Abstract
Acute myelogenous leukemia (AML) is a hematological malignancy marked by the accumulation of large numbers of immature myeloblasts in bone marrow. The overall prognosis in AML is poor; hence, there is a pressing need to improve treatment. Although the sphingolipid (SL) ceramide demonstrates known cancer suppressor properties, it’s mechanism of action is multifaceted. Our studies in leukemia and other cancers have demonstrated that when combined with the antiestrogen, tamoxifen, the apoptosis-inducting effect of ceramide is greatly enhanced. The goal of the present study was to establish whether a ceramide-tamoxifen regimen also affects autophagic-driven cellular responses in leukemia. Using the human AML cell line KG-1, we demonstrate that, unlike exposure to the single agents, combination C6-ceramide-tamoxifen upregulated LC3-II expression, inhibited the mTOR signaling pathway, and synergistically induced KG-1 cell death in an Atg5-dependent manner. In addition, colocalization of autophagosome and mitochondria, indicative of mitophagosome formation and mitophagy, was observed. Versatility of the drug regimen was confirmed by experiments in MV4–11 cells, a FLT3-ITD AML mutant. These results indicate that the C6-ceramide-tamoxifen regimen plays a pivotal role inducing autophagy in AML, and thus constitutes a novel therapeutic design.
Keywords: Ceramide, Autophagy, Mitophagy, Tamoxifen, Leukemia, Sphingolipids
1. Introduction
Acute myeloid leukemia (AML), a cancer of the blood and bone marrow, is characterized by immature myeloid cell (blasts) proliferation and bone marrow failure. AML is a highly heterogeneous malignancy with a poor prognosis and a high mortality rate, and it is the most prevalent leukemia diagnosed in adults. This disease is aggressive, and patients would benefit from novel therapeutic interventions. The incidence of AML increases with age, and most cases occur in patients > 60 years of age. Although considerable progress in understanding the genetic basis of AML has been made, therapeutic options remain limited.
Treatment of AML usually consists of two phases. The first phase or “induction” is directed at targeting leukemic cells in blood and reducing the number of blasts in the bone marrow. In current first-line therapy, patients often receive intravenous anthracyclines given for 3-days combined with a 7-day continuous infusion of cytosine arabinoside (Ara-C). A “consolidation” then follows; its purpose is to eradicate any remaining leukemia cells via the use of more intensive treatment. Maintenance therapy can follow, which usually consists of intermediate doses of Ara-C. Older patients, unfortunately, have difficulty tolerating this intensive regimen [1]; moreover, the cure rate for AML varies. For example, the cure rate in patients who are < 60 years of age ranges from 35 to 40%, and in patients > 60 years of age this decreases to 5–15% [2]. Given the high percentage of morbidity in AML patients, the development of new therapeutic approaches to improve quality of life and durability of the response is critical.
Autophagy, an evolutionarily conserved machinery for bulk breakdown of cytoplasmic components, acts as a lysosomal transport system for degradation of proteins and damaged organelles. Autophagy plays an important role in mammalian cell survival, proliferation, and differentiation, particularly within the hematopoietic system. Diminished autophagic processes are implicated in the initiation of the myeloid-proliferative state, boosting the progression of AML [3,4], and Atg5-dependent autophagy can as well contribute to the development of AML [5]. It is well known that autophagy can be a double-edged sword; it can either serve to drive cancer survival or support cancer cell death. In AML, the induction of autophagy has also been shown to limit cell proliferation and glycolytic capacity [3,6,7], strongly suggesting that targeting autophagy could be useful in AML therapy.
Ceramide, the diradyl backbone of the major sphingolipids (SL), is known to exhibit potent tumor-suppressor properties [8]; however, its efficacy is limited by upregulated SL metabolism that is characteristic in cancer, most prominently in multidrug resistance [9]. Previous studies have shown that ceramide-based intervention with concomitant inhibition of ceramide metabolism/clearance, presents an attractive therapeutic approach in a myriad of cancer types [8,10–13]. Ceramide glycosylation is one of the prominent avenues by which cancer cells “detoxify” ceramide, a metabolic juncture that can be blocked by tamoxifen [11,13–16]. We have recently shown that a regimen comprised of a short-chain ceramide, C6-ceramide and tamoxifen, is cytotoxic in several cancer cell types [11,13,17–19]. These studies showed that the C6-ceramide-tamoxifen regimen triggered the induction of apoptosis and blocked cellular bioenergetics [11]; however, the possibility that additional avenues of cell death were in operation was not investigated. Because autophagy plays a critical role in the anti-leukemic effects of many chemotherapies [3,6,20–22], we sought to investigate whether autophagy contributed to C6-ceramide-tamoxifen responses in AML. Herein we demonstrate that a regimen consisting of C6-ceramide and tamoxifen, unlike exposure to these single agents, induces lethal, Atg5-dependent autophagy and participating mitophagy, as denoted by the formation of mitophagosomes. Cell signal transduction mapping demonstrated LC3-II upregulation in conjugation with AKT, mTOR, and S6K modulation, signals associated with inhibition of cancer cell growth.
2. Materials and methods
2.1. Materials
N-hexanoyl-D-erythro-sphingosine (C6-ceramide) was purchased from Avanti Polar Lipids (Alabaster, Alabama). Tamoxifen-HCl was from Sigma, St. Louis, MO. C6-ceramide and tamoxifen were dissolved in DMSO (10 mM stock solutions) and stored at −20 °C. alamarBlue™ was a product of Thermo Fisher Scientific. RIPA buffer and antibodies specific for LC3-II (catalog # 2775S), p-AKT (catalog # 9271S), p-mTOR (catalog #2971S), p-S6K (catalog #9209S), and β-actin (catalog # 3700), were purchased from Cell Signaling Technology (Danvers, MA). The Cyto-ID kit, including Hoechst stain, was purchased from Enzo Life Sciences (Farmingdale, NY), and Mitotracker was a product of Invitrogen (Carlsbad, CA).
2.2. Cell lines
The human AML cell line, KG-1, and the FLT3-ITD positive, human AML cell line MV4–11 were obtained from the American Type Culture Collection, Manassas, VA. Documentation including antigen expression, DNA profile, short tandem repeat profiling, and cytogenetic analysis was provided by the ATCC. KG-1 Atg5 knockdown cells (KG-1-KD) were established as previously described [23]. GFP-LC3 MEF cells were established in the laboratory of HG Wang, Penn State/Hershey College of Medicine. Cells were maintained in RPMI-1640 medium (Life Technologies, Carlsbad, CA, USA) with 10% fetal bovine serum (FBS) (Atlanta Biologicals, Atlanta, GA, and Peak Serum, Inc, Wellington, CO), 100 units/mL penicillin, and 100 μg/ml streptomycin (Life Technologies, Carlsbad, CA). MV4–11 cells were cultured in IMDM medium (Life Technologies, Carlsbad, CA), supplemented with 10% FBS and antibiotics as above. Cells were cultured in a humidified atmosphere, 95% air, 5% CO2, at 37 °C.
