Abstract
Service members during military actions or combat training are frequently exposed to primary blasts by weaponry. Most studies have investigated moderate or severe brain injuries from blasts generating overpressures >100 kPa, whereas understanding the pathophysiology of low-intensity blast (LIB)-induced mild traumatic brain injury (mTBI) leading to neurological deficits remains elusive. Our recent studies, using an open-field LIB-induced mTBI mouse model with a peak overpressure at 46.6 kPa, demonstrated behavioral impairments and brain nanoscale damages, notably mitochondrial and axonal ultrastructural changes. In this study, we used tandem mass tagged (TMT) quantitative proteomics and bioinformatics analysis to seek insights into the molecular mechanisms underlying ultrastructural pathology. Changes in global- and phospho-proteomes were determined at 3 and 24 h and at 7 and 30 days post injury (DPI), in order to investigate the biochemical and molecular correlates of mitochondrial dysfunction. Results showed striking dynamic changes in a total of 2216 proteins and 459 phosphorylated proteins at vary time points after blast. Disruption of key canonical pathways included evidence of mitochondrial dysfunction, oxidative stress, axonal/cytoskeletal/synaptic dysregulation, and neurodegeneration. Bioinformatic analysis identified blast-induced trends in networks related to cellular growth/development/movement/assembly and cell-to-cell signaling interactions. With observations of proteomic changes, we found LIB-induced oxidative stress associated with mitochondrial dysfunction mainly at 7 and 30 DPI. These dysfunctions included impaired fission-fusion dynamics, diminished mitophagy, decreased oxidative phosphorylation, and compensated respiration-relevant enzyme activities. Insights on the early pathogenesis of primary LIB-induced brain damage provide a template for further characterization of its chronic effects, identification of potential biomarkers, and targets for intervention.
Keywords: mitochondrial dysfunction, open-field LIB, oxidative stress, quantitative proteomics
Introduction
Military personnel frequently exposed to blast waves from explosive weaponry may develop traumatic brain injuries (TBI) of varying severity. Department of Defense and Veterans Administration (DoD /VA) reports indicated that the majority (82.3%) of blast-induced neurotraumas are mild TBI (mTBI)/concussion, which is the “signature wound” of current military conflicts. Subjects with mTBI often do not show detectable signals using conventional imaging techniques, thus missing opportunities for early diagnosis and treatment.1,2 Nevertheless, these subjects are at risk for development of post-traumatic stress disorder, as well as neurodegenerative disorders that result in life-long disabilities.3,4 Although studies have provided insights into TBI mechanisms with moderate-to-high-intensity blast exposures >100 kPa peak overpressure, an urgent need still exists to understand the pathobiology underlying the impact of primary low-intensity blast (LIB)-induced brain injury.
We have recently established an open-field primary LIB murine model to simulate theater combat and military training scenarios.5–8 With exposure to a magnitude of 46.6 kPa and a maximum impulse of 8.7 PSI (aka 60 kPa) × ms blast, in the absence of head motion, animals experienced no mortality or gross bodily injuries. Nevertheless, despite the absence of macroscopic changes or cell necrosis, behavioral deficits and ultrastructural damage including mitochondrial abnormalities and myelinated axonal injury were observed.5 Oxidative stress and mitochondrial dysfunction may decrease mitochondrial fission-fusion activities, and compromise mitophagy and late cellular impairment. Similar signals of mitochondrial dysfunction are found in the pathogenesis of chronic neurodegenerative diseases associated with moderate or severe TBI.9–12 However, the underlying mechanisms in LIB exposure leading to the sustained mitochondrial ultrastructural changes coupled with axonal injury in neuronal cells remain elusive. In the present study, we used systems biology analysis of proteomic profiles to delineate mechanisms associated with mitochondrial ultrastructural changes coupled with axonal injury caused by LIB exposure.
Mass spectrometry (MS)-based proteomics is a specific and sensitive tool for determining protein abundance in brain tissues and predicting biochemical responses.13–15 Quantitative proteomics coupled with bioinformatics have aided the discovery of key proteins, including glial fibrillary acidic protein (GFAP) and ubiquitin carboxyl-terminal hydrolase isozyme L1 (UCH-L1), with severe impact or penetrating TBI.16–19 However, the molecular signatures after LIB exposure remain undefined. In this study, we employed tandem mass tag (TMT) 10-plex peptidyl labeling coupled with liquid chromatography tandem mass spectrometry (LC–MS/MS) to investigate global and phospho-proteomes at different times (3 and 24 h, and 7 and 30 days post-injury [DPI]) after LIB exposure.
Results of this study provide information on specific proteins, signaling pathways/networks, and potential mechanisms related to oxidative-stress-associated mitochondrial dysfunction and dysregulation of axonal/cytoskeletal/synaptic proteins. These insights regarding early effects on mitochondria can offer new strategies for prevention and treatment of primary LIB-induced brain injury.
Methods
Materials
Chemicals were purchased from Sigma-Aldrich (St. Louis, MO). Anesthesia was ketamine hydrochloride injection (100 mg/mL, 1044102, Henry Schein, Melville, NY and xylazine hydrochloride injection (100 mg/mL, 13985-704-10, Vet One, Boise, ID). Reagents for BCA protein assay and sample preparation for LC–MS/MS analysis were purchased from Thermo Scientific (Rockford, IL). TMT reagent 10-plex kits were bought commercially (Thermo Scientific Pierce, Rockford, IL).
Experimental design and statistical rationale
The experimental design incorporated four biological replicates for each group (total five groups: sham control, 3 and 24 h, and 7 and 30 DPI after blast), across two independent TMT 10-plex tagging experiments to examine brain cortex. In order to perform robust quantitative analysis between the two independent TMT 10-plex tagging experiments, we pooled two biological replicates from the sham group together as an index for two TMT tagging cross-batch analysis. All experimental groups in the studies were handled in a randomized, double-blinded manner. For quantification purposes, all proteomics data were normalized across all proteins within each 10-plex batch to eliminate the batch effect. Welch's test and t-distribution were performed with R function “t.test” for proteomics data, and other statistical comparisons were made by student's t test for two groups and by one way analysis of variance (ANOVA) test followed by Tukey test for multiple comparisons. GraphPad Prism (version 6 for Windows; GraphPad Software) were used to calculate the p values. The relative protein abundance ratios (fold changes) between blast and sham groups were calculated. As previously described, the changes in protein levels were considered significant if fold change was >1.0 (upregulated) or <1.0 (downregulated), and the p value was <0.05 in two independent experiments.20 The threshold of this open-field LIB was determined based on peak overpressure (46.6 kPa) as compared with literature reports (majority >100 kPa),6,21 and our previous pathological observations demonstrating the absence of macroscopic damage or necrosis in the presence of nanoscale ultrastructural injuries.5
Animals and open-field blast setting
The protocol for this study has been described previously.5 Twenty male C57BL/6J mice (The Jackson Laboratory, Bar Harbor, ME) at 2 months of age were housed with a 12 h light/dark cycle (lights on at 7:00 a.m.) and given unrestricted access to food and water. Four mice were put in one cage. A cotton pad and an environmental enrichment toy (small plastic house) were placed in each cage. All animals were observed for good health before, during, and after testing. All experimental groups in the studies were handled in a randomized, double-blinded manner. All procedures were performed in accordance to the University of Missouri approved protocols for the Care and Use of Laboratory Animals and the Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines.
Based on the blast exposure setting, animals were randomly grouped as the 3 m (n = 16) blast, and sham control (n = 4). Animals with 3 m blast were randomly grouped into four groups as 3 h (n = 4), 24 h (n = 4), 7 DPI (n = 4), and 30 DPI (n = 4). Mice were anesthetized with intraperitoneal (i.p.) injection of 5 μL/g body weight of ketamine/xylazine mixture (25 mg/mL ketamine and 1.25 mg/mL xylazine). The sham group underwent the identical anesthesia procedures only without blast exposure. Mice were placed in the prone position at the 3 m platforms away from the 350 g C4. No constraint was applied to the animals. Following blast wave exposure, animals were returned to the original cage. After recovery from anesthesia, mice were able to spontaneously move and continuously be monitored for at least 15–30 min. Animals were allowed access to food and water ad libitum before and after blast exposure.
Protein sample preparation, isobaric TMT 10-plex labeling, IMAC enrichment, LC-MS/MS analysis, and protein identification
After blast exposure, cerebral cortices were dissected from the immediately euthanized mouse brains as described previously.5,6 To lyse each brain tissue specimen (100 mg), 600 μL of sample buffer (2% sodium dodecyl sulphate [SDS], 0.5M tetraethylammonium bicarbonate [TEAB]), protease inhibitor cocktail was added and the specimen homogenized by TissueLyser LT (Qiagen, Valencia, CA). Tissue homogenates were centrifuged at 17,000g for 20 min at 4°C. The supernatant was transferred into a new vial for protein concentration measurement by BCA assay. Preparation of tryptic peptides for TMT 10-plex labelling was performed according to manufacturer's instructions. Briefly, 100 μg protein from each sample was transferred into a new vial and adjusted to a final volume of 100 μL with TEAB and reduced with Tris (2-carboxyethyl) phosphine hydrochloride (TCEP) at 55°C for 1 h, and then alkylated with iodoacetamide for 30 min in the dark. Methanol-chloroform precipitation was performed prior to protease digestion. In brief, four volumes of methanol were added to each sample and vortexed, followed by adding one part chloroform and three parts water to the sample, and vortexing. The sample was centrifuged at 14,000g for 4 min at room temperature and the aqueous phase was removed. The organic phases with protein precipitate at the surface were subsequently washed twice with four parts methanol, centrifuged, and subsequently supernatant was removed. After air-drying, precipitated protein pellets were re-suspended with 100 μL of 50 mM TEAB and digested with trypsin overnight at 37°C.
