Abstract
MicroRNA-138 (miR-138) acts as a key regulator in the modulation of carcinogenesis in numerous tumor types. Chemoresistance is common and relevant to the failure of multiple treatment strategies for cervical cancer. However, the biological role of miR-138 in the progression and chemosensitivity of cervical cancer is still unclear. The present study aimed to investigate the expression, function and mechanism of miR-138 in cervical cancer. An miR-138 mimic, inhibitor and negative control were transfected into SiHa and C33A cells. The expression of miR-138 and its target were assessed by reverse transcription-PCR, western blotting and immunohistochemistry. The functional significance of miR-138 in tumor progression and chemosensitivity to cisplatin in vitro was examined by Cell Counting Kit-8, flow cytometry, wound healing and Transwell assays. A tumor xenograft model was used to validate the effects in vivo. These results demonstrated that miR-138 was significantly downregulated in cervical cancer cells. Overexpression of miR-138 suppressed cervical cancer cell proliferation, invasion, increased apoptosis and enhanced chemotherapy sensitivity in vivo and in vitro. Furthermore, bioinformatics analysis and dual luciferase reporter assays demonstrated that H2AX served as a target for miR-138, and the rescue experiment revealed that H2AX was a functional target of miR-138. The protective effects of miR-138 overexpression were dependent on H2AX. This study provides evidence that miR-138/H2AX may be a novel therapeutic target in cervical cancer.
Keywords: microRNA-138, cervical cancer, progression, chemosensitivity, H2AX
Introduction
Cervical cancer has become the fourth most common cause of cancer-related death and the second most common malignancy in women worldwide (1). Despite improved treatment, the number of women succumbing to cervical cancer is increasing. The prognosis of patients with advanced/recurrent cervical cancer is poor and 5-year of survival is only 10–20% (2). Tumor metastasis and recurrence are the main cause of the poor outcome and high mortality. The current standardized treatment for advanced/recurrent cervical cancer is cisplatin-based chemotherapy (3,4). However, numerous patients do not benefit from chemotherapy and tumors often develop chemoresistance (5). Therefore, there is an urgent requirement to uncover the mechanisms of progression and chemoresistance in cervical carcinoma. Notably, recent studies have demonstrated that microRNAs (miRNAs/miRs) may be involved in drug resistance (6–8).
miRNAs are small single stranded non-coding RNA molecules of 17–24 nucleotides, which regulate targeted genes by inducing post-transcriptional degradation or suppressing translation (9). Aberrant miRNA expression has been found to play a vital role in the progression of cervical cancer. It has been reported that miR-150 is overexpressed in patient tissues and promotes cervical cancer cell viability, migration and invasion (10). miR-506 suppresses tumor growth and enhances apoptosis and chemosensitivity in cervical cancer (11). miR-138 has been reported to function as a tumor suppressor, since it plays critical role in certain pathological processes, including tumor progression, metastasis, invasion and apoptosis (12–14). However, the function and chemoresistance mechanism of miR-138 in cervical cancer are poorly understood.
In the current study, the expression and function of miR-138 in cervical cancer were investigated in vitro and in vivo. Furthermore, the target of miR-138 was identified. The results revealed that miR-138 was downregulated in cervical cancer cells. In vitro and in vivo, miR-138 suppressed progression and enhanced chemosensitivity. Dual-luciferase reporter assays and rescue experiments revealed that histone H2AX phosphorylation (H2AX) was the functional target gene of miR-138. The results of the present study indicate that miR-138 may be a novel biomarker of progression and chemoresistance in cervical cancer.
Materials and methods
Cell lines
Cervical cancer cells SiHa and C33A and immortalized normal cervical epithelial squamous cell line H8 were obtained from the Cell Bank of the Chinese Academy of Sciences. SiHa and H8 cells were cultured in Dulbecco's modified Eagle's medium (Thermo Fisher Scientific, Inc.) and C33A cells were cultured in minimum Eagle's medium (Thermo Fisher Scientific, Inc.) each supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, Inc.), in a 37°C incubator with 5% CO2.
Transfection
In total, 50 nM miR-138 mimic (5′-AGCUGGUGUUGUAAUCAGGCCGGCCUGAUUCACAACACCAGCUUU-3′), 100 nM miR-138 inhibitor (5′-CGGCCUGAUUCACAACACCAGCU-3′) or 50 nM miR-negative control (NC; sense, 5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense, 5′-ACGUGACACGUUCGGAGAATT-3′) were transfected into the cells using Lipofectamine® 3000 (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Following 48 h transfection, the cells were subjected to further experimentation. The miR-138 mimic, miR-138 inhibitor and miR-NC were designed by Shanghai GenePharma Co., Ltd. The H2AX expression vector, carrying the human H2AX-coding DNA sequence, was cloned into a pcDNA3.1 vector (Thermo Fisher Scientific, Inc.).
RNA detection
miRNA and total RNA extraction were extracted from cells using miRcute miRNA Isolation Kit (Tiangen Biotech Co., Ltd.) or TRIzol® reagent (Thermo Fisher Scientific, Inc.) according to the manufacturers' protocols. cDNA was synthesized using the miRcute Plus miRNA First-Strand cDNA Kit (Tiangen Biotech Co., Ltd.) according to the manufacturer's protocol. The miRNA cDNA reverse transcription conditions were as follows: 42°C for 60 min and 95°C for 3 min. Total RNA was reverse transcribed into cDNA with the following conditions: 42°C for 60 min and 70°C for 5 min. miRNA and mRNA expression levels were determined using miRcute Plus miRNA qPCR Kit (SYBR® Green; Tiangen Biotech Co., Ltd.) and SYBR® Green Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.), respectively. The thermocycling conditions for miRNA were: Initial denaturation at 95°C for 15 min, followed by 45 cycles of denaturation at 94°C for 20 sec and anneal/extend at 60°C for 34 sec. The thermocycling conditions for mRNA were: Initial denaturation at 95°C for 2 min, followed by 40 cycles of denaturation at 95°C for 15 sec and anneal/extend at 60°C for 1 min. The internal controls were U6 and β-actin. Relative expression was calculated and normalized using the 2−ΔΔCq method (15). The primer sequences are listed in Table I.
