Abstract
Colon cancer is one of the most common types of cancer worldwide. Methotrexate (MTX) is a chemotherapy drug used for the treatment of multiple types of cancer, such as colon and breast cancer. To determine the effects of MTX treatment on colorectal adenocarcinoma cell lines, a microRNA (miRNA) microarray was used to detect miRNA expression profiles of HT-29 colorectal adenocarcinoma MTX-resistant cells and their parental cells. The results demonstrated that 641 genes and 43 miRNAs were differentially expressed between HT-29 MTX-sensitive cells and MTX-resistant cells. In addition, 12 miRNAs and their co-expressed genes were highly correlated in MTX treatment, and one of the identified miRNAs, miR-770-5p, was studied in subsequent experiments. Upregulation of miR-770-5p significantly decreased the sensitivity of HT-29 cells to MTX. Using bioinformatics software, homeodomain-interacting protein kinase 1 (HIPK1) was identified to be a putative target gene of miR-770-5p, which was confirmed by a luciferase reporter assay. Downregulation of miR-770-5p target gene HIPK1 significantly decreased the sensitivity of HT-29 cells to MTX. These results suggest that miR-770-5p may be involved in the regulation of colon cancer resistance to MTX by regulating the expression of the target gene HIPK1.
Keywords: methotrexate, colon carcinoma, drug resistance, microRNA-770-5p, homeodomain-interacting protein kinase 1
Introduction
Methotrexate (MTX) is one of the most commonly used drugs for maintenance therapy in multiple diseases such as cancer (including colon, breast and lung cancer), autoimmune diseases, acute lymphocytic leukemia, rheumatoid arthritis and ectopic pregnancy (1–5). In cancer treatment, Rajagopalan et al (6) reported that MTX functions by inhibiting dihydrofolate reductase, whereas in acute lymphocytic leukemia, MTX exerts its effects through the interaction with folic acid (7). These results suggested that MTX may exhibit different functional mechanisms in different diseases. Colon carcinoma is one of the most common types of cancer in the United States of America and worldwide (8). Multiple treatment methods including chemotherapy, radiotherapy and surgery are used in the treatment of colon cancer (9–11). MTX treatment or combined MTX treatment contributes an important part in chemotherapy in colon cancer (12). Therefore, the mechanism of MTX function in colon cancer is a challenging yet important question in the treatment of colon cancer.
MicroRNAs (miRNAs) are short RNAs that contain ~22 nucleotides and regulate ~30% of human genes by targeting their 3′-untranslated region (3′UTR), which serve essential regulatory roles in the tumorigenesis and tumor development of multiple types of cancer, including colon cancer (13). Several studies have described the function of MTX in the treatment of colon cancer (14,15) or protein targets of MTX in colon cancer; however, a limited number of reports focused on the mechanism of MTX effects at the co-expressed protein, miRNA and network levels.
The present study aimed to investigate the mRNA and miRNA profiles of colon carcinoma using HT29-derived cell lines to explore the MTX-associated mechanisms of action in colon carcinoma. miR-770-5p and its target gene home domain-interacting protein kinase 1 (HIPK1) were identified, and their role in the MTX resistance in colon cancer was studied.
Materials and methods
Cell culture
The human colorectal adenocarcinoma HT-29 cell line was purchased from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences. HT-29 MTX-resistant cells were successfully established from the parental HT-29 cell line by exposing HT-29 cells to gradually increasing concentrations of MTX (Sigma-Aldrich; Merck KGaA). HT-29 MTX-resistant cells were first adapted to grow in the presence of 1×10−8 mol/l MTX. MTX treatment was then performed by exposure to stepwise increasing concentrations of MTX for 6 months. MTX-resistant clones were maintained with 10−6 mol/l MTX. The half-maximal inhibitory concentration (IC50) of MTX in WT HT-29 cells was 3.1×10−8 and 1.0×10−5 mol/l in the MTX-resistant HT-29 cells.
HT-29 cells and HT-29 MTX-resistant cells were cultured in RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc.) medium containing 10% fetal bovine serum (HyClone; GE Healthcare Life Sciences), 2 mM L-glutamine, 10 ng/ml epidermal growth factor (Shanghai PrimeGene Bio-Tech Co., Ltd.), 100 U/ml penicillin and 100 µg/ml streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C with 5% CO2. The medium of HT-29 MTX-resistant cells contained 1 µg/ml MTX.
miRNA and mRNA co-expression analysis
Cells from 3 separate cultures of both the HT-29 MTX sensitive and MTX resistant cell lines were selected for gene and miRNA expression profile analysis. Microarray data were downloaded from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo), which comprised 6 RNA microarray samples (GSE11440) and 6 miRNA microarray samples (GSE28547). Data were downloaded and pre-processed, and differentially expressed genes (DEGs) and miRNAs were identified using R software (https://www.r-project.org). The Limma package in R was used for differential gene and miRNA expression analysis (16). Genes and miRNAs were considered differentially expressed if their |log[fold change (FC)]|>1.2 and adjusted P<0.05. Probes corresponding to multiple genes were removed from the analysis results. When multiple probes corresponded to the same gene, average values were calculated.
