, immunoglobulin G; Ago2, argonaute 2, RISC catalytic component; LOC, locus.
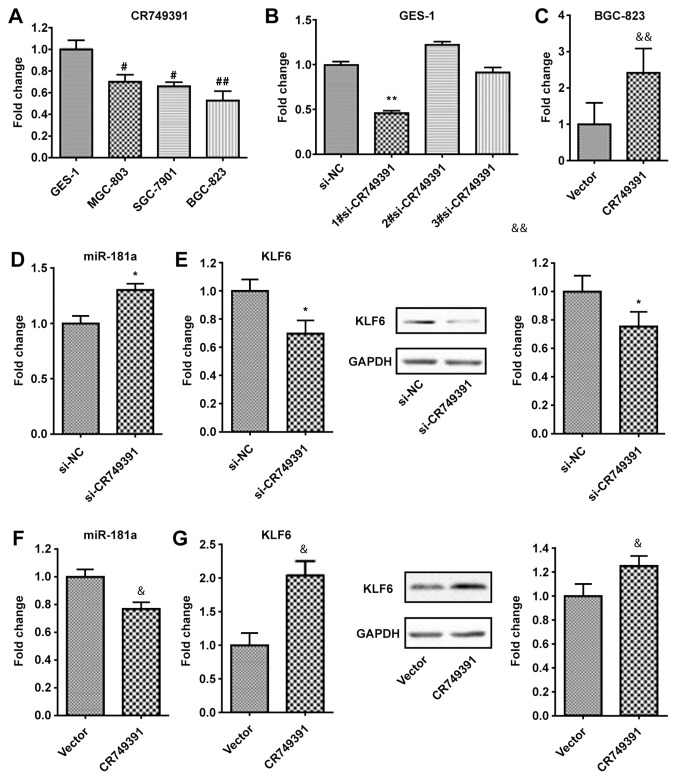
Subsequently, the regulatory interactions among CR749391, KLF6, and miR-181 in GC cells were evaluated. First, the expression of CR749391 was analyzed in several GC cell lines, namely BGC-823, SGC-7901, and MGC-803. Compared with those in the normal gastric cell line GES-1, the levels of CR749391 were significantly reduced, particularly in BGC-823 (Fig. 2A). CR749391 was then knocked down or overexpressed by transfection of CR749391 siRNA and pcDNA3.1/CR749391 vectors, respectively (Fig. 2B and C). Of note, GES-1 cells transfected with si-CR749391 exhibited an increase in miR-181a but a decrease in KLF6 expression (Fig. 2D and E). Conversely, ectopic overexpression of CR749391 in BGC-823 cells resulted in a decrease of miR-181a but an increase in KLF6 expression (Fig. 2F and G). Taken together, these results demonstrated that miR-181a was able to directly bind to CR749391 through its miRNA recognition sites, leading to inhibition of KLF6 expression in GC cells.
Figure 2.
CR749391 regulates KLF6 expression in GC cells. (A) RT-qPCR demonstrated that CR749391 expression levels in the MGC-803, SGC-7901, and BGC-823 GC cell lines were downregulated compared with those in the GES-1 normal gastric epithelial cell line. #P<0.05, ##P<0.01 vs. GES-1. (B and C) RT-qPCR analysis of CR749391 expression levels (B) in GES-1 cells following treatment with siRNAs targeting CR749391 and (C) in BGC-823 cells treatment with the pcDNA3.1/CR749391 vector. (D and E) GES-1 cells were treated with siRNAs targeting CR749391 and the expression of (D) miR-181a and (E) KLF6 mRNA and protein was detected by RT-qPCR and Western blot analysis, respectively. (F and G) BGC-823 cells were treated with pcDNA3.1/CR749391 vector and the expression of (F) miR-181a and (G) KLF6 mRNA and protein was detected by RT-qPCR and Western blot analysis, respectively. Values are expressed as the mean ± standard deviation from three independent experiments. *P<0.05, **P<0.01 vs. si-NC. &P<0.05, &&P<0.01 vs. Vector. KLF, Kruppel-like factor; NC, negative control; GC, gastric cancer; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; siRNA, small interfering RNA; miR, microRNA; pcDNA3.1/CR749391, overexpression plasmid for long non-coding RNA CR749391.
