Abstract
The mammalian brain describes a multiscale system. At the microscale, axonal, dendritic, and synaptic elements ensure neuron-to-neuron communication, and at the macroscale, large-scale projections form the anatomical wiring for communication between cortical areas. Although it is clear that both levels of neural organization play a crucial role in brain functioning, their interaction is not extensively studied. Connectome studies of the mammalian brain in cat, macaque, and human have recently shown that regions with larger and more complex pyramidal cells to have more macroscale corticocortical connections. In this study, we aimed to further validate these cross-scale findings in the human, mouse, and rat brain. We combined neuron reconstructions from the NeuroMorpho.org neuroarchitecture database with macroscale connectivity data derived from connectome mapping by means of tract-tracing (rat, mouse) and in vivo diffusion magnetic resonance imaging (human). Across these three mammalian species, we show cortical variation in neural organization to be associated with features of macroscale connectivity, with cortical variation in neuronal complexity explaining significant proportions of cortical variation in the number of white matter projections of cortical areas. Our findings converge on the notion of a relationship between features of micro- and macroscale neural connectivity to form a central aspect of mammalian neural architecture.
Keywords: connectome organization, cytoarchitecture, human, mouse, rat, structural connectomics
Introduction
Connectome studies of the mammalian brain in cat, macaque, and human have shown that regional variation in microscale structural type (Beul et al., 2015) and layer III pyramidal cell complexity (Scholtens et al., 2014; van den Heuvel et al., 2015c) relates to macroscale projection patterns, with regions with larger and more complex pyramidal cells having more macroscale corticocortical connections. In this study, we aimed to further investigate this relationship in the human brain, as well as to extend the observed micro–macro relationship of neural connectivity organization to the smaller rodent brain.
Cytoarchitectonic variation is one of the cornerstones of mammalian cortex organization. Detailed cortical mappings of early 20th century anatomy pioneers such as Korbinian Brodmann (Brodmann, 1909), Alfred Campbell (Campbell, 1905), Earl Walker (Walker, 1940), Gerhardt Von Bonin (von Bonin and Bailey, 1947), and Constantin Von Economo and George Koskinas (von Economo and Koskinas, 1925) showed a rich and diverse cytoarchitecture of the cortical mantles of mammalian species. Their observations of a profuse variety of cytoarchitectonics across cortical areas led to theories of variation in neuron morphology and complexity across the cortex to play a crucial role in the physiological properties of cortical areas (Mesulam, 1998).
Studies of macaque and human cortex have shown a posterior to anterior gradient in cortical pyramidal complexity, with frontal association areas noted to display larger more branched and more spinous pyramidal neurons as compared with unimodal primary motor, visual, and/or auditory cortex (Elston, 2003; Jacobs et al., 2001) and have accordingly led to the suggestion of frontal regions to be involved in more complex neural processing (Elston, 2003). This posterior–anterior gradient in neuron size has since been reported in many different primate and rodent species (Charvet et al., 2015), showing an increasingly strong gradient with larger brain volume (Finlay and Uchiyama, 2015), a measure related to a species' capacity for cognitive processing (MacLean et al., 2014).
Network studies of the mammalian brain including human (Hagmann et al., 2008), cat (de Reus and van den Heuvel, 2013b; Scannell et al., 1995), macaque (Goulas et al., 2014; Harriger et al., 2012), rat (Bota et al., 2015; van den Heuvel et al., 2015d), and mouse (Oh et al., 2014; Rubinov et al., 2015; van den Heuvel and de Reus, 2014) neural wiring have further shown an overall cost-efficient architecture of the connectome, combining cost-effective local communities with the formation of relatively expensive short communication relays and densely connected hub regions (Bullmore and Sporns, 2009; van den Heuvel and Sporns, 2013b; van den Heuvel et al., 2016a).
High-degree hub regions, in particular, have been noted as expensive attributes of brain network architecture, with their relatively long connections (van den Heuvel et al., 2012), their distinct geometric embedding in the network (Roberts et al., 2016; van den Heuvel and Sporns, 2013a), their cytoarchitecture (Beul et al., 2015; Scholtens et al., 2014), their high metabolic demand (Bullmore and Sporns, 2012; Collin et al., 2013; Vaishnavi et al., 2010), and corresponding gene expression profile (Fulcher and Fornito, 2016). Potentially offsetting this high cost, the formation of highly connected hub regions has been argued to form the anatomical infrastructure for global neural communication and integration (Tomasi and Volkow, 2011; van den Heuvel and Sporns, 2013a; Zamora-López et al., 2010, 2011) and to make an important contribution to high order brain function and behavior in humans (Adelstein et al., 2011; Li et al., 2009; Park and Friston, 2013; Pessoa, 2012; Shanahan, 2012; van den Heuvel et al., 2009). Species adaptations in brain connectivity organization have, therefore, been hypothesized to reflect adaptations in cognition and behavior (Hopkins and Rilling, 2000; Rilling and Insel, 1999; van den Heuvel et al., 2016a).
It is thus evident that properties at both the microscale and macroscale levels of mammalian cortex organization play a critical role in brain function. Recent studies have argued for a potential interplay between these two organizational scales (Beul and Hilgetag, 2017; Beul et al., 2015; Hilgetag and Grant, 2010; Scholtens et al., 2014; van den Heuvel et al., 2015c, 2016b). Cortical structural type and supragranular structure have been hypothesized to form an important factor for the formation of interareal projections (Barbas, 2015; Barbas and Rempel-Clower, 1997; Felleman and Van Essen, 1991; Hilgetag and Grant, 2010; Pandya and Sanides, 1973; Scannell et al., 1995), theories supported by recent observations in the rat, cat, and macaque showing cortical structural type to shape macroscale connectivity (Beul and Hilgetag, 2017; Beul et al., 2015; Goulas et al., 2017). In further support of a microscale–macroscale interaction, microscale layer 3 pyramidal complexity has been linked to the number of white matter corticocortical projections of various regions in the macaque (Scholtens et al., 2014) and human cortex (van den Heuvel et al., 2015c).