Table I.
Reverse transcription-PCR Primer Sequences.
| Primer | Sequence |
|---|---|
| β-actin | 5′-TGTCCACCTTCCAGCAGATGT-3′ (Forward) |
| 5′-GCTCAGTAACAGTCCGCCTAGA-3′ (Reverse) | |
| H2AX | 5′-CGGCAGTGCTGGAGTACCTCA-3′ (Forward) |
| 5′-AGCTCCTCGTCGTTGCGGATG-3′ (Reverse) | |
| GAPDH | 5′-AGAAGGCTGGGGCTCATTTG −3′ (Forward) |
| 5′-AGGGGCCATCCACAGTCTTC −3′ (Reverse) | |
| miR-138 | 5′-AGCUGGUGUUGUGAAUCAGGCCG-3′ (Forward) |
| 5′-TGGTGTCGTGGAGTCG-3′ (Reverse) |
miR, microRNA; H2AX, histone H2A.
Cell proliferation and chemosensitivity assay
To analyze cell proliferation, a Cell Counting Kit-8 assay (CCK-8; Dojindo Molecular Technologies, Inc.) was used according to the manufacturer's protocol. Cells were plated in 96-well plates (2,000 cells/well) following a 24 h transfection. After overnight incubation, the cells were incubated with CCK-8 solution at 37°C for 2 h. Next, viable cell numbers were measured by absorbance at 450 nm using a spectrophotometer. Cells were plated in 96-well plates (4,000 cells/well) and treated with different doses of cisplatin (Sigma-Aldrich; Merck KGaA) following transfection for 24 h. Subsequently, the toxicity of DDP in the cells was determined using a CCK-8 assay.
EdU staining
EdU assays (Guangzhou RiboBio Co., Ltd.) were performed according to the manufacturer's protocol. Transfected cells were incubated with 50 µM of EdU labeling medium for 2 h at 37°C. Then, 4% paraformaldehyde (50 µl/well) was used to fix cells for 20 min at 37°C, prior to incubation with glycine (2 mg/ml). After ~5 min, 200 µl 1X Apollo solution was used to stain cells for 30 min at 37°C in the dark. Triton X-100 (0.5%) in PBS was used to wash cells for 5 min, then three random fields of view per slide were examined using a fluorescence microscope (Olympus Corporation). The number of proliferative cells (EdU positive) was counted.
Apoptosis assay
SiHa and C33A cells (1×106) were seeded in 6-well plates and treated with cisplatin (10 µM/ml) for 24 h prior to harvesting. An Annexin V-FITC Apoptosis Detection kit (BD Biosciences) was used to perform flow cytometry analysis. Cell apoptosis was measured using BD FACSCanto™ II flow cytometer (BD Biosciences) and analyzed using the FlowJo software (version 10.0.7; FlowJo LLC).
Cell migration assay
A 24-well Transwell chamber assay (pore size, 8 µm) was used to perform migration experiments (BD Biosciences). Transfected cervical cancer cells (2×105 cells/well) were placed into the upper chambers. After 24 h of incubation, the cells that had migrated through the membrane were fixed in 4% methanol for 30 min and stained with 0.05% crystal violet for 30 min, both at room temperature. Then, the invaded cells were quantified under a microscope (Olympus Corporation).
Wound-healing assay
Cells (1×106 cells/well) were incubated in a 24-well plate; the cell monolayer was then wounded with a 100 µl pipette tip and cultured in serum-free medium. Cell migration images were captured at 0 and 24 h.
Prediction of miR-138 targets using TargetScan
Potential targets of miR-138 were searched using the TargetScan software (release 7.2; http://www.targetscan.org/vert_72/), and the resulting search resulted in H2AX being a candidate.
Luciferase reporter assay
The wild-type 3′-untranslated region (UTR) sequences of H2AX cDNA (WT-H2AX) were synthesized into the luciferase reporter vector [Obio Technology (Shanghai) Corps., Ltd.]. A mutant H2AX 3′-UTR vector (MUT-H2AX) was also constructed, which contained a mutation in the predicted H2AX-binding sequence. The WT-H2AX or MUT-H2AX and the miR-138 mimic or miR-138 NC, were transfected into cervical cells using Lipofectamine® 3000 reagent (Thermo Fisher Scientific, Inc.). After 48 h transfection, a Luciferase-Reporter Assay System kit (Promega Corporation) was used to measure luciferase activity using Renilla luciferase activity as internal reference.
Western blotting analysis
Total protein was obtained following cell lysis, using RIPA buffer (Beyotime Institute of Biotechnology) supplemented with 1 nM phenylmethylsulfonyl fluoride. Total protein was collected and the concentration was determined with an Enhanced Bicinchoninic Acid Protein Assay kit (Beyotime Institute of Biotechnology). Proteins (30 µg) were subjected to denaturing 10% SDS-polyacrylamide gel electrophoresis and transferred onto a PVDF membrane (EMD Millipore). The following antibodies were used overnight at 4°C: Caspase-3 (1:1,000; Abcam; cat. no. ab13585), bcl-2 (1:1,000; Abcam; cat. no. ab117115), bax (1:1,000; Abcam; cat. no. ab32503), matrix metalloproteinase (MMP) 9 (1:1,000; Abcam; cat. no. ab38898), H2AX (1:1,000; Abcam; cat. no. ab81299) and β-actin (1:1,000; Proteintech Group, Inc.; cat. no. 66009-1-Ig). The membrane was incubated with horseradish peroxidase-conjugated anti-rabbit (1:4,000; cat. no. A0208; Beyotime Institute of Biotechnology) or anti-mouse (1:4,000; cat. no. A0216; Beyotime Institute of Biotechnology) for 2 h at room temperature. An enhanced chemiluminescence system (Thermo Fisher Scientific, Inc.) was used was used to produce signals and Quantity One software (version 3.0; Bio-Rad Laboratories, Inc.) was used for signal detection.
