Abstract
Background: Alone, administrative data poorly identifies patients with palliative care needs.
Objective: To identify patients with uncommon, yet devastating, illnesses using a combination of administrative data and natural language processing (NLP).
Design/Setting: Retrospective cohort study using the electronic medical records of a healthcare network totaling over 2500 hospital beds. We sought to identify patient populations with two unique disease processes associated with a poor prognosis: pneumoperitoneum and leptomeningeal metastases from breast cancer.
Measurements: Patients with pneumoperitoneum or leptomeningeal metastasis from breast cancer were identified through administrative codes and NLP.
Results: Administrative codes alone resulted in identification of 6438 patients with possible pneumoperitoneum and 557 patients with possible leptomeningeal metastasis. Adding NLP to this analysis reduced the number of patients to 869 with pneumoperitoneum and 187 with leptomeningeal metastasis secondary to breast cancer. Administrative codes alone yielded a 13% positive predictive value (PPV) for pneumoperitoneum and 25% PPV for leptomeningeal metastasis. The combination of administrative codes and NLP achieved a PPV of 100%. The entire process was completed within hours.
Conclusions: Adding NLP to the use of administrative codes allows for rapid identification of seriously ill patients with otherwise difficult to detect disease processes and eliminates costly, tedious, and time-intensive manual chart review. This method enables studies to evaluate the effectiveness of treatment, including palliative interventions, for unique populations of seriously ill patients who cannot be identified by administrative codes alone.
Keywords: critical illness, natural language processing, patient identification
Introduction
Palliative care investigators hoping to study the impact of interventions in large populations face challenges in identifying the target population. This difficulty in recognizing patients with serious illness has been termed “the denominator challenge.”1 Researchers remain dependent on extraction specialist to tediously sift through charts isolating relevant information.2–4 Use of structured data codes, such as administrative claims, can obviate this process, but at the cost of a reduction in specificity and accuracy. In a recent report, Kelley and Bollens-Lund1 calculated the sensitivity of standard techniques to be <50%.
The vast majority of patient information (70%–80%) resides in unstructured free-text notes, which are not readily captured by administrative codes.5 Powerful computational methods such as natural language processing (NLP) now make it possible to quickly analyze free-text clinical notes stored within the electronic health record (EHR).6,7 These methods are particularly relevant to the field of palliative medicine, where, as demonstrated above, standard administrative data poorly capture the relevant patient populations. Without a ready ability to identify patients, it is impossible to determine the effectiveness of various interventions. Randomized controlled trials are time consuming, costly, and not always practical, especially in patients with serious illness. Retrospective population-level review can help answer important clinical questions, but only if relevant populations can be identified.8 Unfortunately, palliative care physicians and researchers have not been able to take advantage of these techniques due to a lack of practical methods of identifying these patients.9,10
We sought to validate a novel combination of NLP methods and administrative codes in identification of seriously ill patients. We chose to focus on one surgical diagnosis, pneumoperitoneum, and one medical diagnosis, leptomeningeal metastases, secondary to stage IV breast cancer. Both patient populations have a high associated mortality and many within these groups could benefit from early palliative care referral.11–13 We chose these two examples to highlight how NLP might improve the efficiency of identifying patients with serious illness and facilitate high-quality studies.
Methods
Data source
Our primary data source was the Partners HealthCare Research Patient Data Registry. This registry gathers data from multiple EHR's at Partners HealthCare, a large network in Massachusetts with over 2500 hospital beds serving approximately one-third of all hospitalized patients in the Boston metropolitan area. Administrative data are available for encounters across all hospital and clinic settings within the Partners HealthCare system. Data are linked to EHR notes, including consultation notes, progress notes, procedure notes, and discharge summaries. This study was approved by the Partners Institutional Review Board.