To evaluate the co-expression between mRNAs and miRNAs, Pearson correlation coefficient in R software was used. miRNA-mRNA expression pairs with correlation values <-0.9 were used for further analysis, since miRNAs usually serve a negative regulatory role on mRNAs. miRNA target information was downloaded from miRTarBase database (download date, February 17th, 2016) (17). Cytoscape software (version 3.2.0; http://cytoscape.org/) (18) was used for miRNA-mRNA co-expression network analysis. The network work of each gene was calculated by counting the numbers of upstream genes and downstream genes, which were expressed in the form of in-degree and out-degree.
Prediction of miR-770-5ptarget genes
To search for the targets of miR-770-5p, miRanda (http://www.mocrorna.org) (19), TargetScan (http://www.targetscan.org/) (20) and miRWalk (http://mirwalk.uni-hd.de/) (21) online databases were used. The genes identified in ≥2 databases were considered as targets of DE miRNAs.
Transfection with synthesized oligonucleotides
miRNA mimics, miRNA inhibitor and negative control miRNA oligonucleotides of miR-770-5p were obtained from Guangzhou RiboBio Co. Ltd. The sequences were: miR-770-5p mimic, 5′-UCCAGUACCACGUGUCAGGGCCA-3′; miR-NC mimic, 5′-UCGCUUGGUGCAGGUCGGGAA-3; miR-770-5p inhibitor, 5′-UGGCCCUGACACGUGGUACUGGA-3′; and miR-NC inhibitor, 5′-CAGUACUUUUGUGUAGUACAA-3′. The small interfering RNAs (siRNAs) targeting HIPK1 (genOFF™ h-HIPK1_1999A; cat. no. SIGS0004714-1) were synthesized by Guangzhou RiboBio Co. Ltd. HT-29 cells were cultured to ~70% confluence and transfected with the indicated RNAs using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) at a final concentration of 100 nM. Cells were subjected to subsequent experimentation 24 h following transfection.
Construction of the reporter vector and site-directed mutagenesis
The 3′UTR of HIPK1 was amplified from HT-29 cell cDNA by PCR. Total RNA was extracted from HT-29 cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). cDNA was synthesized by using M-MLV Reverse Transcriptase (Promega Corporation) with random primers and dNTPs (Promega Corporation). The temperature protocol for the reverse transcription reaction consisted of cDNA synthesis at 37°C for 60 min and termination at 80°C for 2 min. qPCR was subsequently performed with GoTaq® Greeen Master Mix (Promega Corporation). Primers for the amplification of HIPK1 3′UTR were as follows: Wild type (WT) forward, 5′-AATCTCGAGGAGGAGTCAAGCCAATATTAAAT-3′ and reverse, 5′-TAAGCGGCCGCTATGGGCAGGAATGTC-3′; mutant forward, 5′-AATCTCGAGGAGGAGTCAAGCCAATATTAAAT-3′ and reverse, 5′-TAAGCGGCCGCAATGTCCCCATCCATCCACTTTTCTAC-3′. The reaction conditions were as follows: Initial denaturation at 95°C for 5 min, followed by 30 cycles of 95°C for 30 sec and 60°C for 30 sec, and final extension at 72°C for 10 min. The amplified products were cloned into the XhoI and NotI sites of the psiCHECK-2 plasmid (Promega Corporation).
Dual luciferase reporter assay
Luciferase reporter assays were performed using the psiCHECK2-3′UTR vector. Cells were cultured to ~70% confluence in 48-well plates and co-transfected with psiCHECK2-3′UTR + miR-770-5p mimics or control mimics using Lipofectamine® 2000 for 24 h followed by the luciferase reporter assay using the Dual Luciferase Assay System (Promega Corporation). Renilla luciferase activity was normalized to firefly luciferase activity. Cell lysates were subjected to luciferase activity measurement according to the manufacturer's instructions.
RNA extraction and semi-quantitative reverse transcription- polymerase chain reaction
Total RNA was extracted from cultured cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). For detection of HIPK1 mRNA, cDNA was synthesized using M-MLV Reverse Transcriptase (Promega Corporation) with random primers (Promega Corporation). The temperature protocol for the reverse transcription reaction consisted of cDNA synthesis at 37°C for 60 min and termination at 80°C for 2 min. qPCR amplification was subsequently performed with GoTaq® Green Master Mix (Promega Corporation) using the following primers: HIPK1 forward, 5′-GCATCCTTTCCCGCCTAAGA-3′ and reverse, 5′-TACATGTGAACCCTCCGATTG-3′; GAPDH forward, 5′-AGCCTTCTCCATGGTGGTGAA-3′ and reverse, 5′-ATCACCATCTTCCAGGAGCGA-3′. The reaction conditions were as follows: Initial denaturation at 95°C for 2 min, followed by 30 cycles 95°C for 20 sec and 60°C for 30 sec, and final extension at 72°C for 30 sec. The PCR products were subsequently subjected to 0.5 µg/ml ethidium bromide-containing 1% agarose gel electrophoresis. The products were visualized using a digital gel image processing system (Tanon 6200 Luminescent Imaging Workstation; Tanon Science and Technology Co., Ltd.). Semi-quantitative-PCR bands results were further analyzed using the Image-Pro Plus 6.0 software (Media Cybernetics, Inc.).