Animal experiment
A total of 20 (4-week old, 15–20 g) female BALB/c nude mice were purchased from the Shanghai Laboratory Animal Center. The animals were housed at 21–24°C with 50–60% relative humidity and a 12-h light/dark cycle. They had free access to food and water. All experiments were approved by the Animal Management Rule of the Chinese Ministry of Health (document 55, 2001) and approved by the Tongji University Animal Ethics Committee. Lentivirus miR-138 vector or empty vector (Obio Technology) was transfected into SiHa cells. A total of 5×106 cells stably transfected with empty vector or miR-138 vector were injected subcutaneously into the right flank region of each mouse. Tumor length and width were measured every 2 days using Vernier calipers, and tumor volume was calculated as: Volume = 0.5 × length × width2. When the tumor became palpable, the mice bearing either miR-138-expressing tumors or empty vector tumors were randomized into 4 groups (5 mice/group) and were intraperitoneally injected with PBS or cisplatin (10 mg/Kg). The mice were humanely sacrificed at 23 days after implantation and the xenograft tumors were collected for weighing and subsequent analysis.
Immunohistochemical (IHC) analysis
For histopathology, paraffin-embedded tissues obtained from xenograft mice were sectioned to a thickness of 4 µm. The expression of caspase-3, cleaved-caspase3, MMP9, H2AX and MDR1 was evaluated, as previously described (16).
Statistical analysis
All experiments were performed at least three times with duplicate or triplicate samples in each assay. Statistical analysis was performed using SPSS 17.0 software (SPSS, Inc.) and GraphPad Prism software 7.0 (GraphPad Software, Inc.). The Student's t-test and one-way analysis of variance followed by Tukey's or Student's t-test were performed to calculate the differences between groups. P<0.05 was considered to indicate a statistically significant difference. Data are presented as the mean ± standard deviation.
Results
miR-138 expression is downregulated in human cervical cancer cell lines
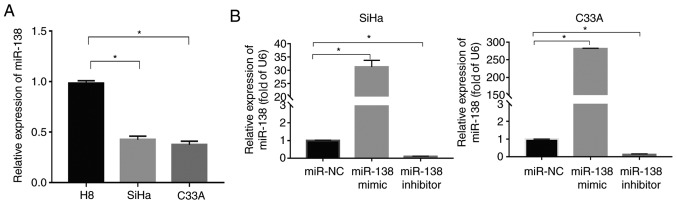
Reverse transcription (RT)-PCR was used to evaluate miR-138 expression in immortalized normal cervical epithelial squamous cell line H8 and cervical squamous cancer cell lines (SiHa and C33A). The results revealed that the miR-138 expression in the SiHa and C33A cells was significantly downregulated compared with in the H8 cells (P<0.05; Fig. 1A).
Figure 1.
Expression of miR-138 in human cervical cancer cell lines and transfection efficiency analysis in SiHa and C33A cells. (A) miR-138 expression is downregulated in human cervical cancer cell lines. Relative miR-138 expression levels of immortalized normal cervical epithelial squamous cell line H8 and two cervical cancer cells were analyzed by RT-PCR, with U6 as the internal control. Data are presented as the mean ± standard deviation. *P<0.05. (B) The relative expression of miR-138 in SiHa and C33A transfected cells was measured by RT-PCR and normalized to U6. Data are presented as the mean ± standard deviation. *P<0.05. RT, reverse transcription; miR, microRNA; NC, negative control.
miR-138 inhibits cell proliferation and induces apoptosis in vitro
To investigate the role of miR-138 in cervical cancer, the miR-138 mimic, miR-138 inhibitor and miR-NC were first transfected into SiHa and C33A cells. RT-PCR was used to detect the transfection efficiency. As shown in Fig. 1B, the level of miR-138 significantly increased in the cells transfected with miR-138 mimic (P<0.05), but the miR-138 inhibitor exhibited the opposite effect in both SiHa and C33A cells.
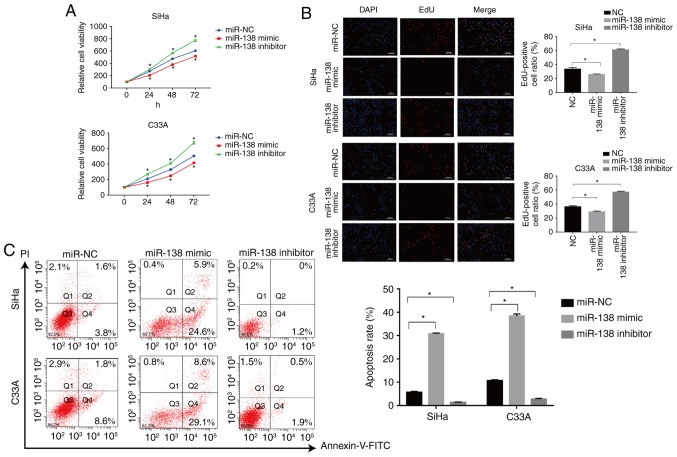
The CCK-8 and EdU assays were used to assess the proliferation of transfected SiHa and C33A cells for the indicated times. The miR-138 mimic significantly suppressed the proliferation of SiHa cells compared with the miR-NC-transfected cells, but miR-138 inhibitor significantly enhanced the proliferation of the cells (P<0.05; Fig. 2A). The effects of miR-138 were also validated with EdU staining. The number of EdU positive cells was significantly decreased in the miR-138 mimic-transfected group (P<0.05), while the number of EdU positive cells in miR-138 inhibitor-transfected group was significantly increased when compared with the miR-NC group in the SiHa cells, as shown in Fig. 2B (P<0.05). Similar results were also found in C33A cells. Flow cytometric analysis was used to assess the apoptosis of SiHa and C33A cells. As shown in Fig. 2C, the miR-138 mimic significantly induced apoptosis of SiHa cells when compared with the miR-NC group (P<0.05), but the miR-138 inhibitor significantly inhibited apoptosis (P<0.05). Similar results were also found in C33A cells. In addition, the expression of important proteins involved in apoptosis were also examined. The miR-138 mimic increased the expression of caspase-3 and Bax, and suppressed the expression of Bcl-2. By contrast, the miR-138 inhibitor significantly inhibited the expression of caspase-3 and Bax but promoted the expression of Bcl-2 (P<0.05; Fig. 3C). Taken together, these findings indicated that miR-138 inhibited proliferation and induced apoptosis in cervical cancer cells.