Population screening
Initial population screening was performed using International Classification of Diseases, 9th Revision (ICD-9) diagnosis codes, which are compiled for every patient encounter in Partners HealthCare. We used ICD-9 diagnosis codes for unspecified peritoneal disorder (568.89) and visceral perforation (596.83) to identify patients with possible pneumoperitoneum between January 1, 2010 and September 30, 2015. We used ICD-9 codes for breast cancer (174.0–174.9, 175.0, and 175.9) and leptomeningeal disease (198.4) to identify patients with possible leptomeningeal metastases between January 1, 2010 and September 30, 2015.
Natural language processing
After the population of patients was screened using diagnostic codes, regular expression of NLP was used to identify patients with a specific disease. Our NLP software, ClinicalRegex, identifies predefined keywords or phrases within the clinical notes, taking into account varieties in language and punctuation. ClinicalRegex also allows for rapid semiautomated review that ensures that keywords have not been taken out of context. For pneumoperitoneum, we used the phrases “free air,” “extraluminal air,” extraluminal gas,” and “pneumoperitoneum” to review radiology reports of patients identified through administrative codes. For leptomeningeal disease, we reviewed radiology reports using the keyword “leptomeningeal.” Notes that contained the above phrases were reviewed to ensure accuracy. The review process was performed in a semiautomated manner, in which the notes were listed with the phrase or word highlighted to facilitate identification. In the case of pneumoperitoneum, the review was performed by a surgical resident. In the case of leptomeningeal disease, the review was performed by an attending neurologist. Semiautomated review ensures that the positive search terms have not been taken out of context. For example, NLP may erroneously identify a note in which the phrase “no evidence of” precedes the desired search term.
Results
Initial population screening, using ICD-9 diagnosis codes, yielded 6438 patients with possible pneumoperitoneum and 557 with metastatic leptomeningeal disease. These patients' hospitalizations were associated with 299,449 and 32,519 radiology reports, respectively. Using NLP, we reduced the number of radiology reports by ∼95%. The remaining reports were reviewed by trained physicians in a semiautomated manner with NLP highlighting key phrases and words to facilitate identification of the relevant patient population. Through this process, we were able to identify 869 patients with pneumoperitoneum and 187 patients with leptomeningeal metastasis. The entire process was completed within hours. Use of administrative codes alone was associated with a positive predictive value of 13% for pneumoperitoneum and 25% for leptomeningeal metastasis from breast cancer. This stepwise progression is summarized in Table 1.
Table 1.
Stepwise Identification of Relevant Patient Populations with Serious Illness
| Pneumoperitoneum | Leptomeningeal metastasis | |
|---|---|---|
| Population screening | ||
| Patients identified using administrative codes | 6438 | 557 |
| Radiology reports associated with patient population | 291,449 | 32,519 |
| Natural language processing | ||
| Radiology reports containing key word(s) | 13,978 | 1602 |
| Confirmed positive radiology reportsa | 1429 | 214 |
| No. of patients in the final denominator | 869 | 187 |
Note that some patients had multiple positive radiology reports.
Discussion
Language can communicate complex ideas and nuanced decisions. In a written document such as the medical record, it is found in text. Yet, the current state of the art methodology is only able to extract “defined field” entries, such as used in coding for administrative databases. Sadly, the extraction of meaning from text relies on humans reading every note then interpreting it—a time-consuming and expensive process. Developments in computer technology yield the potential to gather meaningful nuance from written text in a fraction of time and at a lower cost. This project reports preliminary progress toward that goal.
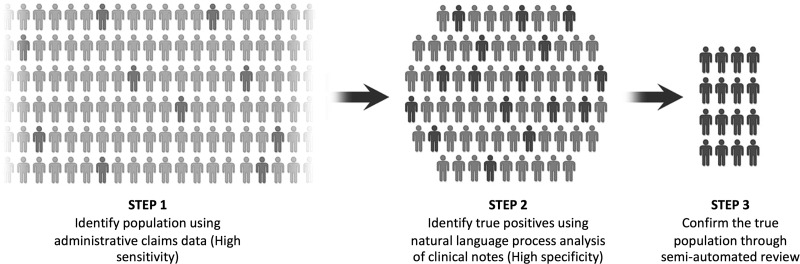
Conceptually, we use a three-step approach to identify patients with serious illness that are otherwise difficult to isolate. In this process, we use ICD-9 diagnostic codes to screen for a potential disease process (step 1). These administrative codes have a high sensitivity, but a low specificity.14 From this pool of potential patients, NLP methods allow for accurate and efficient isolation of the relevant population (step 2). In the examples described in this study, NLP was used to scan through radiology reports. However, NLP methods can be used for any free-text patient data (clinic notes, pathology reports, etc.) to identify keywords or phrases associated with a particular diagnosis. The charts of patients identified through this process can be audited in a semiautomated process to ensure accuracy (step 3). NLP reduces the time required to review charts by more than a 1000-fold compared to standard manual chart review.15 This process is illustrated in Figures 1 and 2.