Effect of CR749391 on cell proliferation and apoptosis
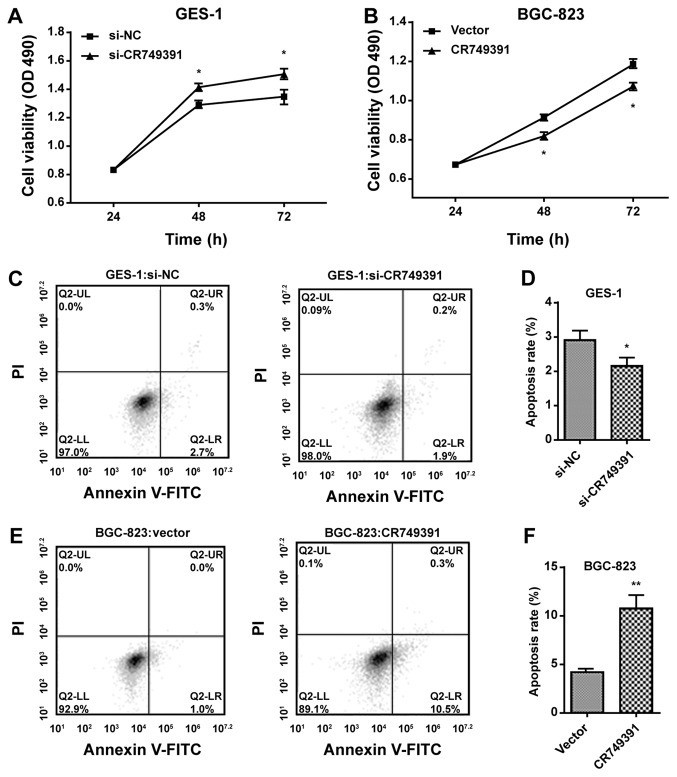
Due to the significant downregulation of CR749391 expression in GC cell lines compared with that in a normal gastric cell line, the possible biological significance of CR749391 in carcinogenesis was investigated. An MTT assay indicated that cell growth was significantly elevated in GES-1 cells transfected with si-CR749391 (Fig. 3A), while the proliferation of BGC-823 cells was significantly reduced in pcDNA3.1/CR749391-transfected cells compared with the respective controls (Fig. 3B). Flow cytometric analysis was also performed in order to determine whether apoptosis is a contributing factor for the growth inhibition observed. The results indicated that the fraction of early apoptotic cells was significantly lower among GES-1 cells treated with si-CR749391 compared with that in scrambled control siRNA-treated cells (Fig. 3C and D). Conversely, BGC-823 cells transfected with pcDNA3.1/CR749391 had a significantly higher apoptotic rate than empty vector-transfected BGC-823 cells (Fig. 3E and F). Collectively, these results indicated thatCR749391 overexpression inhibits GC cell proliferation but induces them to undergo apoptosis in vitro.
Figure 3.
CR749391 expression in gastric cancer cells and its effect on cell proliferation in vitro. (A and B) MTT assays were performed to determine the proliferation of (A) si-CR749391-transfected GES-1 cells or (B) pcDNA3.1/CR749391-transfected BGC-823 cells. (C-F) The apoptotic rates of (C and D) GES-1 cells transfected with si-CR749391 and (E and F) BGC-823 cells transfected with pcDNA3.1/CR749391 vector were detected by flow cytometry. Values are expressed as the mean ± standard deviation from three independent experiments. *P<0.05; **P<0.01. si-CR749391, small interfering RNA targeting long non-coding RNA CR749391; miR, microRNA; pcDNA3.1/CR749391, overexpression plasmid for CR749391; NC, negative control; OD 490, optical density at 490 nm; PI, propidium iodide; FITC, fluorescein isothiocyanate; Q, quadrant; LR, lower right; UL, upper left.