Figure 2.
miR-138 inhibits cervical cancer cell viability and induced apoptosis in vitro. (A) Cell Counting Kit-8 assay was used to detect the growth curves of transfected cells. (B) EdU was used to detect the cell proliferation rate of transfected cells. (C) Flow cytometry was used to evaluate the apoptosis rate of transfected cells. Data are presented as the mean ± standard deviation. *P<0.05. miR, microRNA; NC, negative control; FITC, fluorescein isothiocyanate; PI, propidium iodide.
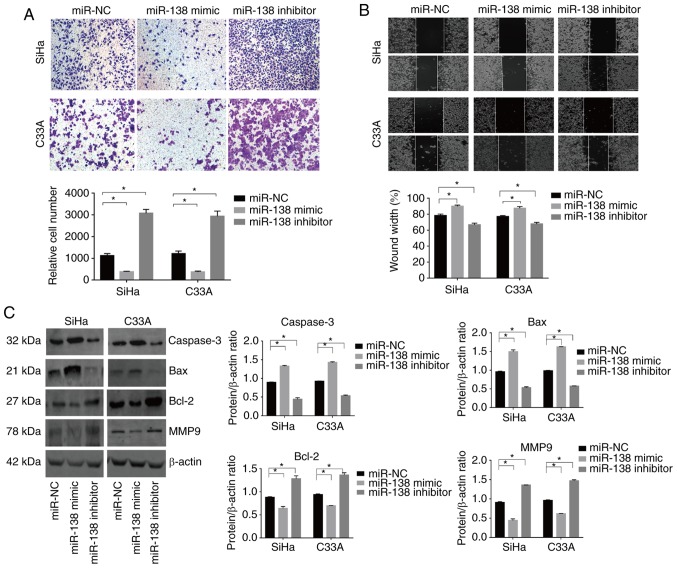
Figure 3.
miR-138 inhibits the cell mobility and the regulated the expression of proteins related to apoptosis and invasion in vitro. (A) A transwell assay and (B) wound-healing assay were used to assess invasion. Magnification, ×200. (C) Caspase-3, Bax, Bcl-2 and MMP9 expression in the transfected cells was evaluated by western blot analysis, and β-actin was used as the control. Data are presented as the mean ± standard deviation. *P<0.05. miR, microRNA; MMP, matrix metalloproteinase; NC, negative control.
miR-138 inhibits cell mobility in vitro
Transwell and wound-healing assays were used to evaluate the effect of miR-138 on the mobility of cervical cancer cells. The miR-138 mimic significantly decreased the number of invading cells in the SiHa and C33A lines. By contrast, the miR-138 inhibitor exhibited the opposite effect (Fig. 3A and B). In addition, the expression of important proteins involved in metastasis were examined. The expression of MMP9 was decreased in the miR-138 mimic-transfected cells, though the miR-138 inhibitor induced the opposite effect (Fig. 3D).
miR-138 inhibits cell proliferation, invasion and induces apoptosis in vivo
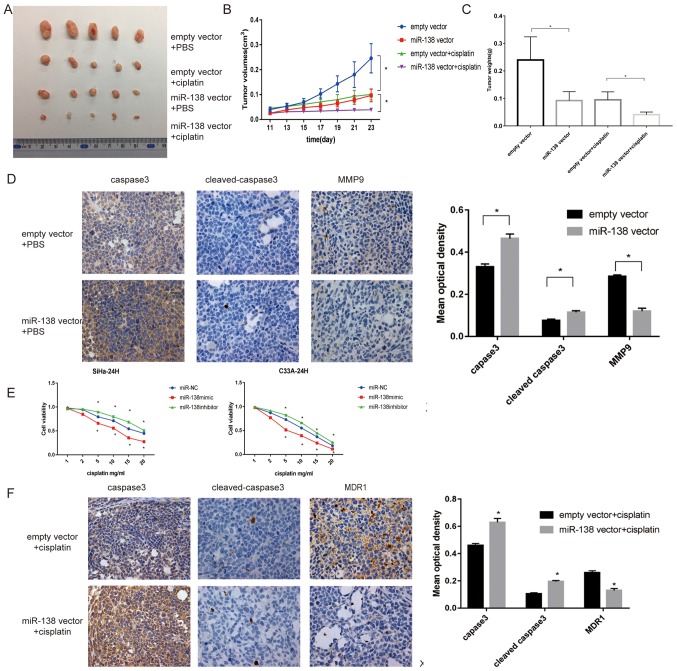
To evaluate whether the inhibitory effect of miR-138 on cell proliferation was also present in vivo, a tumor xenograft study was performed. A lentiviral miR-138 vector or empty vector was transfected into SiHa cells. The tumor volumes in the miR-138 overexpression group were significantly reduced compared with those in the empty vector group (P<0.05; Fig. 4A and B). Tumor weights in the miR-138 overexpression group were also reduced compared with in the empty vector group (Fig. 4C). Furthermore, miR-138-overexpressing tumors showed increased expression levels of caspase-3 and cleaved caspase-3 compared with the empty vector, as determined by IHC. By contrast, the expression of MMP9 was significantly inhibited in tumors with miR-138 overexpression, suggesting that the effect was still present in vivo (P<0.05; Fig. 4D).