Western blot analysis
Cells were lysed using RIPA lysis buffer (Sigma-Aldrich; Merck KGaA) supplemented with protease inhibitor (Invitrogen; Thermo Fisher Scientific, Inc.). The protein concentration was then determined using a Bicinchoninic Acid protein assay kit (Beyotime Institute of Biotechnology). Total proteins (20–50 µg) were subjected to 10% sodiumdodecy1 sulfate-polyacrylamide gel electrophoresis and then transferred onto Immobilon P membranes. The membranes were blocked with TBS supplemented with 5% non-fat dry milk and 0.1% Tween-20 for 1 hat room temperature prior to incubation with primary antibodies against GAPDH (cat. no. HPA061280; 1:1,000; Sigma-Aldrich; Merck KGaA) and HIPK1 (cat. no. ab90103; 1:1,000; Abcam) at 4°C overnight. Following incubation with horseradish peroxidase-conjugated secondary antibody (cat. no. A0208; 1:500; Beyotime Institute of Biotechnology) at room temperature for 1 h, and electrochemiluminescene western blot substrate (Beyotime Institute of Biotechnology) was used for the detection of protein expression. Densitometric analyzes of the protein bands were performed using the Image-Pro Plus 6.0 software (Media Cybernetics, Inc.).
Cell proliferation assay
To examine cell proliferative ability, cells were seeded at the density of 2,000 cells/well and transfected with the indicated RNAs for 24 h as aforementioned. Cell proliferation was evaluated using a Cell Counting Kit-8 (CCK-8) assay. Briefly, each well was supplemented with 10 µl CCK-8 reagent and incubated at 37°C for 4 h. Optical density was measured at 490 nm for each well.
Statistical analysis
All in vitro experiments were performed in triplicate and the data presented were representative of 3 independent experiments. Numerical data are presented as mean ± standard deviation. The Student's t-test was used for the statistical evaluation of the difference between two groups, and one-way analysis of variance with appropriate Bonferroni post-hoc test was performed for comparisons among ≥3 variables. Statistical analysis was performed using SPSS v.16.0 (SPSS, Inc.) statistical software. P<0.05 was considered to indicate a statistically significant difference.
Results
Differential mRNA and miRNA expression profiles in MTX-sensitive vs. MTX-resistant HT29-derived colorectal adenocarcinoma cell lines
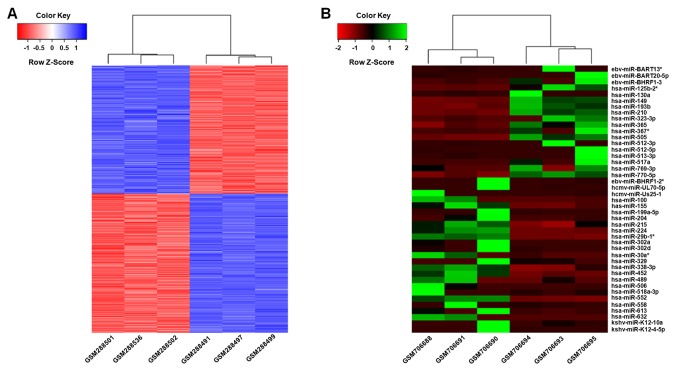
To explore the genes and miRNAs associated with MTX treatment in colorectal adenocarcinoma cell lines, DEGs and miRNAs between 3 MTX-sensitive and 3 MTX-resistant cell lines were determined. A total of 641 genes and 35 miRNAs were differentially expressed between MTX-sensitive and MTX-resistant cell lines, including 305 up-regulated and 336 down-regulated genes (Fig. 1; Tables I and II; Table SI), and 15 upregulated and 20 downregulated miRNAs (Tables III and IV; Table SI).
Figure 1.
Heat map representation of DEGs and miRNAs between MTX-sensitive and MTX-resistant HT-29 cell lines. (A) Heat map of DEGs between 3MTX-sensitive HT-29 cell lines and 3MTX-resistant HT-29 cell lines. The red color represents low mRNA expression level, and the blue color represents high mRNA expression level. (B) Heat map representation of differently expressed miRNAs between MTX-sensitive HT-29 cell lines and MTX-resistant HT-29 cell lines. The red color represents low miRNA expression level, and the green color represents high mRNA expression level. DEGs, differently expressed genes; miRNA, microRNA; MTX, methotrexate.
Table I.
Top 10 upregulated DE genes (adjusted P<0.05) for MTX-resistant vs. MTX-sensitive cell lines as ranked by FC.
| Gene symbol | Genename | logFC | P-value | adj.P-val |
|---|---|---|---|---|
| HAPLN1 | Hyaluronan and proteoglycan link protein 1 | 7.22 | 1.95×10−4 | 2.49×10−2 |
| IGFBP7 | Insulin-like growth factor binding protein 7 | 6.86 | 1.31×10−6 | 8.27×10−3 |
| EDIL3 | EGF-like repeats and discoidin I-like domains 3 | 6.36 | 2.32×104 | 2.80×10−2 |
| ATP6AP1L | ATPase, H+ transporting, lysosomal accessory protein 1-like | 5.85 | 1.76×10−6 | 8.61×10−3 |
| CYBRD1 | Cytochrome b reductase 1 | 5.55 | 1.24×10−5 | 1.13×10−2 |
| GNG2 | G protein subunit gamma 2 | 5.30 | 1.23×10−5 | 1.13×10−2 |
| CD37 | CD37 molecule | 5.14 | 2.00×10−4 | 2.79×10−2 |
| RASSF8 | Ras association domain family member 8 | 4.57 | 1.38×10−4 | 2.54×10−2 |
| SOX8 | SRY (sex determining region Y)-box 8 | 4.49 | 2.10×10−4 | 2.82×10−2 |
| PPP1R14C | Protein phosphatase 1 regulatory inhibitor subunit 14C | 4.47 | 1.12×10−4 | 2.37×10−2 |
FC, fold change; adj. P-val, adjusted P-value.