LncRNA CR749391 inhibits the migration and invasion of GC cells
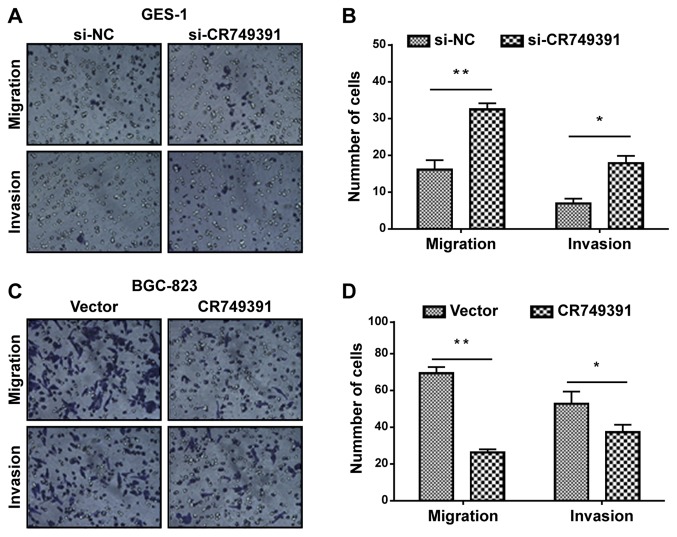
Cell invasion is a marker for the progression of cancer. In the present study, the influence of CR749391 on cancer cell migration and invasion was determined by using Transwell assays. Compared with the scrambled control siRNA, transfection of CR749391 siRNA induced the migratory capacity of GES-1 cells (Fig. 4A and B). An analogous effect on invasiveness was also detected in a parallel invasion assay (Fig. 4A and B). On the contrary, transfection of BGC-823 cells with pcDNA3.1/CR749391 vector reduced cell migration and invasiveness (Fig. 4C and D). These results demonstrated that CR749391 suppresses the migratory and invasive capability of GC cells in vitro.
Figure 4.
Effect of CR749391 on gastric cancer cell migration and invasion in vitro. Transwell assays were performed to investigate changes in cell migration and invasion. (A and B) GES-1 cells were transfected with si-CR749391 or si-NC, and (C and D) BGC-823 cells were transfected with pcDNA3.1/CR749391 vector or empty vector control, and then subjected to Transwell assays. Magnification, ×100. The data are expressed as the mean ± standard deviation of three independent experiments. *P<0.05 and **P<0.01. si-CR749391, small interfering RNA targeting long non-coding RNA CR749391; pcDNA3.1/CR749391, overexpression plasmid for CR749391; NC, negative control.
LncRNA CR749391 overexpression inhibits GC xenograft tumors formation in vivo
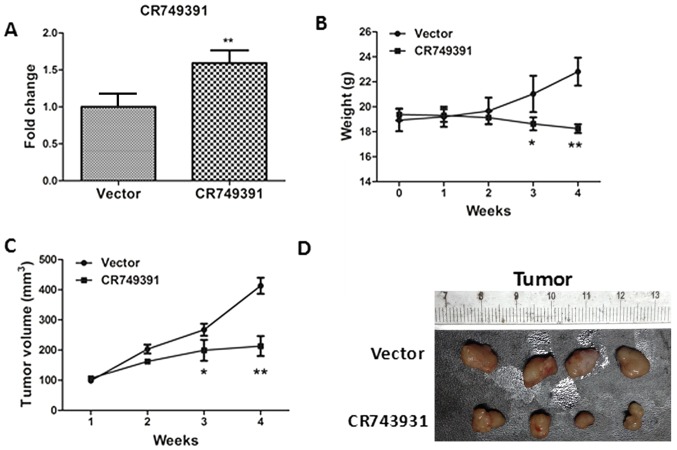
In order to investigate whether CR749391 expression levels affect tumor formation, a xenograft tumor model in nude mice was generated using BGC-823 cells transfected with the pcDNA3.1/CR749391 vector. After 4 weeks, the expression of CR749391 in the tumor tissues was confirmed by RT-qPCR (Fig. 5A). After 3 and 4 weeks, the body weight and tumor volume significantly decreased in the CR749391 group compared with those in the control group (Fig. 5B-D). The above data suggested a significant association between the levels of CR749391 expression and the in vivo proliferation ability of GC cells.
Figure 5.