Figure 4.
miR-138 suppresses cell proliferation and invasion and increases apoptosis in vivo and promoted the sensitivity of cervical cancer cells in vitro and in vivo. (A) Representative images of tumors isolated from nude mice (n=20). (B) Weight of tumors treated with empty vector or miR-138 vector injected with PBS and cisplatin were recorded and presented as the mean ± standard deviation (n=20). (C) Tumor volumes were measured every 2 days, 7 days after injection. (D) Representative images of IHC-stained tumor specimens for apoptotic proteins caspase-3, cleaved caspase-3 and bax, which were markedly upregulated in the miR-138-SiHa group. MMP9, an invasive protein, was markedly decreased in miR-138 vector tumors. Magnification, ×200. (E) Proliferation of transfected cells following treatment with different concentrations of cisplatin, as examined by Cell Counting Kit-8 assay. (F) Representative expression of caspase-3, cleaved caspase-3 and MDR1 in the three groups of tumor xenografts, as determined by IHC. Magnification, ×200. Data are presented as the mean ± standard deviation. *P<0.05. miR, microRNA; MMP, matrix metalloproteinase; IHC, immunohistochemistry.
miR-138 promotes the sensitivity in vitro and in vivo
To investigate whether miR-138 affects the sensitivity of cervical cancer cells, cells were subjected to different doses of cisplatin and a CCK-8 assay was used to measure cell viability at the indicated times. As shown in Fig. 4E, the miR-138 mimic significantly improved the toxicity of cisplatin compared with the miR-NC, while the miR-138 inhibitor exhibited the opposite effect (P<0.05).
Furthermore, whether this effect could also be observed in vivo was investigated. As shown in Fig. 4B and C, following cisplatin treatment, tumor growth derived from miR-138 vector cells was significantly delayed compared with the group that received cisplatin only (P<0.05). IHC revealed that miR-138 increased caspase-3 and cleaved caspase-3 staining, but deceased MDR1 staining, which was consistent with the results in vitro (Fig. 4F).
H2AX is a direct target of miR-138
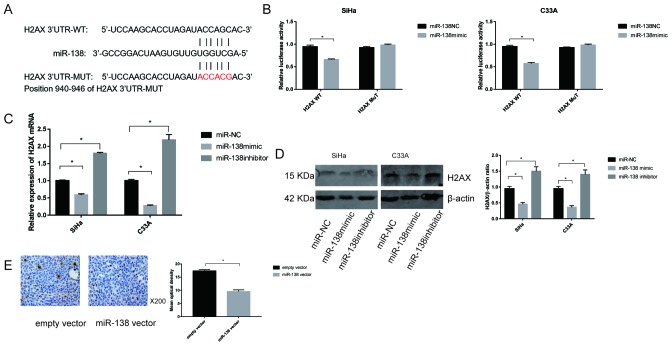
To further clarify the mechanisms of miR-138 in cervical cancer, the miRNA prediction websites, TargetScan software was used to screen for potential target of miR-138, and the H2AX gene was selected as a potential candidate (Fig. 5A). Then, two types of plasmid containing luciferase reporter was constructed to examine whether miR-138 directly binds to H2AX. miR-138 significantly reduced the luciferase activity of the plasmid with WT-H2AX (P<0.05), but did not affect MUT-H2AX in SiHa and C33A cells, suggesting that miR-138 could target H2AX in cervical cancer (Fig. 5B).
Figure 5.
H2AX is a target of miR-138. (A) Predicted binding site between miR-138 and H2AX 3′UTR. (B) Luciferase activity for cells co-transfected with WT-H2AX or MUT-H2AX and miR-138-NC or miR-138 mimic. (C) Relative expression of H2AX mRNA in SiHa and C33A lines transfected with miR-NC, miR-138 mimic and miR-138 inhibitor were determined by reverse transcription-PCR. (D) H2AX protein levels in the transfected cells were evaluated by western blot analysis and β-actin was used as the control. Data are presented as the mean ± standard deviation. (E) Representative expression of H2AX in two groups of xenograft tumors treated with DDP, as determined by IHC. Magnification, ×200. Data are presented as the mean ± standard deviation. *P<0.05. miR, microRNA; IHC, immunohistochemistry; NC, negative control.
To evaluate the association between miR-138 and H2AX, the expression of H2AX was detected by transfection with the miR-138 mimic or inhibitor. The results showed that the mRNA levels of H2AX were downregulated when miR-138 mimic was transfected into both SiHa and C33A cells. In contrast, transfection with miR-138 inhibitor markedly enhanced the expression of H2AX mRNA in SiHa and C33A cells (Fig. 5C). Similar results were also observed for H2AX protein expression (Fig. 5D). In addition, the expression of H2AX in vivo was assessed. As shown in Fig. 5E, IHC revealed that miR-138 significantly decreased the expression of H2AX compared with the empty vector group.