FIG. 1.
Three stage process of patient identification.
FIG. 2.
Schematic of NLP. Sensitive, but nonspecific administrative codes are used to capture thousands of reports, which may be indicative of a disease process (A). NLP rapidly scans through the free-text reports and isolates those that screen positive for the desired search terms (B). In this example, the terms “free air” and “pneumoperitoneum” were used. Semiautomated review allows the researcher to review notes in which the search terms appear. The search terms are highlighted by the NLP to facilitate rapid review. If a note is confirmed positive, the reviewer can place an indicator in the annotated value box. In this case the reviewer used the digit “1” to indicate “free air.” The output from the screening is automatically exported to a spreadsheet (C). In this example, all data have been deidentified. CT, computed tomography; NLP, natural language processing.
The described methods proved successful in both a surgical population (pneumoperitoneum) as well as a medical population (leptomeningeal metastasis). Both disease processes are associated with high short-term mortality and may lead to a reevaluation of treatment priorities.10,11 Despite their high mortality and morbidity, the administrative codes associated with leptomeningeal metastasis and pneumoperitoneum have a low specificity. This has limited previous studies to small single-institution case series, in which manual identification could be performed.11–13,16 The methods described in this work offer a potential mechanism for identifying large numbers of these patients without the aid established registries.
Finally, the methods for identifying seriously ill populations described in this study offer a potential solution to the “denominator” problem within the field of palliative care. As reported by Kelley and Bollens-Lund,1 administrative data poorly capture patients with serious illness, which have limited the optimal treatment of these populations. Palliative care in this population can improve quality and reduce cost.17–20 Understanding which populations may most benefit from palliative care is a critical step to disseminating the service. NLP methods aid in defining the population at risk. While NLP can be used in innumerous research modalities, its potential in the palliative care population is paramount, as there is currently no effective method for efficient identification of patients with palliative care needs at a population level. The methods described in this article can immediately facilitate the identification of palliative care population in retrospective reviews aimed at evaluating the outcomes of various treatment paradigms. Through further work, these methods may be operationalized at a health system level to detect patients with palliative needs in a prospective manner.
While NLP has enormous potential, there are limitations that should be acknowledged. Chief is accuracy of documentation. NLP can efficiently identify patients within the EHR, so long as clinicians produce some form of documentation. If clinicians fail to document their interactions with patients, then NLP will also fail to identify these patients. In addition, as mentioned above, NLP requires semiautomated auditing to ensure that notes with negative modifiers are appropriately categorized. While machine learning techniques are being developed to obviate this requirement, at this time, it is still necessary to ensure specificity.
Conclusion
A combination of administrative codes and NLP rapidly identifies seriously ill patients with otherwise difficult to detect disease processes. NLP methods enable larger retrospective studies evaluating the effectiveness of treatment in patients, who can benefit from palliative interventions. Future work, aimed at operationalizing these methods, may lead to real-time identification of patients with palliative care needs.
Acknowledgments
Dr. Lindvall was supported by a Junior Faculty Career Development Award from the National Palliative Care Research Center (NPCRC), New York City, New York; Pilot Award (NINR U24) from the Palliative Care Research Cooperative Group (PCRC), Denver, Colorado; and Spivak Fund Faculty Development Award, Boston, Massachusetts.
Author Disclosure Statement
No competing financial interests exist.