Effect of CR749391 overexpression on tumor growth in vivo. A xenograft model derived from BGC-823 cells transfected with pcDNA3.1/CR749391 or empty vector was established. (A) CR749391 expression in tissues of resected tumors was determined by reverse transcription-quantitative polymerase chain reaction. (B) Body weight curves of the mice and (C) tumor volume over the course of the experiment. Values are expressed as the mean ± standard deviation (n=4). *P<0.05; **P<0.01. (D) Representative images of the tumors from the two groups at the end of the experiment. si-CR749391, small interfering RNA targeting long non-coding RNA CR749391; pcDNA3.1/CR749391, overexpression plasmid for CR749391; NC, negative control.
LncRNA CR749391 overexpression is positively correlated with KLF6 but negatively correlated with miR-181a in GC tissues
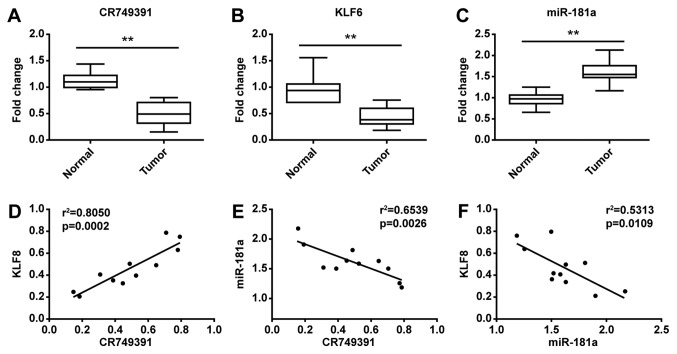
To confirm the association between CR749391 and KLF6 in clinical GC samples, the expression of CR749391, miR-181a, and KLF6 was assessed in 11 paired GC tissues and adjacent non-cancerous gastric tissues by RT-qPCR. The results indicated that the levels of CR749391 and KLF6 were significantly downregulated (Fig. 6A and B), while the levels of miR-181a were upregulated in cancerous tissues compared with adjacent tissues (Fig. 6C). The ratios of CR749391, miR-181a, and KLF6 expression in GC and adjacent tissues were analyzed using the bivariate correlation. A positive association between CR749391 and KLF6 was observed, whereas an inverse association was observed with miR-181a (Fig. 6D and E). Similar to previous data (22), the miR-181a levels were inversely associated with KLF6 expression in GC (Fig. 6F). These results implied that the downregulation of lncRNA CR749391 might be a novel progression-associated marker for GC patients.
Figure 6.
The levels of lncRNA CR749391 were positively correlated with KLF6 but negatively with miR-181a. (A-C) The expression of (A) lncRNA CR749391, (B) KLF6, and (C) miR-181a was determined in 11 gastric cancer tissues and normal gastric tissues using reverse transcription-quantitative PCR. The box indicates the ranges and mean, and the error bars indicate the SD. **P<0.01. (D-F) Analysis of the correlation between (D) lncRNA CR749391 and KLF6, (E) lncRNA CR749391 and miR-181a and (F) miR-181a and KLF6. lncRNA, long non-coding RNA; KLF, Kruppel-like factor; miR, microRNA.
Discussion
LncRNAs may exert their function by binding to DNA or RNA in a sequence-specific manner or by binding to proteins, which may regulate gene expression and protein synthesis in various ways. A vast body of evidence suggests that lncRNAs compete with mRNAs to bind to miRNAs and then enhance the translation of the target mRNA into protein (24–27). However, the precise function of lncRNA and miRNA interactions during the carcinogenesis of GC has remained largely elusive. Understanding this interaction may regulate cancer-associated processes and may promote the development and implementation of lncRNA/miRNA-based diagnostic markers and therapeutic targets for cancer.
In the present study, CR749391 was identified as a novel lncRNA up stream of KLF6, and it was indicated that a cis-regulatory mechanism between CR749391 and KLF6 may exist (28,29). A bioinformatics analysis had indicated that the CR749391 transcript has four putative binding sites of miR-181a, which acts as an oncogene in GC through targeting KLF6, as reported by a previous study by our group (22), and has the same function in clear cell renal cell carcinoma (30). Inspired by the concept of ceRNAs and novel evidence demonstrating that lncRNAs are involved in such regulatory mechanisms, the present study hypothesized that CR749391 may act as a ceRNA to modulate KLF6 through interacting with miR-181a. In order to prove this hypothesis, luciferase assays were performed to prove the direct binding capacity of the predicted miR-181a response elements on the full-length CR749391 transcript. As expected, CR749391 performed complementary base pairing with miR-181a and caused translational repression of an RLuc-KLF6 3′-UTR luciferase reporter gene plasmid. In addition, miR-181a pull-down assays and CR749391-miR-181a co-immunoprecipitation with anti-Ago2 indicated a physical interaction in GC cells, further proving the miRNA sequestering activity of CR749391.
