Abstract
Long-chain polyprenol phosphates feature in membrane-associated glycoconjugate biosynthesis pathways across domains of life. These unique amphiphilic molecules are best known as substrates of polytopic membrane proteins, including polyprenol-phosphate phosphoglycosyl and glycosyl transferases, and as components of more complex substrates. The linear polyprenols are constrained by double bond geometry and lend themselves well to interactions with polytopic membrane proteins, in which multiple transmembrane helices form a rich landscape for interactions. Recently, a new superfamily of monotopic phosphoglycosyl transferase enzymes has been identified that interacts with polyprenol phosphate substrates via a single reentrant membrane helix. Intriguingly, despite the dramatic differences in their membrane-interaction domains, both polytopic and monotopic enzymes similarly favor a unique cis/trans geometry in their polyprenol phosphate substrates.
Herein, we present a multipronged biochemical and biophysical study of PglC, a monotopic phosphoglycosyl transferase that catalyzes the first membrane-committed step in N-linked glycoprotein biosynthesis in Campylobacter jejuni. We probe the significance of polyprenol phosphate geometry both in mediating substrate binding to PglC and in modulating the local membrane environment. Geometry is found to be important for binding to PglC; a conserved proline residue in the reentrant membrane-helix is determined to drive polyprenol phosphate recognition and specificity. Pyrene fluorescence studies show that polyprenol phosphates at physiologically-relevant levels increase the disorder of the local lipid bilayer; however, this effect is confined to polyprenol phosphates with specific isoprene geometries. The molecular insights from this study may shed new light on the interactions of polyprenol phosphates with diverse membrane-associated proteins in glycoconjugate biosynthesis.
Keywords: polyprenol phosphate, reentrant membrane helix, membrane protein interactions, glycoconjugate biosynthesis, SMALP
Graphical Abstract
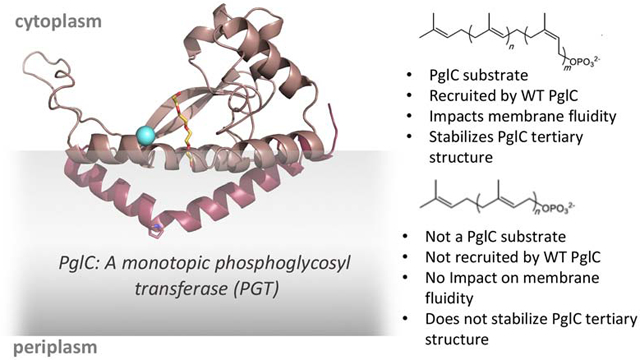
1. Introduction
Glycoconjugates constitute a diverse and complex class of macromolecules that play a wide variety of essential roles in both prokaryotic and eukaryotic biology. The assembly of many complex glycans is associated with cellular membranes (e.g. the endoplasmic reticulum membrane in eukaryotes and the cell membrane in prokaryotes). In such biosynthetic pathways, a series of integral membrane-bound phosphoglycosyl transferases (PGT) and glycosyltransferase (GT) enzymes catalyze the stepwise addition of monosaccharides from glycosyl donor substrates to amphiphilic polyprenol phosphate (PrenP) acceptors in the membrane (Fig. 1A). PrenPs are linear, long-chain products from the same terpene biosynthesis pathway responsible for the biosynthesis of farnesyl- and geranylgeranyl diphosphate for post-translational protein prenylation, as well as cholesterol and other steroids. However, unlike cholesterol, which can make up as much as 30% of the plasma membrane in eukaryotes [1], polyprenol phosphates have been estimated to comprise only ≤1% of bacterial and eukaryotic membranes [2-4].
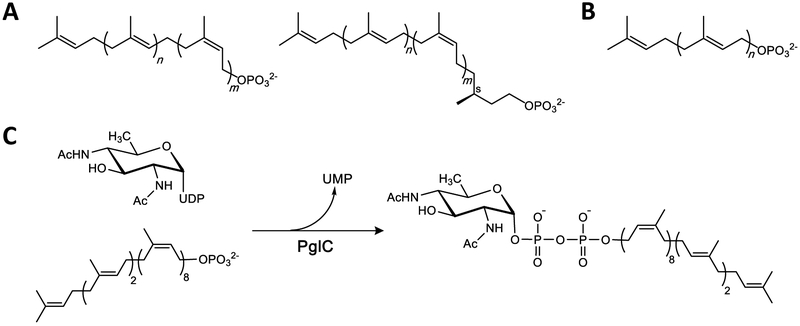
Fig. 1 – Structures of relevant terpenes.
A. Top: Generalized structure of bacterial polyprenol phosphate. For bacterial UndP, n = 2 and m = 8. (Note: the double-bond geometry of the plant-derived UndP used for in vitro assays described herein is n = 3 and m = 7 for bacterial UndP). [17]. Bottom: Generalized structure of dolichol phosphate (DolP); n = 2 and m = 5-17.
B. Structure of the all-trans nonaprenol, solanesol phosphate (SolP); n = 8.
C. The phosphoglycosyl transfer reaction catalyzed by PglC from C. jejuni.
The precise structural features of the PrenPs used in glycan assembly vary by species (Fig. 1A). Archaea and eukaryotes utilize α-saturated polyprenol phosphates, called dolichols, which range in length from as few as eight isoprene units in some archaea, to 20 or more in mammals and plants [5-9]. Archaeal PrenPs can be additionally saturated at other isoprene units [10, 11]. Bacteria, on the other hand, generally feature α-unsaturated polyprenol phosphates. The PrenP found in most bacteria investigated to date, including model organisms Escherichia coli and Staphylococcus aureus, is the 11-isoprene undecaprenol phosphate (UndP) [2, 12], however, a preference for shorter PrenPs has also been reported for some bacterial species [13-15].
Despite the apparent diversity of PrenP structures across domains of life, both the unique linear polyisoprenoid character and the preference for cis (Z) double-bond geometry in the isoprene units proximal to the phosphoryl group are strictly conserved in all glycan assembly pathways. Furthermore, although there exist linear polyprenols composed exclusively of trans (E) isoprene units, such as the nine-isoprene solanesol abundant in tobacco and other solanaceous crops (Fig. 1B) [16], these molecules do not serve as carriers in glycan assembly pathways [17]. The structural conservation of a series of cis configuration isoprene units suggests that the unique geometry of the PrenPs may play a more purposed role in membrane-associated glycoconjugate biosynthesis, beyond that of a hydrophobic membrane anchor on which glycan assembly can occur. In this context, a variety of roles in mediating substrate-enzyme binding and multi-enzyme complex formation, as well as in modulating the biophysical properties of the membrane, have been proposed [18-25].
PrenPs are best known as substrates for polytopic membrane proteins. Prominent examples include PGTs such as MraY and GPT, which catalyze the first membrane-committed step in bacterial peptidoglycan and mammalian N-linked glycoprotein biosynthesis respectively [26, 27], and polyprenol-phosphate GTs, such as Dol-P-mannose synthase, which is important for the biosynthesis of glycosyl donor substrates in the endoplasmic reticulum (ER) [28-30]. Recent structural analyses have beautifully revealed how the constraints imposed by double bond geometry in the PrenPs make them well-suited for interactions with polytopic membrane proteins, in which the multiple transmembrane helices provide complex surfaces for substrate binding [28, 31, 32]. Complementary molecular dynamics simulations of PrenP-linked substrates in lipid bilayer models have also provided further insight into the conformational dynamics of PrenP [20, 33].