Table II.
Top 10 down-regulated differentially expressed genes (adjusted P<0.05) for MTX-resistant vs. MTX-sensitive cell lines as ranked by FC.
| Gene symbol | Gene name | logFC | P-value | adj. P-val |
|---|---|---|---|---|
| IFI27 | Interferon, alpha-inducible protein 27 | −10.37 | 9.70×10−8 | 5.30×10−3 |
| BST2 | Bone marrow stromal cell antigen 2 | −9.02 | 5.81×10−7 | 7.94×10−3 |
| PRSS1 | Serine protease 1 | −7.62 | 1.98×10−6 | 8.61×10−3 |
| ARMCX4 | Armadillo repeat containing X-linked 4 | −6.62 | 1.05×10−6 | 8.61×10−3 |
| PSMB9 | Proteasome subunitbeta 9 | −6.55 | 6.20×10−4 | 4.31×10−2 |
| NETO2 | Neuropilin and tolloid-like 2 | −6.51 | 4.08×10−4 | 3.46×10−2 |
| MX1 | MX dynamin like GTPase 1 | −6.50 | 7.50×10−4 | 4.58×10−2 |
| PDZK1IP1 | PDZK1 interacting protein 1 | −6.14 | 3.63×10−4 | 2.55×10−2 |
| PLIN2 | Perilipin 2 | −5.88 | 6.93×10−4 | 4.50×10−2 |
| IFITM1 | Interferon induced transmembrane protein 1 | −5.84 | 2.08×10−5 | 1.33×10−2 |
FC, fold change; adj. P-val, adjusted P-value.
Table III.
Top 10 upregulated differentially expressed microRNAs (P<0.05) for MTX-resistant vs. MTX-sensitive cell lines as ranked by FC.
| ID | adj. P-val | P-value | logFC | Organism |
|---|---|---|---|---|
| hsa-miR-512-5p | 0.49 | 1.07×10−2 | 8.75 | Homo sapiens |
| hsa-miR-512-3p | 0.57 | 3.08×10−2 | 5.65 | Homo sapiens |
| hsa-miR-130a | 0.57 | 3.63×10−2 | 5.26 | Homo sapiens |
| hsa-miR-513a-3p | 0.57 | 3.89×10−2 | 5.11 | Homo sapiens |
| hsa-miR-125b-2 | 0.49 | 1.04×10−2 | 4.68 | Homo sapiens |
| hsa-miR-517a | 0.57 | 2.65×10−2 | 4.46 | Homo sapiens |
hsa, Homo sapiens; FC, fold change; adj. P-val, adjusted P-value.
Table IV.
Top 10 down-regulated differentially expressed microRNAs (P<0.05) for MTX-resistant vs. MTX-sensitive cell lines as ranked by FC.
| ID | adj. P-val | P-value | logFC | Organism |
|---|---|---|---|---|
| hsa-miR-199a-5p | 0.49 | 9.38×10−3 | −9.21 | Homo sapiens |
| hsa-miR-632 | 0.31 | 4.19×10−3 | −6.31 | Homo sapiens |
| hsa-miR-204 | 0.57 | 3.27×10−3 | −6.08 | Homo sapiens |
| hsa-miR-506 | 0.57 | 2.38×10−2 | −5.09 | Homo sapiens |
| hsa-miR-302d | 0.57 | 4.72×10−2 | −4.86 | Homo sapiens |
| hsa-miR-302a | 0.57 | 4.30×10−2 | −4.87 | Homo sapiens |
hsa, Homo sapiens; FC, fold change; adj. P-val, adjusted P-value.
Differentially expressed miRNA-associated co-expressed genes and target gene networks
To further understand the function of the differentially expressed miRNAs in the effects of MTX in colon carcinoma, the co-expression and target information between differentially expressed miRNAs and DEGs was determined. A total of 12 miRNAs and 690 genes were identified to be negatively co-expressed (Table SII). In the miRNA-mRNA co-expression network, all 12 identified miRNAs were human miRNAs and were divided into two co-expression groups of 5 and 7 miRNAs. The top 5 miRNAs with the highest number of connections in the network were Homo sapiens(hsa)-miR-770-5p, hsa-miR-29b-1, hsa-miR-552, hsa-miR-505 and hsa-miR-224, whereas the top 5 genes with the highest number of connections were Rho GTPase activating protein 24, armadillo repeat containing X-linked 4, epithelial stromal interaction 1, hyaluronan and proteoglycan link protein 1 and Major histocompatibility complex, class I, B (Table SIII). miR-770-5p and its target gene were identified in the miRNA-mRNA co-expression network, which suggested that this miRNA and its target were co-expressed and highly associated with MTX treatment.