Next, the present study demonstrated that the levels of CR749391 were significantly decreased in GC cell lines compared with those in a normal gastric cell line, while vector-mediated changes in CR749391 expression resulted in corresponding changes in miR-181a and KLF6. Specifically, knockdown of CR749391 in the normal gastric cell line increased miR-181a but decreased KLF6 expression, while overexpression of CR749391 in a GC cell line had the opposite effect. Regarding the results of CR749391 interacting with miR-181a, it is suggested that CR749391 may regulate KLF6 by modulating miR-181a in GC cells.
Mounting evidence suggests that certain lncRNAs have a crucial role in regulating various cell functions. For instance, the lncRNA small nucleolar RNA host gene 5 was reported to regulate GC cell proliferation and migration through targeting KLF4 (22). Silencing of lncRNA zinc finger nuclear transcription factor, X-box binding 1-type containing 1 antisense RNA 1 inhibited malignancies by blocking Wnt/β-catenin signaling in GC cells (31). By modulation of the miR-507/forkhead box M1 axis, lncRNA urothelial cancer associated 1 was reported to regulate cell proliferation, invasion, and G0/G1 cell cycle arrest in melanoma (24). Similarly, lncRNA MEG3 upregulated Bcl-2 through its ceRNA activity on miR-181a, regulating cell proliferation, migration, invasion, and apoptosis in GC (18). The results of the present study suggested that inhibition of lncRNA CR749391 induced the proliferation, migration, and invasion and reduced the apoptosis of gastric cells, while lncRNA CR749391 overexpression in GC cells had the opposite effect. Of note, ectopic lncRNA CR749391 overexpression also inhibited the growth of GC-derived tumor cell xenografts in nude mice. The above results suggest that CR749391 may serve as a tumor suppressor or at least possesses an antitumor activity in GC.
To act as an endogenous miRNA ‘sponge’, the abundance of CR749391 should be higher than that of miR-181a. The present results indicated that CR749391 expression was inversely correlated with miR-181a expression in GC tissues, compared with that in adjacent non-cancerous gastric tissues, which is in line with the observation that ectopic overexpression of CR749391 suppressed GC cell invasion and viability, and inhibited GC cell growth in vitro and in vivo. Furthermore, the CR749391 levels were negatively correlated with miR-181a but positively correlated with KLF6, which is also in accordance with the result that an alteration of CR749391 led to a change in miR-181a and KLF6. These results suggested that CR749391 has a direct role in regulating the carcinogenesis and progression of GC and may be considered as a novel therapeutic target in GC. However, the carcinogenic properties and mechanisms of CR749391 are far from fully clarified. The prognostic value of CR749391 may require further studies.
In conclusion, the present study was the first, to the best of our knowledge, to report that lncRNA CR749391 acts as a ceRNA to modulate KLF6 expression by sponging miR-181a and may be a key regulator in GC. Furthermore, the present results imply that the downregulation of lncRNA CR749391 may serve as a novel progression-associated marker in GC.
Acknowledgements
Not applicable.
Funding
This work was supported in part by grants from the National Natural Science Foundation of China (grant no. 81372633) and the Natural Science Foundation of Hunan Province (grant no. 2017JJ3270).
Availability of data and materials
All data generated in the current study are available from the corresponding author upon reasonable request.
Authors' contributions
YN and YL conceived of and designed the experiments. SS and ZF performed the experiments. DL analyzed the data. SS and YN wrote the paper. ZF and YD provided tools used and participated in some data analyses. All authors read and approved the final version of this manuscript.