Recently, a new superfamily of monotopic PGTs, has been defined. Unlike the polytopic PGTs, the superfamily of monotopic PGTs appears to be found exclusively in prokaryotic glycoconjugate biosynthesis pathways [34]. The most extensively studied member of this second superfamily is PglC, a monotopic PGT that initiates the N-linked protein glycosylation (pgl) pathway in the Gram-negative enteropathogen, Campylobacter jejuni. The recently reported structure of the PglC homolog from Campylobacter concisus, which shares 72% sequence identity with the C. jejuni PglC, revealed a unique membrane architecture [35]. An N-terminal hydrophobic domain of PglC forms a reentrant helix-break-helix that penetrates the membrane to a depth of 14 Å and associates with several amphipathic helices to anchor this integral membrane protein into the cytoplasmic face of the inner membrane. The enzyme active site, formed by one of the amphipathic helices and part of the soluble domain, is positioned at the membrane interface, allowing for efficient transfer of the phosphosugar, in this case N,N'-diacetylbacillosamine (diNAcBac) phosphate, from a soluble nucleotide sugar donor to the membrane-anchored PrenP via a covalent phosphosugar intermediate [36] (Fig. 1C). In the structure, the putative PrenP-binding site, located adjacent to the active site, was proposed based on electron density attributed to a co-crystallizing PEG molecule (Fig. 2).
Fig. 2 – Structure of PglC.
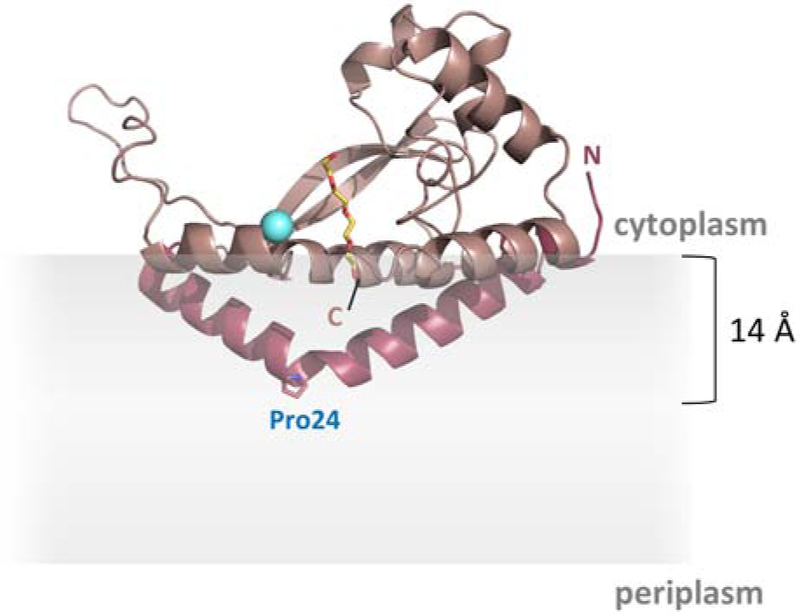
The reported structure of PglC (PDB 5W7L) [35]. The N-terminal reentrant helical domain is shown in dark pink. Pro24 in the membrane-resident domain is shown as sticks. The Mg2+ in the active site is shown in cyan. A co-crystallized PEG molecule marking the putative PrenP-binding site is shown in yellow. The location of the membrane, as calculated in [35], is represented in gray.
Based on extensive homology, all of the PGTs in the monotopic superfamily share the PglC-like functional core with a single, reentrant membrane helix (RMH) domain. However, despite the dramatic differences in their membrane-interaction domains, both polytopic and monotopic enzymes similarly favor a unique cis/trans geometry in their PrenP substrates. Additionally, a conserved proline (Pro24 in C. concisus PglC), located at the break in the reentrant helix domain has been shown to be important for PglC activity (Fig. 2) [34, 37]. Based on the position of Pro24 within the only membrane-resident domain of PglC, and on the known conservation of prolines in polyisoprenol-recognition sequences (PIRSs) in diverse proteins [19, 24, 38], it has been hypothesized that Pro24 may play a key role in recognition and binding of PrenP substrates in the membrane, both in the context of PglC and in other members of the monotopic PGT superfamily [27, 34].
Herein, we apply a range of biochemical and biophysical approaches to identify and quantify PrenPs in C. jejuni membranes, and to assess binding of PrenP to PglC, both in vitro and in a native-like model membrane environment. Further, we probe the importance of proper cis/trans geometry of PrenP both for substrate recognition by PglC and for modulating the local membrane environment. The significance of this work in the broader understanding of PrenPs in glycan assembly across domains is also discussed.
2. Materials and Methods
C. jejuni growth
C. jejuni strain 81-176 from a glycerol stock was streaked on selective blood agar plates (BD, Franklin Lakes, NJ; cat. 221727) at 37 °C under microaerophilic conditions (10% CO2, 5% O2, 85% N2). After 20-24 hr of growth cells were resuspended in a small volume of Mueller-Hinton broth. A 500 μL aliquot of resuspension at an OD600 of 0.5 was used to inoculate a 0.5 L culture of Mueller-Hinton broth + 10 μg/mL trimethoprim (selection antibiotic) in a 1 L Erlenmeyer flask. Cultures were grown at 37 °C under microaerophilic conditions (see above) with shaking at 200 rpm. In preliminary experiments, cultures were sampled periodically to determine a growth curve under these conditions; log-phase growth was found to occur at an OD600 of 0.15-0.35. In subsequent experiments, cultures were harvested at an OD600 within this range and stored at −80 ℃ until analysis.
Total lipid extraction
Lipids were extracted using a modification of the protocol by Bligh and Dyer [39]. Cells from 250 mL of C. jejuni culture were washed once with Tris-buffered saline (25 mM Tris, 150 mM NaCl, pH 7.5) and resuspended in 2 mL of water acidified to a pH of 1-2 by the addition of concentrated HCl (a low pH facilitated the partition of UndP into the lower organic phase during extraction). Next, 4 mL of 2:1 (v/v) CHCl3:MeOH were added to the cell suspension and samples were vortexed for 15-30 sec and centrifuged at 1,000 × g for 5-10 min. The organic phase was removed and the aqueous phase was extracted three additional times with 1-2 mL CHCl3. All organic phases were pooled, concentrated under N2 and washed with an additional 3 mL of water acidified with HCl to a pH of 1-2. Final lipid extracts were dried completely under N2 and stored at −20 °C until analysis.
Quantification and alkaline hydrolysis of total lipid extracts
Lipid extracts were quantified by a modification of the ammonium molybdate method for microdetermination of total phosphate described previously [40] against a phosphorus standard solution (Sigma-Aldrich, St. Louis, MO).
Prior to LC/MS quantification of total PrenP, lipid extracts were subjected to mild alkaline hydrolysis [41] to release PrenP from PrenPP and PrenPP-linked glycans. Dry lipids (~500 nmol) were resuspended in 4 mL of a single-phase Bligh-Dyer mix containing 0.2 M NaOH (1:2:0.8 (v/v) CHCl3:MeOH:0.95M NaOH) and incubated at 60 °C for 1 hr with periodic vortexing. The addition of 1 mL CHCl3 and 0.8 mL 1 M HCl neutralized the solution and converted the solvent mix to two phases. The organic phase was removed and the aqueous phase was extracted three additional times with 1 mL CHCl3. All organic phases were pooled, concentrated under N2 and washed with an additional 2 mL of water acidified with HCl to a pH of 1-2. Total hydrolyzed lipids were divided equally into five aliquots (~100 nmol each) and supplemented with 0-100 pmol of pure UndP standard (see below). Samples were dried under N2 and stored at −20 °C until analysis.