2.3. Cell viability
Cell viability was determined using the alamarBlue™ assay [24], following the suppliers (Thermo Fisher Scientific) instructions. Briefly, cells were seeded in 96-well plates in medium containing 5% FBS and allowed to equilibrate for 2 h in a tissue culture incubator before addition of test agents. At the appropriate times (indicated in figure legends), 10 μl of alamarBlue Reagent was added, an amount equal to 10% of the volume in the well, and the plates were placed at 37 °C, 5% CO2 for 1 h. Plates were removed and fluorescence was measured with excitation wavelength at 530–560 nm and emission wavelength at 590 nm, or absorbance at a wavelength of 570 nm or 600 nm. A negative control for medium only without cells was included to determine background signal, and a positive control of 100 μl of 100% reduced alamarBlue Reagent without cells was also included.
2.4. Flow cytometry
The Cyto-ID Autophagy Detection Kit (Enzo Life Sciences, Farmington, NY) was used to measure autophagy. Cells were plated in a 6-well plates (1 × 106/ml, 5% FBS in RPMI-1640, 2.0 ml final volume), then exposed to C6-ceramide and tamoxifen (dissolved into 5% FBS RPMI-1640 medium from 10 mM DMSO stock solutions) for 18–24 h. Cells were collected, washed three times with phenol red-free RPMI-1640 medium containing 0.2% BSA, resuspended in 500 μl Cyto-ID regent, and incubated in the dark for 30 min at 37 °C. Cells were then collected, washed twice in PBS, re-suspended in assay buffer (provided in the kit), and immediately evaluated by flow cytometry, using an LSRII 4-laser 11-color flow cytometer and FACscan (Becton Dickinson).
2.5. Autophagy and mitophagy detection by fluorescent microscopy
Cell autophagy and mitophagy was assessed by fluorescent microscopy. To determine autophagy, cells were treated and stained with Cyto-ID detection reagent then immediately imaged using an Evos FL auto cell imaging system (Life Technologies AMAFD1000). Mitophagy was determined by staining control and treated cells with a mixture that contained a 1:3000 dilution of Hoechst stain, a 1:3000 dilution of Cyto-ID Green Reagent dye, and a 1:3000 dilution of MitoTracker into 1 × assay buffer. Cyto-ID and Mitotracker were employed according to manufacturer’s instructions. Samples were then imaged on the Evos FL auto cell imaging system, using three different fluorescent light channels (RFP, GFP, DAPI). Samples were imaged at 200× Images of each sample were taken in similar topographical positions for both versions of imaging. Images were then overlaid for assessment.
2.6. Immunoblotting
After treatment, cell lysates were prepared by harvesting cells in RIPA buffer supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF), as previously described [17]. Protein in whole cellular lysates was measured using a BCA Protein Assay Kit (ThermoFisher Scientific, Hudson, NH). Equal amounts of protein were loaded onto 4%–12% NuPAGE gels (Invitrogen) for electrophoresis. Protein was transferred to PVDF membranes (Millipore, Billerica, MA) and probed with antibodies against p-mTOR, p-AKT, p-S6K, LC3-II and β-actin. Blots were imaged using a LI-COR Biosciences Odyssey Infrared Fluorescent scanner (Lincoln, NE).
2.7. Statistics
All statistical analyses were performed using an unpaired two-tailed Student’s t-test, and all data are expressed as the mean ± SEM with three independent experiments. Other statistical tests used are as described in the figure legends. Statistical significance in the figures is indicated as follows: *p < 0.05; **p < 0.01; ***p < 0.001.
3. Results
3.1. C6-ceramide-tamoxifen regimen induces autophagy and autophagic flux in KG-1 cells
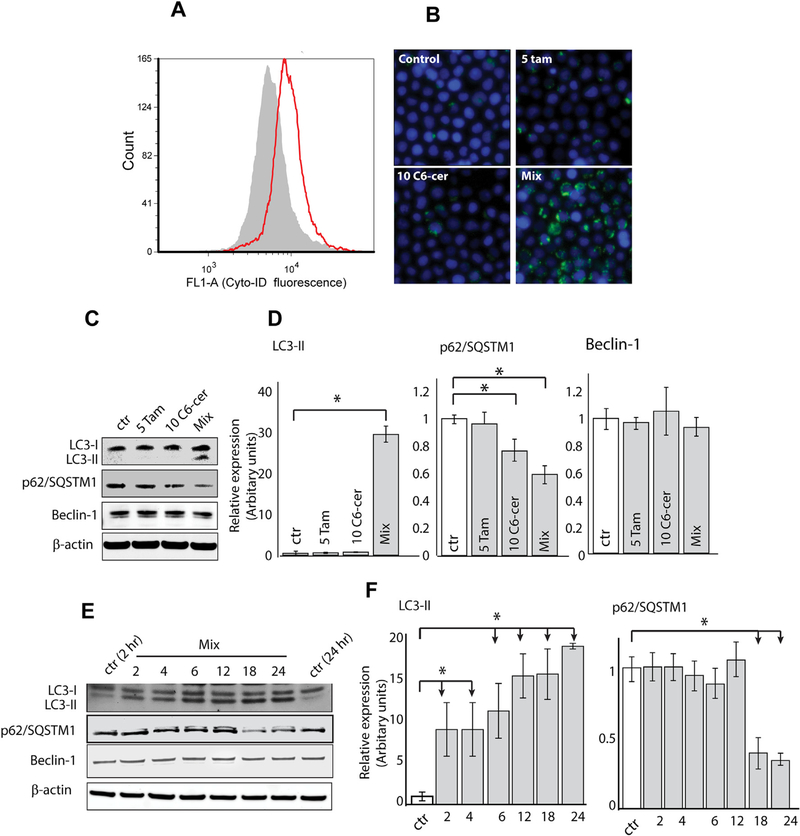
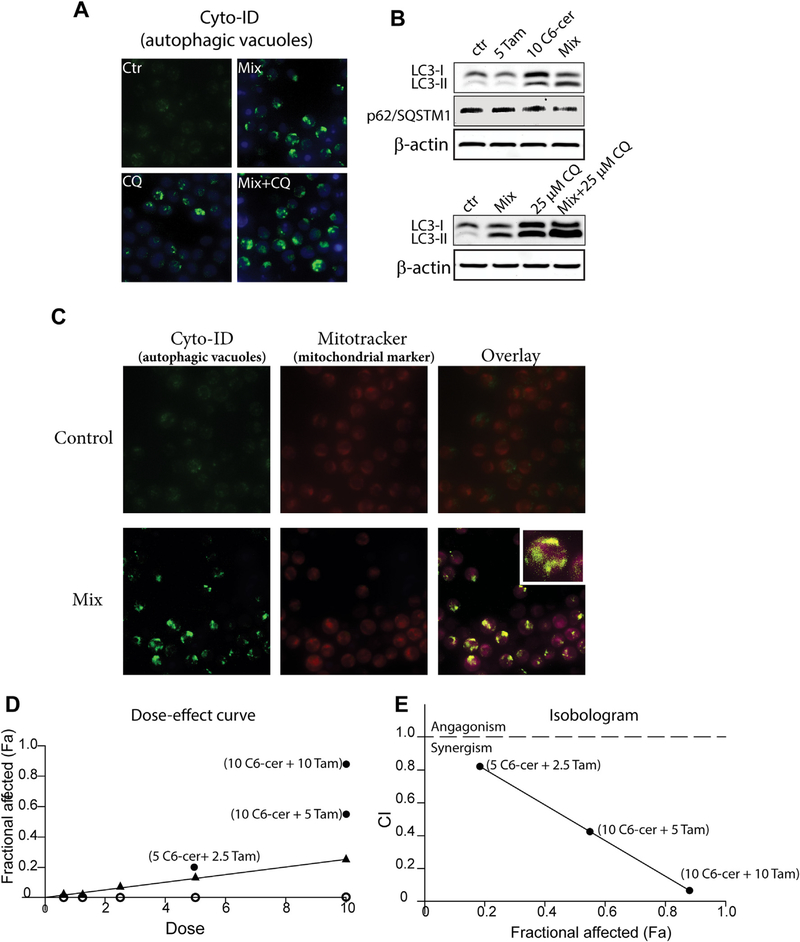
A regimen consisting from C6-ceramide and tamoxifen induces apoptosis in AML cells [11]. Given that autophagy plays an important role in AML cytotoxic responses [3,6,20–22], we were interested to learn whether this C6-ceramide-containing regimen also induced autophagy. Autophagy was assessed by flow cytometry and microscopy using Cyto-ID Green Dye and Western blot analysis. Cyto-ID dye stains autophagosomes and autolysosomes and is used to measure autophagic activity. As shown in Fig. 1A, treatment of KG-1 cells induced significant increases in Cyto-ID dye intensity (right shift), compared to the control, with an autophagic activity factor (see legend) equal 37.78 ± 1.75. Photomicroscopy revealed that the drug combination, unlike treatment with the single agents, increased the accumulation of autophagic vacuoles (Fig. 