miR-770-5p decreases drug sensitivity of HT-29 cells to MTX
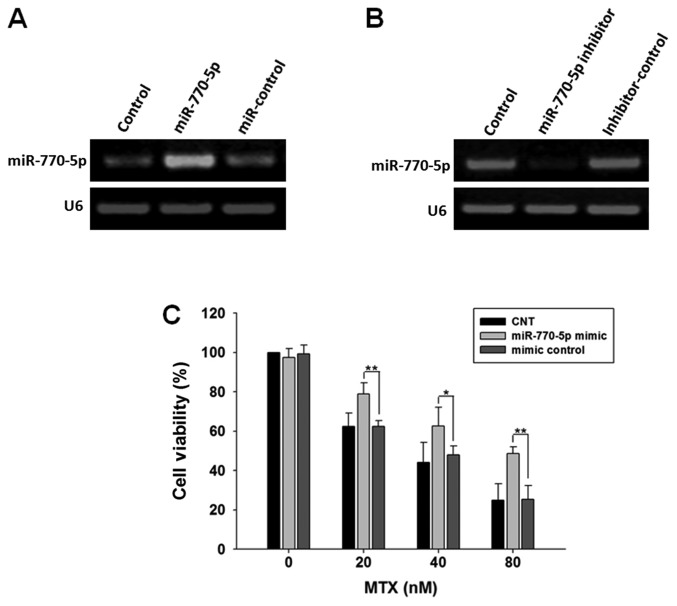
Cells were transfected with miR-770-5p mimic or miR-770-5p inhibitor, respectively. Delivery of miR-770-5p mimic significantly increased the expression level of miR-770-5p, whereas the introduction of the miR-770-5p inhibitor decreased miR-770-5p expression compared with their corresponding RNA-transfected control cells (Fig. 2A and B). The viability of HT-29 cells transfected with miR-770-5p mimic to methotrexate was measured using a CCK-8 assay. The results demonstrated that the viability of HT-29 cells transfected with miR-770-5p mimic treated with MTX was significantly increased compared with that in the control group, suggesting that overexpression of miR-770-5p significantly decreased the sensitivity of HT-29 cells to MTX (Fig. 2C).
Figure 2.
miRNA-770-5p promotes the proliferation of MTX-treated HT-29 cells. (A) PCR results of the measurement of miR-770-5p expression in HT-29 cells transfected with miR-770-5p mimics and mimic control. (B) PCR results of the measurement of miR-770-5p expression in HT-29 cells transfected with miR-770-5p inhibitor or inhibitor control. (C) HT-29 cells were transfected with miR-770-5p mimic or mimic control for 24 h and treated with MTX for 5 days. Cell viability was detected by Cell Counting Kit-8 assay. Results are presented as mean ± standard deviation. *P<0.05 and **P<0.01. MTX, methotrexate; miR, microRNA; PCR, polymerase chain reaction; CNT, control untreated.
HIPK1 is a direct target of miR-770-5p in HT-29
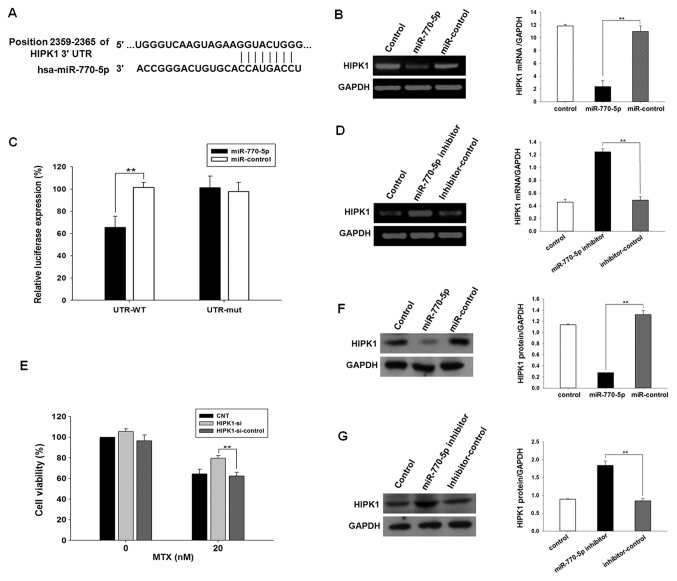
Through computational analysis, the 3′UTR of HIPK1 mRNA was identified to contain binding sites for miR-770-5p. Position 2,359-2,365 of HIPK1 3′UTR was predicted to contain the binding site of the miR-770-5p seed sequence (Fig. 3A). The direct interaction between miR-770-5p and HIPK1 mRNA was determined using a dual luciferase reporter assay. A dual luciferase reporter vector containing the 3′UTR of HIPK1 was introduced into HT-29 cells for detection of luciferase activity. The results demonstrated that the luciferase activity was significantly decreased in the experimental group transfected with miR-770-5p (P<0.01; Fig. 3B). To verify the specificity of the interaction between miR-770-5p and HIPK1 mRNA, a luciferase reporter vector encoding HIPK1 3′UTR with specific base pair mutations at 42–58 was constructed, and luciferase activity was measured in HT-29 cells. No significant difference in luciferase activity was observed in the experimental group transfected with the mutated HIPK1 3′UTR reporter plasmid (P>0.05; Fig. 3B). These results indicated that miR-770-5p specifically bound the 3′UTR of HIPK1 mRNA.