Purification of an UndP standard for LC/MS
Synthesis of UndP by phosphorylation of plant-derived undecaprenol was described previously [42]. This method yields a mixture of C50-C60 PrenP (predominantly UndP, C55) which was used for activity and thermal denaturation assays. For use as a standard for PrenP quantification, this mix was purified further by ion-pair reverse-phase HPLC to give only the UndP [43]. Separation of C50, C55 and C60 PrenP was successfully performed on a YMC-Pack ODS-A RP-18 column (4.6 × 250 mm, 5 μm, 120 Å) under isocratic elution in 76:19:5:2 (v/v) EtOH:CH3CN:hexanes:ion-pair reagent (ion pair-reagent prepared as 8 g 85% H3PO4 + 45 mL 40% aq. tetrabutyl ammonium hydroxide). The fraction containing pure UndP (C55) was quantified by the ammonium molybdate method described above, dried under N2 and stored at - 20 °C until analysis.
Quantification of total polyprenol-P by LC/MS
Each dried lipid extract was resuspended in 0.5 mL 2:1 (v/v) CHCl3:MeOH and 10 μL of sample was injected for normal-phase LC/MS analysis. Normal-phase LC was performed on an Agilent 1200 Quaternary LC system equipped with an Ascentis Silica HPLC column, 5 μm, 25 cm × 2.1 mm (Sigma-Aldrich, St. Louis, MO). Mobile phase A consisted of CHCl3:MeOH:aq. NH4OH (800:195:5, v/v); mobile phase B consisted of CHCl3:MeOH:aq. NH4OH (600:340:50:5, v/v); mobile phase C consisted of CHCl3:MeOH:aq. NH4OH (450:450:95:5, v/v). The elution program consisted of the following: 100% mobile phase A was held isocratically for 2 min, then linearly increased to 100% B over 14 min and held at 100% B for 11 min. The LC gradient was then changed to 100% C over 3 min and held at 100% C for 3 min, and finally returned to 100% A over 0.5 min and held at 100% A for 5 min. The LC eluent (with a total flow rate of 300 μl/min) was introduced into the ESI source of a high resolution TripleTOF5600 mass spectrometer (Sciex, Framingham, MA). Instrumental settings for negative ion ESI and MS/MS analysis of lipid species were as follows: IS= −4500 V; CUR= 20 psi; GSI= 20 psi; DP= −55 V; and FP= −150 V. The MS/MS analysis used nitrogen as the collision gas. Data analysis was performed using Analyst TF1.5 software (Sciex, Framingham, MA).
PrenP abundance is reported as total PrenP (determined by LC/MS) as a percentage of total extracted lipids (determined by total phosphate quantification, described above). Data were plotted using Graphpad Prism 8 (GraphPad Software, La Jolla, CA).
PglC variants and expression
PglC from C. jejuni strain 11168 was cloned into the pET24a vector to insert a C-terminal His6-tag[37] or into the pE-SUMO vector [34]. P24A variants of both constructs were generated using QuikChange II Site-Directed Mutagenesis (Agilent Technologies, Santa Clara, CA) as described previously [37].
PglC-His6 and SUMO-PglC variants were expressed in BL21-CodonPlus (DE3)-RIL E. coli cells (Agilent Technologies) using the Studier auto-induction method.[44] Overnight cultures grown in 3 mL MDG media (0.5% (w/v) glucose, 0.25% (w/v) sodium aspartate, 2 mM MgSO4, 25 mM Na2HPO4, 25 mM KH2PO4, 50 mM NH4Cl, 5 mM Na2SO4 and 0.2x trace metal mix (from 1000x stock, Teknova, Hollister, CA)) with kanamycin and chloramphenicol (30 μg/mL each) were used to inoculate 0.5 L auto-induction media (1% (w/v) tryptone, 0.5% (w/v) yeast extract, 0.5% (v/v) glycerol, 0.05% (w/v) glucose, 0.2% (w/v) α-D-lactose, 2 mM MgSO4, 25 mM Na2HPO4, 25 mM KH2PO4, 50 mM NH4Cl, and 5 mM Na2SO4, 0.2x trace metal mix) containing kanamycin (90 μg/mL) and chloramphenicol (30 μg/mL). Cells were grown for 4 hr at 37 °C followed by an additional 16-18 hr at 16 °C and then harvested at 3700 × g for 30 min. Cells were stored at −80 ℃ until purification.
SMA and DIBMA solubilization and Ni-NTA purification of PglC-His6 variants
All purification steps were carried out at 4 ℃. Cells from 0.5 L of autoinduction culture were resuspended in 20 mL lysis buffer (50 mM Tris, 150 mM NaCl, pH 7.5) and lysed by French Press at 25 psi. Lysate was supplemented with 0.5 mg/mL lysozyme (Research Products International, Mount Prospect, IL), 1:1000 dilution of EDTA-free protease inhibitor cocktail (EMD Millipore, Burlington, MA) and 1 unit/mL DNase I (New England Biolabs, Ipswich, MA) and incubated on ice for 15 min, then centrifuged at 9,000 × g for 45 min. The resulting supernatant was further centrifuged at 140,000 × g for 65 min to yield the cell envelope fraction (CEF), which was homogenized into 20-30 mL solubilization buffer (50 mM Tris, 150 mM NaCl, pH 8.0) to a final protein concentration of 50 mg/mL, as assessed by absorbance at 280 nm.
Homogenized CEF was mixed 1:1 with a 5% (w/v) solution of XIRAN SL40005 styrene-maleic anhydride copolymer (Polyscope, Geleen, Netherlands) or 5% (w/v) of Sokalan CP 9 diisobutylene-maleic acid copolymer (Basf Corporation, Florham Park, NJ) in solubilization buffer (50 mM Tris, 150 mM NaCl, adjusted to a final pH 8.0) and tumbled on a rotating mixer for 3 hr at room temperature. Solubilized lysate was centrifuged at 100,000 × g for 45 min to remove any insoluble material. Approximately 1/10 of the clarified supernatant volume was retained as a “background” sample, while the remaining supernatant was incubated with 5 mL Ni-NTA resin (Thermo Fisher Scientific, Waltham, MA) pre-treated with equilibration buffer (50 mM Tris, 150 mM NaCl, 10 mM imidazole, pH 8.0) overnight at 4 °C on a rotating mixer. Resin was washed with 10 column volumes of wash buffer (50 mM Tris, 150 mM NaCl, 20 mM imidazole, pH 8.0). SMA lipoparticles carrying PglC-His6 were eluted in 2 column volumes of elution buffer (50 mM Tris, 150 mM NaCl, 500 mM imidazole, pH 8.0). The purity of eluted particles was assessed by SDS-PAGE with Coomassie staining. A 30 kDa MWCO centrifugal filter was used to exchange eluted SMA and DIBMA lipoparticles into solubilization buffer (50 mM Tris, 150 mM NaCl, pH 8.0) and to concentrate to ~1 mL. PglC-His6 in DIBMA lipoparticles was quantified by absorbance at 280 nm. PglC-His6 in SMA lipoparticles was quantified by gel densitometry relative to DIBMA lipoparticles.
