To the Editor:
Idiopathic pulmonary fibrosis (IPF) is a progressive fibrotic disease of the lung with increasing incidence and poorly understood pathogenesis (1). The lung microbiome has recently been implicated in IPF pathogenesis, with studies showing that lung microbiota are altered in disease, predictive of disease progression, correlated with variation in alveolar cytokines, and causally involved in animal models (2, 3). Although the radiographic features of IPF have critical diagnostic and prognostic significance (1, 4), the relationship between radiographic findings and lung microbiota is undetermined. To determine the relationship between lung microbiota and the anatomic features of IPF, we compared microbiota in IPF BAL fluid (BALF) with IPF radiographic features. Specifically, we compared microbiota in patients with IPF with and without radiographic evidence of honeycombing detected via high-resolution computed tomography (CT). Honeycombing is a cardinal histopathologic feature of IPF; it increases the diagnostic certainty and independently predicts mortality (4).
To examine lung microbiota and radiographic honeycombing, we studied patients with IPF who were enrolled in the COMET (Correlating Outcomes with Biochemical Markers to Estimate Time to Progression in Idiopathic Pulmonary Fibrosis) study. Bacterial communities in BALF were characterized using 16S rRNA gene sequencing and quantified as previously described (2). Honeycombing was recorded as absent or present on baseline chest high-resolution CT by a thoracic radiologist. Differences in community composition were determined by model‐based analysis of multivariate abundance data (mvabund) and by multivariate ANOVA with permutation testing. Differences in bacterial DNA burden and diversity were determined using Student’s t test. A total of 68 patients with data available for analysis were included. The clinical characteristics and demographics of the subjects have previously been described (2), and differences between patients with and without radiographic honeycombing are reported in Table 1. This was a subanalysis of previously published COMET data. The sequencing data are available via the National Center for Biotechnology Information Sequence Read Archive (accession numbers PRJNA515255 and PRJNA515279). Operational taxonomic unit (OTU), taxonomy, and metadata tables are available for download at https://github.com/dicksonlunglab/murine_pulmonary_fibrosis.
Table 1.
Demographics and Clinical Characteristics of the COMET Idiopathic Pulmonary Fibrosis Cohort
| No Honeycombing | Honeycombing | P Value | |
|---|---|---|---|
| n (%) | 29 (43%) | 39 (57%) | — |
| Age, yr, mean ± SD | 62.7 ± 7.3 | 66.5 ± 7.0 | 0.03 |
| Male, n (%) | 20 (68.9%) | 26 (66.6%) | 0.99 |
| FVC% predicted, mean ± SD | 72.0 ± 16.6 | 69.8 ± 18.3 | 0.61 |
| DlCO% predicted, mean ± SD | 49.3 ± 13.1 | 39.4 ± 13.6 | 0.007 |
| Smoking status | 0.006 | ||
| Non, n (%) | 14 (48%) | 6 (15%) | — |
| Ever, n (%) | 15 (52%) | 33 (85%) | — |
Definition of abbreviation: COMET = Correlating Outcomes with Biochemical Markers to Estimate Time to Progression in Idiopathic Pulmonary Fibrosis.
ANOVA, unpaired t test, or Fisher’s exact test was used when applicable.
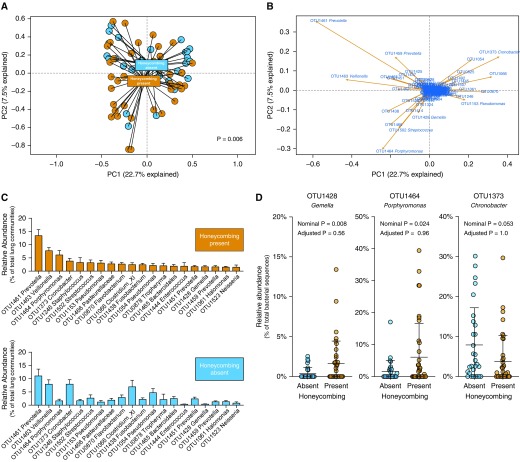
In this hypothesis-generating study, to determine whether the composition of lung bacteria differs among patients with IPF and honeycombing, we first used principal component analysis to visualize the relative similarity or dissimilarity of specimens when clustered by honeycombing status (Figure 1A). Although we observed a considerable overlap in the lung communities of patients with and without honeycombing, we also observed a clustered separation of specimens by honeycombing status. When we compared the communities statistically using mvabund, this collective difference in community composition was significant, robust to the taxonomic level of comparison (e.g., P = 0.006 at the OTU level of taxonomy, and P = 0.037 at the family level). The difference did not meet statistical significance when the communities were compared via multivariate ANOVA with permutation testing (P = 0.07, adjusted for age, baseline DlCO, and smoking status). We thus determined that lung communities in patients with IPF and honeycombing are collectively different from those in patients with IPF and no honeycombing, although a considerable taxonomic overlap exists.