Tagging with TMT multiplex reagents enables relative quantitation of proteins present in multiple samples by labeling peptides with isobaric stable isotope tags. In this study, TMT 10-plex reagents were used for labeling of 10 samples (blast samples and controls) and analyzed simultaneously, in order to avoid run-to-run variation. Tryptic digested peptides from brain samples were labeled with TMT 10-plex reagents (Thermo Fisher Scientific) according to manufacturer's instructions. Briefly, the TMT reagents (0.8 mg) were dissolved in 41 μL of anhydrous acetonitrile. Aliquots of 95 μL of the trypsin-digested peptides from cortical tissue samples were incubated with TMT reagents for 1 h at room temperature. The reactions were quenched by 8 μL of 5% hydroxylamine solution and reacted for 15 min. The combined TMT labelled samples were dried under SpeedVac, and then reconstituted by dilute trifluoroacetic acid solution followed by desalting by Oasis HLB 96-well μElutionplate (Waters) prior to LC-MS/MS analysis.
Immobilized metal affinity chromatography (IMAC) was used as an affinity enrichment technique for the preparation of phosphopeptides for mass spectrometric analysis and sequencing as previously described.22 Briefly, Pierce ferric nitrilotriacetate (Fe-NTA) Phosphopeptide Enrichment Kit (Thermo Fisher Scientific #88300) was used following the manufacture's protocol. This kit enables fast and efficient enrichment of phosphorylated peptides. Each spin column included in the kit contains phosphopeptide-specific resin that offers excellent binding and recovery properties for enriching phosphopeptides. Each trypsin-digested dried sample was re-suspended in 200 μL of binding buffer and was added to a Fe-NTA spin column followed by incubation for 20 min with a slow end-over-end rotation at room temperature. Then, the Fe-NTA spin column was centrifuged at 1,000g for 1 min and washed with wash buffer. The eluted phosphopeptides were applied to a Pierce Graphite Spin column (Thermo Fisher Scientific #88302), which enables fast and efficient capture, concentration, desalting, and elution of hydrophilic peptides. All eluents were quantitated and analyzed, and the phosphorylation sites were localized.
We performed proteomic profiling of global and phospho-proteomes using the established workflow for MS analysis (Fig. 1A). LC-MS/MS was performed on a Q Exactive Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Fisher Scientific) coupled with a Dionex ultimate 3000 HPLC system equipped with a nano-ES ion source.23 The TMT labelled peptides were separated on a C18 reverse-phase capillary column (PepMap, 75 μm × 150 mm, Thermo Fisher) with linear gradients of 2–35% acetonitrile in 0.1% formic acid, at a constant flow rate of 300 nL/min for 220 min. The instrument was operated in the positive-ion mode with the electrospray ionization (ESI) spray voltage set at 1.8 kV. A full scan of MS spectra (300–1800 m/z) was acquired in the Q Exactive at a mass resolution of 70,000 at 200 m/z with an automatic gain control (AGC) target of 3e6. Fifteen peptide ions showing the most intense signal from each scan were selected for a higher energy collision-induced dissociation (HCD)-MS/MS analysis (normalized collision energy 32) in the Q Exactive at a mass resolution of 35,000 and AGC value of 1e5. Maximal filling times were 100 ms in full scans and 120 ms (HCD) for the MS/MS scans. Ions with unassigned charge states and singly charged species were rejected. The dynamic exclusion was set to 50 sec and there was a relative mass window of 10 ppm. Data were acquired using ThermoXcalibur 3.0.63. The phosphopeptides were analyzed under the same conditions as mentioned, except that separation was done with linear gradients of 1–30% acetonitrile in 0.1% formic acid, and full scan MS ranging 300–2000 m/z with NCE at 25.
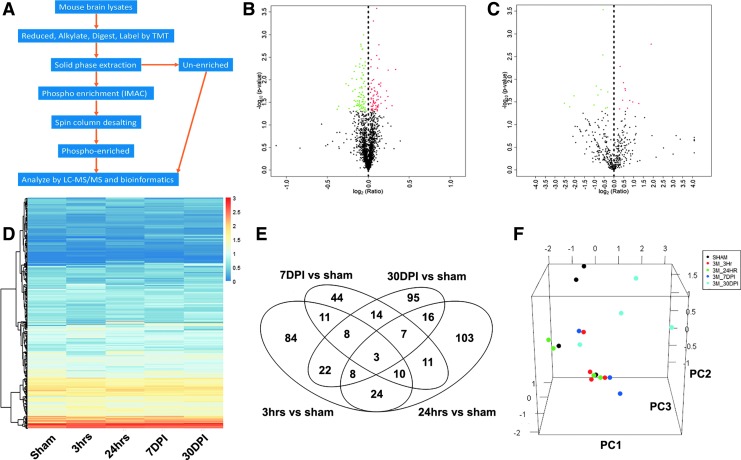
FIG. 1.
Proteomics analyses workflow and identification of differentially expressed global- and phospho- proteins after low-intensity blast (LIB) injury. (A) Workflow for quantitative profiling of mouse brain global and phospho-proteome after blast exposure. (B, C) LIB differentially induced expression of 170 global proteins and 23 phospho-proteins at the 3 h time point, respectively. Volcano plots were generated by R statistical software. Dashed lines indicate the thresholds of log2 (ratio/fold change) and –log10 (p). The identified proteins are displayed as dots in red (upregulation with significant changes), green (downregulation with significant changes), or black (no significant change). (D) Heatmap showing effects of LIB on phospho-protein expression over multiple time points. A large portion of blast-induced up-/downregulated phospho-proteins were dynamically changed along with the time after blast. For visualization, the calculated –log(p value) was shown in red for upregulation and in blue for downregulation. The darkness of the color in each box reflects the absolute value of –log(p value). (E) Venn diagram showing the overlapping differentially expressed global proteins among groups. The overlap portions indicate reoccurring proteins over time. (F) Principal component analysis showed that those differentially expressed global proteins effectively separated blast at 3 h (red), 24 h (green), 7 days post-injury (DPI) (dark blue), and 30 DPI (light blue) from sham control (black). Color image is available online.
Raw data were processed using SEQUEST with Proteome Discoverer (Version 2.1, Thermo Fisher Scientific).24 Data were searched against the mus musculus Universal Protein Resource sequence database (UniProt, August, 2013). The searches were performed with the following parameter settings: trypsin digestion with two missed cleavage allowed; fixed modification, carbamidomethyl of cysteine; variable modification, oxidation of methionine, TMT 10-plex (peptide labeled) for N terminus and Lys; MS tolerance, 5 ppm; MS/MS tolerance, 0.02 Da; false discovery rate (FDR) at peptide and protein levels, <0.01; and required peptide length, ≥6 amino acids. Protein grouping was enabled, meaning if one protein was equal to or completely contained within the set of peptides of another protein, these two proteins were put into the same protein group. At least one unique peptide per protein group was required for identifying proteins. The relative protein abundance ratios (fold changes) between all blast and sham groups were calculated. As previously described, the changes in protein levels were considered significant if fold change was >1.0 (upregulated) or <1.0 (downregulated), and the p value was <0.05 in two independent experiments.20
Analysis of the mitochondrial respiratory chain
Cortical tissues (∼ 20 mg for each sample) were cut into small pieces and homogenized with a Dounce homogenizer in 1 mL of 20 mM phosphate buffered saline (PBS) containing protease and phosphatase inhibitors. The homogenate was diluted fourfold with 20 mM PBS and used to determine the enzymatic activities of Complex I (or nicotinamide adenine dinucleotide [NADH] dehydrogenase), cytochrome C oxidase (COX), and citrate synthase (CS) using an Infinite M200 plate reader (Tecan Group Ltd., Switzerland) as described previously.25 The protein concentration of homogenate was determined using the DC (detergent compatible) protein assay (5000116, Bio-Rad Laboratories, CA). The maximal reaction speed (Vmax) normalized to the amount of protein was used to characterize the activity of each enzyme. For assay of Vmax of Complex I, diluted homogenate was first subjected to three freeze and thaw cycles. Next, 15 mM coenzyme Q1 was added into a cuvette containing 150 μM NADH and 40 μL diluted homogenate. The decrease in NADH absorbance at 340 nm was monitored for 2 min for estimation of Vmax. To measure the Complex I induced NADH oxidation, 10 nM of the Complex I inhibitor rotenone was added and the post-rotenone rate was subtracted from the pre-rotenone rate to yield the rotenone-sensitive NADH oxidation Vmax. For the COX Vmax assay, 25 μM reduced cytochrome C was added into the cuvette containing 20 mM PBS, 5 μL diluted homogenate, and 0.2 mg/mL dodecyl maltoside, to make the total volume of 1 mL. The oxidation of the reduced cytochrome C was monitored for 2 min at 550 nm and the pseudo first-order rate constant was calculated as Vmax. For assay of citrate synthase Vmax activity, the substrates of CS, 100 μM oxaloacetate, and 50 μM acetyl coenzyme A (CoA), were added into the cuvette containing 100 mM Tris-HCl (pH 8.0), 5 μL homogenate, and 100 μM 5,5'-dithiobis-(2-nitrobenzoic acid) (DTNB) (1 mL total volume) and then the formation of 5-mercapto-2-nitrobenzoic acid was monitored for 2 min at 412 nm, as previously described.25
Western blot analysis
Protein samples were added to 4 × SDS sample buffer with 1% 2-mercaptoethanol and resolved on 12%, 1.5mm SDS- polyacrylamide gel electrophoresis (PAGE) gels as previously described.26 Then, proteins in SDS-PAGE gel were transferred to a nitrocellulose membrane. The membrane was first incubated in PBS plus 0.1% Tween-20 (PBST) (pH 7.4) containing 5% non-fat milk for 1 h at room temperature and then incubated with primary antibody, including COX IV (ab16056 Abcam, 1:2000), MnSOD (ab13534 Abcam, 1:3000), GPX4 (ab125066 Abcam, 1:2000), Drp1 (ab184247 Abcam, 1:1000), Fis1 (10956-1-AP Proteintech, 1:1000), OPA1 (cst80471 Cell Signaling Technology, 1:1000), Mitofusin 2 (cst9482 Cell Signaling Technology, 1:1000), VDAC1 (sc390996 Santa Cruz, 1:1000), OXPHOS (containing a cocktail of complex I NDUFB8, complex II SDHB, complex III UQCRC2, complex IV MTCO1, and complex V ATP5A) (45-8099 ThermoFisher Scientific, 1:500), PINK1 (sc33796 Santa Cruz, 1:500), Parkin (cst4211 Cell Signaling Technology, 1:1000), ATG12 (cst4180 Cell Signaling Technology, 1:1000), LC3B (cst2775 Cell Signaling Technology, 1:1000), ULK1 (cst8054 Cell Signaling Technology, 1:1000), and Actin (A3854 Sigma-Aldrich, 1:15000) at 4°C overnight. After washing with PBST three times, the membrane was incubated with corresponding secondary antibody (anti-mouse IgG, A0168 Sigma-Aldrich, 1:2000, anti-rabbit IgG, A0545 Sigma-Aldrich, 1:2000) in PBST for 1 h at room temperature. Immunoreactive bands were detected using Amersham ECL Prime Western Blotting Detection Reagent and LAS-4000 Fujifilm imaging system.