Total lipid extraction from SMA lipoparticles
The extraction protocol for SMA lipoparticles was adapted from Dorr et al [45]. For extraction, 1 mL Ni-NTA enriched or “background” lipoparticles were added to 3.4 mL 10:23:1 (v/v) CHCl3:MeOH:1 M Tris pH 8.0 to make a single phase and vortexed intermittently for 30 min at room temperature. Phase separation was induced by the addition of 0.5 mL 0.1 M Tris, pH 8.0 and 0.5 mL CHCl3; samples were vortexed for 15-30 sec and centrifuged at 1,000 × g for 5-10 min. The organic phase was removed and the aqueous phase was extracted three additional times with 1 mL CHCl3. All organic phases were pooled, concentrated under N2 and washed with 1 mL ion-switch buffer (50 mM Tris, 100 mM NaCl, 100 mM EDTA, pH 8.2). Approximate lipid composition and concentration were estimated by TLC against a commercial E. coli polar lipid extract (Avanti Polar Lipids, Inc., Alabaster, AL), mobile phase 65:25:4 (v/v) CHCl3:MeOH:water, visualization with molybdenum blue stain. Final lipid extracts were dried completely under N2 and stored at −20 °C until analysis.
Purification and activity assays of SUMO-PglC
SUMO-PglC variants were purified by Ni-NTA affinity as described in detail previously [37]. Activity assays were performed using the UMP/CMP-Glo assay (Promega, Madison, WI) on a 10 μL scale using UndP as the PrenP acceptor, as described previously [37]. Data were plotted using Graphpad Prism 8 (GraphPad Software).
Thermal denaturation assay
Thermal denaturation assays were based on a protocol described previously [28]. Aliquots of 3 μM purified SUMO-PglC in assay buffer (50 mM HEPES, 100 mM NaCl, 0.1% Triton X-100, 5 mM MgCl2, pH 7.5) were supplemented with 300 μM UndP or SolP (synthesis and purification described previously) [17] from a 10x stock in DMSO, or with 10% DMSO (vehicle control), and heated for 10 min at 30–99 °C in a thermocycler (MJ Mini Thermal Cycler; Bio-Rad, Hercules, CA). Precipitate was immediately removed by centrifugation at 16,000 × g for 10 min at 4°C. The resulting supernatant, containing protein that remained soluble, was analyzed by SDS-PAGE with Coomassie staining and quantified by gel densitometry using a Molecular Imager Gel Doc XR+ System with Image Lab software (BioRad). Data were fitted using Graphpad Prism 8 (GraphPad Software).
Pyrene fluorescence in liposomes
Lipids (3:1 POPE:POPG, Avanti Lipids, Inc.) from chloroform stocks, supplemented with UndP or SolP as necessary, were dried first under N2 and then overnight under vacuum. Lipids were hydrated in PBS at 42 ℃ for at least two hours, with intermittent vortexing. The final lipid concentration was 50 μM. Pyrene was added to a final concentration of 0.5 μM from a 100x stock in ethanol and allowed to incubate with liposomes for at least 30 min at room temperature.
Fluorescence was measured on a FluoroMax-P (Horiba, Kyoto, Japan), with excitation at 335 nm and emission at 350-500 nm with 4 nm bandwidth in both. Although background fluorescence was low, a blank liposome sample (from which pyrene was omitted) was read and subtracted from each pyrene-containing sample.
3. Results
Quantification of PrenP abundance in C. jejuni
Quantification of PrenP abundance in C. jejuni was performed by NPLC/MS analysis of total lipids extracted from log-phase C. jejuni cultures. Lipid extraction was performed using a modified Bligh-Dyer method [39]. High-resolution MS was performed in the negative ion mode, and showed that the predominant PrenPs in C. jejuni are decaprenol phosphate (DecP) and UndP, whose [M-H]- ions were detected at m/z 777.6 and 845.6, respectively. The total PrenP in extracts was quantified by spiking in an internal UndP standard. The total pool of PrenP in Gram-negative bacteria is distributed between free PrenP and PrenPP-glycoconjugate biosynthetic intermediates, including PrenPP-linked glycans and PrenPP. A preliminary analysis of extracted lipids identified PrenP but no pathway intermediates, suggesting that these moieties might only be present in very low abundance, or they are not extractable by the Bligh-Dyer method. For quantification of the entire PrenP pool, lipid extracts in subsequent analyses were first subjected to a mild alkaline hydrolysis, a step shown previously to release PrenP from PrenPP and PrenPP-linked glycans [41]. The abundance of PrenP in lipid extracts is reported as the percentage of PrenP relative to the total lipids, which were quantified by microdetermination of total phosphate (Fig. 3A).
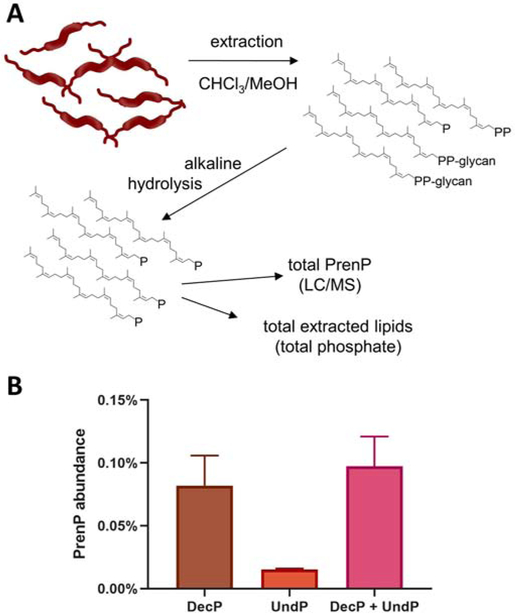
Fig. 3 – Quantification of PrenP in C. jejuni membranes.
A. Schematic showing work flow for extraction, alkaline hydrolysis and quantification of PrenP in C. jejuni membranes.
B. Total abundance of DecP and UndP as a percentage of the total lipidic extract from C. jejuni. Error bars are given for mean ± SD, n = 3.
As UndP is the most common PrenP identified in bacteria studied to date, previous reports have also ascribed UndP the role of glycan carrier in C. jejuni [46, 47], and analyses of PglC catalytic activity and mechanism have successfully utilized the UndP substrate [17, 34, 36]. Indeed, some UndP is identified in the lipidic fraction of C. jejuni extracts. However, the majority of PrenP identified in the extracts was actually DecP, the shorter 10-isoprene PrenP homolog (Fig. 3B). The total abundance of PrenP (DecP + UndP) in C. jejuni lipid extracts was found to be ~0.1%, with DecP accounting for over 80% of the total PrenP.
Enrichment of PrenP in the local membrane around PglC
Quantification of PrenP levels in native extracted lipids provides valuable insight into the identity and average abundance of these species in the bulk C. jejuni membrane, but it does not describe the abundance of PrenP in the membrane local to PglC and other PrenP-binding enzymes. For example, it is possible that specific interactions with such enzymes may result in PrenP enrichment in the local membrane environment. In particular, Pro24 in PglC [34] and similarly conserved prolines in polyisoprenol-recognition sequences in various enzymes were previously proposed to contribute to PrenP binding [19, 24, 38].