In this study, we further explore a possible micro–macro relationship in the mammalian brain by further validation of this phenomenon in the human brain and by expanding investigations to the mouse and rat rodent brain. Recent technological advances have enabled spatially highly detailed tract-tracing mappings of the mouse (Oh et al., 2014) and rat connectome (Bota and Swanson, 2007; Bota et al., 2012, 2015). In particular, this tract-tracing data enables incorporation of information regarding directionality in the brain network, reported to be essential in shaping the brain network (Kale et al., 2018). In addition, high-resolution in vivo neuroimaging data of the Human Connectome Project (Van Essen et al., 2013) allow for detailed connectome reconstructions of human macroscale brain wiring. This, combined with important undertakings in large-scale collation of detailed neuron morphology data across published studies such as in the NeuroMorpho.org database (Ascoli, 1999), allows for a further exploration of a micro-to-macro relationship in neural connectivity organization across mammalian species. Combining detailed connectome mappings with data on pyramidal neuroarchitecture in the human and rodent brain, we show cross-species consistency of a multiscale relationship between microscale pyramidal neuron complexity and macroscale corticocortical connectivity levels, providing converging evidence of architectural aspects of microscale and macroscale connectivity in the mammalian brain to be related.
Materials and Methods
Microscale neuronal data
Data on neuron morphology from the human, rat, and mouse cortex were extracted from the comprehensive NeuroMorpho.org (http://NeuroMorpho.org, version 6.1) database (Ascoli, 1999) and curated by Ascoli and coworkers (Ascoli, 2006; Ascoli et al., 2001a, 2001b). The NeuroMorpho.org database includes a collection of a large number of individual neuron reconstructions as grouped together across multiple experiments presented in the literature. The database includes standardized digital reconstructions of the original reported reconstructed neurons from which (as provided at NeuroMorpho.org) morphological properties can be derived (Ascoli et al., 2001a, 2001b), as well as detailed meta information on the original literature source, staining procedures, and the examined cortical areas and layers. In our study, data from healthy control individuals of experiments examining adult human, rat, and mouse cortical areas were extracted. To exclude effects resulting from differences in reconstruction methods, neuron reconstructions made using Neurolucida software were included for analysis.
Based on consistent micro–macro associations reported in previous work on micro–macro associations in the macaque (Scholtens et al., 2014) and human cerebral cortex (van den Heuvel et al., 2016b), total neuron length, number of dendritic branches, and soma surface were selected as main properties of examination for correlation analysis in all three included species. Other cytoarchitectonic properties included metrics of total neuron volume, total neuron surface, number of stems, number of bifurcations, overall width, overall height, overall depth, average diameter, max path distance, and max branch order. Summary statistics for all microscale measures are shown in Supplementary Figures S1–S3 (Supplementary Data are available online at www.liebertpub.com/brain).
Human neuron morphology
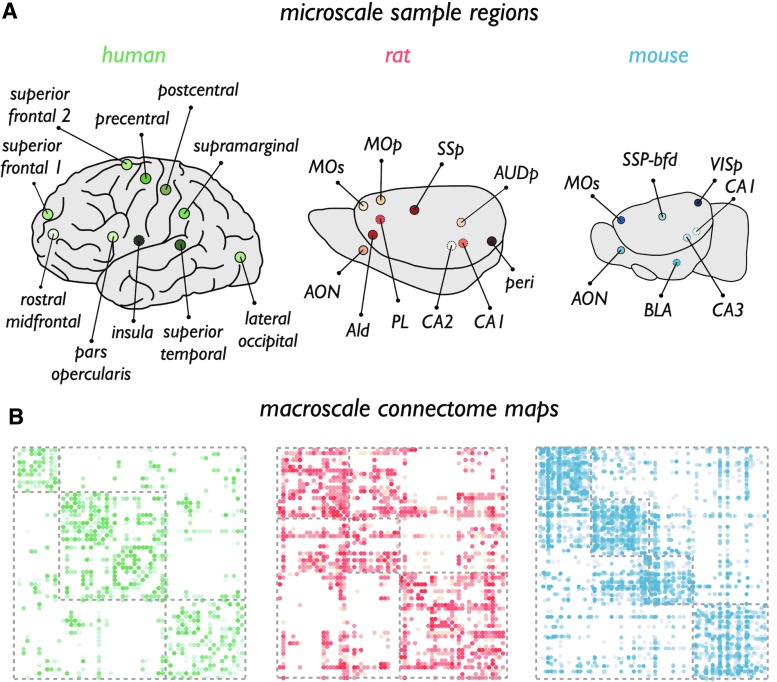
NeuroMorpho.org extraction of human data included experiments using Golgi, rapid-Golgi, and Golgi-Scheibel staining procedures, and described a total of 1916 neuron reconstructions across 10 cortical areas, all from the left hemisphere. Information on neuronal cytoarchitectonic properties (3 primary and 10 secondary properties) was extracted from these recordings, and averaged for each cortical area. Next, the reported regions were manually mapped to a 2 × 57 regions subdivision of the Desikan–Killiany cortical atlas of the FreeSurfer software suite (Cammoun et al., 2012; Desikan et al., 2006; Fischl and Dale, 2000; Scholtens et al., 2015; van den Heuvel et al., 2015c) (DK-57, describing 57 distinct cortical areas per hemisphere) for comparison with macroscale connectivity (Fig. 1A shows the mapping of the included cortical regions). Supplementary Table S1 describes the data of the individual experiments, their original reported cortical site, and their mapping to regions of the DK-57 cortical atlas. This resulted in morphological data of in total 10 areas of the human cortex (mean/SD: 185/147 [range: 70–594] recordings of neuron metrics per cortical area were present in the database), including left hemispheric subareas of lateral occipital, pars opercularis, postcentral, precentral, rostral middle frontal, superior frontal (two subareas), superior temporal, supramarginal, and insular cortex. Regions and their mappings are shown in Figure 1A and listed in Supplementary Table S1.
FIG. 1.
