Abstract
Lung adenocarcinoma (LACA) is the leading cause of cancer-associated death worldwide. The present study intended to identify DNA methylation patterns that may serve as diagnostic and prognostic biomarkers for LACA. Data on DNA methylation and the survival data of the patients of LACA were obtained from The Cancer Genome Atlas. Kaplan-Meier curves and receiver operating characteristic curve analysis were utilized to build diagnostic and prognostic models. A total of 13 CpG sites were identified and validated as the optimal diagnostic and prognostic signature for overall survival. It was concluded that the CpG-based signature is a reliable predictor for the diagnosis and prognosis of patients with LACA.
Keywords: lung adenocarcinoma, methylation, prognostic signature
Introduction
Lung cancer, including small cell lung cancer (SCLC) and non-SCLC (NSCLC) (1,2), may be regarded as the most common tumor type and major contributor to the high tumor-associated mortality rate worldwide (3). Recent epidemiological data have revealed that it affected 1.8 million patients and resulted in 1.6 million deaths in 2012 (4). As one of the most frequent histological subtypes of lung cancer, lung adenocarcinoma (LACA) is a major cause of tumor-associated death (5,6). Despite recent advances in surgical techniques, radiotherapeutic interventions and combined chemotherapy strategies, the long-term survival rate of patients diagnosed with primary LACA has not significantly improved (7,8). Due to tumor heterogeneity factors and different molecular subtypes of LACA, its treatment faces large challenges. In this light, it is significant to identify specific details regarding characteristic molecules in LACA tissues to delineate the heterogeneity of LACA and develop strategies for personalized therapy.
In recent years, epigenetics, which has a critical role in carcinogenesis, has gained attention (9). Aberrant DNA methylation, as the core element of epigenetic modification, influences certain tumor suppressor genes and regulates gene functions (10,11). Increasing studies also demonstrated that DNA methylation is associated with genome stability, gene imprinting and cell differentiation (12,13). Thus, the methylation level is deemed a molecular biomarker for the diagnosis and prognostication of patients with tumors. However, the current expertise on the association between the epigenetic modifications and the clinically predicted outcomes of LACA is limited. Thus, in the present study, distinctive DNA methylation data for LACA vs. control tissues were acquired to evaluate the prognostic significance of distinctive DNA methylation patterns and provide insight regarding survival prediction for patients with LACA.
Materials and methods
Data processing
Original, publicly available and open-access genetic representation data of LACA samples and relevant clinical information of the patients obtained from The Cancer Genome Atlas (TCGA) database (http://cancergenome.nih.gov/) were included in the present study. The clinical information included the following attributes: Age, sex, ethnicity, stage and histological type of LACA. The exclusion criteria were as follows: i) Samples without clinical information, ii) samples without survival data, and iii) samples from patients that survived for <1 month. Ultimately, 447 LACA samples with DNA methylation data and clinical information were screened for further testing. The data were provided by TCGA and the study was performed in compliance with the TCGA publication guidelines (14).
Selection of differential DNA methylation sites
In the present study, aggregation and collection of DNA methylation information was performed by using R. First, the data were normalized by log2 transformation. The Limma package was employed for analyzing the differential DNA methylation sites between LACA tumor tissues and normal tissues. The fold changes (FCs) of DNA methylation were also calculated and significant aberrations in gene methylation were defined as those having a log2|FC|>2.0, P-value <0.01 and beta value >0.1. The least absolute shrinkage and selection operator (LASSO) method, which is suitable for regression of high-dimensional data (15), was used to select the most useful predictive features from the primary data set. The potential association of the CpG-based signature with LACA status was first assessed in the primary cohort and then validated in the validation cohort using a Mann-Whitney U-test. With this CpG-based signature, patients in each dataset were classified into a high-risk group and a low-risk group by using the median risk score. Kaplan-Meier curves and log-rank analysis were then performed to calculate the association between the CpG-based signature and patient's OS in the two groups with high-risk and low-risk CpG-based signatures. The CpG-based signature, which was significantly associated with OS (P<0.001), was then subjected to receiver operating characteristic (ROC) curve analysis to evaluate the predictive accuracy and sensitivity of the prognostic model. The area under the ROC curve (AUC) was also calculated. In the Kaplan-Meier curve, log-rank test and ROC analysis, the significance was defined as P<0.05.