The local concentration of the PrenP substrate in the membrane surrounding PglC was investigated using styrene maleic acid lipoparticle (SMALP) technology [48-50], which exploits a styrene-maleic acid polymer to solubilize membrane proteins and lipids directly from cell membranes to make SMALPs. The application of SMA-based solubilization obviates the need for detergent or other non-native surfactants, and importantly, solubilizes proteins together with phospholipids and other lipidic species from their native membrane environment. For this reason, extraction and quantification of the lipidic fraction from SMALPs has been used to provide insight into the lipid composition of the local membrane environment surrounding membrane proteins [45, 51]. To this end, PglC from C. jejuni with a C-terminal His6 tag was heterologously overexpressed in E. coli and solubilized into SMALPs; particles containing PglC-His6 were then isolated by Ni-NTA affinity chromatography (Fig. 4A-B). Lipids extracted both from the Ni-NTA-purified PglC-SMALPs and from SMA-solubilized membranes not enriched on Ni-NTA for PglC-His6 (“background”) were assessed qualitatively by TLC, and determined to be congruent with commercially-available E. coli lipid extracts (Fig. 4C). The extracted lipids were subjected to mild alkaline hydrolysis and quantified by LC/MS against a pure UndP standard as described above; notably, in this case only UndP was quantified, as this is the only PrenP found in E. coli.
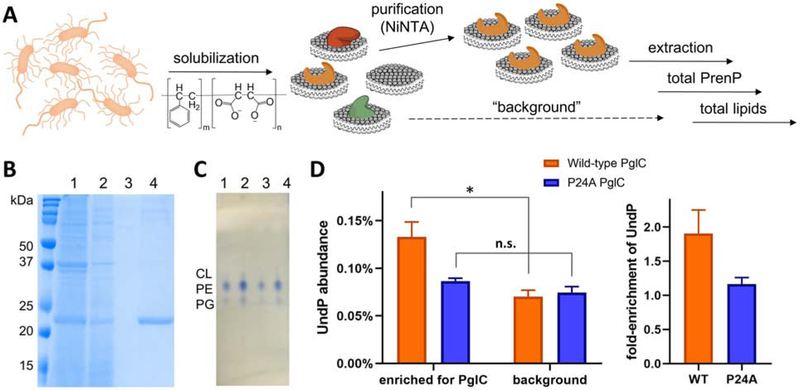
Fig. 4 – Quantification of PrenP enrichment around SMA-solubilized PglC.
A. Schematic showing work-flow for solubilization and purification of SMA-solubilized PglC-His6 and extraction and quantification of the surrounding lipids.
B. Coomassie-stained SDS-PAGE gel showing purification of SMA-lipoparticles with PglC-His6, purified by Ni-NTA affinity chromatography. Lanes: 1 = load, 2 = flow-through, 3 = wash, 4 = elution.
C. Thin-layer chromatography analysis of extracted lipids. Lanes: 1, 2 = E. coli lipid commercial standard, 5 nmol and 10 nmol respectively. Lanes 3, 4 = extracted lipids from SMA-lipoparticles, ~4 nmol and ~8 nmol respectively. CL = cardiolipin, PE = phosphotidylethanolamine, PG = phosphotidylglycerol.
D. Left: Quantification of UndP in lipid extracts from PglC- and background SMA-lipoparticles. * p = 0.02; n.s. = not significant. Right: Fold-enrichment of UndP, calculated as the UndP abundance in PglC SMA-lipoparticles over UndP abundance in background SMA-lipoparticles. Error bars are given for mean ± SD, n = 3.
The UndP abundance in background lipid extracts was ~0.075%, similar to the bulk abundance of PrenP determined in native lipid extracts from C. jejuni membrane (Fig. 4D). Importantly, UndP was found to be 1.91±0.35-fold more abundant in PglC-SMALPs, relative to SMA-solubilized membranes alone, indicating an enrichment of UndP around PglC in the local membrane environment. In these experiments, PglC-His6 concentration in SMALPs could not be quantified directly by absorbance at 280 nm due to absorbance of the SMA copolymer. Therefore, PglC-His6 was first quantified by Coomassie gel densitometry, using PglC-His6 lipoparticles isolated with a non-absorbing, diisobutylene-maleic acid (DIBMA) copolymer [52] to create a standard curve using absorbance at 280 nm. This standard curve was then used to rigorously quantify PglC-His6 in SMALPs. polyisoprenol-recognition sequences (PIRSs) in SMALPs. The ratio of total recovered UndP to PglC-His6 in purified SMALPs was less than unity, supporting that some SMALPs contained PglC-His6 not bound to UndP substrate. This finding reflects the low overall native abundance of free PrenP in the bacterial membrane relative to the total PglC-His6 overexpressed in the E. coli, and recapitulates the native membrane environment, in which enzymes must compete for a single pool of PrenP.
To probe the role of conserved PglC Pro24 in the observed localization of UndP, the same experimental protocol was applied to a P24A PglC-His6 variant. P24A PglC-His6 SMALPs of similar purity and quantity to the wild-type PglC-His6 variant were obtained (SI Fig. 2) and protein quantification was carried out as described for wild-type PglC-His6. In this case, UndP was found not to be significantly more abundant in PglC-SMALPs relative to background. Thus, the enrichment of UndP around PglC is largely abrogated by the mutation of Pro24 to Ala, supporting a role for this conserved residue in binding PrenP in the native membrane environment.
The role of Pro24 in recognition and binding of PrenP substrate
The putative role played by Pro24 in binding PrenP was investigated further in vitro by comparing the stabilities of wild-type and P24A SUMO-PglC variants. Previously, incorporation of an N-terminal SUMO-tag was shown to significantly improve detergent solubilization, stability and purification of PglC [34, 35], and SUMO-PglC was shown to adopt a native membrane topology [37] and be catalytically active [34, 36]. The interactions between SUMO-PglC variants and the UndP substrate were assessed both by circular dichroism (CD) spectroscopy and by a thermal denaturation assay without removal of the coexpression tag.
CD studies revealed that wild-type and P24A SUMO-PglCs adopt very similar, α-helical secondary structures (Fig. 5A), and show similar cooperative unfolding upon heating, when monitoring ellipticity at 222 nm (Fig. 5B). Additionally, the two variants demonstrated comparable thermal transitions in the presence of 300 μM of UndP substrate, indicating secondary structure stabilization by UndP.
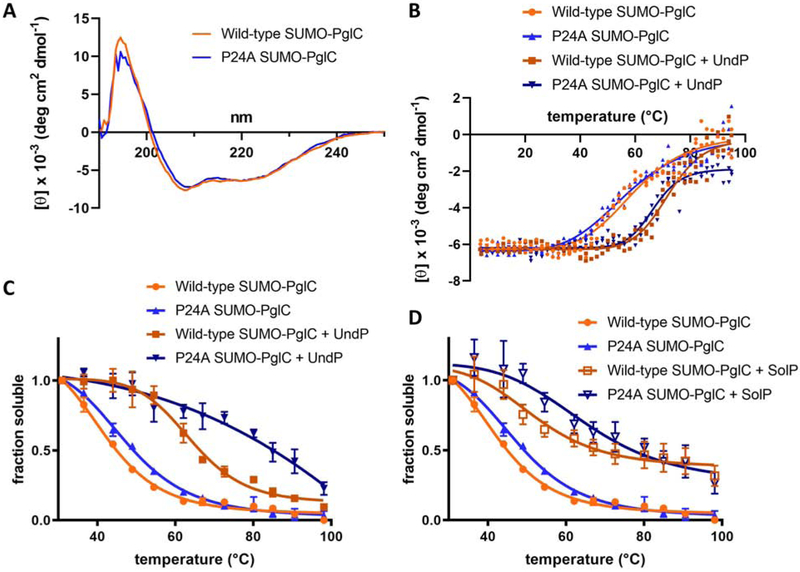
Fig. 5 – Thermal stability studies to support PrenP substrate binding.
A. CD spectroscopy scans of wild-type and P24A SUMO-PglC show that wild-type and P24A SUMO-PglC adopt very similar secondary structures. Mean residue ellipticity is reported.