References
- 1. Kelley AS, Bollens-Lund E: Identifying the population with serious illness: The “denominator” challenge. J Palliat Med 2018;21:S7–S16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bennett S, Agyepong IA, Sheikh K, et al. : Building the field of health policy and systems research: An agenda for action. PLoS Med 2011;8:e1001081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gilson L, Hanson K, Sheikh K, et al. : Building the field of health policy and systems research: Social science matters. PLoS Med 2011;8:e1001079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sheikh K, Gilson L, Agyepong IA, et al. : Building the field of health policy and systems research: Framing the questions. PLoS Med 2011;8:e1001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Murdoch TB, Detsky AS: The inevitable application of big data to health care. JAMA 2013;309:1351–1352 [DOI] [PubMed] [Google Scholar]
- 6. Bates DW, Saria S, Ohno-Machado L, et al. : Big data in health care: Using analytics to identify and manage high-risk and high-cost patients. Health Aff (Millwood) 2014;33:1123–1131 [DOI] [PubMed] [Google Scholar]
- 7. Krumholz HM: Big data and new knowledge in medicine: The thinking, training, and tools needed for a learning health system. Health Aff (Millwood) 2014;33:1163–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kelley AS, Ettner SL, Morrison RS, et al. : Determinants of medical expenditures in the last 6 months of life. Ann Intern Med 2011;154:235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aldridge MD, Meier DE: It is possible: Quality measurement during serious illness. JAMA Intern Med 2013;173:2080–2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Badgwell B, Krouse R, Cormier J, et al. : Frequent and early death limits quality of life assessment in patients with advanced malignancies evaluated for palliative surgical intervention. Ann Surg Oncol 2012;19:3651–3658 [DOI] [PubMed] [Google Scholar]
- 11. Badgwell B, Feig BW, Ross MI, et al. : Pneumoperitoneum in the cancer patient. Ann Surg Oncol 2007;14:3141–3147 [DOI] [PubMed] [Google Scholar]
- 12. Shinall MC, Jr., Ehrenfeld JM, Gunter OL, Jr.: Nonoperative management of perforated hollow viscera in a palliative care unit. Ann Surg 2018;268:e1–e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Niwinska A, Pogoda K, Michalski W, et al. : Determinants of prolonged survival for breast cancer patient groups with leptomeningeal metastasis (LM). J Neurooncol 2018;138:191–198 [DOI] [PubMed] [Google Scholar]
- 14. Chang DC, Evans C, Parker J: Conceptualizing administrative databases as screening tools for health system quality: Rethinking the issue of data accuracy. JAMA Surg 2015;150:5–6 [DOI] [PubMed] [Google Scholar]
- 15. Lilley EJ, Lindvall C, Lillemoe KD, et al. : Measuring processes of care in palliative surgery: A novel approach using natural language processing. Ann Surg 2018;267:823–825 [DOI] [PubMed] [Google Scholar]
- 16. Kingston B, Kayhanian H, Brooks C, et al. : Treatment and prognosis of leptomeningeal disease secondary to metastatic breast cancer: A single-centre experience. Breast 2017;36:54–59 [DOI] [PubMed] [Google Scholar]
- 17. Blumenthal D, Chernof B, Fulmer T, et al. : Caring for high-need, high-cost patients—An urgent priority. N Engl J Med 2016;375:909–911 [DOI] [PubMed] [Google Scholar]
- 18. Teno JM, Gozalo PL, Bynum JP, et al. : Change in end-of-life care for Medicare beneficiaries: Site of death, place of care, and health care transitions in 2000, 2005, and 2009. JAMA 2013;309:470–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bakitas M, Lyons KD, Hegel MT, et al. : Effects of a palliative care intervention on clinical outcomes in patients with advanced cancer: The Project ENABLE II randomized controlled trial. JAMA 2009;302:741–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brumley R, Enguidanos S, Jamison P, et al. : Increased satisfaction with care and lower costs: Results of a randomized trial of in-home palliative care. J Am Geriatr Soc 2007;55:993–1000 [DOI] [PubMed] [Google Scholar]