Bioinformatics and statistical analysis
All quantitative proteomics data were normalized across all proteins within each 10-plex batch to eliminate the batch effect. Heatmap is generated by R package “pheatmap.” Welch's test and t-distribution were performed with R function “t.test.” Principal component analysis (PCA) plots were generated with R package “prcomp” and the first three components were plotted with R package “rgl.” Numbers of significantly changed protein levels among different groups after blast (3 h, 24 h, 7 DPI, and 30 DPI) versus sham control, were shown by Venn diagram with R package “VennDiagram.”27 Pathway and gene ontology (GO) enrichment analysis was performed by R package “clusterProfiler.” Proteins that had significant change in levels were enriched with Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database with function “enrichKEGG,” and cellular component enrichment analysis was performed with function “enrichGO.”28 The circularly composition analysis was performed with GO plot R packages as previously described.29 All cutoffs for p value were set to 0.05 for enrichment analysis.
Machine learning-based bioinformatic proteome analysis QIAGEN's Ingenuity Pathway Analysis (IPA, QIAGEN Redwood City, www.qiagen.com/ingenuity) was used to examine protein functions and molecular pathways associated with blast injury, as well as to predict protein–protein interaction networks, as previously described.30 Z score for IPA analysis was a statistical measure of the match between expected relationship direction and observed protein expression. Statistical analyses were performed with GraphPad Prism (version 6 for Windows, GraphPad Software). All data were expressed as means ± standard error of the mean (SEM). Statistical comparisons were made by student's t test for two groups, and a one way ANOVA test followed by Tukey test for multiple comparisons. Differences were considered significant at p < 0.05 for all analyses, with *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Results
Quantitative proteomic analysis of cortical global and phospho-proteome profiles after primary LIB
In order to provide a comprehensive understanding of the molecular events associated with dynamic changes occurring after primary LIB exposure, profiles of global and phospho-proteomes in mouse cortex with or without blast injury were analyzed utilizing the TMT 10-plex tagging approach. For global proteome analysis, with criteria of FDR <1% for both peptide and protein identification, we identified 2216 proteins in brain tissues after a 46.6 kPa blast exposure (Table S1). Among these, there were 170, 182, 108, and 173 differentially expressed proteins in 3 h, 24 h, 7 DPI, and 30 DPI groups as compared wth sham control, respectively (Fig. 1B, Fig. S1, and Table S2). Using the phospho-proteome analysis by IMAC pull-down of phosphopeptides along with LC-MS/MS, we further identified 459 phosphorylated proteins (Table S3). Specifically, there were 23, 15, 36, and 41 differentially expressed phosphorylated proteins at 3 h, 24 h, 7 DPI, and 30 DPI after the blast, respectively (Fig. 1C and 1D, Fig. S1, and Table S4). The Venn diagrams (Fig. 1E and Fig. S2) illustrated the changes in expression levels of individual proteins, especially some proteins reoccurring or unique at various time points in the context of a primary LIB exposure. Expression levels of some proteins, such as NADH ubiquinone oxidoreductase subunits (NDUFA), inner membrane mitochondrial protein (IMMT), and voltage dependent anion channel 1 (VDAC1), changed at 3 h after blast and remained altered up to 30 DPI (see online supplementary material at http://www.liebertpub.com).
To delineate relationships between sham and blast proteomes at different time points and to visualize the convergence and/or divergence among the proteomes, we performed PCA on the normalized expression values of the differentially expressed proteins. Results demonstrated consistency of these effects both in global and phospho-proteomes (Fig. 1F, Fig. S3, and Table S5). Among the PCA components, the blast groups were distinct from the sham control group. Nevertheless, various degrees of differences were found in individual blast groups, supporting the notion that even a single LIB exposure can drive significant impacts on protein expressions at different time points (see online supplementary material at http://www.liebertpub.com).
GO annotation and canonical pathways associated with mitochondrial dysfunction after primary LIB
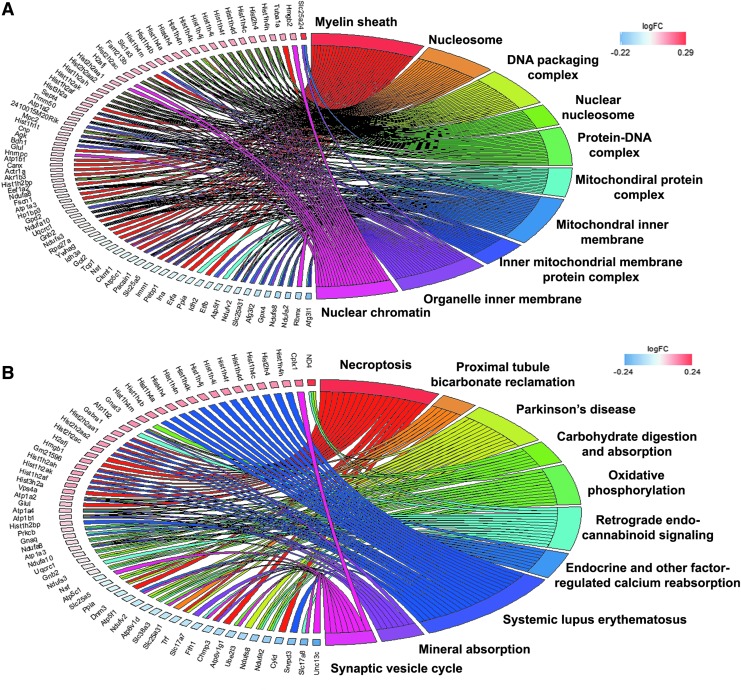
In our previous studies, we observed persistent changes in brain mitochondria ultrastructure up to 30 DPI after LIB exposure.5 In order to understand effects of primary LIB on dynamic molecular events, we mapped both global and phospho-proteome identities to the GO cellular components, canonical signaling pathways, and regulatory networks potentially affected by blast at different time points, by applying KEGG pathways annotation and machine learning based IPA analysis.30 In the cellular components of KEGG GO, the top 10 represented categories include mitochondria (mitochondrial protein complex and mitochondrial inner membrane), myelin sheath, actin cytoskeleton, and synapse (pre-synapse, post-synapse, post-synaptic density, neuron to neuron synapse, and asymmetric synapse) (Fig. 2A). Figures S4–S7 show other top 10 shared or distinct cellular component details, when comparing among blast groups at different time points in global and phospho-proteomes (see online supplementary material at http://www.liebertpub.com).
FIG. 2.
Cellular components and signaling pathways enriched in global proteome at 3 h after low-intensity blast (LIB) injury. (A) Global proteins enriched in the top shared cellular components. (B) Global proteins enriched in the top shared functional pathways. Each cluster of cellular components and canonical pathways in the plot is assigned a unique color. Each connection between a protein and the cellular component or canonical pathway represents the fold change. Color image is available online.
KEGG annotation and IPA pathway analysis showed that the top 10 canonical pathways in blast affected global and phospho-proteomes were categorized into mitochondrial dysfunction, oxidative stress (included oxidative phosphorylation, fatty acid degradation, glutathione metabolism, chemokine and mechanistic target of rapamycin (mTOR) signaling pathways, nuclear factor (erythroid-derived 2)-like 2 (Nrf2) mediated oxidative stress response, synaptic dysfunction, axonal guidance signaling, regulation of cytoskeleton and gap junction, and neurodegeneration (related to Parkinson's diseases and Alzheimer's disease) (Fig. 2B). Detailed data about signaling pathways commonly shared among blast groups at different time points are depicted in Figures S6–S9 and Tables S6 and S7. The dynamics of involved signaling pathways, including oxidative stress and mitochondrial dysfunction, myelin sheath and axonal injury, and synaptic dysregulation pathways, changed along with different blast time points in both global and phospho-proteomes (Fig. 3A and Table S8). From these signaling pathways, we observed various degrees of temporal changes in differentially expressed protein levels from 3 h up to 30 DPI related to mitochondrial dysfunction, axonal injury, and synaptic dysregulation (Tables 1–3 and Tables S9–S11). Particularly, these changes included multiple proteins associated with oxidative stress and mitochondrial dysfunction pathways. As shown in Table 1 these included NDUFA, ubiquinol-cytochrome c reductase core protein 2 (UQCRC2), adenosine triphosphate (ATP) synthase (ATP synthase peripheral stalk-membrane subunit B [ATP5PB] and ATP synthase peripheral stalk subunit D [ATP5PD]) (oxidative phosphorylation), mitochondrial isocitrate dehydrogenase (IDH2) (peroxisome), and mitofusin 2 (MFN2) (fission-fusion and mitophagy) (see online supplementary material at http://www.liebertpub.com).
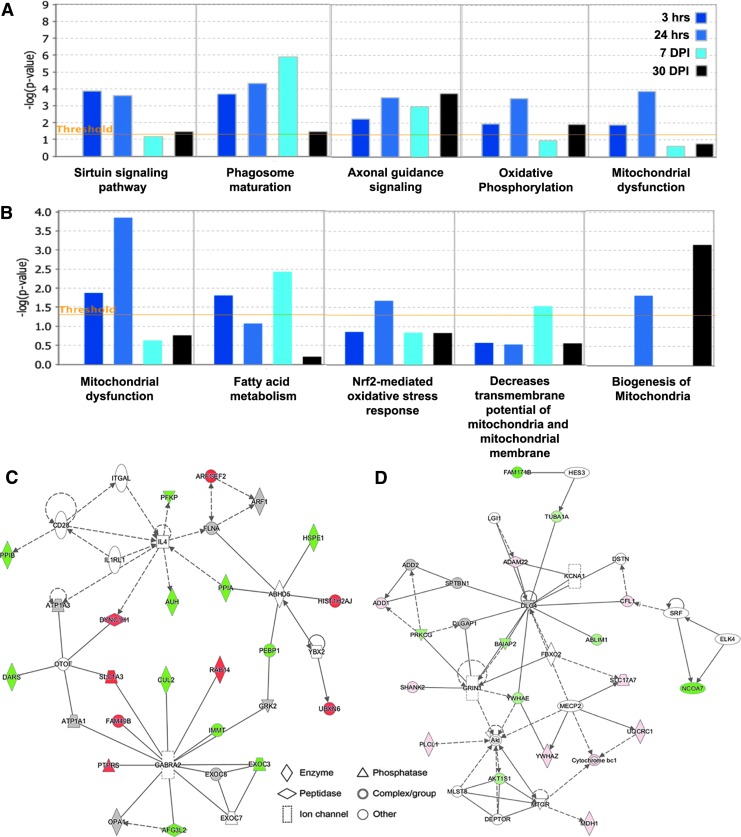
FIG. 3.