B. CD spectroscopy thermal melts of wild-type and P24A SUMO-PglC with and without UndP. Tm,WT = 59.7±1.5 ℃; Tm,P24A = 57.4±1.5 ℃; Tm,WT+UndP = 72.2±0.9 ℃; Tm,P24A+UndP = 66.7±0.7 ℃. Mean residue ellipticity at 222 nm is reported.
C. Thermal denaturation assays of wild-type and P24A SUMO-PglC variants with and without UndP. Tm,WT = 41.9±1.3 ℃; Tm,P24A = 47.4±1.1 ℃; Tm,WT+UndP = 64.2±1.4 ℃; Tm,P24A+UndP not determined. Error bars are given for mean ± SEM, n = 4.
D. Thermal denaturation assays of wild-type and P24A SUMO-PglC variants with and without SolP. Tm,WT = 41.9±1.3 ℃; Tm,P24A = 47.4±1.1 ℃; Tm,WT+UndP and Tm,P24A+UndP not determined. Error bars are given for mean ± SEM, n = 4.
As it has been reported that unfolding of α-helical membrane proteins is often driven through loss of tertiary contacts rather than secondary structure [53, 54], the effect of UndP on PglC tertiary structure stability was also investigated, using a thermal denaturation assay reported previously to assess the thermal stability of PglC [37] and the membrane protein, DPMS [28]. In this assay, thermal stability was measured as a function of resistance to denaturation and precipitation upon heating: following heating, precipitated protein is removed by centrifugation and the remaining soluble fraction quantified by gel densitometry. Both wild-type and P24A SUMO-PglC exhibited complete, cooperative denaturation in a similar temperature range (Fig. 5C). However, the two variants were affected differently by the presence of UndP. Wild-type SUMO-PglC showed cooperative denaturation in the presence of 300 μM UndP, with a significant (22.3±1.9 ℃) increase in thermal stability indicating stabilization of the native fold via substrate binding. In contrast, although P24A SUMO-PglC showed apparent stabilization at higher temperatures in 300 μM UndP, this variant no longer exhibited cooperative denaturation and remained partially soluble even at elevated temperatures. This atypical thermal denaturation behavior suggests the observed stabilization is not due to native substrate binding, but rather results from non-specific stabilization of PglC in non-native, likely partially denatured, tertiary conformations.
Indeed, a very similar behavior was observed for both wild-type and P24A SUMO-PglC in the presence the non-substrate all-trans nonaprenol, solanesol phosphate (SolP) [17]. Again, although wild-type and P24A SUMO-PglC both appeared more stable in the presence of 300 μM SolP, both variants no longer exhibited cooperative denaturation, and remained partially soluble at high temperatures (Fig. 5D), suggesting that interaction with SolP stabilized non-native or partially denatured conformations.
PrenP modulation of lipid bilayer fluidity depends on isoprene geometry
NMR and molecular modeling studies of PrenPs in membranes have previously suggested that these amphiphilic molecules modulate the fluidity of the membrane and induce changes in membrane structure [19, 21-23], similarly to the known effects of “kinked” unsaturated fatty acid acyl chains [55, 56]. We investigated this effect further using pyrene, an aromatic membrane probe that embeds in the lipid bilayer proximal to the head-group region [57]. The characteristic fluorescence spectra of pyrene are reported to change with the polarity of the surrounding environment, with an increase in the ratio between two vibrionic peak intensities, I1/I3, corresponding to an increase in polarity. Pyrene fluorescence has previously been used to characterize changes in membrane fluidity as a function of membrane composition, for example, as a result of lipid acyl chain saturation in bacterial membranes [55] and cholesterol depletion in hippocampal membranes [58]. Within the context of the lipid bilayer, an increase in fluidity is thought to allow greater penetration of water molecules into the membrane interior, resulting in an increase in polarity that is sensed by the pyrene probe.
The ratio between the first (372 nm) and third (384 nm) emission peaks of pyrene was measured in liposomes composed of 3:1 POPE:POPG and increasing amounts of UndP (Fig. 6A). The I1/I3 ratio in the liposomes was found to increase with UndP concentration, indicating that UndP caused an increase in polarity in the local membrane environment. The evident trend was approximately linear up to 10% UndP, but a modest effect was observable even at 0.5% UndP, the lowest concentration studied. Pyrene fluorescence was also measured in POPE/POPG liposomes with increasing amounts of the all-trans SolP (Fig. 6B). In this case, no change in membrane polarity was observable, even at 10% SolP, the highest concentration tested.
Fig. 6 – Pyrene fluorescence reflects changes in membrane environment.
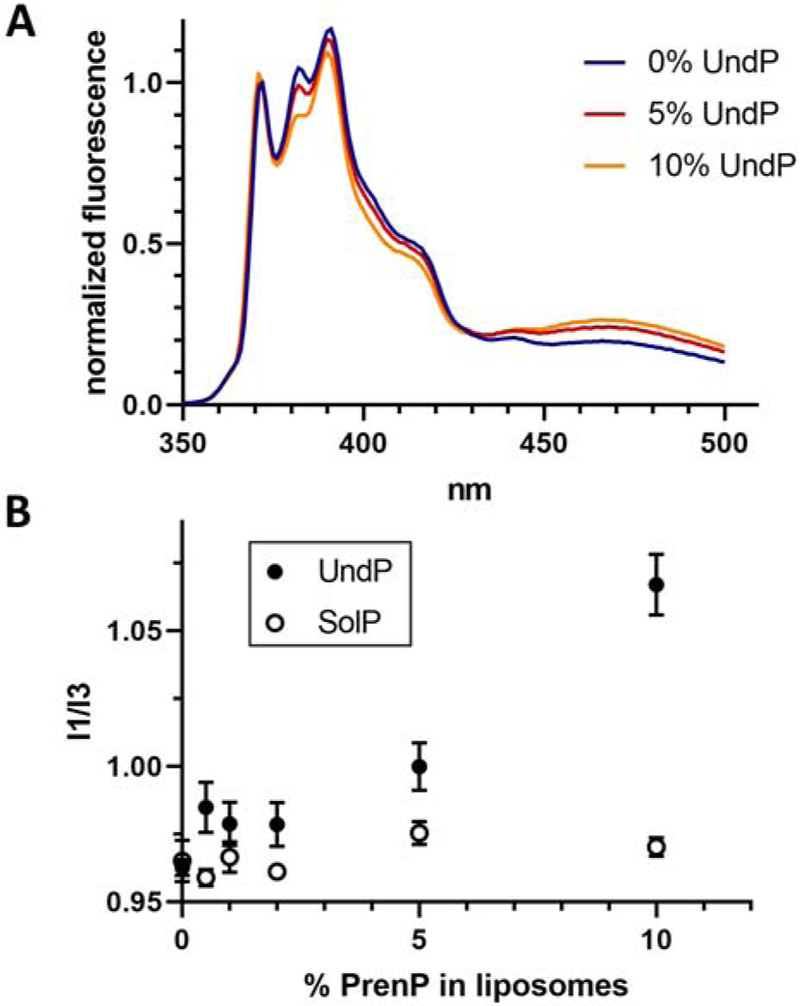
A. Fluorescence spectra of pyrene in liposomes composed of POPE/POPG and increasing concentrations of UndP. Representative data is shown normalized to fluorescence at 372 nm.
B. The ratio of between the first (372 nm) and third (382 nm) vibrionic peaks of pyrene increases with increasing UndP concentration but is not affected by the presence of SolP. Error bars are given for mean ± SEM, n = 4-8.