Figure 3.
miR-770-5p downregulates endogenous HIPK1 expression in HT-29 cells. (A) Sites at 2,359-2,365 of the HIPK1 3′UTR were predicted to be the potential binding sites of miR-770-5p by the MiRanda, MiRwalk, and TargetScan software. (B) PCR results of the measurement of HIPK1 mRNA expression in HT-29 cells transfected with miR-770-5p mimics or mimic control. (C) Luciferase activity detected in miR-770-5p-transfected HT-29 cells co-transfected with psiCHECK2-3′UTR or psiCHECK2-3′UTR-mutation. (D) PCR results of the measurement of HIPK1 mRNA expression in HT-29 cells transfected with miR-770-5p inhibitors or inhibitor control. (E) HT-29 cells were transfected with siRNA targeting HIPK1 or siRNA control for 24 h and treated with MTX for 5 days. Cell viability was detected by Cell Counting Kit-8 assay. (F) Western blot analysis of the protein expression levels of HIPK1 in HT-29 cells transfected with miR-770-5p mimics or mimic control. (G) Western blot analysis of the protein expression levels of HIPK1 in HT-29 cells transfected with miR-770-5p inhibitors or inhibitor control. Results are presented as the mean ± standard deviation. s**P<0.01. MTX, methotrexate; 3′UTR, 3′-untranslated region; HIPK1, homeodomain-interacting protein kinase 1; siRNA, small interfering RNA; miR, microRNA; WT, wild type; mut, mutant; hsa, Homo sapiens; CNT, control untreated.
miR-770-5p downregulates endogenous HIPK1 expression in HT-29
To investigate the regulation of endogenous HIPK1 expression by miR-770-5p in HT-29 cells, the mRNA and protein expression levels of HIPK1 were analyzed by semi-quantitative PCR and western blot analysis Compared with the negative control group, an ~80% decrease in HIPK1 mRNA levels and an 80% decrease in HIPK1 protein levels were observed when HT-29 cells were transfected with the miR-770-5p mimic (P<0.01; Fig. 3C and E). By contrast, the expression levels of HIPK1 mRNA and protein were increased by 2.6- and 2.2-fold, respectively, when HT-29 cells were transfected with miR-770-5p inhibitor (P<0.01; Fig. 3D and F). Although the number of templates used was consistent, the digital gel image processing system (Tanon) has an automatic exposure function, which automatically adjusted the brightest bands to a certain fixed value. Therefore, the mRNA expression levels of HIPK1 in control and inhibitor-control would look different between the two groups of data. These data suggested that endogenous HIPK1 expression was negatively regulated by miR-770-5p in HT-29 cells.
Downregulation of the miR-770-5p target gene decreases the sensitivity of HT-29 cells to MTX
HIPK1 siRNA was transfected into HT-29 cells, and the CCK-8 assay was used to detect the sensitivity of cells to MTX. The results demonstrated that the sensitivity of HT-29 cells transfected with HIPK1 siRNA to MTX was significantly decreased compared with that of the control group (Fig. 3G).
Discussion
The present study focused on the miRNAs and mRNAs associated with MTX treatment in the colorectal adenocarcinoma HT29 cell line and identified miRNAs and their co-expressed genes that were associated with MTX sensitivity. Zhang et al (22) previously demonstrated that miR-770-5p was upregulated in the in vitro model of diabetic nephropathy (DN) and that it may promote the development of DN by regulating podocyte apoptosis through targeting tp53 regulated inhibitor of apoptosis 1. Another previous study revealed that miR-770 inhibits glioma cell proliferation and induces apoptosis through the suppression of the Wnt/β-catenin signaling pathway by targeting cyclin-dependent kinase 8, suggesting that miR-770 may serve a crucial role in glioma progression and may be used as a potential novel target for glioma therapy (23). A recent study demonstrated that when the expression of mir-770-5p is inhibited, cell radiosensitivity is decreased, which suggested that miR-770-5p may be a useful therapeutic target miRNA that sensitizes tumors to radiation through negative regulation of PBK (24). In addition, miR-770-5p expression is decreased in patients with platinum-resistant malignancies, whereas the overexpression of miR-770-5p in vitro decreased the survival rate of chemoresistant cell lines following cisplatin treatment; suggesting that miR-770-5p may therefore be a useful biomarker for predicting chemosensitivity to cisplatin in patients with ovarian cancer and improve the selection of effective personalized treatment strategies (25). These 2 studies suggested that miR-770-5p may serve a crucial role in the drug resistance of tumors, which is consistent with the results of the present study. The present study demonstrated that has-miR-770-5p and its target gene HIPK1 were highly associated with MTX treatment in HT-29-resistant cells, suggesting that it may serve a role in modulating drug resistance. Therefore, miR-770-5p was overexpressed using miRNA mimics in HT29 cells, and the sensitivity of HT-29 cells to MTX was significantly decreased.
miRNAs serve an important role in energy homeostasis, particularly in intestinal homeostasis regulation, cell cycle, apoptosis, cell migration, proliferation and neuron function (26–28). A recent study investigated the functional involvement of maternally expressed 3 (MEG3) and its intron miR-770-5p in Hirschsprung's disease (HSCR) progression and identified the mechanism of pathophysiological roles and the potential association between long non-coding (lnc)RNA MEG3 and miR-770-5p, indicating that the lncRNA MEG3/miR-770-5p/SLIT-ROBO Rho GTPase-activating protein 1 pathway may serve important roles in the pathogenesis of HSCR (29). However, there are a limited number of studies on the role of miR-770 in colon cancer. HIPK is one of the 4 members of a closely associated serine/threonine kinase family with similar structure and function (HIPK1 to 4). HIPK1 regulates the activity of a broad range of transcription factors (30–33). Rey et al (34) identified that HIPK1 is significantly overexpressed in colorectal cancer and the level of HIPK1 gradually decreased as tumor stage progressed. However, the role of HIPK1 in colon cancer HT-29 cell resistance to MTX remains unclear. In the present study, the knockdown of HIPK1 in HT-29 cells significantly decreased the sensitivity of cells to MTX, indicating that HIPK1 may serve an important role in colon cancer HT-29 cell resistance to MTX.