Low-intensity blast (LIB) effects at different times on alterations of canonical pathways, toxic annotation, and disease/function networks. (A) Top five canonical pathways predicted by an IPA analysis of the differentially expressed global and phospho-proteins affected by the LIB exposure at different time points (color coded). (B) Top five toxic lists predicted by IPA. The canonical pathways and toxic list annotations were ranked according to the -log (p value). A ratio (height) indicates the number of proteins that were differentially expressed in each pathway or list over the total number of proteins in that specific pathway. (C) The top disease/function network in global proteome is associated with cellular development, cellular growth and proliferation, cellular movement, and cellular assembly and organization corresponding to the LIB. (D) The top disease/function network in phospho-proteome is associated with neurological disease, organismal injury and abnormalities, cell-to-cell signaling and interaction, and cell morphology corresponding to the LIB. The identified genes involved in the networks are displayed in red (upregulation) and green (downregulation). The color intensity indicates the degree of regulation. Solid lines in the network imply direct interactions between genes, and dashed lines indicate indirect interactions. Geometric shapes represent different general functional families of gene regulation (diamond for enzyme, oval for transcription regulator, trapezoid for transporter, inverted triangle for kinase, double circle for complex/group, and circle for others). Color image is available online.
Table 1.
Canonical Pathways Annotation from Global Proteome Associated with Oxidative Stress and Mitochondrial Dysfunction after LIB Injury
| Canonical pathway | Differentially expressed proteins | ||||
|---|---|---|---|---|---|
| ID | Name | 3 h | 24 h | 7 DPI | 30 DPI |
| mmu00190 | Oxidative phosphorylation | NDUFA2, NDUFS8 | UQCRC1, NDUFS3, NDUFA2, UQCRC2, ATP5H, ATP5F1, PPA1 | ATP5F1 | ATP5C1, NDUFA2 |
| 2.90E-05 | 5.81E-04 | 4.93E-05 | 3.52E-03 | ||
| mmu04137 | Mitophagy | NA | Ubb | MFN2, RRAS2 | NA |
| 1.13E-01 | 8.50E-01 | 8.16E-01 | 4.02E-01 | ||
| mmu04140 | Autophagy | NA | LAMP1, RRAGB, PRKCQ, MAPK3, CAMKK2 | RRAS2 | RRAGB |
| 6.90E-02 | 3.01E-02 | 8.41E-01 | 5.05E-01 | ||
| mmu04142 | Lysosome | NA | LAMP1, AP3B2 | AP3B2 | CLTB |
| 4.89E-01 | 5.01E-01 | 4.14E-01 | 5.09E-01 | ||
| mmu04145 | Phagosome | TUBA1A, DYNC1H1, Tuba3a | LAMP1, TUBA1B, RAB5A, RAB5C | DYNC1H1, RAB5C | NA |
| 6.89E-03 | 1.98E-02 | 2.52E-02 | 4.30E-05 | ||
| mmu04146 | Peroxisome | ECI2, IDH2 | ECI2 | ECI2 | IDH2 |
| 1.52E-01 | 1.58E-01 | 2.88E-01 | 2.88E-01 | ||
| mmu04150 | mTOR signaling pathway | NA | RRAGB, MAPK3 | NA | RRAGB |
| 1.47E-01 | 7.60E-02 | 3.60E-01 | 2.27E-01 | ||
| mmu04210 | Apoptosis | TUBA1A, Tuba3a | TUBA1B, MAPK3 | SPTA1 | NA |
| 6.06E-02 | 4.25E-01 | 3.33E-01 | 6.13E-01 | ||
p values for each specific canonical pathway at different time points are presented directly under each pathway at different time points. NA, no differentially expressed protein is identified.
LIB, low-intensity blast; DPI, days post-injury; mTOR, mechanistic target of rapamycin.
Table 2.
Canonical Pathways Annotation from Global Proteome Associated with Myelin Sheath and Axonal Injury after LIB Injury
| Canonical pathway | Differentially expressed proteins | ||||
|---|---|---|---|---|---|
| ID | Name | 3 h | 24 h | 7 DPI | 30 DPI |
| mmu04360 | Axon guidance | CDK5 | CDK5, GNAI3, MAPK3, NTNG1 | NA | NA |
| 3.55E-01 | 3.68E-01 | 9.37E-01 | 9.75E-01 | ||
| mmu04810 | Regulation of actin cytoskeleton | NCKAP1 | MYH9, WASL, NCKAP1, MAPK3, WASF2 | RRAS2 | NCKAP1 |
| 2.86E-02 | 2.03E-05 | 5.94E-01 | 4.25E-01 | ||
| mmu04071 | Sphingolipid signaling pathway | NA | GNAI3, MAPK3 | GNAQ | GNAQ |
| 2.86E-02 | 2.03E-05 | 5.94E-01 | 4.25E-01 | ||
| mmu04514 | Cell adhesion molecules (CAMs) | NA | NRXN3, NEGR1, NTNG1 | NA | NA |
| 6.38E-01 | 6.49E-01 | 1.00E+00 | 8.41E-01 | ||
LIB, low-intensity blast; DPI, days post-injury.
Table 3.
Canonical Pathways Annotation from Global Proteome Associated with Synaptic Dysregulation after LIB Injury
| Canonical pathway | Differentially expressed proteins | ||||
|---|---|---|---|---|---|
| ID | Name | 3 h | 24 h | 7 DPI | 30 DPI |
| mmu04721 | Synaptic vesicle cycle | NA | NA | VAMP2 | CLTB |
| 6.24E-03 | 1.44E-03 | 1.24E-04 | 3.18E-05 | ||
| mmu04727 | GABAergic synapse | GNB2, GLUL | GNB2, GNAI3, GLUL, SLC38A3 | GNB2 | GLUL, SLC38A3 |
| 1.97E-03 | 1.88E-05 | 1.71E-02 | 2.95E-04 | ||
| mmu04724 | Glutamatergic synapse | GNB2, GLUL, SHANK3 | GNB2, GNAI3, GLUL, MAPK3, SLC38A3, SHANK3 | GNB2, GNAQ, GRK2, SHANK3 | GNAQ, GLUL, SLC38A3, GRK2 |
| 1.78E-05 | 9.48E-05 | 1.48E-04 | 9.88E-03 | ||
| mmu04720 | Long-term potentiation | GNB2, GLUL, SHANK3 | GNB2, GNAI3, GLUL, MAPK3, SLC38A3, SHANK3 | GNB2, GNAQ, GRK2, SHANK3 | GNAQ, GLUL, SLC38A3, GRK2 |
| 4.46E-02 | 1.39E-02 | 2.56E-01 | 1.76E-01 | ||
| mmu04730 | Long-term depression | NA | GNAI3, MAPK3 | GNAQ | GNAQ |
| 1.08E-02 | 1.16E-02 | 2.38E-01 | 7.55E-01 | ||
LIB, low-intensity blast; DPI, days post-injury.
Based upon the analysis of canonical pathways, we further characterized the blast effects relating to brain injuries at the molecular level by IPA to predict the toxic annotation. Bioinformatic analysis indicated that this level of LIB exposure caused mitochondrial dysfunction (associated with decreased depolarization and transmembrane potential of mitochondria and mitochondrial membrane, biogenesis of mitochondria) and damage caused by oxidative stress (involved with fatty acid metabolism and Nrf2 mediated oxidative stress response). These findings were reflected by up- and downregulation of specific mitochondrial proteins, including glutathione peroxidase 4 (GPX4), NDUFA, ATP5PB and ATP5PD, UQCRC2, IMMT, and protein kinase C (PRKC); and by upregulation of phosphorylated proteins, such as VDAC1, stathmin 1 (STMN1), and ATPase sarcoplasmic/endoplasmic reticulum calcium transporting 2 (ATP2A2) (Fig. 3B). The differential expression of multiple key mitochondrial proteins related to oxidative phosphorylation, respiration, and electron transport chain (ETC) function demonstrates that LIB exposure causes bioenergetic failure.
Annotation of disease and functional networks regulated after primary LIB
To further investigate functional links among the differentially expressed proteins as well as related biological processes in context of the LIB exposure, IPA was used to map the global and phospho-proteome identities to reveal signaling pathways and regulatory networks. The top disease/function networks induced by the blast injury in global proteomes were involved with cellular development, cellular growth and proliferation, cellular movement, and cellular assembly and organization (Fig. 3C). This network was associated with downregulation of IMMT, mitochondrial dynamin-related GTPase (OPA1), cullin-2 (CUL2) (core component of E3 ubiquitin-protein ligase complexes), synaptosomal-associated protein 25 (SNAP25); and upregulation of mitogen-activated protein (MAP) kinase-activated protein kinase 3 (MAPK3), peroxiredoxin-6 (PRDX6) (redox activity) at various time points after blast. By contrast, the top networks, associated with blast signaled downregulation of leucine-rich repeat-containing protein 7 (LRRC7) (required for normal synaptic spine architecture and function), tropomodulin-2 (TMOD2) (regulating membrane skeleton), and ubiquitin-like modifier-activating enzyme 1 (UBA1); upregulated STMN1 (destabilizing microtubules) and VDAC1, related to phospho-proteome include neurological disease, organismal injury and abnormalities, cell-to-cell signaling and interaction, and cell morphology (Fig. 3D). Importantly, consistent with the canonical pathway findings, the disease/functional network data further confirm mitochondrial dysfunction after LIB-induced brain injury.