4. Discussion
PrenP abundance in C. jejuni is consistent with that of E. coli and eukaryotes
Previous work to quantify PrenP levels in bacteria has focused on model bacteria such as E. coli. One such study determined levels of ~350 ng UndP per gram of dry cell weight in E. coli; we estimate that this corresponds to ~0.3% of the total lipid pool [59]. Thus, the current quantification of ~0.1% PrenP abundance in C. jejuni approximately agrees with what has been determined for E. coli. Differences in methodology and interspecies variability likely account for deviations in the numbers. The total dolichol phosphate content of eukaryotic membranes has similarly been estimated to be ~0.1% [3, 4], suggesting that the abundance of polyprenol phosphate in membranes may be similar across domains of life.
Although the PrenP utilized for glycoconjugate assembly in C. jejuni was previously presumed to be UndP, the use of alternate PrenP lipid carriers is not unprecedented: DecP is the preferred PrenP in Mycobacterium tuberculosis, and even shorter PrenPs are observed in other prokaryotes [13-15]. A previous study of PrenP substrate specificity in PglC and other enzymes from the pgl pathway demonstrated that while a series of cis double bonds and unsaturation at the α-isoprene unit of PrenP play important roles in substrate recognition, the length of the chain (e.g. 8-11 isoprenes) plays a minor role [17]. Thus, the bacterial PglC reacts with DecP and UndP with similar efficiency and, use of UndP as the more available PrenP for biochemical and biophysical studies is well supported.
Enrichment of PrenP in the local membrane environment
Quantification of PrenP abundance in C. jejuni and other bacteria are an important step towards understanding the role of PrenP in glycan assembly. However, several lines of evidence suggest that PrenP is not uniformly distributed throughout the membrane, but instead enriched in the local membrane environment of enzymes involved in glycoconjugate assembly. PrenP is utilized in biosynthetic pathways leading to different bacterial glycoconjugates including lipopolysaccharide (LPS) and peptidoglycan within the same organism; thus, multiple pathways must compete for a single pool of PrenP in the membrane [60, 61]. Indeed, the occurrence of both polytopic and monotopic PGTs at the inception of different glycoconjugate biosynthesis pathways may play a role in the coordinated use of the limited PrenP pool. Early studies of bacterial glycoconjugate biosynthesis pathways suggested that these pathways function in large multi-enzyme complexes that assemble completed glycoconjugates without releasing pathway intermediates into the membrane [62-64]. Such a strategy would capitalize on limited PrenP resources and facilitate efficient flux of PrenP-linked intermediates through biosynthetic pathway as needed.
Prior studies have suggested that enzymes in the C. jejuni pgl pathway associate into a macromolecular complex [42]. Additionally, significant levels of PrenPP-glycoconjugate intermediates were not observed in the current analysis of membrane extracts, consistent with a model of glycoconjugate assembly in which enzymes in close proximity function sequentially to minimize the release of intermediates. In such a model, the local concentration of PrenP-linked substrates is expected to be higher in the membrane surrounding the glycoconjugate biosynthesis machinery than in the bulk membrane. This effect is expected to be particularly significant with respect to PGTs, due to their pivotal role in the initiation of glycan biosynthesis. Furthermore, PglC and other members of the monotopic PGT superfamily feature a reentrant membrane helix with a conserved proline, previously predicted to be a key player in PrenP substrate recognition. Using the SMA copolymer to solubilize PglC together with the local lipid constituents, we quantified the abundance of PrenP in the membrane surrounding PglC relative to the rest of the solubilized membrane (Fig. 4). We note that the current study is the first to reconstitute a PGT into SMA lipoparticles, although reconstitution of PglC into lipid Nanodiscs was reported by our group previously [42].
For wild-type PglC that was heterologously expressed in E. coli membranes, we observed an almost two-fold enrichment of UndP relative to background. This finding supports a model of glycan assembly in which PrenP becomes enriched in areas of the membrane where glycan biosynthesis is occurring. In light of the otherwise low abundance of PrenP in the membrane, local enrichment of PrenP substrate likely promotes flux through downstream pathway enzymes. In addition, local enrichment of PrenP has the potential to modulate the biophysical properties of the surrounding membrane.
We note that in this E. coli model, the C. jejuni-specific phosphosugar substrate for PglC, UDP-diNAcBac, as well as the other enzymes of the pgl pathway, are not present, allowing us to study enrichment of PrenP around PglC separately from catalysis and pathway flux. In a native C. jejuni system, with the all of the pgl pathway enzymes participating in glycan assembly present, we would anticipate an even greater enrichment of PrenP in the local membrane environment.
Pro24 in PglC plays a key role in PrenP binding and specificity
Whereas UndP was found to be almost two-fold enriched around SMA-solubilized wild-type PglC relative to background, the lipids surrounding a P24A PglC variant did not show significant enrichment in UndP (Fig. 4D), strongly supporting a critical role for Pro24 in PrenP binding. Importantly, although Pro24 is part of a conserved two-residue motif that was previously found to drive folding of the reentrant membrane domain of PglC, the P24A PglC variant was found to adopt a native-like reentrant topology [37]. In addition, both CD spectroscopy and thermal denaturation assays of wild-type and P24A SUMO-PglC in the absence of UndP indicate that the two variants have very similar intrinsic thermal stabilities (Fig. 5A-C). Thus, although minor changes in structure between the wild-type and P24A PglC variants cannot be ruled out, the available data suggest that loss of UndP binding in P24A PglC reflects an inability of the variant to recognize the PrenP substrate, rather than a global misfolding event.
In vitro thermal stability studies provide additional insights into key molecular determinants of PrenP recognition and binding by PglC. Thermal denaturation assays of wild-type and P24A SUMO-PglC in the presence of 300 μM UndP revealed two very different behaviors (Fig. 5C). Wild-type SUMO-PglC exhibited a complete, cooperative denaturation in the presence of UndP, and was significantly more stabilized, corresponding to a ΔTm >20 °C, indicating stabilization due to substrate binding. In contrast, P24A SUMO-PglC in the presence of UndP no longer exhibited cooperative denaturation, and remained partially soluble even at elevated temperatures, suggesting non-specific stabilization of non-native states of PglC.
The thermal stability studies suggest that P24A SUMO-PglC does not bind the PrenP substrate in the active site; instead, interaction with UndP may stabilize P24A SUMO-PglC in a non-native conformation upon heating. Due to the amphiphilic nature of PrenPs, it is possible that such molecules have a detergent-like solubilizing effect in solution, stabilizing the hydrophobic portion of the PglC fold even as tertiary structure is lost during thermal denaturation. Indeed, CD spectroscopy studies of wild-type and P24A SUMO-PglC indicate that the secondary structures of both variants are significantly stabilized by the presence of UndP (Fig. 5B). Importantly, unfolding of α-helical membrane proteins has been reported to occur through loss of tertiary, rather than secondary, structure [53, 54]. Thus, while UndP stabilizes the secondary structure of both wild-type and P24A SUMO-PglC, and contributes to increasing the solubility of both, only the wild-type PglC tertiary structure appears to be stabilized in a native, UndP-binding conformation. Such a model agrees well with the observation that the P24A SUMO-PglC variant is inactive (SI Fig. 1) and further supports a key role for Pro24 in mediating PrenP substrate binding.