1B, mix). Beclin 1 and LC3-II, two specific markers of autophagy, are involved in the autophagic process, particularly in the early stages. Therefore, we investigated the levels of LC3-II and beclin 1 after treatment. The data in Fig. 1C and D (immunoblot quantitation) show that the drug combination, denoted by “mix”, significantly increased the production of LC3-II but had no effect on beclin 1 expression. We then explored earlier time points to decipher the temporal pattern of the autophagic process. Results demonstrated that the drug regimen increased the level of LC3-II as early as 2 h after treatment whereas Beclin 1 levels remained steady throughout the 24 h time course (Fig E, F immunoblot quantitation). Autophagosome accumulation, as indicated by Cyto-ID (Fig. 1A and B), and the levels of LC3-II (Fig. 1C–F), could be due to either induction of autophagy, or alternatively, suppression of steps in the autophagic pathway downstream of autophagosome formation, such as inefficient fusion or decreased lysosomal degradation [25,26]. Thus, we thought it important to assess autophagic flux. Autophagic flux is a term used to identify the dynamic process of autophagosome synthesis, delivery of autophagic substrates to the lysosome, and degradation of autophagic substrates inside the lysosome. Thus, autophagic flux is a more reliable indicator of autophagic activity than measurements of autophagosome numbers. p62/SQSTM1 (p62), a molecule that targets cargo for autophagic degradation, is a key molecule managing autophagic clearance of proteins. p62 is selectively incorporated into autophagosomes and is, itself, efficiently degraded by autophagy, thus it is an appropriate candidate to monitor autophagic flux [25,26]. Analysis of KG-1 cells treated with single agents and the mix for 24 h revealed diminished p62 levels when exposed to the drug combination (Fig. 1C and D), indicative of p62 degradation. Time course experiments conducted with the regimen showed that downregulation of p62 levels were apparent by 18 h (Fig. 1E and F). Thus, the accumulation of LC3-II and downregulation of p62 coincide in KG-1 cells, a strong indication that the drug combination induces autophagic flux.
Fig. 1. Effect of single agent and co-administration of C6-ceramide and tamoxifen on induction of autophagy in KG-1 cells.
A) Cells were exposed to combination C6-ceramide-tamoxifen for 24 h, after which the presence of autophagosomes (noted by the right-shift in fluorescence intensity) was determined using Cyto-ID and flow cytometry. B) Cells were exposed to agents indicated for 24 h. Autophagic vesicles were detected by Cyto-ID and fluorescence microscopy. C) Effect of C6-ceramide and tamoxifen on LC3-II, p62/SQSTM1 (p62), and Beclin-1 expression. Cells were exposed to the agents shown for 24 h. Levels of target proteins were analyzed by Western blot. D) Time course for expression of LC3-II, p62, and Beclin-1 in response to combination C6-ceramide-tamoxifen exposure. Autophagic activity factor (AAF). AAF = 100 × (MFI treated − MFI control)/MFI treated. Mean Fluorescence Intensity (MFI).
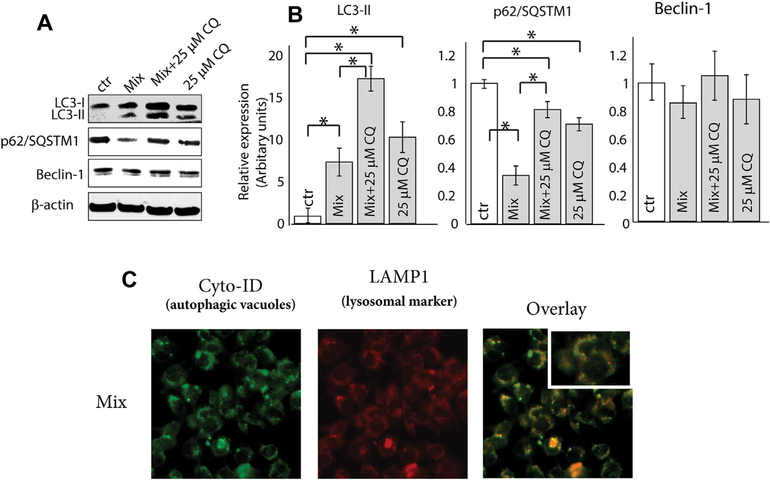
To determine whether autophagy elicited by the C6-ceramide-containing regimen interferes with downstream autophagosome formation, we utilized an autophagic inhibitor, chloroquine, a lysosomotropic agent that inhibits both fusion of autophagosomes with lysosomes and lysosomal protein degradation. As expected, when combined with chloroquine, the C6-ceramide-tamoxifen-induced depression (mix) in p62 levels was reversed, and the levels of LC3-II increased (Fig. 2A and B), confirming induction of autophagic flux. Next, we investigated whether the C6-ceramide-containing regimen impacted the fusion of lysosome with autophagosome. In this series of experiments, we used MEF cells transfected with lysosomal marker LAMP1-RFP (red fluorescent protein) (Fig. 2C). After treatment with the drug mix, MEF LAMP1+/+ cells were stained with Cyo-ID to identify autophagosomes. Overlay co-localization analysis revealed that the drug combination did not interfere with the fusion of lysosome with autophagosome, indicating the formation of autolysosomes and the induction autophagic flux. Although the results in murine fibroblasts suggest the potential for cytotoxicity in normal cells, we have previously demonstrated that the C6-ceramide-tamoxifen combination is not toxic in human peripheral blood mononuclear cells [27].
Fig. 2. Effect of co-administration of C6-ceramide and tamoxifen on induction of autophagic flux in KG-1 cells.