Figure 1.
In patients with idiopathic pulmonary fibrosis, radiographic honeycombing is associated with changes in lung microbiota. Sixty-eight patients with idiopathic pulmonary fibrosis were stratified by the presence or absence of honeycombing on computed tomography scan. Lung microbiota were analyzed using amplicon sequencing of the 16S rRNA gene. Operational taxonomic unit (OTU) data in the principal component (PC) analysis were Hellinger transformed to allow for better behaviors with longer species gradients. (A) Ordination of specimens by community composition (principal component analysis) revealed clustering by honeycombing status, although considerable overlap was observed. Overall community composition was significantly distinct across groups (P = 0.006, mvabund). (B) BiPlot analysis of this ordination identified candidate bacterial taxa responsible for driving the separation of honeycombing specimens (e.g., OTU1428 Gemella and OTU1464 Porphyromonas). (C) Rank abundance analysis revealed taxonomic differences between groups. The 20 most abundant taxa in honeycombing-present specimens are shown in decreasing order of mean relative abundance (mean ± SEM). (D) Specific bacterial taxa were then compared across groups using direct comparison of mean relative abundance. Although select taxa were nominally significant when compared directly (e.g., OTU1428 Gemella and OTU1373 Chronobacter), no specific taxa were significant when adjusted for multiple comparisons by mvabund. Significance was determined via mvabund, and the nominal P values presented were obtained before adjustment for multiple comparisons (A and D).
To better understand these collective differences in community composition, we next used complementary techniques to compare specific bacterial taxa. We used BiPlot analysis (Figure 1B) to determine which bacterial taxa could explain the separation of specimens in our principal component analysis plot. This revealed several candidate taxa that explained variation in specimens toward the honeycombing-present cluster (e.g., OTU1464 Porphyromonas and OTU1428 Gemella), as well as several that explained variation toward the honeycombing-absent cluster (e.g., OTU1373 Cronobacter). We next used rank abundance analysis to visualize differences in the relative abundance of prominent taxa across groups (Figure 1C). Although the most abundant taxa were common across groups (e.g., OTU1461 Prevotella and OTU1463 Veillonella), several prominent taxa differed visibly across the comparison (e.g., OTU1464 Porphyromonas, OTU1373 Cronobacter, and OTU1428 Gemella). We directly compared the relative abundance of candidate taxa identified by BiPlot and rank abundance analysis (Figure 1D). We found that although OTU1428 Gemella and OTU1464 Porphyomonas were nominally enriched in specimens from patients with honeycombing (P = 0.008 and 0.024, respectively; mvabund), these comparisons were not significant when controlled for multiple comparisons (P > 0.05 for both). Similarly, although OTU1373 Cronobacter was nominally enriched among honeycombing-absent specimens, it was not significantly enriched when adjusted for multiple comparisons (P > 0.05). Cronobacter OTU1373 was detectable at a low signal (2.63% of reads) in negative controls. Both Gemella OTU1428 and Porphyromonas OTU1464 were not detectable in negative controls. We then compared the burden of bacterial DNA and the diversity of bacterial communities (Shannon diversity index) in BALF from patients with IPF with and without honeycombing. We found no significant difference in either feature of the microbiome (P > 0.05). We concluded that although honeycombing alters community composition, it is not associated with differences in bacterial burden or diversity.
Here, we demonstrate that the honeycombed lung is associated with alterations in the community composition of lung microbiota. These taxonomic differences are appreciable at both the species/genus and family levels of taxonomy and may not be directly attributable to individual taxa but rather to collective differences in community composition. Several studies have demonstrated the importance of lung dysbiosis in IPF. Molyneaux and colleagues have shown that a greater bacterial burden is predictive of increased mortality (5), a finding we recently validated (2). Studies have linked changes in the expression of host defense genes in blood with lung microbiota in IPF (3). Immunosuppression has a deleterious effect on clinical outcomes in patients with IPF, although antibiotic therapy in IPF may convey a clinical benefit (6, 7). A recent study reported minimal bacterial signal in excised lung tissue from patients with IPF (8), which may suggest that the host–microbiota interface can be more readily sampled via the airway mucosa (BALF) than from peripheral parenchymal tissue, which is largely comprised of extracellular matrix.
Honeycombing is a feature of IPF with pathologic and prognostic significance. It is believed to derive from mucin dysfunction in the distal airway, whereby epithelial cells exhibit increased expression of Muc5b, a mucin that is essential for normal mucocillary clearance (9). Overexpression of mucin in the distal airway may alter the ecological conditions of the lungs, selectively promoting dysbiosis. We observed a potential association between honeycombing and a member of the Gemella genus. Gemella belong to the Firmicutes phylum, are found at the mucosal surface of the aerodigestive tract, and have been implicated in exacerbations of chronic lung disease (10). We speculate that honeycombing results in ecological changes to the distal airways, fostering the growth of taxa such as Gemella spp., altering community composition, and contributing to injury from mucin overexpression and defective mucocillary clearance.