H2AX is a functional target of miR-138
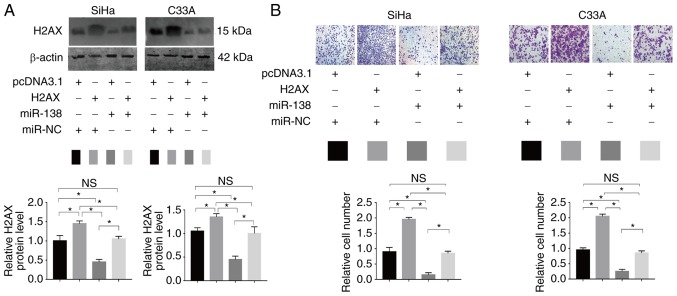
To assess whether H2AX has functional relevance for miR-138 expression, a rescue assay was generated. PcDNA 3.1-H2AX or pcDNA3.1 and miR-138 mimic or NC were co-introduced into SiHa and C33A cells (Fig. 6A). A Transwell assay revealed that the enforced expression of H2AX partially restored the migratory abilities of SiHa and C33A cells (Fig. 6B). The results suggested that H2AX is a functional target of miR-138 in human cervical cancer.
Figure 6.
Interaction between miR-138 and H2AX. (A) H2AX alterations following pcDNA3.1-H2AX and/or miR-138 mimic co-transfection. miR-138 mimic and pcDNA3.1-H2AX were co-transfected into SiHa and C33A cells. The western blotting results showed that miR-138 could inhibit H2AX expression. (B) Transwell assay after miR-138 and pcDNA3.1-H2AX co-transfection. Pc-DNA3.1-H2AX transfection could counteract miR-138-induced invasion inhibition. *P<0.05. Data are presented as the mean ± standard deviation. NS, no significance; miR, microRNA.
Discussion
Uncontrolled tumor progression and chemoresistance are the leading causes of poor treatment outcomes in cervical cancer. Clarifying the mechanisms involved in cervical cancer cell proliferation and chemoresistance is urgently required in order to improve the survival rates of patients with cervical cancer. miR-138 was previously reported to be dysregulated in various malignancies, including non-small cell lung cancer (17) and colorectal cancer (18). In the present study, the results revealed that miR-138 was downregulated in cervical cancer cell lines. To investigate what role the downregulation of miR-138 plays in cervical cancer, miR-138 mimic or inhibitor was transfected into the cells. miR-138 suppressed cell viability, metastasis and invasion, and enhanced apoptosis in cervical cancer cells. By contrast, downregulation of miR-138 increased proliferation, metastasis and invasion, and inhibited apoptosis. Xenograft experiments also confirmed these findings in vivo. The results were consistent with a previous report, which stated that miR-138 functions as a tumor suppressor (19).
Downregulation of miR-138 has been demonstrated to be involved in the chemoresistance of several cancer types (20). Prior studies have demonstrated that miR-138 modulate sensitization in prostate and non-small cell lung cancer, multiple myeloma, renal cell carcinoma, and osteosarcoma (14,21–25). Consistent with previous studies, the results of the present study showed that miR-138 enhanced the cisplatin-induced decrease in cervical cancer cell viability. To characterize the effect of miR-138 more precisely, xenograft models transfected with lentiviral miR-138 vector or empty vector (negative control group) were used. Tumor sizes and volumes were decreased following miR-138 overexpression and cisplatin treatment, as compared with in the group that received cisplatin only, indicating that upregulation of miR-138 sensitized cervical cancer cells to cisplatin. MDR1, which has been investigated closely in relation to drug resistance, was used to measure chemosensitivity (26). IHC was used to analyze apoptosis and drug resistance. These results revealed that miR-138 induced apoptosis and reversed drug resistance.
Identification of miRNA gene targets helps to further understand the function of miRNA in carcinogenesis. Several target genes of miR-138 had been revealed, including GIT1, PDK1, Survivin and SIRT1 (27–30). The present data demonstrated that H2AX acted as a functional target of miR-138 in cervical cancer. H2AX, histone H2A variant phosphorylation, is an early marker of DNA double-strand break (DSB) formation and regulates molecules needed for DNA repair (31,32). DSBs signify genomic instability and contribute to cancer initiation and progression. DNA damage occurs in a variety of events, such as treatment with radiomimetic agents, drugs. DNA damage can active three phosphatidylinositol 3-kinase-like kinases, which in turn phosphorylated H2AX at Ser 139. H2AX phosphorylated at the sites of DNA damage and facilitated repair proteins, including the MRN complex, 53BP1, MDC1 and RAD51. In addition to this function role as response to DSBs, H2AX also play a structural role, by regulating cell proliferation (33), angiogenesis (34) and other biological processes. H2AX can serve as an indicator of clonogenic survival and induction of apoptosis. Radio-resistant tumor cells can be made sensitive to radiation through blocking H2AX (35,36).
Previous studies had revealed that miR-138 negatively regulates the expression of H2AX in osteosarcoma (37) and in small cell lung cancer (38). miR-138 enhances cellular sensitivity to DNA damaging agents by inhibiting H2AX. However, the specific molecular mechanisms in cervical cancer have been unclear and few studies have explored the relationship between H2AX and chemosensitivity in cervical cancer. Consistent with previous results, the present study revealed that miR-138 targets H2AX in cervical cancer. Furthermore, a xenograft model was used to confirm these findings. These results provide further evidence that H2AX is a target of miR-138 in cervical cancer, not just in osteosarcoma and small lung cancer.
In the current study, a luciferase reporter assay, mRNA quantification, western blot analysis and animal experiments demonstrated that miR-138 targets H2AX, and that an inverse relationship exists between miR-138 and H2AX. Furthermore, a functional study showed that H2AX expression was directly inhibited by miR-138. Ectogenic overexpression of H2AX was able to rescue the invasive abilities induced by miR-138, confirming that miR-138 targets H2AX. Moreover, functional studies revealed that H2AX silencing induced miR-138 overexpression in cervical cancer, resulting in the suppression of cell viability and migration. These results suggested that H2AX is a functional mediator of miR-138.