A) Impact of treatment on expression of LC3-II, p62, and Beclin-1. Cells were treated with C6-ceramide-tamoxifen for 18 h and then assessed for LC3-II, p62, and Beclin-1 expression by Western blot. B) Densitometric quantitation using ratios of LC3-II, p62, Beclin-1, to β-actin. C) Effect of treatment regimen on fusion of lysosome with autophagosome. MEF cells transfected with lysosomal marker LAMP1-RFP (red fluorescent protein) were used. After treatment (18 h), MEF LAMP1+/+ cells were stained with Cyo-ID to identify autophagosomes. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. Drug regimen inhibits mTOR signaling pathway
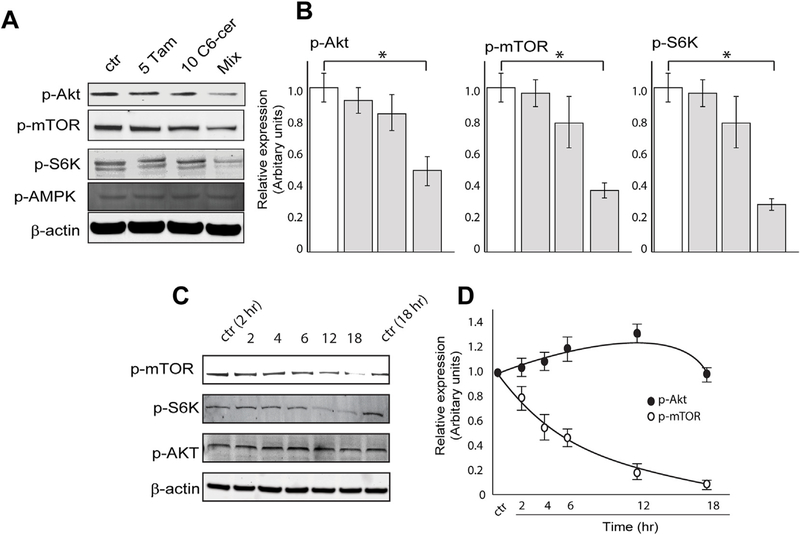
The mTOR signaling pathway plays a critical role in regulating autophagy [28]. Additionally, targeting mTOR has been shown to be effective in leukemia therapy [21,29]. Therefore, we extended our analysis to examine the effect of the drug combination on the mTOR signaling pathway, including mTOR upstream regulators, AKT and AMPK, and the downstream effector of mTOR, P70S6K. As shown in Fig. 3A and B (immunoblot quantitation), cells treated with C6-ceramide-tamoxifen (mix) for 24 h demonstrated downregulated expression of p-AKT, p-mTOR, and p-70S6K, with no effect on pAMPK. To distinguish whether the drug regimen inhibits AKT phosphorylation and subsequently blocks mTOR activity or whether mTOR is the primary target, cells were treated for different time periods to assess the temporal profile of AKT/mTOR signaling (Fig. 3C). Analysis revealed that the regimen downregulated the expression of p-mTOR as early as 2–4 h and p-70S6K by 12 h, whereas little effect on p-AKT expression was noted by 12 h. Immunoblot quantitation for p-AKT and p-mTOR is shown in Fig. 3D. These data indicate that mTOR is the primary target of the C6-ceramide-tamoxifen regimen, with possible albeit slight upregulation of p-AKT as a feedback signaling mechanism [30–32].
Fig. 3. Effect of C6-ceramide and tamoxifen on the mTOR signaling pathway in KG-1 cells.
A) Effect of single and combination agents on p-mTOR, p-70S6K (p-S6K), and p-AMPK expression. KG-1 cells were exposed to the agents shown for 24 h, after which protein expression was determined by Western blot. B) Effect of C6-ceramide-tamoxifen exposure time on expression of p-mTOR, p-S6K, and p-AKT. KG-1 cells were treated with the drug combination for the times shown. C) Densitometric quantitation of p-AKT and p-mTOR from Western blot in B.
3.3. Induction of mitophagy
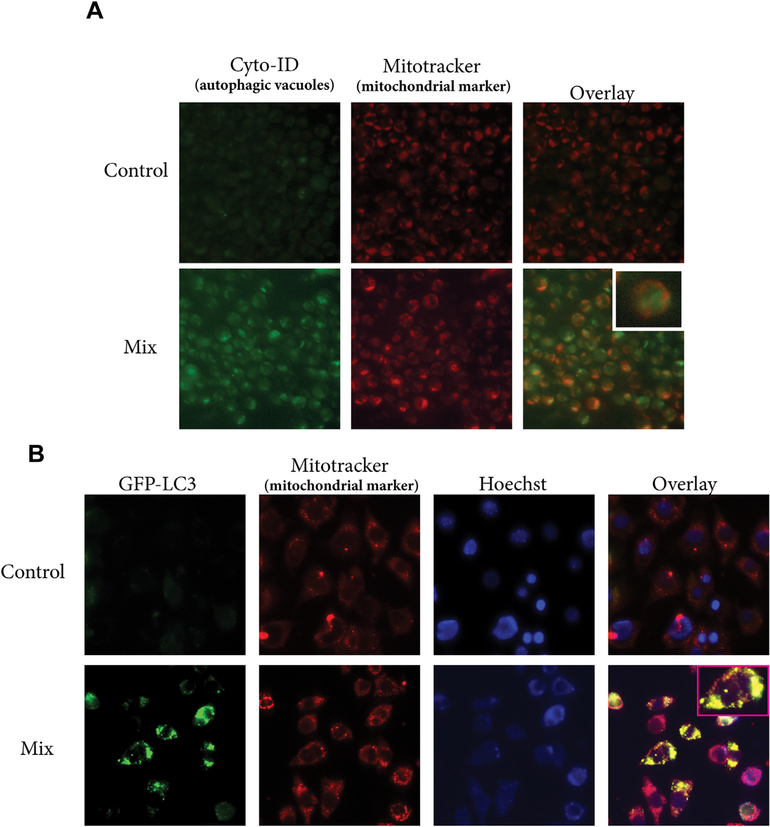
Mitophagy, or autophagy of the mitochondria, is an essential process to monitor mitochondrial quality control and considered a novel cell death mechanism [33,34]. We have shown that the C6-ceramide-tamoxifen regimen targets cellular bioenergetics in AML [11]. This targeting was associated with perturbation of mitochondrial function, indicating the possible interplay of mitophagy. We therefore investigated the impact of the drug regimen on the induction of mitophagy. Co-localization of autophagosome and mitochondria was monitored by staining cells with Cyto-ID (autophagosomal marker) and Mitotracker Red (mitochondrial marker). Co-localization of autophagosme and mitochondria was observed in KG-1 cells exposed to C6-ceramide-tamoxifen, (Fig. 4A, overlay), indicative of the formation of mitophagosomes. Co-localization of GFP-LC3, a marker of autophagy, with mitochondria has also been used as a measure of mitophagy [35]. In these experiments, we treated GFP-LC3 MEF’s with vehicle control or the C6-ceramide-tamoxifen regimen, followed by staining with Mito-tracker Red dye. As shown in Fig. 4B, treated cells (mix) demonstrated co-localization of GFP-LC3 puncta with mitochondria, compared to vehicle-treated controls, demonstrative of mitochondrial autophagy. Fragmented nuclei noted in Fig. 4B (GFP-LC3 Mix) result from exposure to the drug regimen, which also induces apoptosis [11].