Functional and pathway enrichment analysis
To further elucidate the biological functions of the mapped genes and the molecular mechanisms, a functional enrichment analysis was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (16). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and Gene Ontology (GO) enrichment analyses with P<0.05 were identified and biological process terms were further clustered using the Enrichment Map Plugin of Cytoscape (17).
Statistical analyses
Fundamental characteristics of the sample in the study were summarized by using descriptive statistics. Data for continuous variables, which were expressed as the mean ± standard deviation, were compared using the Student's t-test, Mann-Whitney U-test, Kruskal-Wallis H-test or one-way ANOVA with post hoc Student-Newman-Keuls tests, depending on the normality of data distribution as tested by Kolmogorov-Smirnov tests; data for categorical variables, which were presented as percentages, were compared using the Chi-square test. Statistical analyses were performed using SPSS 17.0 (SPSS Inc.) and R version 3.5.1 software (http://www.r-project.org/) (18). P<0.05 was considered to indicate a statistically significant difference.
Results
Patient characteristics
The data of all 447 samples with clinical information and methylation data available were obtained from the TCGA database. The samples included 440 LACA tissues and 7 normal tissues. Table I lists the detailed clinical characteristics of patients in the initial stage and specific groups (primary and validation cohort), including age at diagnosis, sex, ethnicity, disease stage and survival status. The results revealed that there were no major distinctions between the two groups in terms of these five clinical characteristics.
Table I.
Baseline clinical characteristics of the patients in the primary and validation cohorts.
| Characteristic | Primary cohort (n=220) | Validation cohort (n=440) | P-value |
|---|---|---|---|
| Age at diagnosis (years) | 66.6±9.9 | 65.4±10.1 | 0.86 |
| Sex | 1.00 | ||
| Female | 120 (54.5) | 240 (54.5) | |
| Male | 100 (45.5) | 200 (45.5) | |
| Ethnicity | 0.08 | ||
| Caucasian | 176 (80.0) | 340 (77.3) | |
| Of African descent | 22 (10.0) | 50 (11.4) | |
| Othersa | 22 (10.0) | 50 (11.4) | |
| Stage | 0.45 | ||
| I/II | 175 (77.3) | 336 (76.4) | |
| III/IV | 40 (18.2) | 97 (22.0) | |
| NA | 5 (2.3) | 7 (1.6) | |
| Survival status | 0.86 | ||
| Alive | 139 (63.2) | 275 (62.5) | |
| Dead | 81 (36.8) | 165 (37.5) |
Others includes patients of Native American, Asian and unknown descent/ethnicity. Values are expressed as the mean ± standard deviation or n (%). NA, not available.
Differential methylation sites in LACA
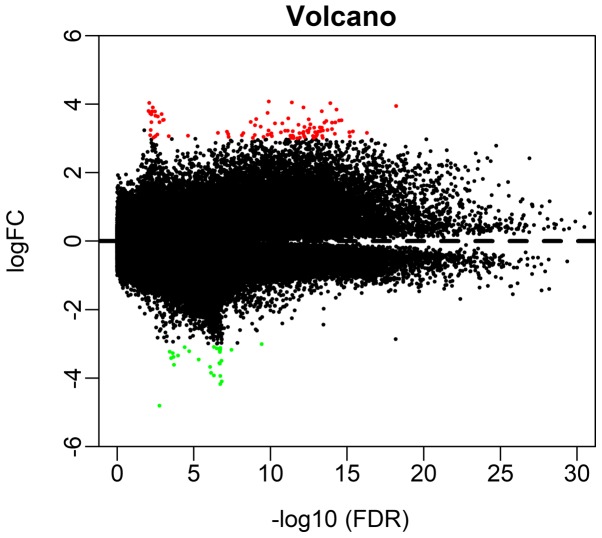
A total of 209 differential methylation sites were recognized between the LACA and regular tissue samples in the present study, including 133 hypermethylation and 76 hypomethylation sites. The distribution of hypermethylated and hypomethylated sites were visualized in Fig. 1. Furthermore, the methylation data were analyzed using the Limma incremental model. The five most hypermethylated sites (cg16306898, cg00648301, cg01869632, cg18837178 and cg22449330) and hypomethylated sites (cg05100666, cg15998127, cg07764932, cg12581354 and cg27649653) between LACA and normal tissue samples are listed in Table II.
Figure 1.
Volcano plot of differentially methylated loci. The red dots represent hypermethylated and the green dots represent hypomethylated loci. FC, fold change; FDR, false discovery rate.