(A) Cortical areas reported in the NeuroMorpho.org data mapped to regions of the adopted human (DK-57 atlas), rat [Swanson atlas (Swanson, 1992)], and mouse (Allen Mouse Brain Atlas) brain atlases. Figures highlight the regions for which neuronal morphological data could be extracted from the NeuroMorpho.org database, approximate location of medial regions depicted with a dotted outline. From left to right: human, rat, and mouse data. Included regions in the rat are AId, rostral agranular insular cortex; AON, anterior olfactory nucleus; AUDp, primary auditory cortex; CA1 and CA2 of the hippocampus; MOp, primary motor cortex; MOs, secondary motor cortex; peri, perirhinal cortex; PL, prelimbic cortex; SSp, primary sensory cortex. Included mouse regions are AON, anterior olfactory nucleus; BLA, basolateral amygdala, CA1 and CA3 of the hippocampus; MOs, secondary motor cortex; SSP-bfd, barrel field of the primary sensory cortex; VISp, primary visual cortex. (B) Extracted connectivity matrices of the human [as derived from in vivo DWI data from the Human Connectome Project (Van Essen et al., 2013)], rat [as derived from the BAMS-II rat connectome database (Bota and Swanson, 2007)], and mouse [as derived from the Allen Mouse Brain Connectivity Atlas (Oh et al., 2014)] connectome. DWI, diffusion weighted imaging. Color images available online at www.liebertpub.com/brain
Rat neuron morphology
NeuroMorpho.org extraction from studies on the rat brain included experiments using Golgi, Golgi-Cox, Biocytin, Lucifer Yellow, Neurobiotin, biotinylated dextran amine, and horseradish peroxidase staining procedures, and described 875 neuron reconstructions across 10 different cortical sites. Similar to the human extraction, data (now including data from both left and right hemispheres) were collected from the NeuroMorpho.org reconstructions and averaged for each cortical area included in the unihemispheric connectome data. Regions included areas AId (rostral agranular insular cortex); AON (anterior olfactory nucleus); AUDp (primary auditory cortex); CA1 and CA2 of the hippocampus; MOp (primary motor cortex); MOs (secondary motor cortex); peri (perirhinal cortex); PL (prelimbic cortex); and SSp (primary sensory cortex) (mean/SD: 83/97 [range: 1–283] recordings per cortical area) (Fig. 1). Using the detailed Swanson rat cortical atlas (Swanson, 1992), these sites were manually mapped to regions of which macroscale connectivity was available in the highly detailed collated mesoscale BAMS-II rat cortical connectome map as presented by Swanson and coworkers (Bota et al., 2015) (see Mouse Connectome section). Supplementary Table S2 lists the individual source publications, their original reported cortical site in the rat cortex, and their mapping to regions of the Swanson rat brain atlas.
Mouse neuron morphology
NeuroMorpho.org extraction on the mouse brain included experiments using Golgi, Golgi-Cox, and rapid-Golgi staining procedures, and described a total of 408 neuron recordings across seven brain areas. Similar to the human and rat data set, morphological data of the reconstructed neurons in the database were extracted, mapped to the unihemispheric connectome, and values subsequently averaged across cortical areas. Using the Allen Mouse Brain Atlas (http://mouse.brain-map.org), these cortical areas were manually mapped to cortical areas as examined by Oh and coworkers (2014) (see Fig. 1A for a visual representation of the included regions), resulting in neuron information for seven brain regions (mean/SD: 58/58 [range: 1–167] recordings per cortical area): AON (anterior olfactory nucleus); BLA (basolateral amygdala); CA1 and CA3 of the hippocampus;MOs (secondarymotor cortex); SSP-bfd (barrel field of the primary sensory cortex); and VISp (primary visual cortex). Supplementary Table S3 describes the data of the individual experiments, their original reported cortical site and layer (as present in the NeuroMorpho.org database), and their mapping to the regions of the Allen Mouse Brain Atlas.
Macroscale connectome data
Human connectome
Data on macroscale connectivity of the human brain were derived from the high-resolution diffusion weighted imaging (DWI) data from the Human Connectome Project (Van Essen et al., 2013), including data of 215 subjects (Q3 release, voxel size 1.25 mm isotropic, TR/TE 5520/89.5 msec, 3 × 90 diffusion directions with diffusion weighting 1000, 2000, and 3000 sec/mm2). Preprocessing involved correction for eddy current and susceptibility distortions and realignment of the diffusion-weighted and B = 0 images (Glasser et al., 2013). Next, for each individual data set, white matter, gray matter, and cortical spinal fluid classification was performed based on the T1 anatomical image (Q3, voxel size 0.7 mm isotropic) using FreeSurfer (Fischl and Dale, 2000), followed by a three-dimensional (3D) reconstruction of the cortical mantle and parcellation of the cortex into 114 distinct cortical areas (57 unique regions per hemisphere) using a subdivision of FreeSurfer's Desikan–Killiany atlas (Cammoun et al., 2012; Desikan et al., 2006; Scholtens et al., 2015). This resulted in 57 cortical areas to which the reported cortical regions of the NeuroMorpho.org data set were mapped (Fig. 1A). Subsequently, for each voxel, the diffusion profile was computed by means of generalized q-sampling imaging, allowing for the reconstruction of complex fiber configurations (e.g., crossing fibers) (de Reus and van den Heuvel, 2014; Yeh et al., 2010).
Streamline tractography was used to reconstruct white matter pathways, starting a streamline in each white matter voxel, following the best matching diffusion direction from voxel to voxel. A streamline was stopped when it exited the brain tissue mask, made a sharp turn of 45° or more, or reached a voxel with a low fractional anisotropy (a threshold of 0.1 was used). Combination of the 114 parcellated cortical regions and the tractography streamlines resulted in the formation of a weighted structural connectivity (SC) matrix of size N × N by selecting the subsets of reconstructed tractography streamlines that touched both regions i and j, for all pairs (i, j) of the N = 114 cortical regions. To reduce potential false positive reconstructions, pathways consisting of >5 streamlines were included for further analysis (Verstraete et al., 2014). Next, the resulting individual connectivity matrices of the 215 subjects were combined into a group-averaged SC matrix (Fig. 1B) by taking the nonzero mean of the individual matrices, including a region-to-region connection when a pathway was found in at least 71 of the 215 subjects (i.e., 33%) (de Reus and van den Heuvel, 2013a; Verstraete et al., 2014).
Graph theoretical analysis was used to compute the number of pathways (in DWI, due to the absence of information on projection directionality, data on efferent and afferent pathways were combined) of each of the cortical areas, reflecting a region's network degree. To match the single hemisphere data sets of the mouse and rat connectome (see Rat Connectome and Mouse Connectome sections), nodal degree values of regions from the left hemisphere were used for further analysis. [Taking nodal degree data from the right hemisphere revealed similar findings].
Rat connectome
Data on macroscale connectivity of the rat brain were taken from the recent rat connectome mapping of the BAMS-II database by Swanson and Bota and colleagues, providing a comprehensive collation of data from multiple tract-tracing experiments in the rat brain. The unique detailed BAMS-II database of Swanson and Bota [http://brancusi1.usc.edu/connectome/; (Bota and Swanson, 2007; Bota et al., 2015)] describes macroscale connectivity between 67 regions of the rat brain [described and analyzed in Bota et al. (2015)]. For 1662 region pairs (37.6% of total number of pairs), information on the nonexistence, existence, and/or strength (1424 of 1487 projections) of region-to-region macroscale pathways is present. In total, these data describe a single hemisphere connectome of 67 regions and 1487 pathways, represented as a directed 67 × 67 connectivity matrix. Region pairs for which no information on connectivity was present (i.e., NaNs in the matrix) were taken as nonconnected regions (i.e., represented as a 0) (Bota et al., 2015; van den Heuvel et al., 2015c), resulting in a connectivity matrix with a density of 31.6% (Fig. 1B). For each of the cortical regions of the rat brain, the network degree was computed as the total number of efferent or afferent pathways of a cortical area.