Fig. 4. C6-ceramide-tamoxifen combination induces mitophagy.
A) Effect of drug mix on co-localization of autophagosome and mitochondria. KG-1 cells were exposed to DMSO (control) or to the drug mix (18 h), after which, cells were stained with Cyto-ID and Mitotracker Red. The fluorescence photomicroscopy overlay denotes co-incidental staining between autophagosome and mitochondria in cells exposed to mix (C6-ceramide-tamoxifen). B) GFP-LC3-MEF cells were exposed to vehicle control (DMSO) or the drug mix for 18 h, followed by staining with Mitotracker Red and Hoechst (nucleus staining). Images were obtained by fluorescence photomicroscopy. The mix overlay denotes co-localization of GFP-LC3 with mitochondria. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.4. C6-ceramide-tamoxifen regimen induces Atg5-dependent autophagic cell death
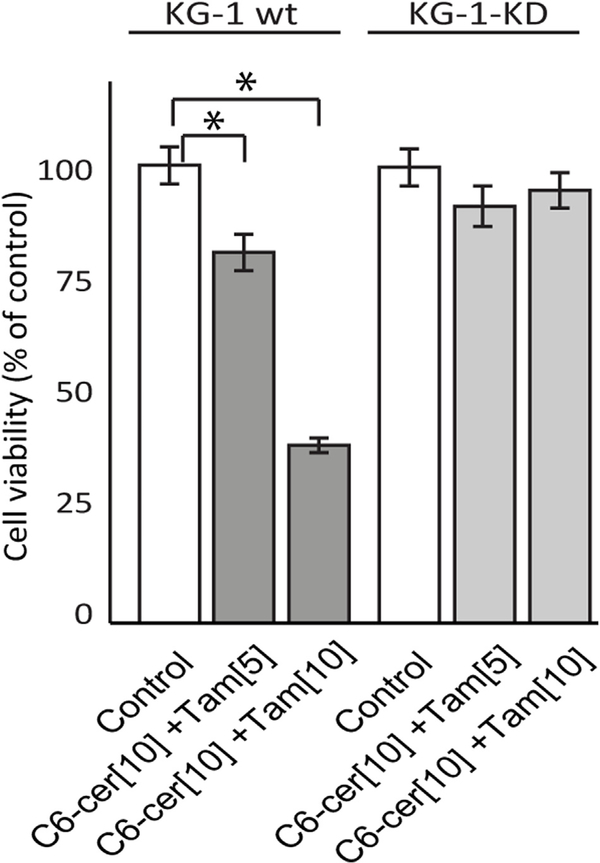
Autophagy maintains cell hemostasis but can also drive cell death, in extreme conditions. To investigate the role of autophagy in cell death, we blocked autophagy by depleting Atg5, an essential player in autophagosome formation. In these specific experiments, we employed KG-1 wild-type and counterpart Atg5 knockdown, KG-1-KD. The data in Fig. 5 clearly demonstrate sensitivity to C6-ceramide-tamoxifen in KG-1 wild-type with near-total insensitivity in the Atg5 knockdown, KG-1-KD.
Fig. 5. Atg5 expression impacts sensitivity to C6-ceramide-tamoxifen.
KG-1 wild-type and Atg5-knockdown, KG-1-KD, were seeded in 96-well plates (50,000 cells/well). After 2 h equilibration in a tissue culture incubator, cells were treated as indicated in the figure, for 24 h. Viability was determined using alamarBlue.
3.5. Drug regimen induces mitophagy and is synergistic in FLT3-ITD AML cells
It has recently been shown that induction of mitophagy comprises a promising approach to regulate AML cell death [7,33], particularly in FLT3-ITD AML, a mutation that occurs in about 30% of patients. With this, we endeavored to determine the impact of the C6-ceramide-containing regimen on mitophagy in MV4–11 cells, AML cells that harbor the FLT3-ITD mutation. Microscopic analysis using Cyto-ID as an autophagy indicator revealed that exposure of MV4–11 cells to C6-ceramide-tamoxifen (mix) induced the accumulation of autophagic vacuoles, indicative of autophagy (Fig. 6A, top right). Immunoblot analysis showed that treatment of MV4–11 cells with the mix lead to increased LC3-II production (Fig. 6B, top). Interestingly, compared with KG-1 cells, Fig. 6B also shows that C6-ceramide, alone, is major player in the induction of autophagy in MV4–11 cells (LC3-II increase), reinforcing an essential role for ceramide in the initiation of autophagy [7]. Similar with KG-1 cells, the drug mix downregulated the expression of p62 in MV4–11 cells (Fig. 6B, top), indicative of the increase in autophagic flux. Addition of the autophagy inhibitor, chloroquine, to the C6-ceramide-tamoxifen regimen, resulted in increased accumulation of LC3-II over that of mix alone and chloroquine alone (Fig. 6B, bottom). To assess mitophagy, we again monitored co-localization of autophagosome and mitochondria, and as shown in Fig. 6C, comparing control with mix-treated cells, the overlay clearly denotes co-localization of autophagosme and mitochondria (mitophagosomes) in the treated group. Mitophagy is a lethal pathway in AML [36,37], particularly in FLT3-ITD AML [7], thus we sought to determine whether the C6-ceramide-tamoxifen regimen was cytotoxic in MV4–11 cells. The data in Fig. 6D show that MV4–11 cells were largely refractory to C6-ceramide at the concentrations tested, with viability maintained at nearly 80% of control at the highest concentration, 10 μM. In addition, tamoxifen alone had no effect on MV4–11 cell viability. Strikingly, however, co-administration of these agents synergistically reduced cell viability, as shown by increases in the fraction affected (Fa) with increased concentrations of the drug regimen (Fig. 6D, solid black points). Chou-Talalay analysis [38] was used to determine synergy. By this method, a drug combination index (CI) is determined wherein a CI of 1 indicates an additive effect, and CI’s of 0.7–0.85 and 0.3–0.7 indicate moderate synergism and synergism, respectively. Fig. 6E shows the correlation between the CI and Fa in response exposure to the drug regimen. Strong synergy was observed with exposure to C6-ceramide-tamoxifen at 10 μM each agent, a combination that yielded a CI = 0.05 and Fa of 0.89. Synergy was also observed with 10 and 5 μM C6-ceramide and tamoxifen, respectively (CI = 0.42; Fa = 0.58). The combination of 10 μM C6-ceramide and 2.5 μM tamoxifen was also synergistic (CI = 0.8; Fa = 0.19).
Fig. 6.