Table II.
Top 5 hyper- and hypomethylated sites of differential methylation.
| A, Hypermethylated sites | |||
|---|---|---|---|
| Composite | Log FC | adj.P-val | Gene |
| cg16306898 | 4.07515 | 1.28×10−10 | TMEM240 |
| cg00648301 | 4.04696 | 3.99×10−12 | INSM1 |
| cg01869632 | 4.034999 | 8.17×10−3 | DUSP26 |
| cg18837178 | 4.034999 | 8.17×10−3 | LINC01194 |
| cg22449330 | 4.034999 | 8.17×10−3 | WDPCP |
| B, Hypomethylated sites | |||
| Composite | Log FC | adj.P-val | Gene |
| cg05100666 | −4.80454 | 1.76×10−3 | BRD9 |
| cg15998127 | −4.80454 | 1.76×10−3 | HDAC4 |
| cg07764932 | −4.17089 | 1.87×10−7 | ARHGAP6 |
| cg12581354 | −4.17089 | 1.87×10−7 | RP11-175P13.2 |
| cg27649653 | −4.17089 | 1.87×10−7 | AC010642.1 |
FC, fold change; adj.P-val, adjusted P-value. TMEM240, transmembrane protein 240; INSM1, INSM transcriptional repressor 1; DUSP26, dual specificity phosphatase 26; LINC01194, long intergenic non-protein coding RNA 1194; WDPCP, WD repeat containing planar cell polarity effector; BRD9, bromodomain containing 9; HDAC4, histone deacetylase 4; ARHGAP6, Rho GTPase activating protein 6.
Methylated loci signature
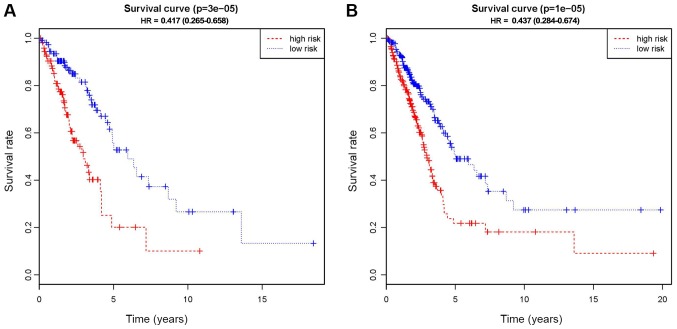
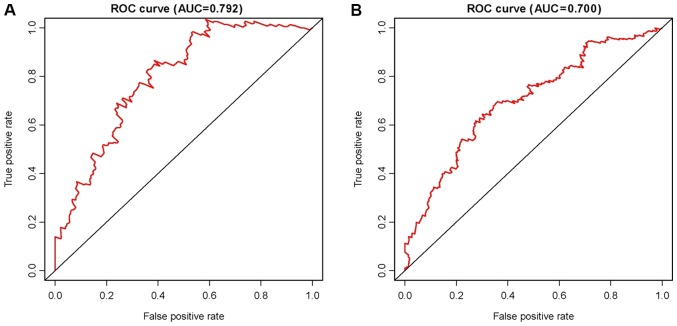
Based on the LASSO regression, 13 potential predictors were showed in the primary cohort (Fig. S1). As indicated in Fig. 2, Kaplan-Meier curves were drawn and the log-rank test was performed to evaluate the association between the CpG-based signals and the survival rates of patients with LUAD. The 13 methylated CpG signature, including cg00002719, cg02769743, cg05239163, cg05507908, cg07918170, cg08213398, cg08516516, cg08623223, cg12748948, cg14904034, cg16007456, ch.6.2958553R and cg19868631, was a significant predictor of OS in the primary cohort (P<0.01; Fig. 2A). The risk-score formula is as follows: (0.93× cg00002719) + (1.15× cg02769743) + (0.24× cg05239163) + (−1.20× cg05507908) + (1.31× cg07918170) + (0.83× cg08213398) + (0.54× cg08516516) + (0.06× cg08623223) + (0.12× cg12748948) + (0.03× cg14904034) + (−0.36× cg16007456) + (−0.36× ch.6.2958553R) + (−0.21× cg19868631). The mapped genes of the methylation sites are also provided in Table III. In addition, for identifying LACA, the approach of ROC curve analysis was pursued (Fig. 3). The AUC was 0.79, and the optimal cut-off value was 0.11 (Fig. 3A).
Figure 2.