Mouse connectome
Data on macroscale connectivity of the mouse brain were taken from the recently mapped mesoscale mouse connectome from the Allen Institute for Brain Science [as published by http://connectivity.brain-map.org (Oh et al., 2014)]. This detailed connectome data set comprises a fully mapped mesoscale connectome of 213 regions describing a single hemisphere of the full mouse brain, including an initial weighted and directed connectivity matrix of 213 regions with 16,954 interareal pathways and 37.5% density. To eliminate very weak projections [reflecting potential false positive mappings, as reported by the curators of this connectome data set (Oh et al., 2014)], a strength threshold of 0.75 (other thresholds, e.g., 0.05–0.1, revealed similar findings) was applied, resulting in a directed connectivity matrix of 3433 pathways and 7.6% density (depicted in Fig. 1B). Next, for each of the cortical and subcortical areas, the network degree was computed, taken as the total number of efferent and afferent pathways of each area.
Cross-scale analysis
For all three data sets (i.e., human, rat, and mouse), data on regional microscale neuron morphology (describing 13 micrometrics, of in total 10 cortical regions for the human data set, 9 for the rat, and 7 regions for the mouse brain) were examined in the context of levels of regional variation in macroscale connectivity (i.e., nodal network degree) by means of Pearson's correlation analyses. Per species data set, correction for multiple testing (3 primary metrics and 10 secondary metrics = 13 micro–macro correlations assessed per data set) was performed by computing the actual number of independent tests performed through principal component analysis (PCA) (Scholtens et al., 2014). Per data set, a PCA was performed on the microscale data, determining the number of principal components that explained >95% of the variance in the data. In all three data sets, this resulted in two components. Next, using the extracted components, a partial Bonferroni alpha of 0.05/2 = 0.025 was computed for each data set, and effects reaching this partial Bonferroni alpha were taken as significant.
Results
Micro–macro correlations
Human
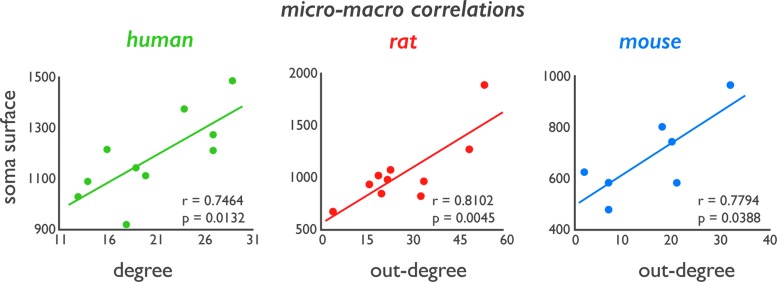
In the human data set, regional variation in soma surface (r = 0.7706, p = 0.0091) (Fig. 2) and total neuron length (r = 0.7015, p = 0.0238) were significantly correlated with regional variation in number of reconstructed white matter pathways (macroscale degree), with regions displaying a dense macroscale connectivity profile overlapping with those cortical areas showing larger neurons. No clear associations were observed between macroscale degree and dendritic branching (p = 0.2290; Supplementary Fig. S4).
FIG. 2.
Panels show cross-scale correlation plots of regional variation in neuron morphology and macroscale connectivity. Left panel shows the association between regional variation in neuron soma surface and macroscale connectivity in the human data set (correlation over 10 cortical regions of which data in the NeuroMorpho.org database were present). Middle panel shows the association between regional variation in total soma surface and number of efferent pathways (i.e., network out-degree) in the rat brain. Right panel shows the association between regional variation in soma surface and number of efferent pathways (out-degree) in the mouse data set. Color images available online at www.liebertpub.com/brain
Rat
Validating human findings, cortical variation in soma surface was found to be significantly associated with regional variation in the number of efferent macroscale pathways (i.e., out-degree, r = 0.8102, p = 0.0045; Fig. 2). This effect was observed to be the strongest for out-degree, with no such effect observed for in-degree (p = 0.1864; Supplementary Fig. S4).
Mouse
Cross-scale analysis in the mouse revealed a trend-level relationship of microscale soma surface (r = 0.7794, p = 0.0388; Fig. 2). In addition, within the set of remaining metrics, a significant relationship was observed between out-degree and total neuron volume (r = 0.8754, p = 0.0059; Supplementary Fig. S4). Similar as in the rat data set, no correlations were observed with network in-degree (i.e., the number of afferent projections of a cortical area) (Supplementary Fig. S4).
Discussion
Cross-scale analysis across three data sets on collated neuron morphology and mesoscale and macroscale connectivity provides further evidence that aspects of neural connectivity across different scales of mammalian cortical organization are related. Recent findings of an association between microscale and macroscale features of brain connectivity in the cat, macaque, and human cortex include observations of a predictive role of cytoarchitectonic classes on the existence of corticocortical axonal pathways (Barbas, 2015; Beul et al., 2015; Hilgetag and Grant, 2010), as well as an association between macroscale connectivity and layer 3 pyramidal complexity of macaque (Scholtens et al., 2014) and human cortical areas (van den Heuvel et al., 2015c, 2016b). Our findings also confer on recent observations showing granular structure of the cortex to be related to the level of between-network functional connectivity (Wylie et al., 2015). We now further extend findings to the rodent brain, providing evidence of a micro–macro association in two of the most highly detailed tract-tracing connectome mappings present for mammalian species. Findings in the human, mouse, and rat converge on an association and potential interplay between architectonic features of neural connectivity at the microscale and macroscale of brain organization in mammalian neural systems.
What may be the etiology of such a potential micro–macro relationship? We note that our observations—and those of the mentioned previous studies—are inferential of nature, meaning that they show a correlation between microscale and macroscale metrics of brain organization. Such correlation analyses cannot answer the important question of whether one effect is driving the other. Whether neuronal morphological or cytoarchitectonic structure allows for the existence of large-scale macroscale pathways and connectivity profiles, or whether the cytoarchitectonic organization of a cortical region is shaped by the amount and type of neural information from large-scale macroscale axonal projections remains unknown.