C6-ceramide-tamoxifen regimen induces mitophagy and is synergistic in FLT3-ITDmut MV4–11 cells. A) Effect of drug combination and chloroquine on autophagic vesicle formation in MV4–11 cells. Cells were seeded into 6-well plates as described Materials and methods and exposed to combination C6-ceramide-tamoxifen (10 and 5 μM, respectively) for 18 h, washed and stained with Cyto-ID and immediately imaged. Chloroquine (CQ) was used at a concentration of 25 μM. B) Western blot depicting effect of single agents and mix on expression of LC3-II and p62, and the effect of chloroquine. Cells were treated with C6-ceramide, tamoxifen, and CQ as in A. C) Evaluation of mitophagy by co-localization of autophagosome and mitochondria. Cells were exposed to DMSO vehicle (control) or the mix (10 μM C6-ceramide, 5 μM tamoxifen) for 18 h, and stained as indicated. D) Impact of combinatorial dosage on cell viability. Cells were treated with the single agents (o, tamoxifen; ▲, C6-ceramide) or the combinations (•) indicated for 18 h, and viability was assessed by alamarBlue assay. y-axis refers to fraction of cells affected (Fa), where 1.0 is equal to 100%. E) Synergy plots for the drug combination. CI indicates combination index. The lower the CI, the higher the synergy, as shown by increase in fraction affected (Fa). All doses are in μM.
4. Discussion
This study was conducted to investigate the effect of combination C6-ceramide-tamoxifen on autophagy and its role in AML cell fate. Our previous studies have demonstrated the anticancer efficacy of short-chain ceramides in combination with agents that block ceramide glycosylation [18,19,39], wherein endpoint cell death was attributed to apoptosis [11]. Tamoxifen and its metabolite, N-desmethyltamoxifen have been shown to be effective inhibitors of C6-ceramide glycosylation [11,13]. This off-target property of the popular antiestrogen serves to enhance ceramide-driven responses.
Autophagy is a dynamic process in which the cell degrades and turns over cellular proteins and cytoplasmic organelles. The primary goal of this turnover is to maintain cellular homeostasis in response to unsuitable conditions. However, autophagy can result in cancer cell survival or culminate in cell death, and autophagy can protect cancer cells against chemotherapy. At the early stage of tumor progression, autophagy acts as tumor suppressor, and in advanced stages, autophagy promotes cancer survival [28]. The engulfment of damaged mitochondria by the autophagosome is called selective macroautophagy or mitophagy. Many studies have stressed the importance of mitophagy in cancer with evidence pointing to therapeutic benefit in promoting cancer cell death [7,33,34,36,37]. Thus, the biological role of autophagy in cancer is dynamic and multi-faceted.
In the present work, we show that the same regimen that induces apoptosis in AML cells elicits autophagy in parallel. In addition, we show that the elevation in autophagic markers is due to an increase in autophagic flux and not associated with inhibition of lysosomal function. Increases in autophagic flux, wherein there is active synthesis and degradation of autophagasomes, indicates that the cells are undergoing autophagy-mediated cell death [40,41]. We identified the participation of autophagic cell death by knockdown techniques focusing on Atg5, an essential protein involved in the early stages of autophagosome formation. In viability assays, the Atg5 knockdown cell line, KG-1-KD, demonstrated clear resistance to the C6-ceramide-tamoxifen combination, whereas in the wild-type, autophagy contributed to loss of cell viability. These results are consistent with studies demonstrating the potential role of autophagy as a cell death executioner in AML [4,7,20,22]. Moreover, the diminishing of autophagic flux promotes AML oncogenesis [4]. Thus, our findings complement the expanding evidence on the importance of targeting autophagy in AML with upregulation of autophagic flux.
Autophagy can be triggered by a myriad of factors such as starvation, increased intracellular protein aggresomes, ER stress, DNA damage, and activation of signaling pathways. One of these signaling avenues is comprised of the AKT/mTORC1 pathway. In AML, the PI3K/AKT and mTORC1 pathways play a critical role in regulating autophagy, cell survival, and resistance to chemotherapy [42–45]. Thus, targeting PI3K/AKT and mTORC1 pathways is a promising direction in AML therapeutics [46,47]. Decreased phosphorylation of AKT, mTOR, and p70S6K was evident in C6-ceramide-tamoxifen treated AML cells. Moreover, we showed that downregulation of p-mTOR was an early event, with little impact on AMPK, an upstream regulator of mTOR. Thus, we propose that the C6-ceramide-containing regimen regulates autophagy via mTOR targeting. Given that the AKT/mTORC1 pathway plays an essential role in the regulation of chemotherapy resistance in AML [44,46], the C6-ceramide-tamoxifen regimen could potentially counter cytarabine and daunorubicin resistance in AML. Lastly, it may be important to consider that Atg5-dependent iDISC-mediated (intracellular death-inducing signaling complex) apoptosis contributes to cell death induced by the C6-ceramide-tamoxifen regimen [23].
It has recently been shown that C18:0-ceramide plays a role in induction of mitophagy in AML [7]. The authors showed that C18:0-ceramide localizes to mitochondria and binds to LC3-II to recruit autophagosomes to mitochondria. In line with their findings, it is possible that C6-ceramide-tamoxifen-driven mitophagy is regulated by C18:0 ceramide derived via metabolism of C6-ceramide, because we have shown that C6-ceramide is metabolized to C16:0, C18:0, and C24:1 ceramide molecular species in KG-1 cells [11].
In summary, C6-ceramide in combination with tamoxifen, comprises a versatile regimen capable of inducing both apoptosis [11] and autophagy, inclusive of lethal mitophagy. The pathway for induction is Atg5-dependent and downregulates cell-proliferative, cell cycle, and nutrient-sensing signaling elements. This ceramide-containing, multitargeted drug regimen is a promising therapeutic that could off-set both survival autophagy as well as anti-apoptotic pathways in cancer.
Acknowledgements
This work was supported by a grant from the National Institutes of Health (NCI) P01 CA171983, and by a grant from the Brody Brothers Foundation, Kinston, NC.
Abbreviations
- AKT
protein kinase B
- AML
acute myelogenous leukemia
- AMPK
5’ adenosine monophosphate-activated protein
- Ara-C
cytosine arabinoside
- Atg
autophagy-related genes
- C6-ceramide
N-hexanoyl-D-erythro-sphingosine
- CQ
Chloroquine
- FLT3-ITD
fms-like tyrosine kinase 3-internal tandem duplication
- GFP-LC3
LC3 taq with green fluorescent protein
- KG-1-KD
KG-1 Atg5 knockdown cells
- LAMP
lysosome-associated membrane protein
- LC3
microtubule-associated protein
- mTOR
the mammalian target of rapamycin
- P62 (p62/SQSTM1
ubiquitin-binding protein/sequestosome-1
- PMSF
phenylmethylsulfonyl fluoride
- RIPA
radioimmunoprecipitation assay buffer
- S6K (70S6K)
ribosomal protein S6 kinase
- SL
sphingolipid
Footnotes
Declaration of interest
Authors declare that they have no conflicts of interest with the contents of this article. MK is Chief Medical Officer and cofounder of Keystone Nano, Inc.