Kaplan-Meier survival curves of the methylated sites in (A) the primary cohort and (B) the validation cohort. The red data-points represent high risk group and the blue data-points represent low risk group. HR, hazard ratio.
Table III.
Methylation loci significantly associated with survival.
| Composite | Chromosome | Start | End | Gene |
|---|---|---|---|---|
| cg00002719 | 1 | 169427468 | 169427469 | CCDC181 |
| cg02769743 | 1 | 9608344 | 9608345 | TMEM201 |
| cg05239163 | 1 | 154218790 | 154218791 | C1orf43 |
| cg05507908 | 5 | 75237454 | 75237455 | ANKRD31 |
| cg07918170 | 17 | 82932272 | 82932273 | TBCD |
| cg08213398 | 11 | 9722579 | 9722580 | SWAP70 |
| cg08516516 | 5 | 115816795 | 115816796 | CDO1 |
| cg08623223 | 8 | 11283906 | 11283907 | AF131216.1 |
| cg12748948 | 1 | 21779802 | 21779803 | USP48 |
| cg14904034 | 2 | 10389150 | 10389151 | HPCAL1 |
| cg16007456 | 1 | 100539082 | 100539083 | GPR88 |
| ch.6.2958553R | 6 | 152198831 | 152198831 | SYNE1 |
| cg19868631 | 7 | 54542083 | 54542084 | VSTM2A |
CCDC181, coiled-coil domain containing 181; TMEM201, transmembrane protein 201; C1orf43, chromosome 1 open reading frame 43; ANKRD31, ankyrin repeat domain 31; TBCD, tubulin folding cofactor D; SWAP70, switching B cell complex subunit SWAP70; CDO1, cysteine dioxygenase type 1; USP48, ubiquitin specific peptidase 48; HPCAL1, hippocalcin like 1; GPR88, G protein-coupled receptor 88; SYNE1, spectrin repeat containing nuclear envelope protein 1; VSTM2A, V-set and transmembrane domain containing 2A.
Figure 3.
ROC curve analysis for the survival prediction by methylated sites in (A) the primary cohort and (B) the validation cohort. ROC, receiver operating characteristic; AUC, area under the ROC curve.
Diagnostic and prognostic validation of the signature
To assess the utility of the 13 CpG-based signature in the diagnosis and prognosis of LUAD, the above-mentioned analyses were performed using the validation cohort. There was a marked distinction between the high-risk and low-risk groups in the primary cohort (P<0.01), which successfully provided confirmation in this process of validation (Fig. 2B). Subsequently, the signatures were tested using an ROC analysis of the validation cohort and the results revealed the AUC was 0.70, indicating that the signature was an effective predictor for LACA, although the AUC was lower than that in the primary cohort (Fig. 3B).
Functional enrichment analysis
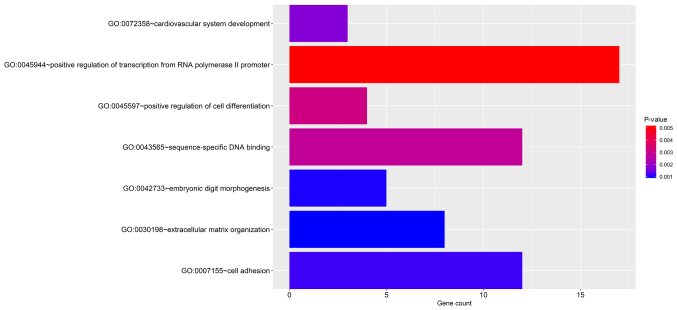
The significant terms of the GO enrichment analysis performed by DAVID and the KEGG pathways are provided in Fig. 4 and Fig. S2. The genes were significantly enriched KEGG pathways including extracellular matrix (ECM)-receptor interaction, cell adhesion molecules, PI3K-Akt signaling pathway, dilated cardiomyopathy and cysteine and methionine metabolism. In addition, the GO biological process terms were mainly enriched in ECM organization, embryonic digit morphogenesis, cell adhesion, cardiovascular system development, sequence-specific DNA binding, positive regulation of transcription from RNA polymerase II promoter and positive regulation of cell differentiation.
Figure 4.
Clustering analyses of significantly enriched biological process terms. The x-axis represents the number of genes accumulated in the pathways. Darker color indicates higher significance (smaller P-value). GO, Gene Ontology.