Cortical development may form a strong influential factor on this relationship, with both organizational scales of connectivity simultaneously shaped by developmental processes. Postmortem observations in young developing animals have suggested initial widespread axonal growth followed by a long period of pruning and local specialization of long-range axonal projections (Innocenti and Price, 2005; LaMantia and Rakic, 1994; Price and Ferrer, 1993), thus suggesting a potential interaction between local neuronal activation and the formation and tuning of interareal communication patterns (Innocenti and Price, 2005). As such, with a reductionist view, one could argue that processes of dendritic specialization and synaptic pruning may lead to the formation and pruning of long-range axonal pathways that connect to these pruned synapses. Vice versa, variation in axonal pathways of cortical regions has also been suggested to have direct consequences for regional differentiation in cytoarchitecture (Kaas et al., 2002), from which one could speculate on the notion of macroscale anatomical and functional patterns to play a role in developmental patterns in the formation, growth, and pruning of dendritic branches and synapses.
Examination of macaque developmental gene expression and cytoarchitecture showed a late formation of adult-like cortical regional and laminar molecular phenotypes, pointing toward an important role of functional cell–cell interaction in forming mature cellular phenotypes (Bakken et al., 2016). From the presented micro–macro findings, we speculate on a similar—and intertwined—developmental pattern of macroscale connectivity. Indeed, in humans, studies have shown clear developmental change of large-scale patterns of anatomical and functional connectivity (Betzel et al., 2014; Dosenbach et al., 2010; Fair et al., 2008, 2009; Hagmann et al., 2010; van den Heuvel et al., 2015b). Furthermore, previous observations have shown differential developmental time windows between cortical regions on both the microscale [e.g., cellular proliferation, migration, and elimination (Dombrowski et al., 2001)] and the macroscale [e.g., myelination of large axonal pathways as observed by Flechsig (1920)], suggesting that the timing of the different interacting processes may serve as an important modulator in establishing the observed adult region-wise micro–macro associations. Future studies examining how (the timing of) developmental changes at the microscale level relate to developmental changes at the macroscale level are, therefore, of high interest.
Studies have reported both overlap (e.g., Brodmann, 1909; Campbell, 1905; Goulas et al., 2014) and cross-species differences in pyramidal and cytoarchitectonic organization of cortical regions (Elston et al., 2001, 2011) and macroscopic connectome formation (Li et al., 2013; Miranda-Dominguez et al., 2014). Contrasts in pyramidal complexity between unimodal and multimodal association cortex have been suggested to be significantly different across primate species, arguing for both an absolute and a relative increase in neuronal size and complexity of pyramidal neurons from unimodal regions to higher order association cortex in humans as compared with primates (Elston et al., 2001, 2011).
Interestingly, in parallel, studies examining cross-species differences in macroscale connectivity of cortical areas have shown evidence of frontal cortical areas to display differences in macroscale connectivity patterns from primates to humans (Sallet et al., 2013). The frontal cortex is among those brain regions showing the largest evolutionary differences in developmental timing, with an extended developmental period in humans compared with other primates on both the microscale [e.g., synaptogenesis (Petanjek et al., 2011)] and the macroscale [e.g., myelination (Miller et al., 2012)]. Future studies could examine whether, and if so how, such cross-species differences in microscale and macroscale connectivity are related.
Several points need to be taken into account when interpreting the reported findings. First, as a direct result of our study design, our examination is limited by the range of microscale data (and, in particular, the number of areas) available in the NeuroMorpho.org database, which can lead to inflated correlation values (Yarkoni, 2009). Second, the currently included set of microscale data gives detailed information on proximal neuron morphology, including soma and dendritic branching, but does not provide information on axonal measurements (e.g., axon length and axon volume). Soma size—the microscale measure most strongly related to macroscale connectivity in the current comparison—has been reported to be related both with larger basal dendritic tree length (for in-going connections) (Jacobs et al., 2001) and with larger axon caliber (out-going connections) (Sloper and Powell, 1979). Given the observed association between soma size and out-degree, future micro–macro comparisons including data on axon morphology could be of interest in further exploring the relationship between microscale neuron morphology and macroscale in-degree and out-degree. Third, it should be noted that the examined microscale data involves a collation of data across multiple experiments and across different studies of different groups, thus involving a collation of data acquired across multiple research conditions, study designs, specimens with varying age and gender, varying mouse/rat strains, and data obtained across different measurement methodologies. Different staining methods have, for example, different sensitivity profiles, potentially leading to subjective measurements across research groups. To overcome these differences, the NeuroMorpho.org database contains detailed information on the original source and applied experimental procedures and, importantly, includes standardized 3D reconstructions of the individual neurons examined in the reported studies, which allows for a standardized extraction of morphological metrics. Fourth, even though the mouse and rat connectome data sets contain highly detailed information on mesoscale connectivity, the included connectome data sets are formed by collating data from a large number of specimens (Bota et al., 2015; Oh et al., 2014), involving a group-averaged consensus connectome map (de Reus and van den Heuvel, 2013a). Concerning the reconstructed human connectome map, diffusion weighted magnetic resonance imaging (MRI) depends on the estimation of the diffusion profile of water molecules and thus involves an indirect measurement of axonal pathways. As a result, diffusion MRI is known to include several limitations and caveats, with, in particular, a difficulty in resolving pathway orientation in white matter areas with complex fiber architecture, leading to argued over- and underestimations of classes of white matter pathways (Jones, 2008; van den Heuvel et al., 2015a). As a fifth remark, we once again stress that our study involves a meta-regression type of analysis in which data of several histology reports and data on tract reconstruction of multiple specimens are collated into one single data set. The microscale and macroscale data are thus acquired across different specimens and we note that this means that we could only examine variation across different areas of the cortex, but not across individual specimens. A data set including data on more comprehensive or even cortex-wide microscale and macroscale connectivity of single specimens, including information on for instance axon size, gene expression, and spine density, would allow for a much more detailed examination. Integration of data on resting-state functional connectivity or dynamics [reported to be related to in-degree in the tract-tracing structural mouse connectome (Sethi et al., 2017)] could be of additional interest in forming a more complete multiscale connectome framework. Such multiscale and multimodal studies would allow for the examination of influences of external factors on a micro–macro interplay of connectivity and as such could provide new insights into how brain properties such as learning or complex cognitive behavior can emerge from the smallest and largest aspects of neural organization.
Our findings across the human, rat, and mouse cortex provide converging evidence for a relationship between aspects of neural organization at the smallest and largest organizational scales of the mammalian brain. With many neurological and psychiatric brain disorders showing alterations in neural connectivity and disrupted connectome formation at both the microscale and macroscale level of brain organization, studying this micro–macro interplay may help to better comprehend underlying disease mechanisms of neurodegenerative and neurodevelopmental brain disorders (van den Heuvel et al., 2016b).