References
- [1].Ossenkoppele G, Lowenberg B, How I treat the older patient with acute myeloid leukemia, Blood 125 (2015) 767–774. [DOI] [PubMed] [Google Scholar]
- [2].Dohner H, Weisdorf DJ, Bloomfield CD, Acute myeloid leukemia, N. Engl. J. Med. 373 (2015) 1136–1152. [DOI] [PubMed] [Google Scholar]
- [3].Watson AS, Riffelmacher T, Stranks A, Williams O, De Boer J, Cain K, MacFarlane M, McGouran J, Kessler B, Khandwala S, Chowdhury O, Puleston D, Phadwal K, Mortensen M, Ferguson D, Soilleux E, Woll P, Jacobsen SE, Simon AK, Autophagy limits proliferation and glycolytic metabolism in acute myeloid leukemia, Cell Death Dis. 1 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lalaoui N, Johnstone R, Ekert PG, Autophagy and AML-food for thought, Cell Death Differ. 23 (2016) 5–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Liu Q, Chen L, Atkinson JM, Claxton DF, Wang HG, Atg5-dependent autophagy contributes to the development of acute myeloid leukemia in an MLL-AF9-driven mouse model, Cell Death Dis. 7 (2016) e2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Tao YF, Li ZH, Du WW, Xu LX, Ren JL, Li XL, Fang F, Xie Y, Li M, Qian GH, Li YH, Li YP, Li G, Wu Y, Feng X, Wang J, He WQ, Hu SY, Lu J, Pan J, Inhibiting PLK1 induces autophagy of acute myeloid leukemia cells via mammalian target of rapamycin pathway dephosphorylation, Oncol. Rep. 37 (2017) 1419–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Dany M, Gencer S, Nganga R, Thomas RJ, Oleinik N, Baron KD, Szulc ZM, Ruvolo P, Kornblau S, Andreeff M, Ogretmen B, Targeting FLT3-ITD signaling mediates ceramide-dependent mitophagy and attenuates drug resistance in AML, Blood 128 (2016) 1944–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Morad SA, Cabot MC, Ceramide-orchestrated signalling in cancer cells, Nature reviews, Cancer 13 (2013) 51–65. [DOI] [PubMed] [Google Scholar]
- [9].Morad SAF, Cabot MC, The onus of sphingolipid enzymes in cancer drug resistance, Adv. Cancer Res. 140 (2018) 235–263. [DOI] [PubMed] [Google Scholar]
- [10].Dick TE, Hengst JA, Fox TE, Colledge AL, Kale VP, Sung SS, Sharma A, Amin S, Loughran TP Jr., Kester M, Wang HG, Yun JK , The apoptotic mechanism of action of the sphingosine kinase 1 selective inhibitor SKI-178 in human acute myeloid leukemia cell lines, J. Pharmacol. Exp. Ther. 352 (2015) 494–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Tan SF, Liu X, Fox TE, Barth BM, Sharma A, Turner SD, Awwad A, Dewey A, Doi K, Spitzer B, Shah MV, Morad SA, Desai D, Amin S, Zhu J, Liao J, Yun J, Kester M, Claxton DF, Wang HG, Cabot MC, Schuchman EH, Levine RL, Feith DJ, Loughran TP Jr., Acid ceramidase is upregulated in AML and represents a novel therapeutic target, Oncotarget 7 (2016) 83208–83222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Morad SA, Messner MC, Levin JC, Abdelmageed N, Park H, Merrill AH Jr., Cabot MC, Potential role of acid ceramidase in conversion of cytostatic to cytotoxic end-point in pancreatic cancer cells, Cancer Chemother. Pharmacol. 71 (2013) 635–645. [DOI] [PubMed] [Google Scholar]
- [13].Morad SA, Tan SF, Feith DJ, Kester M, Claxton DF, Loughran TP Jr., Barth BM, Fox TE, Cabot MC , Modification of sphingolipid metabolism by tamoxifen and N-desmethyltamoxifen in acute myelogenous leukemia–Impact on enzyme activity and response to cytotoxics, Biochim. Biophys. Acta 1851 (2015) 919–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Morad SA, Cabot MC, Tamoxifen regulation of sphingolipid metabolism–Therapeutic implications, Biochim. Biophys. Acta 1851 (2015) 1134–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lavie Y, Cao H, Volner A, Lucci A, Han TY, Geffen V, Giuliano AE, Cabot MC, Agents that reverse multidrug resistance, tamoxifen, verapamil, and cyclosporin A, block glycosphingolipid metabolism by inhibiting ceramide glycosylation in human cancer cells, J. Biol. Chem. 272 (1997) 1682–1687. [DOI] [PubMed] [Google Scholar]
- [16].Cabot MC, Giuliano AE, Volner A, Han TY, Tamoxifen retards glycosphingolipid metabolism in human cancer cells, FEBS Lett. 394 (1996) 129–131. [DOI] [PubMed] [Google Scholar]
- [17].Morad SA, Levin JC, Tan SF, Fox TE, Feith DJ, Cabot MC, Novel off-target effect of tamoxifen-inhibition of acid ceramidase activity in cancer cells, Biochim. Biophys. Acta 1831 (2013) 1657–1664. [DOI] [PubMed] [Google Scholar]
- [18].Morad SA, Madigan JP, Levin JC, Abdelmageed N, Karimi R, Rosenberg DW, Kester M, Shanmugavelandy SS, Cabot MC, Tamoxifen magnifies therapeutic impact of ceramide in human colorectal cancer cells independent of p53, Biochem. Pharmacol. 85 (2013) 1057–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Morad SA, Levin JC, Shanmugavelandy SS, Kester M, Fabrias G, Bedia C, Cabot MC, Ceramide-antiestrogen nanoliposomal combinations-novel impact of hormonal therapy in hormone-insensitive breast cancer, Mol. Cancer Ther. 11 (2012) 2352–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Fang J, Rhyasen G, Bolanos L, Rasch C, Varney M, Wunderlich M, Goyama S, Jansen G, Cloos J, Rigolino C, Cortelezzi A, Mulloy JC, Oliva EN, Cuzzola M, Starczynowski DT, Cytotoxic effects of bortezomib in myelodysplastic syndrome/acute myeloid leukemia depend on autophagy-mediated lysosomal degradation of TRAF6 and repression of PSMA1, Blood 120 (2012) 858–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wu J, Hu G, Dong Y, Ma R, Yu Z, Jiang S, Han Y, Yu K, Zhang S, Matrine induces Akt/mTOR signalling inhibition-mediated autophagy and apoptosis in acute myeloid leukaemia cells, J. Cell Mol. Med. 21 (2017) 1171–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wei Y, Kadia T, Tong W, Zhang M, Jia Y, Yang H, Hu Y, Tambaro FP, Viallet J, O’Brien S, Garcia-Manero G, The combination of a histone deacetylase inhibitor with the Bcl-2 homology domain-3 mimetic GX15–070 has synergistic antileukemia activity by activating both apoptosis and autophagy, Clin. Cancer Res. 16 (2010) 3923–3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Young MM, Takahashi Y, Khan O, Park S, Hori T, Yun J, Sharma AK, Amin S, Hu CD, Zhang J, Kester M, Wang HG, Autophagosomal membrane serves as platform for intracellular death-inducing signaling complex (iDISC)-mediated caspase-8 activation and apoptosis, J. Biol. Chem. 287 (2012) 12455–12468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Uzunoglu S, Karaca B, Atmaca H, Kisim A, Sezgin C, Karabulut B, Uslu R, Comparison of XTT and Alamar blue assays in the assessment of the viability of various human cancer cell lines by AT-101 (−/− gossypol), Toxicol. Mech. Methods 20 (2010) 482–486. [DOI] [PubMed] [Google Scholar]