Discussion
Due to high invasion and poor prognosis, the outcome for patients with LACA remains unsatisfactory, with a 5-year OS rate of 4–17%, depending on the stage and regional differences (19,20). Most lung cancer patients are in the advanced stages at the time of diagnosis. It is achievable to enhance the efficiency of diagnosing and prognosticating LACA patients once the indication of the tumor's presence is able to be detected and explored at an early stage. Thus, in-depth studies on the aetiological elements and progressive mechanisms, early detection of the prognostic markers and identification of specific methylation CpG sites are urgently required.
For the purpose of clear classification of different secondary types of cancer, making use of CpG methylation locations is probably more efficient than it would be to collect detailed information on genetic representation based on covalent chemical alterations and steady hysterogenic markers of conjugated duplication (21). Hence, in the present study, a signature of 13 CpG sites with differential methylation was established, which were as follows: cg00002719, cg02769743, cg05239163, cg05507908, cg07918170, cg08213398, cg08516516, cg08623223, cg12748948, cg14904034, cg16007456, ch.6.2958553R and cg19868631. The 13 CpG signature, which was significantly associated with the OS of patients with LACA, was also recognized as an independent element for diagnosing LACA and predicting the prognosis of the patients. Furthermore, compared with that of the high-risk patients, low-risk patients had better OS, and the 5-year survival rate in low-risk patients was also higher than that in high-risk patients. The functional enrichment analysis for the mapped genes of the CpG methylation sites were all presented by means of approaches of the fields of biology and information technology. The outcomes indicate that the methylation sites included in the 13-CpG-based prognostic signature may have a role in the molecular pathogenetic mechanisms and clinical progression in LACA patients, and this provides novel information for survival prediction and personalized treatment of patients with LACA.
As an important epigenetic mechanism in tumors, DNA methylation has a critical role in carcinogenesis (22). It regulates the extent of gene expression to control the function of the biomolecules encoded by those genes (23,24). Numerous studies have revealed that DNA methylation has a significant role in the initiation, progression and metastasis of cancer by controlling different aspects, including DNA repair, cell cycle regulation, angiogenesis and apoptosis (25). In the present study, 13 mapped genes were identified to be aberrantly methylated in LACA, which were as follows: Coiled-coil domain containing 181 (CCDC181), transmembrane protein 201 (TMEM201), chromosome 1 open reading frame 43 (C1orf43), ankyrin repeat domain 31 (ANKRD31), tubulin folding cofactor D (TBCD), switching B cell complex subunit SWAP70 (SWAP70), cysteine dioxygenase type 1 (CDO1), AF131216.1, ubiquitin specific peptidase 48 (USP48), hippocalcin like 1 (HPCAL1), G protein-coupled receptor 88 (GPR88), spectrin repeat containing nuclear envelope protein 1 (SYNE1) and V-set and transmembrane domain containing 2A (VSTM2A). Previous studies have directly shown that CCDC181, CDO1 and SYNE1 are associated with LACA. Gao et al (26) stated that CCDC181 could be a provisional prognostic biomarker of LACA. Moreover, Diaz-Lagares et al (27) indicated that there is a possibility for the cancer-specific methylation of CDO1 to enhance the diagnosis approach at the early stage and also achievements for patients. The methylation status of SYNE1 has also been valuable in estimating the sporadic lung cancer prognosis (28). However, no previous studies have reported on the association of TMEM201, C1orf43, ANKRD31, TBCD, SWAP70, AF131216.1, USP48, HPCAL1, GPR88 or VSTM2A and LACA. Hence, it is necessary to perform further studies on the methylation of these 10 genes and LACA.
In addition, GO annotations and KEGG pathways were established to provide detailed information regarding the molecular functions of the genes. The differentially methylated genes were mainly enriched in ECM, embryonic morphogenesis, cell adhesion and vascular system development; these are why high-risk tumors are more biologically aggressive. Generated through direct diffusion, lymphatic and vascular metastases are the common metastases in LACA. Once metastasis occurs, the prognosis of patients with LACA is poor. Increasing evidence has indicated that the ECM has a significant role in tumor occurrence and progression (29). Lim et al (30) developed a 29-gene ECM-associated signature to predict the prognosis of the patients at the early stage of NSCLC. Furthermore, it has been well established that the ECM is involved in regulating metastasis and invasion of lung cancer (31,32). Thus, it is necessary to perform in-depth research on these molecules to confirm these predictions, and simultaneously develop novel therapeutic interventions for LACA.