Ethics approval and consent to participate
All procedures involving human participants and specimens were in accordance with the ethical standards of the Medical Ethics Committee of Guangzhou Medical University (Guangzhou, China) and with the declaration of Helsinki from 1964 and its later amendments or comparable ethical standards. All tissues were obtained with informed consent from the patients. The present study of human subjects was approved by the Medical Ethics Committee of Guangzhou Medical University (Guangzhou, China), while the animal experiment was approved by the Animal Care and Use Committee of Guangzhou Medical University (Guangzhou, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD, Kong R, Xia R, Lu KH, Li JH, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92. doi: 10.1186/1476-4598-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jema A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Miller KD, Nogueira L, Mariotto AB, Rowland JH, Yabroff KR, Alfano CM, Jemal A, Kramer JL, Siegel RL. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin. 2019;69:363–385. doi: 10.3322/caac.21565. [DOI] [PubMed] [Google Scholar]
- 4.Yang L. Incidence and mortality of gastric cancer in China. World J Gastroenterol. 2006;12:17–20. doi: 10.3748/wjg.v12.i1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crew KD, Neugut AI. Epidemiology of gastric cancer. World J Gastroenterol. 2006;12:354–362. doi: 10.3748/wjg.v12.i3.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartgrink HH, Jansen EP, van Grieken NC, van de Velde CJ. Gastric cancer. Lancet. 2009;374:477–490. doi: 10.1016/S0140-6736(09)61948-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W, et al. Relation between microRNA expression and progression and prognosis of gastric cancer: A microRNA expression analysis. Lancet Oncol. 2010;11:136–146. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Arita T, Ichikawa D, Konishi H, Komatsu S, Shiozaki A, Shoda K, Kawaguchi T, Hirajima S, Nagata H, Kubota T, et al. Circulating long non-coding RNAs in plasma of patients with gastric cancer. Anticancer Res. 2013;33:3185–3193. [PubMed] [Google Scholar]
- 10.Ergun S, Oztuzcu S. Oncocers: ceRNA-mediated cross-talk by sponging miRNAs in oncogenic pathways. Tumor Biol. 2015;36:3129–3136. doi: 10.1007/s13277-015-3346-x. [DOI] [PubMed] [Google Scholar]
- 11.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen L, Wang W, Cao L, Li Z, Wang X. Long non-coding RNA CCAT1 acts as a competing endogenous RNA to regulate cell growth and differentiation in acute myeloid leukemia. Mol Cells. 2016;39:330–336. doi: 10.14348/molcells.2016.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lü MH, Tang B, Zeng S, Hu CJ, Xie R, Wu YY, Wang SM, He FT, Yang SM. Long noncoding RNA BC032469, a novel competing endogenous RNA, upregulates hTERT expression by sponging miR-1207-5p and promotes proliferation in gastric cancer. Oncogene. 2016;35:3524–3534. doi: 10.1038/onc.2015.413. [DOI] [PubMed] [Google Scholar]
- 14.Chen DL, Ju HQ, Lu YX, Chen LZ, Zeng ZL, Zhang DS, Luo HY, Wang F, Qiu MZ, Wang DS, et al. Long non-coding RNA XIST regulates gastric cancer progression by acting as a molecular sponge of miR-101 to modulate EZH2 expression. J Exp Clin Cancer Res. 2016;35:142. doi: 10.1186/s13046-016-0420-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song B, Guan Z, Liu F, Sun D, Wang K, Qu H. Long non-coding RNA HOTAIR promotes HLA-G expression via inhibiting miR-152 in gastric cancer cells. Biochem Biophys Res Commun. 2015;464:807–813. doi: 10.1016/j.bbrc.2015.07.040. [DOI] [PubMed] [Google Scholar]