Primary LIB-induced oxidative stress associated with mitochondrial dysfunction
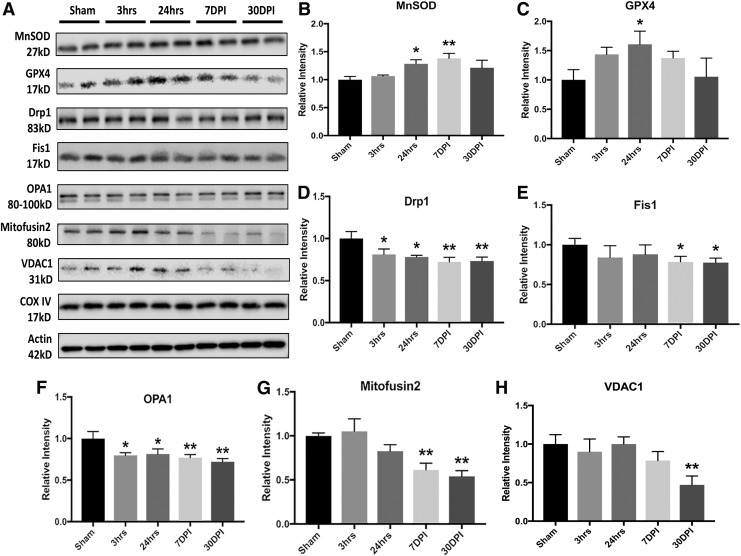
Driven by quantitative proteomics and machine learning based bioinformatics analysis, we investigated specific mitochondrial related molecular and biochemical events over time in mice exposed to primary LIB. Mitochondrial superoxide dismutase 2 (MnSOD), a well-documented mitochondrial oxidative stress marker, was progressively and significantly elevated at 24 h and 7 DPI, with an increased trend at 30 DPI after blast exposure (Fig. 4A and B and by immunostaining shown in Fig. S10A and B)(see online supplementary material at http://www.liebertpub.com). Additionally, we observed that GPX4 level was significantly increased at 24 h after LIB injury (Fig. 4C). GPX4 expression returned to sham control level at 30 DPI. These results suggest that primary LIB blast exposure increases levels of oxidative stress associated with mitochondrial dysfunction.
FIG. 4.
Low-intensity blast (LIB)-induced mitochondrial dysfunctions associated with oxidative stress and fission-fusion dynamics. (A) Western blot and analysis of mitochondrial markers associated with oxidative stress and fission-fusion dynamics after LIB. (B) Significant increase of mitochondrial superoxide dismutase 2 (MnSOD) levels was found at 24 h (p < 0.05) and 7 days post-injury (DPI) (p < 0.01) after blast. (C) Glutathione peroxidase 4 (GPX4) level was significantly elevated at 24 h (p < 0.05) after blast. (D) Dynamin-1-like protein (Drp1) expression was significantly decreased at all time points after blast as compared with sham (p < 0.05). (E) Fis1 expression was significantly decreased at 7 and 30 DPI (p < 0.05). (F) OPA1 level was significantly decreased at all time points after blast (p < 0.05). (G) Significant decrease of Mitofusin2 levels was observed at 7 and 30 DPI (p < 0.01). (H) Significant decrease of voltage dependent anion channel 1 (VDAC1) was identified at 30 DPI (p < 0.01). n = 4 for each individual group for comparison. All data are expressed as mean ± standard error of the mean (SEM) and were analyzed by one way analysis of variance (ANOVA) test followed by Tukey test for multiple comparisons. Differences were considered significant at p < 0.05 for all analyses, *p < 0.05; **p < 0.01.
Primary LIB altered mitochondrial dynamics and bioenergetics, and compromised mitophagy
Next, we investigated molecular changes after LIB associated with mitochondrial fission-fusion dynamics, mitophagy, and autophagy. In this experiment, corroborated with proteomics identification, changes in fission and fusion activities were evidenced by significantly decreased dynamin-1-like protein (Drp1, mitochondrial fission), Fis1 (mitochondrial fission and recruitment/association of Drp1), OPA1 (inner-membrane fusion), and mitofusin 2 (outer-membrane fusion) levels mainly at 7 and 30 DPI (Fig. 4D–G and by immunostaining shown in Fig. S10C–E) (see online supplementary material at http://www.liebertpub.com). Levels of VDAC1, associated with mitochondrial volume regulation, was also reduced at 30 DPI (Fig. 4H).
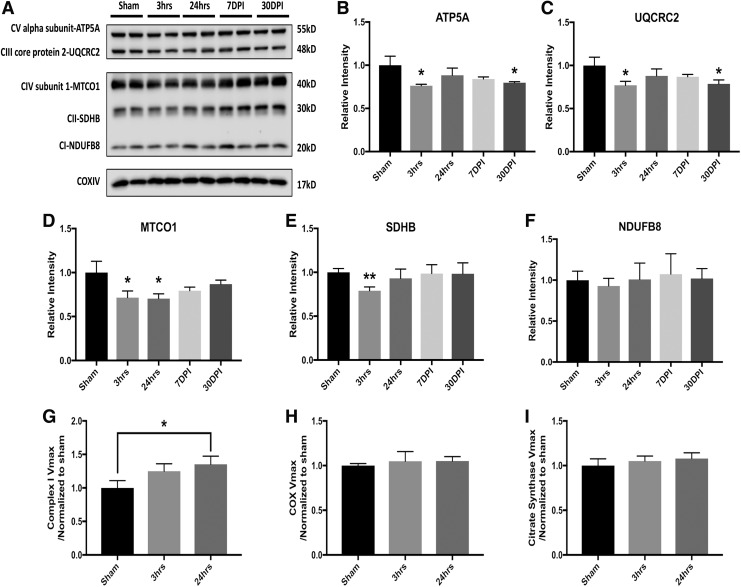
We further identified mitochondrial bioenergetic failure after LIB exposure by assessing mitochondrial oxidative phosphorylation (OXPHOS) essential for ATP production, and measuring mitochondrial respiration chain enzymatic activity. We found significantly decreased levels of the complex V ATP5A and complex III UQCRC2 subunits at 3 h and 30 DPI (Fig. 5A–C). The results validated the proteomics identifications. The Complex IV MTCO1 subunit was also reduced at 3 and 24 h after blast (Fig. 5D). Additionally, we observed that complex II succinate dehydrogenase subunit B (SDHB) subunit expression significantly decreased at 24 h (Fig. 5E). Although no significant change was identified in the complex I NDUFB8 subunit protein (Fig. 5F), the blast at 24 h post- injury led to a significant increase of complex I activity (Fig. 5G). Activity of COX and CS (a matrix-soluble enzyme whose levels tend to reflect mitochondrial mass) showed similar trends in mice exposed to blast as compared with sham (Fig. 5H and I). Decreased levels of multiple OXPHOS subunit proteins and increased complex I Vmax activity suggest that LIB alters mitochondria, which secondarily compensate for either LIB-induced dysfunction or for increased energy needs.
FIG. 5.
Low-intensity blast (LIB)-induced mitochondrial oxidative phosphorylation and bioenergetic failure. (A) Western blot and analysis of mitochondrial markers associated with oxidative phosphorylation after LIB. (B) Significant decreases of complex V adenosine triphosphate (ATP) synthase peripheral stalk-membrane subunit A (ATP5A) levels were found at 3 h (p < 0.05) and 30 days post-injury (DPI) (p < 0.05) after blast. (C) Complex III ubiquinol-cytochrome c reductase core protein 2 (UQCRC2) levels were significantly reduced at 3 h (p < 0.05) and 30 DPI (p < 0.05) after blast. (D) Complex IV MTCO1 expressions were significantly decreased at 3 and 24 h after blast as compared with sham (p < 0.05). (E) Complex II succinate dehydrogenase subunit B (SDHB) expression was significantly decreased at 24 h (p < 0.01). (F) No significant change was identified for complex I NDUFB8 protein expression level after blast. (G) However, significant elevated complex I enzyme activity was found at 24 h after blast (p < 0.05). (H–I) Cytochrome C oxidase (COX) and citrate synthase (CS) activities assessed after blast injury (no significant differences among comparisons). n = 4 for each individual group for comparison. All data are expressed as mean ± standard error of the mean (SEM) and were analyzed by one way analysis of variance (ANOVA) test followed by Tukey test for multiple comparisons. Differences were considered significant at p < 0.05 for all analyses, *p < 0.05; **p < 0.01.
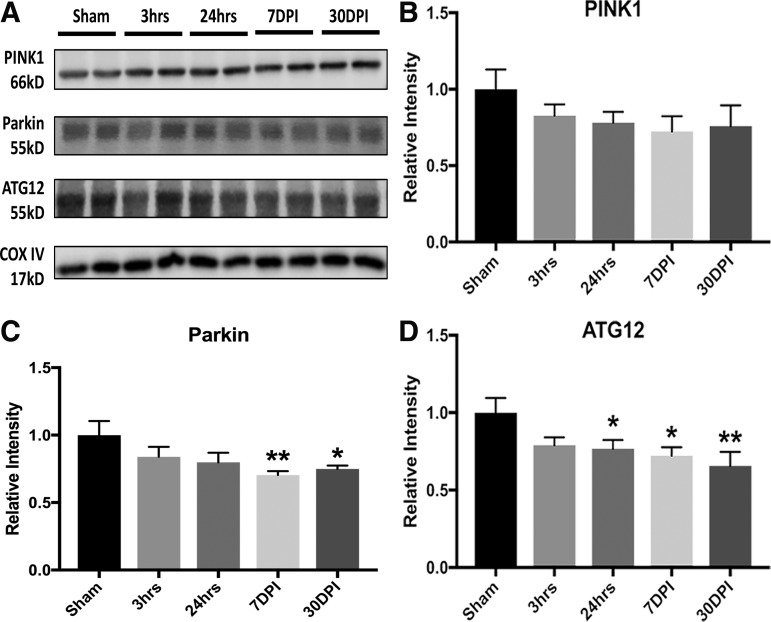
In order to further examine changes in markers related to mitophagy and autophagy, results showed significantly decreased levels of parkin, and with a trend for phosphatase and tensin homolog (PTEN)-induced kinase 1 (PINK1) at 7 and 30 DPI (Fig. 6A–C). These changes suggest efforts to eliminate damaged mitochondria. Significant decreased levels of autophagy-related protein 12 (ATG12, involved in the processes of autophagic initiation and elongation to engulf cytoplasmic components), with lower trends for autophagy activating kinase 1 (ULK1)/ATG1 and microtubule-associated protein 1B-light chain 3 (LC3B), from 24 h to 30 DPI, suggested impaired autophagy by reducing the maturation of functional autophagosome and subsequent attenuation of autophagic flux after blast injury (Fig. 6D). Taken together, these results are in agreement with proteomics and bioinformatics data demonstrating primary LIB-induced mitochondrial dysfunction, particularly involved with increased oxidative stress, impaired fission-fusion dynamics, diminished mitophagy, and acute compensatory respiration activity.