Supplementary Material
Acknowledgments
We thank Ruben Schmidt, Suus van Noort, Siemon C. de Lange, and Marcel de Reus for help with the data collection and analysis. M.P.v.d.H. is an MQ fellow and was supported by an Innovational Research Incentives Scheme Vidi grant (Grant No. VIDI-452-16-015) of the Netherlands Organisation of Scientific Research (Nederlandse Organisatie voor Wetenschappelijk Onderzoek). M.P.v.d.H. and L.H.S. were supported by an ALW Open grant (Grant No. ALWOP.179) of the Netherlands Organisation of Scientific Research. LFB was funded by the National Institute of Mental Health (R01 MH113234, R01 MH109464) and the National Cancer Institute (U01 CA193632).
Data were provided in part by the Human Connectome Project, WU-Minn Consortium (Principal Investigators: David Van Essen and Kamil Ugurbil; 1U54MH091657) funded by the 16 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research; and by the McDonnell Center for Systems Neuroscience at Washington University.
Author Disclosure Statement
No competing financial interests exist.
References
- Adelstein JS, Shehzad Z, Mennes M, DeYoung CG, Zuo X-N, Kelly C, et al. 2011. Personality is reflected in the brain's intrinsic functional architecture. PLoS One 6:e27633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascoli GA. 1999. Progress and perspectives in computational neuroanatomy. Anat Rec 257:195–207 [DOI] [PubMed] [Google Scholar]
- Ascoli GA. 2006. Mobilizing the base of neuroscience data: the case of neuronal morphologies. Nat Rev Neurosci 7:318. [DOI] [PubMed] [Google Scholar]
- Ascoli GA, Krichmar JL, Nasuto SJ, Senft SL. 2001a. Generation, description and storage of dendritic morphology data. Philos Trans R Soc Lond B Biol Sci 356:1131–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascoli GA, Krichmar JL, Scorcioni R, Nasuto SJ, Senft SL, Krichmar G. 2001b. Computer generation and quantitative morphometric analysis of virtual neurons. Anat Embryol 204:283–301 [DOI] [PubMed] [Google Scholar]
- Bakken TE, Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L, et al. 2016. A comprehensive transcriptional map of primate brain development. Nature 535:367–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbas H. 2015. General cortical and special prefrontal connections: principles from structure to function. Annu Rev Neurosci 38:269–289 [DOI] [PubMed] [Google Scholar]
- Barbas H, Rempel-Clower N. 1997. Cortical structure predicts the pattern of corticocortical connections. Cereb Cortex 7:635–646 [DOI] [PubMed] [Google Scholar]
- Betzel RF, Byrge L, He Y, Goni J, Zuo XN, Sporns O. 2014. Changes in structural and functional connectivity among resting-state networks across the human lifespan. Neuroimage 102 Pt 2:345–357 [DOI] [PubMed] [Google Scholar]
- Beul SF, Grant S, Hilgetag CC. 2015. A predictive model of the cat cortical connectome based on cytoarchitecture and distance. Brain Struct Funct 220:3167–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beul SF, Hilgetag CC. 2017. Neuron density is a fundamental determinant of structural connectivity in the primate cerebral cortex. bioRxiv DOI: 10.1101/117051 [DOI]
- Bota M, Dong H-W, Swanson LW. 2012. Combining collation and annotation efforts toward completion of the rat and mouse connectomes in BAMS. Front Neuroinform 6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bota M, Sporns O, Swanson LW. 2015. Architecture of the cerebral cortical association connectome underlying cognition. Proc Natl Acad Sci U S A 112:E2093–E2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bota M, Swanson LW. 2007. Online workbenches for neural network connections. J Comp Neurol 500:807–814 [DOI] [PubMed] [Google Scholar]
- Brodmann K. 1909. Localization in the cerebral cortex: the principles of comparative localization in the cerebral cortex based on cytoarchitectonics [in German]. Leipzig, Germany: Barth [Google Scholar]
- Bullmore E, Sporns O. 2009. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci 10:186–198 [DOI] [PubMed] [Google Scholar]
- Bullmore E, Sporns O. 2012. The economy of brain network organization. Nat Rev Neurosci 13:336–349 [DOI] [PubMed] [Google Scholar]
- Cammoun L, Gigandet X, Meskaldji D, Thiran JP, Sporns O, Do KQ, et al. 2012. Mapping the human connectome at multiple scales with diffusion spectrum MRI. J Neurosci Methods 203:386–397 [DOI] [PubMed] [Google Scholar]
- Campbell AW. 1905. Histological Studies on the Localisation of Cerebral Function. Cambridge, United Kingdom: Cambridge University Press [Google Scholar]
- Charvet CJ, Cahalane DJ, Finlay BL. 2015. Systematic, cross-cortex variation in neuron numbers in rodents and primates. Cereb Cortex 25:147–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin G, Sporns O, Mandl RC, van den Heuvel MP. 2013. Structural and functional aspects relating to cost and benefit of rich club organization in the human cerebral cortex. Cereb Cortex 24:2258–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Reus M, van den Heuvel M. 2013a. Estimating false positives and negatives in brain networks. Neuroimage 70:402–409 [DOI] [PubMed] [Google Scholar]
- de Reus MA, van den Heuvel MP. 2013b. Rich club organization and intermodule communication in the cat connectome. J Neurosci 33:12929–12939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Reus MA, van den Heuvel MP. 2014. Simulated rich club lesioning in brain networks: a scaffold for communication and integration? Front Hum Neurosci 8:647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan RS, Segonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, et al. 2006. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 31:968–980 [DOI] [PubMed] [Google Scholar]
- Dombrowski SM, Hilgetag CC, Barbas H. 2001. Quantitative architecture distinguishes prefrontal cortical systems in the rhesus monkey. Cereb Cortex 11:975–988 [DOI] [PubMed] [Google Scholar]
- Dosenbach NU, Nardos B, Cohen AL, Fair DA, Power JD, Church JA, et al. 2010. Prediction of individual brain maturity using fMRI. Science 329:1358–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elston GN. 2003. Cortex, cognition and the cell: new insights into the pyramidal neuron and prefrontal function. Cereb Cortex 13:1124–1138 [DOI] [PubMed] [Google Scholar]