Our work has several limitations. We were unable to examine the topographical extent and severity of honeycombing in our study cohort. BALF was acquired from the right middle lobe or lingular segment based on the extent of comparative disease between these two locations, but the severity of honeycombing was not recorded or incorporated into our analysis. It may be that graded relationships exist between honeycombing and lung microbiota. Furthermore, interobserver variability exists in the reporting of CT honeycombing. It is possible that confounding is present in lung microbiome studies (e.g., due to medications or disease severity) that cannot be known or accounted for in our analysis. The size of our study cohort and the power to detect differences were limited. Therefore, these observations should be further examined in other cohorts of patients with IPF.
Our exploratory findings suggest a bidirectional interaction between lung microbiota and the anatomic disruption of IPF. Future studies should interrogate further the causal role of dysbiosis in the progressive tissue distortion and honeycombing of IPF to determine whether the lung microbiome represents a therapeutic target in IPF.
Footnotes
Supported by NHLBI grants K99HLI39996 (D.N.O’D.), R01AI117229 (B.B.M.), R01HL127805 (B.B.M.), K23HL130641 (R.P.D.), and R21AI137669 (R.P.D.).
Author Contributions: R.P.D., J.R.E.-D., B.B.M., and D.N.O’D. conceived the work, analyzed and interpreted data, and wrote and revised the manuscript. K.R.F., E.S.W., and F.J.M. acquired data. G.B.H. revised the manuscript. All authors approved the final version of the manuscript.
Originally Published in Press as DOI: 10.1164/rccm.201903-0680LE on August 16, 2019
Author disclosures are available with the text of this letter at www.atsjournals.org.
References
- 1.Raghu G, Collard HR, Egan JJ, Martinez FJ, Behr J, Brown KK, et al. ATS/ERS/JRS/ALAT Committee on Idiopathic Pulmonary Fibrosis. An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med. 2011;183:788–824. doi: 10.1164/rccm.2009-040GL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Dwyer DN, Ashley SL, Gurczynski SJ, Xia M, Wilke C, Falkowski NR, et al. Lung microbiota contribute to pulmonary inflammation and disease progression in pulmonary fibrosis. Am J Respir Crit Care Med. 2019;199:1127–1138. doi: 10.1164/rccm.201809-1650OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Molyneaux PL, Willis-Owen SAG, Cox MJ, James P, Cowman S, Loebinger M, et al. Host-microbial interactions in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2017;195:1640–1650. doi: 10.1164/rccm.201607-1408OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lynch DA, Godwin JD, Safrin S, Starko KM, Hormel P, Brown KK, et al. Idiopathic Pulmonary Fibrosis Study Group. High-resolution computed tomography in idiopathic pulmonary fibrosis: diagnosis and prognosis. Am J Respir Crit Care Med. 2005;172:488–493. doi: 10.1164/rccm.200412-1756OC. [DOI] [PubMed] [Google Scholar]
- 5.Molyneaux PL, Cox MJ, Willis-Owen SA, Mallia P, Russell KE, Russell AM, et al. The role of bacteria in the pathogenesis and progression of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2014;190:906–913. doi: 10.1164/rccm.201403-0541OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raghu G, Anstrom KJ, King TE, Jr, Lasky JA, Martinez FJ Idiopathic Pulmonary Fibrosis Clinical Research Network. Prednisone, azathioprine, and N-acetylcysteine for pulmonary fibrosis. N Engl J Med. 2012;366:1968–1977. doi: 10.1056/NEJMoa1113354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Macaluso C, Maritano Furcada J, Alzaher O, Chaube R, Chua F, Wells AU, et al. The potential impact of azithromycin in idiopathic pulmonary fibrosis. Eur Respir J. 2019;53:1800628. doi: 10.1183/13993003.00628-2018. [DOI] [PubMed] [Google Scholar]
- 8.Kitsios GD, Rojas M, Kass DJ, Fitch A, Sembrat JC, Qin S, et al. Microbiome in lung explants of idiopathic pulmonary fibrosis: a case-control study in patients with end-stage fibrosis. Thorax. 2018;73:481–484. doi: 10.1136/thoraxjnl-2017-210537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seibold MA, Smith RW, Urbanek C, Groshong SD, Cosgrove GP, Brown KK, et al. The idiopathic pulmonary fibrosis honeycomb cyst contains a mucocilary pseudostratified epithelium. PLoS One. 2013;8:e58658. doi: 10.1371/journal.pone.0058658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carmody LA, Zhao J, Schloss PD, Petrosino JF, Murray S, Young VB, et al. Changes in cystic fibrosis airway microbiota at pulmonary exacerbation. Ann Am Thorac Soc. 2013;10:179–187. doi: 10.1513/AnnalsATS.201211-107OC. [DOI] [PMC free article] [PubMed] [Google Scholar]