In conclusion, the present data revealed that miR-138 was downregulated in cervical cancer. miR-138 inhibited cell proliferation, invasion and migration, and enhanced drug sensitivity by targeting H2AX in cervical cancer. The present results not only provide insight into the molecular mechanisms that regulate human cervical cancer, but also offer an explanation for the challenges associated with chemoresistance. The study provides evidence that miR-138 may have potential as a novel therapeutic target in cervical cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by National Natural Science Foundation of China (grant number 81360380).
Authors' contributions
QW and JC conceived and designed the experiments. MY and SZ performed the experiments and constructed the table and figures. RC, GW, YB contributed reagents/materials/analysis tools. SZ wrote the paper. All authors reviewed the manuscript.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
All experiments were approved by the Animal Management Rule of the Chinese Ministry of Health (document 55, 2001) and approved by the Tongji University Animal Ethics Committee (Shanghai, China). No human data were applicable in the present manuscript.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Diaz-Padilla I, Monk BJ, Mackay HJ, Oaknin A. Treatment of metastatic cervical cancer: Future directions involving targeted agents. Crit Rev Oncol Hematol. 2013;85:303–314. doi: 10.1016/j.critrevonc.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 3.Lukka H, Johnston M. Concurrent cisplatin-based chemotherapy plus radiotherapy for cervical cancer: A meta-analysis. Clin Oncol (R Coll Radiol) 2004;16:160–161. doi: 10.1016/j.clon.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Pfaendler KS, Tewari KS. Changing paradigms in the systemic treatment of advanced cervical cancer. Am J Obstet Gynecol. 2016;214:22–30. doi: 10.1016/j.ajog.2015.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dasari S, Tchounwou PB. Cisplatin in cancer therapy: Molecular mechanisms of action. Eur J Pharmacol. 2014;740:364–378. doi: 10.1016/j.ejphar.2014.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bayraktar R, Van Roosbroeck K. miR-155 in cancer drug resistance and as target for miRNA-based therapeutics. Cancer Metastasis Rev. 2018;37:33–44. doi: 10.1007/s10555-017-9724-7. [DOI] [PubMed] [Google Scholar]
- 7.Huang YA, Hu P, Chan KCC, You ZH. Graph convolution for predicting associations between miRNA and drug resistance. Bioinformatics: 2019;(pii):btz621. doi: 10.1093/bioinformatics/btz621. [DOI] [PubMed] [Google Scholar]
- 8.Tung SL, Huang WC, Hsu FC, Yang ZP, Jang TH, Chang JW, Chuang CM, Lai CR, Wang LH. miRNA-34c-5p inhibits amphiregulin-induced ovarian cancer stemness and drug resistance via downregulation of the AREG-EGFR-ERK pathway. Oncogenesis. 2017;6:e326. doi: 10.1038/oncsis.2017.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Z, Wang J, Li J, Wang X, Song W. MicroRNA-150 promotes cell proliferation, migration and invasion of cervical cancer through targeting PDCD4. Biomed Pharmacother. 2018;97:511–517. doi: 10.1016/j.biopha.2017.09.143. [DOI] [PubMed] [Google Scholar]
- 11.Wen SY, Lin Y, Yu YQ, Cao SJ, Zhang R, Yang XM, Li J, Zhang YL, Wang YH, Ma MZ, et al. miR-506 acts as a tumor suppressor by directly targeting the hedgehog pathway transcription factor Gli3 in human cervical cancer. Oncogene. 2015;34:717–725. doi: 10.1038/onc.2014.9. [DOI] [PubMed] [Google Scholar]
- 12.Xu R, Zeng G, Gao J, Ren Y, Zhang Z, Zhang Q, Zhao J, Tao H, Li D. miR-138 suppresses the proliferation of oral squamous cell carcinoma cells by targeting Yes-associated protein 1. Oncol Rep. 2015;34:2171–2178. doi: 10.3892/or.2015.4144. [DOI] [PubMed] [Google Scholar]
- 13.Ye XW, Yu H, Jin YK, Jing XT, Xu M, Wan ZF, Zhang XY. miR-138 inhibits proliferation by targeting 3-phosphoinositide-dependent protein kinase-1 in non-small cell lung cancer cells. Clin Respir J. 2015;9:27–33. doi: 10.1111/crj.12100. [DOI] [PubMed] [Google Scholar]
- 14.Stojcheva N, Schechtmann G, Sass S, Roth P, Florea AM, Stefanski A, Stühler K, Wolter M, Müller NS, Theis FJ, et al. MicroRNA-138 promotes acquired alkylator resistance in glioblastoma by targeting the Bcl-2-interacting mediator BIM. Oncotarget. 2016;7:12937–12950. doi: 10.18632/oncotarget.7346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Nie W, Huang W, Zhang W, Xu J, Song W, Wang Y, Zhu A, Luo J, Huang G, Wang Y, Guan X. TXNIP interaction with the Her-1/2 pathway contributes to overall survival in breast cancer. Oncotarget. 2015;6:3003–3012. doi: 10.18632/oncotarget.3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao Y, Fan X, Li W, Ping W, Deng Y, Fu X. miR-138-5p reverses gefitinib resistance in non-small cell lung cancer cells via negatively regulating G protein-coupled receptor 124. Biochem Biophys Res Commun. 2014;446:179–186. doi: 10.1016/j.bbrc.2014.02.073. [DOI] [PubMed] [Google Scholar]