- Elston GN, Benavides-Piccione R, DeFelipe J. 2001. The pyramidal cell in cognition: a comparative study in human and monkey. J Neurosci 21:RC163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elston GN, Benavides-Piccione R, Elston A, Manger PR, Defelipe J. 2011. Pyramidal cells in prefrontal cortex of primates: marked differences in neuronal structure among species. Front Neuroanat 5:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair DA, Cohen AL, Dosenbach NUF, Church JA, Miezin FM, Barch DM, et al. 2008. The maturing architecture of the brain's default network. Proc Natl Acad Sci U S A 105:4028–4032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair DA, Cohen AL, Power JD, Dosenbach NU, Church JA, Miezin FM, et al. 2009. Functional brain networks develop from a “local to distributed” organization. PLoS Comput Biol 5:e1000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felleman DJ, Van Essen DC. 1991. Distributed hierarchical processing in the primate cerebral cortex. Cereb Cortex 1:1–47 [DOI] [PubMed] [Google Scholar]
- Finlay BL, Uchiyama R. 2015. Developmental mechanisms channeling cortical evolution. Trends Neurosci 38:69–76 [DOI] [PubMed] [Google Scholar]
- Fischl B, Dale AM. 2000. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci U S A 97:11050–11055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flechsig PE. 1920. Anatomy of the human brain and spinal cord based on myelogenesis [in German]. Leipzig, Germany: Thieme [Google Scholar]
- Fulcher BD, Fornito A. 2016. A transcriptional signature of hub connectivity in the mouse connectome. Proc Natl Acad Sci U S A 113:1435–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Sotiropoulos SN, Wilson JA, Coalson TS, Fischl B, Andersson JL, et al. 2013. The minimal preprocessing pipelines for the human connectome project. Neuroimage 80:105–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulas A, Bastiani M, Bezgin G, Uylings HBM, Roebroeck A, Stiers P. 2014. Comparative analysis of the macroscale structural connectivity in the macaque and human brain. PLoS Comput Biol 10:e1003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulas A, Uylings HBM, Hilgetag CC. 2017. Principles of ipsilateral and contralateral cortico-cortical connectivity in the mouse. Brain Struct Funct 222:1281–1295 [DOI] [PubMed] [Google Scholar]
- Hagmann P, Cammoun L, Gigandet X. 2008. Mapping the structural core of human cerebral cortex. PLoS Biol 6:e159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann P, Sporns O, Madan N, Cammoun L, Pienaar R, Wedeen VJ, et al. 2010. White matter maturation reshapes structural connectivity in the late developing human brain. Proc Natl Acad Sci U S A 107:19067–19072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harriger L, van den Heuvel MP, Sporns O. 2012. Rich club organization of macaque cerebral cortex and its role in network communication. PLoS One 7:e46497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgetag CC, Grant S. 2010. Cytoarchitectural differences are a key determinant of laminar projection origins in the visual cortex. Neuroimage 51:1006–1017 [DOI] [PubMed] [Google Scholar]
- Hopkins WD, Rilling JK. 2000. A comparative MRI study of the relationship between neuroanatomical asymmetry and interhemispheric connectivity in primates: implication for the evolution of functional asymmetries. Behav Neurosci 114:739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innocenti GM, Price DJ. 2005. Exuberance in the development of cortical networks. Nat Rev Neurosci 6:955–965 [DOI] [PubMed] [Google Scholar]
- Jacobs B, Schall M, Prather M, Kapler E, Driscoll L, Baca S, et al. 2001. Regional dendritic and spine variation in human cerebral cortex: a quantitative Golgi study. Cereb Cortex 11:558–571 [DOI] [PubMed] [Google Scholar]
- Jones DK. 2008. Studying connections in the living human brain with diffusion MRI. Cortex 44:936–952 [DOI] [PubMed] [Google Scholar]
- Kaas JH, Schüz A, Miller R. 2002. Cortical areas and patterns of cortico-cortical connections. In: Schüz A, Miller R (eds.) Cortical Areas: Unity and Diversity. London, United Kingdom: Taylor & Francis, pp. 179–191 [Google Scholar]
- Kale P, Zalesky A, Gollo LL. 2018. Estimating the impact of structural directionality: how reliable are undirected connectomes? Netw Neurosci 2:259–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMantia AS, Rakic P. 1994. Axon overproduction and elimination in the anterior commissure of the developing rhesus monkey. J Comp Neurol 340:328–336 [DOI] [PubMed] [Google Scholar]
- Li L, Hu X, Preuss TM, Glasser MF, Damen FW, Qiu Y, Rilling J. 2013. Mapping putative hubs in human, chimpanzee and rhesus macaque connectomes via diffusion tractography. Neuroimage 80:462–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Liu Y, Li J, Qin W, Li K, Yu C, Jiang T. 2009. Brain anatomical network and intelligence. PLoS Comput Biol 5:e1000395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean EL, Hare B, Nunn CL, Addessi E, Amici F, Anderson RC, et al. 2014. The evolution of self-control. Proc Natl Acad Sci U S A 111:E2140–E2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesulam M-M. 1998. From sensation to cognition. Brain 121:1013–1052 [DOI] [PubMed] [Google Scholar]
- Miller DJ, Duka T, Stimpson CD, Schapiro SJ, Baze WB, McArthur MJ, et al. 2012. Prolonged myelination in human neocortical evolution. Proc Natl Acad Sci U S A 109:16480–16485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Dominguez O, Mills BD, Grayson D, Woodall A, Grant KA, Kroenke CD, Fair DA. 2014. Bridging the gap between the human and macaque connectome: a quantitative comparison of global interspecies structure-function relationships and network topology. J Neurosci 34:5552–5563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SW, Harris JA, Ng L, Winslow B, Cain N, Mihalas S, et al. 2014. A mesoscale connectome of the mouse brain. Nature 508:207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya DN, Sanides F. 1973. Architectonic parcellation of the temporal operculum in rhesus monkey and its projection pattern. Anat Embryol 139:127–161 [DOI] [PubMed] [Google Scholar]
- Park H-J, Friston K. 2013. Structural and functional brain networks: from connections to cognition. Science 342:1238411. [DOI] [PubMed] [Google Scholar]
- Pessoa L. 2012. Beyond brain regions: network perspective of cognition–emotion interactions. Behav Brain Sci 35:158–159 [DOI] [PubMed] [Google Scholar]
- Petanjek Z, Judaš M, Šimić G, Rašin MR, Uylings HBM, Rakic P, Kostović I. 2011. Extraordinary neoteny of synaptic spines in the human prefrontal cortex. Proc Natl Acad Sci U S A 108:13281–13286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DJ, Ferrer JM. 1993. The incidence of bifurcation among corticocortical connections from area 17 in the developing visual cortex of the cat. Eur J Neurosci 5:223–231 [DOI] [PubMed] [Google Scholar]