FIG. 6.
Low-intensity blast (LIB)-induced mitochondrial dysfunctions associated with mitophagy and autophagy. (A) Western blot and analysis of mitochondrial markers associated with mitophagy and autophagy after LIB. (B) Phosphatase and tensin homolog (PTEN)-induced kinase 1 (PINK1) expression trended lower after blast as compared with sham. (C) Parkin expression was significantly decreased at 7 (p < 0.01) and 30 days post-injury (DPI) (p < 0.05). (D) Significant decrease of autophagy-related protein 12 (ATG12) levels, with a trend toward microtubule-associated protein 1B-light chain 3 (LC3B), was found at 24 h (p < 0.05), and 7 (p < 0.05) and 30 days post-injury (DPI) (p < 0.01). n = 4 for each individual group for comparison. All data are expressed as mean ± standard error of the mean (SEM) and were analyzed by one way analysis of variance (ANOVA) test followed by Tukey test for multiple comparisons. Differences were considered significant at p < 0.05 for all analyses, *p < 0.05; **p < 0.01.
Discussion
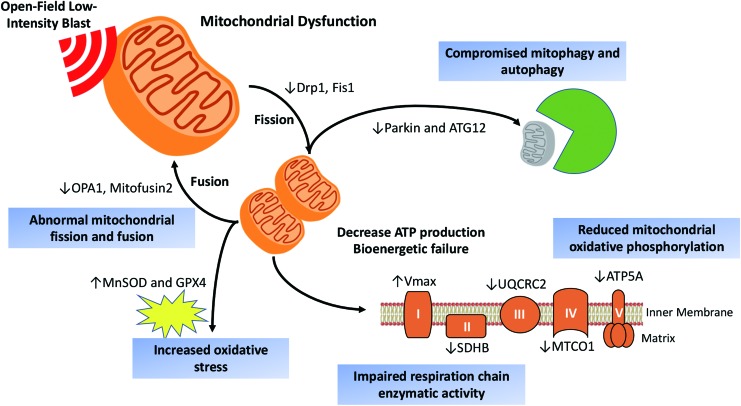
Explosive blast-induced mTBI, a “signature wound” of the recent military conflicts, remains a serious concern. Although experimental animal studies of high intensity blast exposures with >100 kPa peak overpressure have provided insights into neurological damage caused by moderate or severe TBI, the pathobiology and underlying mechanisms of mild brain injury by primary blast wave require further investigation. In this context, we have characterized details of mitochondrial dysfunction involved with increased oxidative stress, impaired fission-fusion dynamics, diminished mitophagy, decreased oxidative phosphorylation, and acute compensated respiration activity by using quantitative proteomic analyses of the effects of primary LIB up to 30 days after exposure, as depicted by a scheme in Figure 7. We used this military-relevant mouse model of LIB brain injury with a magnitude of 46.6 kPa peak overpressure and 8.7 PSI × ms maximum impulse.5,6 The temporal profiles of global and phospho- proteome changes at acute (3 and 24 h), subacute (7 DPI), and longer term (30 DPI) time points after LIB exposure exhibited striking sequential changes, some of which persisted up to 30 DPI.
FIG. 7.
A scheme depicting effects of open-field low-intensity primary blast on mitochondrial dysfunction and canonical pathways. Quantitative proteomics and biochemical analysis reveal the mitochondrial dysfunction following low-intensity primary blast. We identified blast-induced mitochondrial damages associated with impaired fission-fusion dynamics, diminished mitophagy, increased oxidative stress, decreased oxidative phosphorylation, and compensated respiration activity. Color image is available online.
mTBI usually shows no visible damage using routine imaging techniques. Understanding molecular and ultrastructural mechanisms of LIB-induced brain injury requires better identification of specific biomarker surrogates.16,21 Proteins, currently including UCH-L1,18 neuron-specific enolase (NSE),31 gangliosides/ceramides,32 and S100 calcium-binding protein B (S100B),33 have been identified as biomarkers for clinical research in patients with severe impact-induced TBI. However, the underlying pathobiology of neurological deficits caused by LIB remains unclear. In this study, we examined the effects of primary LIB on temporal protein changes in both global and phospho-proteomes by comparing the proteins associated with the cellular components and functional pathways. We first identified a total of 2216 proteins and 459 phosphorylated proteins in brain cortical tissues that were affected by primary blast. Among these, temporal profiles of differentially expressed proteins were characterized at 3 h, 24 h, 7 DPI, and 30 DPI after LIB exposure (Fig. 1). Changes in specific protein and phosphorylated protein targets at 3 h after blast remained significantly altered up to 30 DPI, such as NDUFA, IMMT, and VDAC1. Some differentially expressed proteins were significantly enriched in the shared cellular components of mitochondria, myelin sheath, actin cytoskeleton, and synapse, as well as in the shared canonical pathways related to mitochondrial dysfunction, axonal/cytoskeletal dysregulation, and synaptic abnormalities in corresponding to their cellular components, and neurodegeneration (Fig. 2). Distinct proteins with differential expression in the cellular components and signaling pathways were also identified. These findings are consistent with other reports suggesting compromised biological processes, including myelination and cytoskeletal damage caused by moderate or severe TBI.34,35 In agreement with our previous observation on LIB-induced damages in mitochondrial ultrastructure,5 results here uncovered changes in mitochondrial injury pathways (Fig. 3A and B and Table 1). By regulating proteins including GPX4, UQCRC2, IMMT, VDAC1, and STMN1, in both global and phospho-proteome levels, these signal pathways involve membrane depolarization and biogenesis of mitochondria, as well as fatty acid metabolism and Nrf2-mediated response related to oxidative stress. Further, the IPA disease/function network analysis showed disordered cell-to-cell signaling suggesting interactional damage by mitochondrial and other proteins, including IMMT, OPA1, SNAP25, and STMN1 (Fig. 3C and D). Most importantly, a group of identified mitochondrial related proteins from the proteomic analysis (UQCRC2, IMMT, VDAC1, OPA1) appear to play key roles in regulating mitochondrial integrity (fission and fusion), mitophagy, oxidative phosphorylation, and respiration.36,37 The global and phospho-proteomes suggest that mitochondrial dysfunction peaks between 24 h and 7 DPI (Fig. 3), but some mitochondrial proteins still are persistently altered up to 30 DPI.
Mitochondrial dysfunction and bioenergetic failure have been implicated in moderate to severe TBI and in certain neurodegenerative diseases.38–40 Impairment of mitochondrial bioenergetics links to neuronal excitotoxicity, disruption of Ca2+ homeostasis, production of reactive oxygen species (ROS), and ATP depletion.39 Upregulation of antioxidant MnSOD in TBI study with blast exposure >100 kPa has been reported previously.41 SODs allow conversion of free radicals to hydrogen peroxide and GPX. The present study demonstrated increased MnSOD and GPX4 levels, suggesting that LIB may elicit an early-occurring compensatory upregulation of mitochondrial antioxidant defenses target (Fig. 7).
We have characterized the molecular effects of primary LIB by regulating various mitochondrial proteins on its functions including fusion-fission dynamics, oxidative stress, and bioenergetic infrastructure. Of interest is the mitochondrial fission protein Drp1, which mediates mitochondrial membrane dynamics and is involved with ROS/reactive nitrogen species (RNS) and impaired bioenergetics-induced synaptic damage and subsequent neuronal loss.42,43 Fis1 is involved with mitochondrial fission and recruitment/association of Drp1.42 Conversely, mitochondrial fusion is regulated by mitofusin 2 (outer-membrane fusion) and OPA1 (inner-membrane fusion).44 We first looked at the mitochondrial fusion and fission activities. We found that levels of Drp1, Fis1, OPA1, and mitofusin 2 were significantly reduced to various degrees up to 30 DPI after blast (Fig. 4D–G and Fig. S10C–E) (see online supplementary material at http://www.liebertpub.com). For outer-membrane functions, we identified a significantly reduced level of VDAC1, which is associated with mitochondrial volume regulation (Fig. 4H). Both mitochondrial fission and fusion play a role in helping to traffic abnormal mitochondria toward autophagic processing and in maintaining normal mitochondrial numbers throughout the neuron. Impaired fission and fusion reportedly interferes with the cell's ability to target dysfunctional mitochondria for autophagic removal.45 Mitochondrial fission-fusion dynamics have been implicated in neurodegeneration and synaptic plasticity.46 Our findings on loss of mitochondrial fission and fusion proteins and consequently compromised activities suggest a causal role in the pathogenesis of LIB brain injury and possibly initiation of later neurodegeneration (Fig. 7).
Structurally damaged mitochondria are most likely bioenergetically impaired.42 In the nervous system, mitochondria provide a large proportion of ATP required for neuronal function. The oxidative phosphorylation system OXPHOS, embedded in the inner membrane, contains the respiratory chain that utilizes substrates derived from glucose, fatty acids, and amino acids to produce ATP. The electron transport enzymes of the respiratory chain consists of four protein complexes (complexes I–IV), three of which (I, III, and IV) couple electron transfer to proton pumping across the mitochondrial inner membrane to generate a transmembrane electrochemical potential.46 Inefficient electron transfer through complexes I–IV interferes with energy metabolism and produces of excessive ROS with toxic effects. OXPHOS dysfunction occurs in many neurodegenerative diseases.47 However, the relationship between OXPHOS and blast injury remain to be fully elucidated.48 Our previous transmission electron microscopy (TEM) study demonstrated extensive mitochondrial abnormalities with swollen and clear cristae morphology.5 In this study, we have identified mitochondrial respiration chain enzyme alterations after LIB exposure by measuring the Vmaxes and subunit levels of key respiratory chain enzymes including complex I and COX, which enzymes contain mitochondrial DNA (mtDNA)-encoded subunits (Fig. 5). We observed that multiple OXPHOS protein levels decreased, which could affect cell respiration and ATP production. OXPHOS changes likely also contributed to excessive ROS production (evidenced by the findings of MnSOD and GPX4). Still, it remains to be conclusively determined whether the OXPHOS and respiration enzyme activity are the cause of LIB-induced injury or if they are affected by downstream defects in mitochondrial dynamics, structure, and quality control. Further investigation is required to understand these mitochondrial processes.