- 16.Yan J, Dang Y, Liu S, Zhang Y, Zhang G. LncRNA HOTAIR promotes cisplatin resistance in gastric cancer by targeting miR-126 to activate the PI3K/AKT/MRP1 genes. Tumour Biol. 2016 doi: 10.1007/s13277-016-5448-5. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 17.Yan J. LncRNA HOTAIR acts as a competing endogenous RNA to promote cisplatin resistance in gastric cancer via miR-126/PI3K/AKT/MRP1 pathway. J Gastroenterol Hepatol. 2016;31:53–53. [Google Scholar]
- 18.Peng W, Si S, Zhang Q, Li C, Zhao F, Wang F, Yu J, Ma R. Long non-coding RNA MEG3 functions as a competing endogenous RNA to regulate gastric cancer progression. J Exp Clin Cancer Res. 2015;34:79. doi: 10.1186/s13046-015-0197-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sangodkar J, Shi J, DiFeo A, Schwartz R, Bromberg R, Choudhri A, McClinch K, Hatami R, Scheer E, Kremer-Tal S, et al. Functional role of the KLF6 tumour suppressor gene in gastric cancer. Eur J Cancer. 2009;45:666–676. doi: 10.1016/j.ejca.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Q, Tan XP, Yuan YS, Hu CM, He CH, Wang WZ, Li JC, Zhao Q, Liu NZ. Decreased expression of KLF6 and its significance in gastric carcinoma. Med Oncol. 2010;27:1295–1302. doi: 10.1007/s12032-009-9377-7. [DOI] [PubMed] [Google Scholar]
- 21.Cho YG, Kim CJ, Park CH, Yang YM, Kim SY, Nam SW, Lee SH, Yoo NJ, Lee JY, Park WS. Genetic alterations of the KLF6 gene in gastric cancer. Oncogene. 2005;24:4588–4590. doi: 10.1038/sj.onc.1208670. [DOI] [PubMed] [Google Scholar]
- 22.Zhang X, Nie Y, Du Y, Cao J, Shen B, Li Y. MicroRNA-181a promotes gastric cancer by negatively regulating tumor suppressor KLF6. Tumor Biol. 2012;33:1589–1597. doi: 10.1007/s13277-012-0414-3. [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Zhao L, Han T, Li Y, Sun J, Zhang S, Liu Y, Shan B, Zheng D, Shi J. The lncRNA SNHG5/miR-32 axis regulates gastric cancer cell proliferation and migration by targeting KLF4. FASEB J. 2017;31:893–903. doi: 10.1096/fj.201600994R. [DOI] [PubMed] [Google Scholar]
- 25.Zhao H, Peng R, Liu Q, Liu D, Du P, Yuan J, Peng G, Liao Y. The lncRNA H19 interacts with miR-140 to modulate glioma growth by targeting iASPP. Arch Biochem Biophys. 2016;610:1–7. doi: 10.1016/j.abb.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 26.Wang SH, Wu XC, Zhang MD, Weng MZ, Zhou D, Quan ZW. Long noncoding RNA H19 contributes to gallbladder cancer cell proliferation by modulated miR-194-5p targeting AKT2. Tumor Biol. 2016;37:9721–9730. doi: 10.1007/s13277-016-4852-1. [DOI] [PubMed] [Google Scholar]
- 27.Wei Y, Sun Q, Zhao L, Wu J, Chen X, Wang Y, Zang W, Zhao G. LncRNA UCA1-miR-507-FOXM1 axis is involved in cell proliferation, invasion and G0/G1 cell cycle arrest in melanoma. Med Oncol. 2016;33:88. doi: 10.1007/s12032-016-0804-2. [DOI] [PubMed] [Google Scholar]
- 28.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guil S, Esteller M. Cis-acting noncoding RNAs: Friends and foes. Nature Struct Mol Biol. 2012;19:1068–1075. doi: 10.1038/nsmb.2428. [DOI] [PubMed] [Google Scholar]
- 30.Lei Z, Ma X, Li H, Zhang Y, Gao Y, Fan Y, Li X, Chen L, Xie Y, Chen J, et al. Up-regulation of miR-181a in clear cell renal cell carcinoma is associated with lower KLF6 expression, enhanced cell proliferation, accelerated cell cycle transition, and diminished apoptosis. Urol Oncol. 2018;36:93.e23–93.e37. doi: 10.1016/j.urolonc.2017.09.019. [DOI] [PubMed] [Google Scholar]
- 31.Xu W, He L, Li Y, Tan Y, Zhang F, Xu H. Silencing of lncRNA ZFAS1 inhibits malignancies by blocking Wnt/β-catenin signaling in gastric cancer cells. Biosci Biotechnol Biochem. 2018;82:456–465. doi: 10.1080/09168451.2018.1431518. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated in the current study are available from the corresponding author upon reasonable request.