- [25].Zhang XJ, Chen S, Huang KX, Le WD, Why should autophagic flux be assessed? Acta Pharmacol. Sin. 34 (2013) 595–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Mizushima N, Yoshimori T, Levine B, Methods in mammalian autophagy research, Cell 140 (2010) 313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Morad SA, Ryan TE, Neufer PD, Zeczycki TN, Davis TS, MacDougall MR, Fox TE, Tan SF, Feith DJ, Loughran TP Jr.,Kester M, Claxton DF, Barth BM, Deering TG, Cabot MC, Ceramide-tamoxifen regimen targets bioenergetic elements in acute myelogenous leukemia, J. Lipid Res. 57 (2016) 1231–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kondo Y, Kanzawa T, Sawaya R, Kondo S, The role of autophagy in cancer development and response to therapy, Nat. Rev. Canc. 5 (2005) 726–734. [DOI] [PubMed] [Google Scholar]
- [29].Martelli AM, Evangelisti C, Chappell W, Abrams SL, Basecke J, Stivala F, Donia M, Fagone P, Nicoletti F, Libra M, Ruvolo V, Ruvolo P, Kempf CR, Steelman LS, McCubrey JA, Targeting the translational apparatus to improve leukemia therapy: roles of the PI3K/PTEN/Akt/mTOR pathway, Leukemia 25 (2011) 1064–1079. [DOI] [PubMed] [Google Scholar]
- [30].Shi Y, Yan H, Frost P, Gera J, Lichtenstein A, Mammalian target of rapamycin inhibitors activate the AKT kinase in multiple myeloma cells by up-regulating the insulin-like growth factor receptor/insulin receptor substrate-1/phosphatidylinositol 3-kinase cascade, Mol. Cancer Ther. 4 (2005) 1533–1540. [DOI] [PubMed] [Google Scholar]
- [31].Wan X, Harkavy B, Shen N, Grohar P, Helman LJ, Rapamycin induces feedback activation of Akt signaling through an IGF-1R-dependent mechanism, Oncogene 26 (2007) 1932–1940. [DOI] [PubMed] [Google Scholar]
- [32].Chen XG, Liu F, Song XF, Wang ZH, Dong ZQ, Hu ZQ, Lan RZ, Guan W, Zhou TG, Xu XM, Lei H, Ye ZQ, Peng EJ, Du LH, Zhuang QY, Rapamycin regulates Akt and ERK phosphorylation through mTORC1 and mTORC2 signaling pathways, Mol. Carcinog. 49 (2010) 603–610. [DOI] [PubMed] [Google Scholar]
- [33].Dany M, Ogretmen B, Ceramide mediated lethal mitophagy: a novel cell death mechanism in FLT3 targeted therapy for acute myeloid leukemia, FASEB J. 29 (2015). [Google Scholar]
- [34].Hamacher-Brady A, Brady NR, Mitophagy programs: mechanisms and physiological implications of mitochondrial targeting by autophagy, Cell. Mol. Life Sci. 73 (2016) 775–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zhu J, Dagda RK, Chu CT, Monitoring mitophagy in neuronal cell cultures, Methods Mol. Biol. 793 (2011) 325–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Jiang W, Ogretmen B, Ceramide stress in survival versus lethal autophagy paradox: ceramide targets autophagosomes to mitochondria and induces lethal mitophagy, Autophagy 9 (2013) 258–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Sentelle RD, Senkal CE, Jiang W, Ponnusamy S, Gencer S, Selvam SP, Ramshesh VK, Peterson YK, Lemasters JJ, Szulc ZM, Bielawski J, Ogretmen B, Ceramide targets autophagosomes to mitochondria and induces lethal mitophagy, Nat. Chem. Biol. 8 (2012) 831–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Chou TC, Talalay P, Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors, Adv. Enzym. Regul. 22 (1984) 27–55. [DOI] [PubMed] [Google Scholar]
- [39].Watters RJ, Fox TE, Tan SF, Shanmugavelandy S, Choby JE, Broeg K, Liao J, Kester M, Cabot MC, Loughran TP, Liu X, Targeting glucosylceramide synthase synergizes with C6-ceramide nanoliposomes to induce apoptosis in natural killer cell leukemia, Leuk. Lymphoma 54 (2013) 1288–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Gurpinar E, Grizzle WE, Shacka JJ, Mader BJ, Li N, Piazza NA, Russo S, Keeton AB, Piazza GA, A novel sulindac derivative inhibits lung adenocarcinoma cell growth through suppression of Akt/mTOR signaling and induction of autophagy, Mol. Cancer Ther. 12 (2013) 663–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Denton D, Nicolson S, Kumar S, Cell death by autophagy: facts and apparent artefacts, Cell Death Differ. 19 (2012) 87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zeng Z, Wang RY, Qiu YH, Mak DH, Coombes K, Yoo SY, Zhang Q, Jessen K, Liu Y, Rommel C, Fruman DA, Kantarjian HM, Kornblau SM, Andreeff M, Konopleva M, MLN0128, a novel mTOR kinase inhibitor, disrupts survival signaling and triggers apoptosis in AML and AML stem/progenitor cells, Oncotarget 7 (2016) 55083–55097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Mohammadi S, Ghaffari SH, Shaiegan M, Zarif MN, Nikbakht M, Akbari Birgani S, Alimoghadam K, Ghavamzadeh A, Acquired expression of osteopontin selectively promotes enrichment of leukemia stem cells through AKT/mTOR/PTEN/beta-catenin pathways in AML cells, Life Sci. 152 (2016) 190–198. [DOI] [PubMed] [Google Scholar]
- [44].Lindblad O, Cordero E, Puissant A, Macaulay L, Ramos A, Kabir NN, Sun J, Vallon-Christersson J, Haraldsson K, Hemann MT, Borg A, Levander F, Stegmaier K, Pietras K, Ronnstrand L, Kazi JU, Aberrant activation of the PI3K/mTOR pathway promotes resistance to sorafenib in AML, Oncogene 35 (2016) 5119–5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Park S, Chapuis N, Tamburini J, Bardet V, Cornillet-Lefebvre P, Willems L, Green A, Mayeux P, Lacombe C, Bouscary D, Role of the PI3K/AKT and mTOR signaling pathways in acute myeloid leukemia, Haematologica 95 (2010) 819–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Amadori S, Stasi R, Martelli AM, Venditti A, Meloni G, Pane F, Martinelli G, Lunghi M, Pagano L, Cilloni D, Rossetti E, Di Raimondo F, Fozza C, Annino L, Chiarini F, Ricci F, Ammatuna E, La Sala E, Fazi P, Vignetti M, Temsirolimus, an mTOR inhibitor, in combination with lower-dose clofarabine as salvage therapy for older patients with acute myeloid leukaemia: results of a phase II GIMEMA study (AML-1107), Br. J. Haematol. 156 (2012) 205–212. [DOI] [PubMed] [Google Scholar]
- [47].Altman JK, Sassano A, Platanias LC, Targeting mTOR for the treatment of AML. New agents and new directions, Oncotarget 2 (2011) 510–517. [DOI] [PMC free article] [PubMed] [Google Scholar]