Mitophagy and autophagy are important cellular processes responsible for breaking down damaged organelles, thereby protecting against accumulation of damaged and aggregated biomolecules. Mitophagy is the selective removal of damaged mitochondria by autophagosomes and lysosomes. Both PINK1 and parkin are known involved with mitophagy.47 Upon mitochondrial stress, PINK1 and parkin remove damaged mitochondria. PINK1, the upstream kinase, accumulates on the mitochondrial surface and recruits the E3 ubiquitin ligase parkin on site to ubiquitylate substrate proteins, which in turn induce the recruitment of receptors, followed by autophagy.48 Loss of PINK1 or parkin after blast disrupts mitophagy from preventing oxidative stress damage (shown as the MnSOD data), and is found to be associated with neurodegeneration.49 Mitochondrial autophagy is also important for mitochondrial quality control. Poor quality mitochondria may enhance oxidative stress and induce cell death.50 In this study, we identified that primary LIB exposure significantly reduced the parkin levels at 7 and 30 DPI, and with a trend towards reduced PINK1 (Fig. 6A–C). Parkin substrates including mitofusin 2 and VDAC1, which are embedded in the outer mitochondrial membrane,51 were significantly reduced from 7 to 30 DPI. During autophagy, conjugation of ATG12 to ATG5 is essential for LC3 lipidation and autophagosome formation. In addition, ATG12 functions in diverse processes including mitochondrial fusion and mitochondrial-dependent apoptosis.52 We observed a significantly decreased level of ubiquitin-like protein ATG12, and a trend toward reduced ULK1/ATG1 and LC3B after LIB exposure (Fig. 6D). These results suggest a decreased level of ATG12-ATG5 covalent binding, which further leads to insufficient autophagosome maturation and subsequent attenuation of autophagic flux.53 Together, our findings indicated that LIB resulted in compromised mitophagy and autophagy (Fig. 7).
Mitochondrial damage after blast may further lead to impaired neuronal cell function and contribute to the development or progression of neurodegeneration.10,38 Defects in mitochondrial bioenergetics and associated metabolic function are known to link with alterations of mitostatic processes, including mitochondrial clearance/dynamics, and these changes would impact myelination, axonal transport, and synaptic regulation.54 From our previous report and others, myelin sheaths defects exhibit multiple features, including sheath disruption, detachment from the axonemal, splitting of the myelin lamellae, and dense degeneration with vacuole inclusions within the myelin lamellae.5,55 This study identified canonical pathways associated with axonal guidance, regulation of cytoskeleton, and synaptic vesicle cycle regulation (Tables 2 and 3). Interestingly, we observed downregulation of SNAP25 and upregulation of STMN1 phosphorylation after LIB exposure. It has been reported that STMN1 phosphorylation regulates microtubule disassembly affecting axonal injury after blast at 74.5 kPa.56 SNAP 25, a component of synapses, is also important to synaptic functions, and is altered in diffuse axonal injury.57
Overall, our findings demonstrate that primary LIB exposure at 46.6 kPa causes striking mitochondrial dysfunction. Resulting dynamic global and phospho-proteome alterations and underlying mechanisms involved with mitochondrial dysfunction, oxidative stress, and axonal/cytoskeleton regulation are further revealed in the LIB-exposed cortical brain tissues. It is a logical extension of the effects of blast intensities that LIB-induced ultrastructural mitochondrial damage leads to prolonged dysregulation of cytoskeletal and synaptic proteins. Future studies with isolated mitochondrial proteome will permit further characterization of LIB-induced mitochondrial changes in relation to mitochondrial permeability, mitochondrial membrane potential, and ATP production leading to neurodegeneration. In addition, LIB-induced injury affecting other brain regions, such as hippocampus, striatum, and cerebellum, will be surveyed. The chronic effects of LIB-induced brain injury will also be investigated.7 Nevertheless, these studies have documented the key relationships of blast exposures to oxidative stress and mitochondrial dysfunction. Insights into the acute pathogenesis of LIB-induced mTBI may indicate links between early biochemical changes and later brain degeneration. It remains to be determined whether highly specific target characterization at molecular levels may enable more sensitive biomarker development, as well as particular target proteins and/or time lines for prevention and treatment of primary blast injuries.
Supplementary Material
Acknowledgments
The authors thank Jie Hou, Tina Ndam, and Runting Li for the technical support, and Dr. Grace Y. Sun of the University of Missouri for manuscript editing. This publication was made possible by funding from the DoD Congressionally Directed Medical Research Programs (CDMRP) for the Peer Reviewed Alzheimer's Research Program Convergence Science Research Award (PRARP-CSRA; AZ140109) and the research funds of the University of Missouri (Z.G.). This study was also supported by the awards I21BX002215, I01BX003527, and I21BX003807 from the Biomedical Laboratory Research and Development Service of the Veterans Affairs Office of Research and Development and the Cure Alzheimer's Fund (W.X.). Components of the mitochondrial analysis were performed by the University of Kansas Alzheimer's Disease Center Mitochondrial Genomics and Metabolism Core (P30AG035982). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the United States government, the DoD, the United States Army, or the Department of Veterans Affairs.
Author Disclosure Statement
No competing financial interests exist.
References
- 1. DeKosky S.T., Ikonomovic M.D., and Gandy S. (2010). Traumatic brain injury—football, warfare, and long-term effects. N. Engl. J. Med. 363, 1293–1296 [DOI] [PubMed] [Google Scholar]
- 2. Aertker B.M., Bedi S., and Cox Jr C.S. (2016). Strategies for CNS repair following TBI. Exp. Neurol. 275, 411–426 [DOI] [PubMed] [Google Scholar]
- 3. DePalma R.G., Burris D.G., Champion H.R., and Hodgson M.J. (2005). Blast injuries. N. Engl. J. Med. 352, 1335–1342 [DOI] [PubMed] [Google Scholar]
- 4. Cernak I., and Noble-Haeusslein L.J. (2010). Traumatic brain injury: an overview of pathobiology with emphasis on military populations. J. Cereb. Blood Flow Metab. 30, 255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Song H., Konan L.M., Cui J., Johnson C.E., Langenderfer M., Grant D., Ndam T., Simonyi A., White T., Demirci U., Mott D.R., Schwer D., Hubler G.K., Cernak I., DePalma R.G., and Gu Z. (2018). Ultrastructural brain abnormalities and associated behavioral changes in mice after low-intensity blast exposure. Behav. Brain Res. 347, 148–157 [DOI] [PubMed] [Google Scholar]
- 6. Song H., Cui J., Simonyi A., Johnson C.E., Hubler G.K., DePalma R.G., and Gu Z. (2018). Linking blast physics to biological outcomes in mild traumatic brain injury: narrative review and preliminary report of an open-field blast model. Behav. Brain Res. 340, 147–158 [DOI] [PubMed] [Google Scholar]
- 7. Song H., Konan L.M., Cui J., Johnson C.E., Hubler G.K., DePalma R.G., and Gu Z. (2018). Nanometer ultrastructural brain damage following low intensity primary blast wave exposure. Neural Regen. Res. 13, 1516–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen M., Song H., Cui J., Johnson C., Hubler G., DePalma R.G., Gu Z., and Xia W. (2018). Proteomic profiling of mouse brains exposed to blast-induced mild traumatic brain injury reveals changes in axonal proteins and phosphorylated tau. J. Alzheimers Dis. 66, 751–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lin M.T., and Beal M.F. (2006). Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 443, 787. [DOI] [PubMed] [Google Scholar]
- 10. Hiebert J.B., Shen Q., Thimmesch A.R., and Pierce J.D. (2015). Traumatic brain injury and mitochondrial dysfunction. Am. J. Med. Sci. 350, 132–138 [DOI] [PubMed] [Google Scholar]
- 11. Bertholet A., Delerue T., Millet A., Moulis M., David C., Daloyau M., Arnaune-Pelloquin L., Davezac N., Mils V., and Miquel M. (2016). Mitochondrial fusion/fission dynamics in neurodegeneration and neuronal plasticity. Neurobiol Dis 90, 3–19 [DOI] [PubMed] [Google Scholar]
- 12. Xiao M., Zhong H., Xia L., Tao Y., and Yin H. (2017). Pathophysiology of mitochondrial lipid oxidation: role of 4-hydroxynonenal (4-HNE) and other bioactive lipids in mitochondria. Free Radic. Biol. Med. 111, 316–327 [DOI] [PubMed] [Google Scholar]
- 13. Swan A.L., Mobasheri A., Allaway D., Liddell S., and Bacardit J. (2013). Application of machine learning to proteomics data: classification and biomarker identification in postgenomics biology. Omics 17, 595–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sarkis G.A., Mangaonkar M.D., Moghieb A., Lelling B., Guertin M., Yadikar H., Yang Z., Kobeissy F., and Wang K.K. (2017). The application of proteomics to traumatic brain and spinal cord injuries. Curr. Neurol. Neurosci. Rep. 17, 23. [DOI] [PubMed] [Google Scholar]
- 15. Kim D.K., Park J., Han D., Yang J., Kim A., Woo J., Kim Y., and Mook-Jung I. (2018). Molecular and functional signatures in a novel Alzheimer's disease mouse model assessed by quantitative proteomics. Mol. Neurodegener. 13, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kobeissy F.H., Ottens A.K., Zhang Z., Liu M.C., Denslow N.D., Dave J.R., Tortella F.C., Hayes R.L., and Wang K.K. (2006). Novel differential neuroproteomics analysis of traumatic brain injury in rats. Mol. Cell Proteomics 5, 1887–1898 [DOI] [PubMed] [Google Scholar]