In conclusion, the present study demonstrated the regulatory association between miR-770-5p and HIPK1 in colorectal adenocarcinoma cells and demonstrated that miR-770-5p may regulate cell sensitivity to MTX, suggesting that the miR-770-5p-HIPK1 regulatory axis may serve an important role in colorectal adenocarcinoma cell MTX resistance.
Supplementary Material
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
DZ, YL and PS conceived and designed the experiments. DZ and YL performed the experiments. PS analyzed the data. DZ, YL and PS wrote the paper. All authors approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Mol F, Mol BW, Ankum WM, van der Veen F, Hajenius PJ. Current evidence on surgery, systemic methotrexate and expectant management in the treatment of tubal ectopic pregnancy: A systematic review and meta-analysis. Hum Reprod Update. 2008;14:309–319. doi: 10.1093/humupd/dmn012. [DOI] [PubMed] [Google Scholar]
- 2.Herfarth HH, Long MD, Isaacs KL. Methotrexate: Underused and ignored? Dig Dis. 2012;30(Suppl 3):S112–S118. doi: 10.1159/000342735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cronstein BN. Low-dose methotrexate: A mainstay in the treatment of rheumatoid arthritis. Pharmacol Rev. 2005;57:163–172. doi: 10.1124/pr.57.2.3. [DOI] [PubMed] [Google Scholar]
- 4.Tisman G, Wu SJ. Effectiveness of intermediate-dose methotrexate and high-dose 5-fluorouracil as sequential combination chemotherapy in refractory breast cancer and as primary therapy in metastatic adenocarcinoma of the colon. Cancer Treat Rep. 1980;64:829–835. [PubMed] [Google Scholar]
- 5.Bleyer WA. Methotrexate: Clinical pharmacology, current status and therapeutic guidelines. Cancer Treat Rev. 1977;4:87–101. doi: 10.1016/S0305-7372(77)80007-8. [DOI] [PubMed] [Google Scholar]
- 6.Rajagopalan PT, Zhang Z, McCourt L, Dwyer M, Benkovic SJ, Hammes GG. Interaction of dihydrofolate reductase with methotrexate: Ensemble and single-molecule kinetics. Proc Natl Acad Sci USA. 2002;99:13481–13486. doi: 10.1073/pnas.172501499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morgan SL, Baggott JE, Vaughn WH, Young PK, Austin JV, Krumdieck CL, Alarcón GS. The effect of folic acid supplementation on the toxicity of low-dose methotrexate in patients with rheumatoid arthritis. Arthritis Rheum. 1990;33:9–18. doi: 10.1002/art.1780330102. [DOI] [PubMed] [Google Scholar]
- 8.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 9.Schrag D, Cramer LD, Bach PB, Begg CB. Age and adjuvant chemotherapy use after surgery for stage III colon cancer. J Natl Cancer Inst. 2001;93:850–857. doi: 10.1093/jnci/93.11.850. [DOI] [PubMed] [Google Scholar]
- 10.Cammà C, Giunta M, Fiorica F, Pagliaro L, Craxì A, Cottone M. Preoperative radiotherapy for resectable rectal cancer: A meta-analysis. JAMA. 2000;284:1008–1015. doi: 10.1001/jama.284.8.1008. [DOI] [PubMed] [Google Scholar]
- 11.Nelson H, Petrelli N, Carlin A, Couture J, Fleshman J, Guillem J, Miedema B, Ota D, Sargent D, National Cancer Institute Expert Panel Guidelines 2000 for colon and rectal cancer surgery. J Natl Cancer Inst. 2001;93:583–596. doi: 10.1093/jnci/93.8.583. [DOI] [PubMed] [Google Scholar]
- 12.Marsh JC, Bertino JR, Katz KH, Davis CA, Durivage HJ, Rome LS, Richards F, II, Capizzi RL, Farber LR, Pasquale DN, et al. The influence of drug interval on the effect of methotrexate and fluorouracil in the treatment of advanced colorectal cancer. J Clin Oncol. 1991;9:371–380. doi: 10.1200/JCO.1991.9.3.371. [DOI] [PubMed] [Google Scholar]
- 13.Cai Y, Yu X, Hu S, Yu J. A brief review on the mechanisms of miRNA regulation. Genomics Proteomics Bioinformatics. 2009;7:147–154. doi: 10.1016/S1672-0229(08)60044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weber G, Ichikawa S, Nagai M, Natsumeda Y. Azidothymidine inhibition of thymidine kinase and synergistic cytotoxicity with methotrexate and 5-fluorouracil in rat hepatoma and human colon cancer cells. Cancer Commun. 1990;2:129–133. doi: 10.3727/095535490820874498. [DOI] [PubMed] [Google Scholar]
- 15.Van Mouwerik TJ, Pangallo CA, Willson JK, Fischer PH. Augmentation of methotrexate cytotoxicity in human colon cancer cells achieved through inhibition of thymidine salvage by dipyridamole. Biochem Pharmacol. 1987;36:809–814. doi: 10.1016/0006-2952(87)90168-7. [DOI] [PubMed] [Google Scholar]