- 18.Zhao L, Yu H, Yi S, Peng X, Su P, Xiao Z, Liu R, Tang A, Li X, Liu F, Shen S. The tumor suppressor miR-138-5p targets PD-L1 in colorectal cancer. Oncotarget. 2016;7:45370–45384. doi: 10.18632/oncotarget.9659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu D, Gu L, Li Z, Jin W, Lu Q, Ren T. MiR-138-5p suppresses lung adenocarcinoma cell epithelial-mesenchymal transition, proliferation and metastasis by targeting ZEB2. Pathol Res Pract. 2019;215:861–872. doi: 10.1016/j.prp.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 20.Yu C, Wang M, Chen M, Huang Y, Jiang J. Upregulation of microRNA1385p inhibits pancreatic cancer cell migration and increases chemotherapy sensitivity. Mol Med Rep. 2015;12:5135–5140. doi: 10.3892/mmr.2015.4031. [DOI] [PubMed] [Google Scholar]
- 21.Sossey-Alaoui K, Plow EF. miR-138-mediated regulation of KINDLIN-2 expression modulates sensitivity to chemotherapeutics. Mol Cancer Res. 2016;14:228–238. doi: 10.1158/1541-7786.MCR-15-0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang X, Jiang J, Zhu J, He N, Tan J. HOXA4-regulated miR-138 suppresses proliferation and gefitinib resistance in non-small cell lung cancer. Mol Genet Genomics. 2019;294:85–93. doi: 10.1007/s00438-018-1489-3. [DOI] [PubMed] [Google Scholar]
- 23.Rastgoo N, Pourabdollah M, Abdi J, Reece D, Chang H. Dysregulation of EZH2/miR-138 axis contributes to drug resistance in multiple myeloma by downregulating RBPMS. Leukemia. 2018;32:2471–2482. doi: 10.1038/s41375-018-0140-y. [DOI] [PubMed] [Google Scholar]
- 24.Yun EJ, Zhou J, Lin CJ, Xu S, Santoyo J, Hernandez E, Lai CH, Lin H, He D, Hsieh JT. The network of DAB2IP-miR-138 in regulating drug resistance of renal cell carcinoma associated with stem-like phenotypes. Oncotarget. 2017;8:66975–66986. doi: 10.18632/oncotarget.17756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu Z, Tang J, Wang J, Duan G, Zhou L, Zhou X. MiR-138 acts as a tumor suppressor by targeting EZH2 and enhances cisplatin-induced apoptosis in osteosarcoma cells. PLoS One. 2016;11:e150026. doi: 10.1371/journal.pone.0150026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen KG, Sikic BI. Molecular pathways: Regulation and therapeutic implications of multidrug resistance. Clin Cancer Res. 2012;18:1863–1869. doi: 10.1158/1078-0432.CCR-11-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang B, Wang D, Yan T, Yuan H. MiR-138-5p promotes TNF-α-induced apoptosis in human intervertebral disc degeneration by targeting SIRT1 through PTEN/PI3K/Akt signaling. Exp Cell Res. 2016;345:199–205. doi: 10.1016/j.yexcr.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 28.Yang R, Liu M, Liang H, Guo S, Guo X, Yuan M, Lian H, Yan X, Zhang S, Chen X, et al. miR-138-5p contributes to cell proliferation and invasion by targeting Survivin in bladder cancer cells. Mol Cancer. 2016;15:82. doi: 10.1186/s12943-016-0569-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y, Yang K, Sun X, Fang P, Shi H, Xu J, Xie M, Li M. MiR-138 suppresses airway smooth muscle cell proliferation through the PI3K/AKT signaling pathway by targeting PDK1. Exp Lung Res. 2015;41:363–369. doi: 10.3109/01902148.2015.1041581. [DOI] [PubMed] [Google Scholar]
- 30.Li J, Wang Q, Wen R, Liang J, Zhong X, Yang W, Su D, Tang J. MiR-138 inhibits cell proliferation and reverses epithelial-mesenchymal transition in non-small cell lung cancer cells by targeting GIT1 and SEMA4C. J Cell Mol Med. 2015;19:2793–2805. doi: 10.1111/jcmm.12666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siddiqui MS, Francois M, Fenech MF, Leifert WR. Persistent γH2AX: A promising molecular marker of DNA damage and aging. Mutat Res Rev Mutat Res. 2015;766:1–19. doi: 10.1016/j.mrrev.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 32.Turinetto V, Giachino C. Multiple facets of histone variant H2AX: A DNA double-strand-break marker with several biological functions. Nucleic Acids Res. 2015;43:2489–2498. doi: 10.1093/nar/gkv061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seo J, Kim K, Chang DY, Kang HB, Shin EC, Kwon J, Choi JK. Genome-wide reorganization of histone H2AX toward particular fragile sites on cell activation. Nucleic Acids Res. 2014;42:1016–1025. doi: 10.1093/nar/gkt951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao H, Tong R, Ding C, Lv Z, Du C, Peng C, Cheng S, Xie H, Zhou L, Wu J, Zheng S. γ-H2AX promotes hepatocellular carcinoma angiogenesis via EGFR/HIF-1α-/VEGF pathways under hypoxic condition. Oncotarget. 2015;6:2180–2192. doi: 10.18632/oncotarget.2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bassing CH, Chua KF, Sekiguchi J, Suh H, Whitlow SR, Fleming JC, Monroe BC, Ciccone DN, Yan C, Vlasakova K, et al. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc Natl Acad Sci USA. 2002;99:8173–8178. doi: 10.1073/pnas.122228699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Banáth JP, Macphail SH, Olive PL. Radiation sensitivity, H2AX phosphorylation and kinetics of repair of DNA strand breaks in irradiated cervical cancer cell lines. Cancer Res. 2004;64:7144–7149. doi: 10.1158/0008-5472.CAN-04-1433. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y, Huang JW, Li M, Cavenee WK, Mitchell PS, Zhou X, Tewari M, Furnari FB, Taniguchi T. MicroRNA-138 modulates DNA damage response by repressing histone H2AX expression. Mol Cancer Res. 2011;9:1100–1111. doi: 10.1158/1541-7786.MCR-11-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang H, Luo J, Liu Z, Zhou R, Luo H. MicroRNA-138 regulates DNA damage response in small cell lung cancer cells by directly targeting H2AX. Cancer Invest. 2015;33:126–136. doi: 10.3109/07357907.2015.1006329. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.