- 17. Liu T., Qian W.-J., Gritsenko M.A., Xiao W., Moldawer L.L., Kaushal A., Monroe M.E., Varnum S.M., Moore R.J., and Purvine S.O. (2006). High dynamic range characterization of the trauma patient plasma proteome. Mol. Cell Proteomics 5, 1899–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Boutté A.M., Yao C., Kobeissy F., May Lu X.C., Zhang Z., Wang K.K., Schmid K., Tortella F.C., and Dave J.R. (2012). Proteomic analysis and brain‐specific systems biology in a rodent model of penetrating ballistic‐like brain injury. Electrophoresis 33, 3693–3704 [DOI] [PubMed] [Google Scholar]
- 19. Peters M.E., Rao V., Bechtold K.T., Roy D., Sair H.I., Leoutsakos J.-M., Diaz-Arrastia R., Stevens R.D., Batty Jr D.S., and Falk H. (2017). Head injury serum markers for assessing response to trauma: design of the HeadSMART study. Brain Inj. 31, 370–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ning M., Sarracino D.A., Kho A.T., Guo S., Lee S.-R., Krastins B., Buonanno F.S., Vizcaíno J.A., Orchard S., McMullin D., Wang X., and Lo E.H. (2011). Proteomic temporal profile of human brain endothelium after oxidative stress. Stroke 42, 37–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Saljo A., Svensson B., Mayorga M., Hamberger A., and Bolouri H. (2009). Low-level blasts raise intracranial pressure and impair cognitive function in rats. J. Neurotrauma 26, 1345–1353 [DOI] [PubMed] [Google Scholar]
- 22. Villén J., and Gygi S.P. (2008). The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat Protoc 3, 1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang F.-K., Zhang G., Lawlor K., Nazarian A., Philip J., Tempst P., Dephoure N., and Neubert T.A. (2017). Deep coverage of global protein expression and phosphorylation in breast tumor cell lines using TMT 10-plex isobaric labeling. J. Proteome Res. 16, 1121–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Elias J.E. and Gygi S.P. (2007). Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207. [DOI] [PubMed] [Google Scholar]
- 25. Wang X., Zhang Q., Bao R., Zhang N., Wang Y., Polo-Parada L., Tarim A., Alemifar A., Han X., Wilkins H.M., Swerdlow R.H., Wang X., and Ding S. (2017). Deletion of nampt in projection neurons of adult mice leads to motor dysfunction, neurodegeneration, and death. Cell Rep. 20, 2184–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Qu Z., Meng F., Bomgarden R.D., Viner R.I., Li J., Rogers J.C., Cheng J., Greenlief C.M., Cui J., Lubahn D.B., Sun G.Y., and Gu Z. (2014). Proteomic quantification and site-mapping of S-nitrosylated proteins using isobaric iodoTMT reagents. J. Proteome Res. 13, 3200–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen H., and Boutros P.C. (2011). VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics 12, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yu G., Wang L.-G., Han Y., and He Q.-Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 16, 284–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Walter W., Sánchez-Cabo F., and Ricote M. (2015). GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics 31, 2912–2914 [DOI] [PubMed] [Google Scholar]
- 30. Song H., Lu Y., Qu Z., Mossine V.V., Martin M.B., Hou J., Cui J., Peculis B.A., Mawhinney T.P., and Cheng J. (2016). Effects of aged garlic extract and FruArg on gene expression and signaling pathways in lipopolysaccharide-activated microglial cells. Sci. Rep. 6, 35323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mondello S., Muller U., Jeromin A., Streeter J., Hayes R.L., and Wang K.K. (2011). Blood-based diagnostics of traumatic brain injuries. Expert Rev. Mol. Diagn. 11, 65–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Woods A.S., Colsch B., Jackson S.N., Post J., Baldwin K., Roux A., Hoffer B., Cox B.M., Hoffer M., and Rubovitch V. (2013). Gangliosides and ceramides change in a mouse model of blast induced traumatic brain injury. ACS Chem. Neurosci. 4, 594–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rodríguez-Rodríguez A., Egea-Guerrero J.J., León-Justel A., Gordillo-Escobar E., Revuelto-Rey J., Vilches-Arenas Á., Carrillo-Vico A., Domínguez-Roldán J.M., Murillo-Cabezas F., and Guerrero J.M. (2012). Role of S100B protein in urine and serum as an early predictor of mortality after severe traumatic brain injury in adults. Clin. Chim. Acta 414, 228–233 [DOI] [PubMed] [Google Scholar]
- 34. Yap Y.C., King A.E., Guijt R.M., Jiang T., Blizzard C.A., Breadmore M.C., and Dickson T.C. (2017). Mild and repetitive very mild axonal stretch injury triggers cystoskeletal mislocalization and growth cone collapse. PloS One 12, e0176997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Song H., Fang S., Gao J., Wang J., Cao Z., Guo Z., Huang Q., Qu Y., Zhou H., and Yu J. (2018). Quantitative proteomic study reveals up-regulation of cAMP signaling pathway-related proteins in mild traumatic brain injury. J. Proteome Res. 17, 858–869 [DOI] [PubMed] [Google Scholar]
- 36. Dai W., Cheng H.-l., Huang R.-q., Zhuang Z., and Shi J.-X. (2009). Quantitative detection of the expression of mitochondrial cytochrome c oxidase subunits mRNA in the cerebral cortex after experimental traumatic brain injury. Brain Res. 1251, 287–295 [DOI] [PubMed] [Google Scholar]
- 37. Di Pietro V., Lazzarino G., Amorini A.M., Signoretti S., Hill L.J., Porto E., Tavazzi B., Lazzarino G., and Belli A. (2017). Fusion or fission: the destiny of mitochondria in traumatic brain injury of different severities. Sci. Rep. 7, 9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Swerdlow R.H. (2018). Mitochondria and mitochondrial cascades in Alzheimer's disease. J. Alzheimers Dis. 62, 1403–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kulbe J., and Hall E. (2017). Chronic traumatic encephalopathy-integration of canonical traumatic brain injury secondary injury mechanisms with tau pathology. Prog. Neurobiol. 158, 15–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Harmon J.L., Gibbs W.S., Whitaker R.M., Schnellmann R.G., and Adkins D.L. (2017). Striatal mitochondrial disruption following severe traumatic brain injury. J Neurotrauma 34, 487–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Huber B.R., Meabon J.S., Martin T.J., Mourad P.D., Bennett R., Kraemer B.C., Cernak I., Petrie E.C., Emery M.J., and Swenson E.R. (2013). Blast exposure causes early and persistent aberrant phospho-and cleaved-tau expression in a murine model of mild blast-induced traumatic brain injury. J. Alzheimers Dis. 37, 309–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Barsoum M.J., Yuan H., Gerencser A.A., Liot G., Kushnareva Y., Gräber S., Kovacs I., Lee W.D., Waggoner J., and Cui J. (2006). Nitric oxide‐induced mitochondrial fission is regulated by dynamin‐related GTPases in neurons. EMBO J. 25, 3900–3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cho D.-H., Nakamura T., Fang J., Cieplak P., Godzik A., Gu Z., and Lipton S.A. (2009). S-nitrosylation of Drp1 mediates β-amyloid-related mitochondrial fission and neuronal injury. Science 324, 102–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Selfridge J.E., Lezi E., Lu J., and Swerdlow R.H. (2013). Role of mitochondrial homeostasis and dynamics in Alzheimer's disease. Neurobiol. Dis. 51, 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Twig G., Hyde B., and Shirihai O.S. (2008). Mitochondrial fusion, fission and autophagy as a quality control axis: the bioenergetic view. Biochim. Biophys. Acta 1777, 1092–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Balog J., Mehta S.L. and Vemuganti R. (2016). Mitochondrial fission and fusion in secondary brain damage after CNS insults. J. Cereb. Blood Flow Metab. 36, 2022–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nguyen T.N., Padman B.S., and Lazarou M. (2016). Deciphering the molecular signals of PINK1/Parkin mitophagy. Trends Cell Biol. 26, 733–744 [DOI] [PubMed] [Google Scholar]
- 48. Matsuda N., Sato S., Shiba K., Okatsu K., Saisho K., Gautier C.A., Sou Y.-s., Saiki S., Kawajiri S., and Sato F. (2010). PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 189, 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Itoh K., Nakamura K., Iijima M., and Sesaki H. (2013). Mitochondrial dynamics in neurodegeneration. Trends Cell Biol. 23, 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang J. (2013). Autophagy and mitophagy in cellular damage control. Redox Biol. 1, 19–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Youle R.J., and Narendra D.P. (2011). Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 12, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Radoshevich L., Murrow L., Chen N., Fernandez E., Roy S., Fung C., and Debnath J. (2010). ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death. Cell 142, 590–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sarkar C., Zhao Z., Aungst S., Sabirzhanov B., Faden A.I., and Lipinski M.M. (2014). Impaired autophagy flux is associated with neuronal cell death after traumatic brain injury. Autophagy 10, 2208–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Misgeld T., and Schwarz T.L. (2017). Mitostasis in neurons: maintaining mitochondria in an extended cellular architecture. Neuron 96, 651–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ojo J.O., Bachmeier C., Mouzon B.C., Tzekov R., Mullan M., Davies H., Stewart M.G., and Crawford F. (2015). Ultrastructural changes in the white and gray matter of mice at chronic time points after repeated concussive head injury. J. Neuropathol. Exp. Neurol. 74, 1012–1035 [DOI] [PubMed] [Google Scholar]
- 56. Elder G.A., Dorr N.P., De Gasperi R., Gama Sosa M.A., Shaughness M.C., Maudlin-Jeronimo E., Hall A.A., McCarron R.M., and Ahlers S.T. (2012). Blast exposure induces post-traumatic stress disorder-related traits in a rat model of mild traumatic brain injury. J. Neurotrauma 29, 2564–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang P., Zhu S., Li Y., Zhao M., Liu M., Gao J., Ding S., and Li J. (2016). Quantitative proteomics analysis to identify diffuse axonal injury biomarkers in rats using iTRAQ coupled LC–MS/MS. J. Proteom. 133, 93–99 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.