- 16.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu SD, Lin FM, Wu WY, Liang C, Huang WC, Chan WL, Tsai WT, Chen GZ, Lee CJ, Chiu CM, et al. miRTarBase: A database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 2011;39((Database Issue)):D163–D169. doi: 10.1093/nar/gkq1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;11:2498–5504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 21.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk-database: Prediction of possible miRNA binding sites by ‘walking’ the genes of three genomes. J Biomed Inform. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 22.Zhang SZ, Qiu XJ, Dong SS, Zhou LN, Zhu Y, Wang MD, Jin LW. MicroRNA-770-5p is involved in the development of diabetic nephropathy through regulating podocyte apoptosis by targeting TP53 regulated inhibitor of apoptosis 1. Eur Rev Med Pharmacol Sci. 2019;23:1248–1256. doi: 10.26355/eurrev_201902_17018. [DOI] [PubMed] [Google Scholar]
- 23.Zhang JF, Zhang JS, Zhao ZH, Yang PB, Ji SF, Li N, Shi QD, Tan J, Xu X, Xu CB, Zhao LY. MicroRNA-770 affects proliferation and cell cycle transition by directly targeting CDK8 in glioma. Cancer Cell Int. 2018;18:195. doi: 10.1186/s12935-018-0694-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee HC, Her NG, Kang D, Jung SH, Shin J, Lee M, Bae IH, Kim YN, Park HJ, Ko YG, Lee JS. Radiation-inducible miR-770-5p sensitizes tumors to radiation through direct targeting of PDZ-binding kinase. Cell Death Dis. 2017;8:e2693. doi: 10.1038/cddis.2017.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao H, Yu X, Ding Y, Zhao J, Wang G, Wu X, Jiang J, Peng C, Guo GZ, Cui S. MiR-770-5p inhibits cisplatin chemoresistance in human ovarian cancer by targeting ERCC2. Oncotarget. 2016;7:53254–53268. doi: 10.18632/oncotarget.10736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belcheva A. MicroRNAs at the epicenter of intestinal homeostasis. Bioessays. 2017;39 doi: 10.1002/bies.201600200. [DOI] [PubMed] [Google Scholar]
- 27.Song H, Xu W, Song J, Liang Y, Fu W, Zhu XC, Li C, Peng JS, Zheng JN. Overexpression of Lin28 inhibits the proliferation, migration and cell cycle progression and induces apoptosis of BGC-823 gastric cancer cells. Oncol Rep. 2015;33:997–1003. doi: 10.3892/or.2014.3674. [DOI] [PubMed] [Google Scholar]
- 28.Im HI, Kenny PJ. MicroRNAs in neuronal function and dysfunction. Trends Neurosci. 2012;35:325–334. doi: 10.1016/j.tins.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Li B, Zhu D, Xie H, Du C, Xia Y, Tang W. Downregulation of lncRNA MEG3 and miR-770-5p inhibit cell migration and proliferation in Hirschsprung's disease. Oncotarget. 2017;8:69722–69730. doi: 10.18632/oncotarget.19207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi CY, Kim YH, Kim YO, Park SJ, Kim EA, Riemenschneider W, Gajewski K, Schulz RA, Kim Y. Phosphorylation by the DHIPK2 protein kinase modulates the corepressor activity of Groucho. J Biol Chem. 2005;280:21427–21436. doi: 10.1074/jbc.M500496200. [DOI] [PubMed] [Google Scholar]
- 31.Kim YH, Choi CY, Lee SJ, Conti MA, Kim Y. Homeodomain-interacting protein kinases, a novel family of co-repressors for homeodomain transcription factors. J Biol Chem. 1998;273:25875–25879. doi: 10.1074/jbc.273.40.25875. [DOI] [PubMed] [Google Scholar]
- 32.Sung KS, Go YY, Ahn JH, Kim YH, Kim Y, Choi CY. Differential interactions of the homeodomain-interacting protein kinase 2 (HIPK2) by phosphorylation-dependent sumoylation. FEBS Lett. 2005;579:3001–3008. doi: 10.1016/j.febslet.2005.04.053. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Q, Yoshimatsu Y, Hildebrand J, Frisch SM, Goodman RH. Homeodomain interacting protein kinase 2 promotes apoptosis by downregulating the transcriptional corepressor CtBP. Cell. 2003;115:177–186. doi: 10.1016/S0092-8674(03)00802-X. [DOI] [PubMed] [Google Scholar]
- 34.Rey C, Soubeyran I, Mahouche I, Pedeboscq S, Bessede A, Ichas F, De Giorgi F, Lartigue L. HIPK1 drives p53 activation to limit colorectal cancer cell growth. Cell Cycle. 2013;12:1879–1891. doi: 10.4161/cc.24927. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.