The present study does have certain limitations. First, in view of the LACA cohort exhibiting a reasonably high censored rate, this probably had an effect on the credibility of the Kaplan-Meier evaluation. Furthermore, as all of the samples analyzed in the present study were acquired from TCGA only, in-depth verification should be performed using independent datasets. In addition, the mechanistic role of each components of the prognostic signature remains to be investigated. Therefore, experimental research on cancer cell lines may provide significant information to further the understanding of their functional roles.
In conclusion, a 13 CpG-based prognostic signature for OS prediction in patients with LACA was obtained through comprehensively analyzing DNA methylation. The present results suggest that further research is required to validate the diagnostic ability of the novel diagnostic model in LACA. Retrospective and prospective studies may be performed to verify the prognostic utility of the CpG-based signature model.
Supplementary Material
Acknowledgements
Not applicable.
Glossary
Abbreviations
- NSCLC
non-small cell lung carcinoma
- LACA
lung adenocarcinoma
- TCGA
The Cancer Genome Atlas
- FC
fold change
- OS
overall survival
- ROC
receiver operating characteristic
- LASSO
least absolute shrinkage and selection operator
- DAVID
Database for Annotation, Visualization and Integrated Discovery
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- AUC
area under the ROC curve
- ECM
extracellular matrix
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
RZ, HX, WM, ZD and MH participated in the study design, analysis and interpretation of data and drafting of the manuscript. MW performed the research and revised the paper. XG contributed to the acquisition of data, collection of relevant literature and drafting of the article together. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Locher C, Debieuvre D, Coëtmeur D, Goupil F, Molinier O, Collon T, Dayen C, Le Treut J, Asselain B, Martin F, et al. Major changes in lung cancer over the last ten years in France: The KBP-CPHG studies. Lung Cancer. 2013;81:32–38. doi: 10.1016/j.lungcan.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 2.B'chir F, Laouani A, Ksibi S, Arnaud MJ, Saguem S. Cigarette filter and the incidence of lung adenocarcinoma among Tunisian population. Lung Cancer. 2007;57:26–33. doi: 10.1016/j.lungcan.2007.01.034. [DOI] [PubMed] [Google Scholar]
- 3.Woods LM, Coleman MP, Lawrence G, Rashbass J, Berrino F, Rachet B. Evidence against the proposition that ‘UK cancer survival statistics are misleading’: Simulation study with National Cancer Registry data. BMJ. 2011;342:3399. doi: 10.1136/bmj.d3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 5.Lemjabbar-Alaoui H, Hassan OU, Yang YW, Buchanan P. Lung cancer: Biology and treatment options. Biochim Biophys Acta. 2015;1856:189–210. doi: 10.1016/j.bbcan.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Travis WD. Pathology of lung cancer. Clin Chest Med. 2011;32:669–692. doi: 10.1016/j.ccm.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 7.Ganti AK, Mulshine JL. Lung cancer screening: Panacea or Pipe dream? Ann Oncol. 2005;16:215–219. doi: 10.1093/annonc/mdi723. [DOI] [PubMed] [Google Scholar]
- 8.Field JK, Raji OY. The potential for using risk models in future lung cancer screening trials. F1000 Med Rep. 2010;24:38. doi: 10.3410/M2-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Momparler RL. Cancer epigenetics. Oncogene. 2003;22:6479–6483. doi: 10.1038/sj.onc.1206774. [DOI] [PubMed] [Google Scholar]
- 10.Meng H, Cao Y, Qin J, Song X, Zhang Q, Shi Y, Cao L. DNA methylation, its mediators and genome integrity. Int J Biol Sci. 2015;11:604–617. doi: 10.7150/ijbs.11218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klutstein M, Nejman D, Greenfield R, Cedar H. DNA methylation in cancer and aging. Cancer Res. 2016;76:3446–3450. doi: 10.1158/0008-5472.CAN-15-3278. [DOI] [PubMed] [Google Scholar]
- 12.Berdasco M, Esteller M. Aberrant epigenetic landscape in cancer: How cellular identity goes awry. Dev Cell. 2010;19:698–711. doi: 10.1016/j.devcel.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 13.Senner CE. The role of DNA methylation in mammalian development. Reprod Biomed Online. 2011;22:529–535. doi: 10.1016/j.rbmo.2011.02.016. [DOI] [PubMed] [Google Scholar]