- Rilling JK, Insel TR. 1999. Differential expansion of neural projection systems in primate brain evolution. Neuroreport 10:1453–1459 [DOI] [PubMed] [Google Scholar]
- Roberts JA, Perry A, Lord AR, Roberts G, Mitchell PB, Smith RE, et al. 2016. The contribution of geometry to the human connectome. Neuroimage 124:379–393 [DOI] [PubMed] [Google Scholar]
- Rubinov M, Ypma RJF, Watson C, Bullmore ET. 2015. Wiring cost and topological participation of the mouse brain connectome. Proc Natl Acad Sci U S A 112:10032–10037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallet J, Mars RB, Noonan MP, Neubert FX, Jbabdi S, O'Reilly JX, et al. 2013. The organization of dorsal frontal cortex in humans and macaques. J Neurosci 33:12255–12274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell JW, Blakemore C, Young MP. 1995. Analysis of connectivity in the cat cerebral cortex. J Neurosci 15:1463–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholtens LH, de Reus MA, van den Heuvel MP. 2015. Linking contemporary high resolution magnetic resonance imaging to the von economo legacy: a study on the comparison of MRI cortical thickness and histological measurements of cortical structure. Hum Brain Mapp 36:3038–3046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholtens LH, Schmidt R, de Reus MA, van den Heuvel MP. 2014. Linking macroscale graph analytical organization to microscale neuroarchitectonics in the macaque connectome. J Neurosci 34:12192–12205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi SS, Zerbi V, Wenderoth N, Fornito A, Fulcher BD. 2017. Structural connectome topology relates to regional BOLD signal dynamics in the mouse brain. Chaos 27:047405. [DOI] [PubMed] [Google Scholar]
- Shanahan M. 2012. The brain's connective core and its role in animal cognition. Philos Trans R Soc Lond B Biol Sci 367:2704–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloper JJ, Powell TP. 1979. A study of the axon initial segment and proximal axon of neurons in the primate motor and somatic sensory cortices. Philos Trans R Soc Lond B Biol Sci 285:173–197 [DOI] [PubMed] [Google Scholar]
- Swanson L. 1992. Brain Maps: Structure of the Rat Brain. Amsterdam: Elsevier [Google Scholar]
- Tomasi D, Volkow ND. 2011. Association between functional connectivity hubs and brain networks. Cereb Cortex 21:2003–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnavi SN, Vlassenko AG, Rundle MM, Snyder AZ, Mintun MA, Raichle ME. 2010. Regional aerobic glycolysis in the human brain. Proc Natl Acad Sci U S A 107:17757–17762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Bullmore ET, Sporns O. 2016a. Comparative connectomics. Trends Cogn Sci 20:345–361 [DOI] [PubMed] [Google Scholar]
- van den Heuvel MP, de Reus MA. 2014. Chasing the dreams of early connectionists. ACS Chem Neurosci 5:491–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, de Reus MA, Feldman Barrett L, Scholtens LH, Coopmans FMT, Schmidt R, et al. 2015a. Comparison of diffusion tractography and tract-tracing measures of connectivity strength in rhesus macaque connectome. Hum Brain Mapp 36:3064–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Kahn RS, Goñi J, Sporns O. 2012. High-cost, high-capacity backbone for global brain communication. Proc Natl Acad Sci U S A 109:11372–11377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Kersbergen KJ, de Reus MA, Keunen K, Kahn RS, Groenendaal F, et al. 2015b. The neonatal connectome during preterm brain development. Cereb Cortex 25:3000–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Scholtens LH, de Reus MA, Kahn RS. 2016b. Associated microscale spine density and macroscale connectivity disruptions in schizophrenia. Biol Psychiatry 80:293–301 [DOI] [PubMed] [Google Scholar]
- van den Heuvel MP, Scholtens LH, Feldman Barrett L, Hilgetag CC, de Reus MA. 2015c. Bridging cytoarchitectonics and connectomics in human cerebral cortex. J Neurosci 35:13943–13948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Scholtens LH, Reus MA. 2015d. Topological organization of connectivity strength in the rat connectome. Brain Struct Funct 221:1719–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Sporns O. 2013a. An anatomical substrate for integration among functional networks in human cortex. J Neurosci 33:14489–14500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Sporns O. 2013b. Network hubs in the human brain. Trends Cogn Sci 17:683–696 [DOI] [PubMed] [Google Scholar]
- van den Heuvel MP, Stam CJ, Kahn RS, Hulshoff Pol HE. 2009. Efficiency of functional brain networks and intellectual performance. J Neurosci 29:7619–7624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Smith SM, Barch DM, Behrens TE, Yacoub E, Ugurbil K. 2013. The WU-Minn Human Connectome Project: an overview. Neuroimage 80:62–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstraete E, Veldink JH, van den Berg LH, van den Heuvel MP. 2014. Structural brain network imaging shows expanding disconnection of the motor system in amyotrophic lateral sclerosis. Hum Brain Mapp 35:1351–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bonin G, Bailey P. 1947. The Neocortex of Macaca Mulatta. Urbana: Univ. of Illinois Press [Google Scholar]
- von Economo CF, Koskinas GN. 1925. The cytoarchitectonics of the human cerebral cortex [in German]. Berlin, Germany: J. Springer [Google Scholar]
- Walker E. 1940. A cytoarchitectural study of the prefrontal area of the macaque monkey. J Comp Neurol 73:59–86 [Google Scholar]
- Wylie KP, Kronberg E, Maharajh K, Smucny J, Cornier M-A, Tregellas JR. 2015. Between-network connectivity occurs in brain regions lacking layer IV input. Neuroimage 116:50–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarkoni T. 2009. Big Correlations in little studies: inflated fMRI correlations reflect low statistical power-commentary on Vul et al. (2009). Perspect Psychol Sci 4:294–298 [DOI] [PubMed] [Google Scholar]
- Yeh F-C, Wedeen VJ, Tseng W-YI. 2010. Generalized q-sampling imaging. IEEE Trans Med Imaging 29:1626–1635 [DOI] [PubMed] [Google Scholar]
- Zamora-López G, Zhou C, Kurths J. 2010. Cortical hubs form a module for multisensory integration on top of the hierarchy of cortical networks. Front Neuroinform 4:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamora-López G, Zhou C, Kurths J. 2011. Exploring brain function from anatomical connectivity. Front Neurosci 5:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.