- 14.National Cancer Institute, corp-author. The Cancer Genome Atlas Program. http://cancergenome.nih.gov/publications/publicationguidelines. [Dec 21;2015 ];
- 15.Sauerbrei W, Royston P, Binder H. Selection of important variables and determination of functional form for continuous predictors in multivariable model building. Stat Med. 2007;26:5512–5528. doi: 10.1002/sim.3148. [DOI] [PubMed] [Google Scholar]
- 16.Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID Bioinformatics Resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:169–175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: A network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5:13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.R Core Team, corp-author. R Foundation for Statistical Computing; Vienna: 2014. R: A language and environment for statistical computing. [Google Scholar]
- 19.American Cancer Society, corp-author. American Cancer Society; Atlanta, GA: 2015. Cancer facts & figures 2015. [Google Scholar]
- 20.Grinberg-Rashi H, Ofek E, Perelman M, Skarda J, Yaron P, Hajdúch M, Jacob-Hirsch J, Amariglio N, Krupsky M, Simansky DA, et al. The expression of three genes in primary non-small cell lung cancer is associated with metastatic spread to the brain. Clin Cancer Res. 2009;15:1755–1761. doi: 10.1158/1078-0432.CCR-08-2124. [DOI] [PubMed] [Google Scholar]
- 21.Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38:23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lorincz MC, Schübeler D. Evidence for converging DNA methylation pathways in placenta and cancer. Dev Cell. 2017;43:257–258. doi: 10.1016/j.devcel.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 23.Ding YX, Cui H. Integrated analysis of genome-wide DNA methylation and gene expression data provide a regulatory network in intrauterine growth restriction. Life Sci. 2017;179:60–65. doi: 10.1016/j.lfs.2017.04.020. [DOI] [PubMed] [Google Scholar]
- 24.Lim JH, Kim SY, Han JY, Kim MY, Park SY, Ryu HM. Comprehensive investigation of DNA methylation and gene expression in trisomy 21 placenta. Placenta. 2016;42:17–24. doi: 10.1016/j.placenta.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 25.Zhang X, Feng H, Li D, Liu S, Amizuka N, Li M. Identification of differentially expressed genes induced by aberrant methylation in oral squamous cell carcinomas using integrated bioinformatic analysis. Int J Mol Sci. 2018;19:1698. doi: 10.3390/ijms19061698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao C, Zhuang J, Li H, Liu C, Zhou C, Liu L, Sun C. Exploration of methylation-driven genes for monitoring and prognosis of patients with lung adenocarcinoma. Cancer Cell Int. 2018;18:194. doi: 10.1186/s12935-018-0691-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diaz-Lagares A, Mendez-Gonzalez J, Hervas D, Saigi M, Pajares MJ, Garcia D, Crujerias AB, Pio R, Montuenga LM, Zulueta J, et al. A novel epigenetic signature for early diagnosis in lung cancer. Clin Cancer Res. 2016;22:3361–3371. doi: 10.1158/1078-0432.CCR-15-2346. [DOI] [PubMed] [Google Scholar]
- 28.Tessema M, Belinsky SA. Mining the epigenome for methylated genes in lung cancer. Proc Am Thorac Soc. 2008;5:806–810. doi: 10.1513/pats.200805-045TH. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Multhaupt HA, Leitinger B, Gullberg D, Couchman JR. Extracellular matrix component signaling in cancer. Adv Drug Deliv Rev. 2016;97:28–40. doi: 10.1016/j.addr.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 30.Lim SB, Tan SJ, Lim WT, Lim CT. An extracellular matrix-related prognostic and predictive indicator for early-stage non-small cell lung cancer. Nat Commun. 2017;8:1734. doi: 10.1038/s41467-017-01430-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peng DH, Ungewiss C, Tong P, Byers LA, Wang J, Canales JR, Villalobos PA, Uraoka N, Mino B, Behrens C, et al. ZEB1 induces LOXL2-mediated collagen stabilization and deposition in the extracellular matrix to drive lung cancer invasion and metastasis. Oncogene. 2017;36:1925–1938. doi: 10.1038/onc.2016.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stevens LE, Cheung WKC, Adua SJ, Arnal-Estapé A, Zhao M, Liu Z, Brewer K, Herbst RS, Nguyen DX. Extracellular matrix receptor expression in subtypes of lung adenocarcinoma potentiates outgrowth of micrometastases. Cancer Res. 2017;77:1905–1917. doi: 10.1158/0008-5472.CAN-16-1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.