Non-cooperative and incomplete denaturation was also observed for both wild-type and P24A SUMO-PglC in the presence of 300 μM SolP (Fig. 5D), indicating that both variants are similarly unable to bind the all-trans PrenP. These findings are corroborated by previous reports of the importance of the geometry of the internal cis double-bonds of PrenP for PglC activity [17]. Thus, only interaction between wild-type SUMO-PglC and the UndP substrate resulted in stabilization of a native conformation of PglC. These findings further support the significance of Pro24 as a key mediator of PrenP recognition and binding, and suggest an additional role in discerning substrate PrenP from phospholipid acyl tails and non-substrate isoprenoids in the membrane, on the basis of double-bond geometry.
A structural basis for substrate specificity mediated by Pro24
NMR and molecular modeling studies of polyprenol phosphates have previously suggested that the cis-isoprene units of UndP and dolichol phosphate chains create a coiled structure within the membrane, while the trans-isoprene units form a non-coiled “tail” [19]. Structural models predict that peptides containing polyisoprenol-recognition sequences (PIRS) bind to the coiled, cis-isoprene region of PrenPs [19, 65]. In addition, as PIRS often contain conserved proline residues, it has been hypothesized that the ability of these sequences to distinguish PrenP substrate is due in part to the unique structure of the pyrrolidine side chain of proline. Proline residues are known to disrupt α-helices by perturbing peptide-backbone hydrogen bonding [66]; thus, it has been suggested that the change in peptide-backbone hydrogen bonding around conserved prolines in polyisoprenol-recognition sequences facilitates interactions between PrenP and PrenP-binding proteins [65]. Indeed, recent structural and molecular dynamics analyses of PglC show an extensive and dynamic network of modified hydrogen bonds in the peptide backbone surrounding Pro24 in the hydrophobic domain of PglC [35, 37]. The current work similarly supports key roles in substrate recognition for both Pro24 and the cis PrenP geometry, and provides a first account of how monotopic PGTs use a single reentrant helix to recognize this scare substrate in the membrane. A more precise understanding of the molecular mechanisms underlying PglC substrate specificity remain to be elucidated by later structural and modeling studies.
In the reported structure of PglC, a co-crystallized PEG molecule reveals the putative PrenP-binding site, which is near the active site and directly above Pro24 (Fig. 2). Thus, we can envision a model of PrenP binding wherein Pro24, buried within the membrane, associates with the unique coiled structure of the PrenP substrate and facilitates positioning of the phosphate group of PrenP at the active site, which is located at the membrane interface, for catalysis. Notably, unlike members of the polytopic PGT superfamily, which have 10- or 11- transmembrane helices that may provide an interaction surface for PrenP, members of the monotopic PGT superfamily rely on a single reentrant helix, carrying the conserved Pro24, to facilitate PrenP binding.
Increased concentrations of PrenP modulate the local membrane environment
The exact consequences of PrenP enrichment in the membrane are unclear, although several hypotheses have been advanced. It has been proposed that PrenP may serve as a structural scaffold for the organization of complexes with downstream glycosyl transferases [19, 65]. In addition, it has been found that PrenP alters the biophysical properties of the local membrane, increasing fluidity and modifying membrane structure [19, 21-23]; such local changes may also be important for promoting multi-enzyme complex formation or for increasing substrate flux through the pathway [18]. Analogous roles in modulation of local membrane fluidity and organization of membrane proteins have been proposed for other membrane components, including cis-geometry unsaturated fatty acid acyl chains [56] and cholesterol [1, 67].
Our studies using the membrane polarity probe, pyrene, confirm earlier reports that UndP modulates the biophysical properties of the membrane (Fig. 6). However, whereas prior studies were carried out with high levels (5-10%) of PrenP, the effect observed in the pyrene fluorescence studies is observed at much lower, and more physiologically relevant, UndP concentrations. As little as 0.5% UndP in liposomes is found to increase the polarity of the local membrane environment resulting from increased water penetration into the membrane due to loss of membrane order surrounding UndP [58]. Our findings support the proposed model wherein increased local concentrations of PrenP modulate the biophysical properties of the membrane to facilitate multi-enzyme complex formation and support substrate flux through the pathway.
In contrast, pyrene fluorescence studies show that polarity in the membrane environment does not increase in the presence of up to 10% of the all-trans SolP, indicating that double-bond geometry plays a key role in the reported ability of PrenP to disrupt lipid membranes. Notably, the all-trans SolP cannot adopt the unique coiled conformation predicted for the cis-isoprene portion of UndP; it may be that this coiled structure of UndP is more disruptive to local lipid packing than the all-trans linear structure of SolP. Thus, in addition to promoting recognition by PglC, the specific cis-trans geometry of the isoprene units of UndP allows this PrenP to play a distinct secondary role in modulating the local membrane environment.
5. Conclusion
The elusive role of PrenP in glycan assembly in the bacterial pgl pathway is explored with respect to both its effect on the local membrane environment and interactions with PglC, a monotopic PGT that acts at the initiation of N-glycan assembly. The specific cis-trans double - bond geometry of PrenP is found to be essential both to interactions with the local membrane and with PglC. We also confirm a role for the conserved Pro24 in PglC in binding to the PrenP substrate, and propose an additional role in driving specificity for the unique cis-isoprene geometry of the PrenP substrate. Therefore, although the membrane topology of PglC and the broader monotopic PGT superfamily is distinct from that of the various polytopic PGTs, both enzyme superfamilies favor PrenPs with comparable structural features. This key observation is intriguing and suggests that the two distinct PGT superfamilies, may be the result of convergent evolution achieving two different structural and mechanistic solutions to the common goal of initiating glycan assembly on a common membrane resident amphiphilic PrenP.
Finally, it is poorly understood how a single limited pool of PrenP in bacterial membranes simultaneously supports multiple glycoconjugate biosynthesis pathways including those leading to lipopolysaccharide (LPS), peptidoglycan and glycoprotein biosynthesis and the occurrence of both polytopic and monotopic PGT superfamilies at the inception of different glycoconjugate biosynthesis pathways may be important in the coordinated use of the limited PrenP pool.
Supplementary Material
Highlights.
Polyprenol phosphates make up ~0.1% of total C. jejuni bacterial membrane lipids
Phosphoglycosyl transferases (PGTs) enrich polyprenol substrates in the membrane
The PGT reentrant membrane helix is critical for substrate recognition and specificity
Polyprenol phosphates modulate the fluidity of the local membrane environment
Polyprenol phosphate geometry dictates functional interactions
Acknowledgements
Dr. Jean-Marie Swiecicki is gratefully acknowledged for assistance with SMA-solubilization of PglC-His6 and associated lipids. Dr. Jerry Eichler is thanked for many valuable discussions.
Funding
This work was supported by the National Institutes of Health [R01-GM039334 to BI, GM069338 and EY023666 to ZG] and the Predoctoral Training Program in the Biological Sciences [T32-GM007287 to SE].
Abbreviations
- CD
circular dichroism
- diNAcBac
N,N'-diacetylbacillosamine
- DecP
decaprenol phosphate
- DIBMA
diisobutylene maleic acid
- Dol-P-DPMS
mannose synthase
- NPLC/MS
normal phase liquid chromatography/mass spectrometry
- OST
oligosaccharyl transferase
- PEG
polyethylene glycol
- PGT
phosphoglycosyl transferase
- PIRS
polyisoprenol-recognition sequences
- PrenP
polyprenol phosphate
- PrenPP
polyprenol diphosphate
- RMH
reentrant membrane helix
- SMA
styrene-maleic acid copolymer
- SMALP
styrene-maleic acid lipoparticle
- UndP
undecaprenol phosphate
- SolP
solanesol phosphate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Krause MR, Regen SL, The structural role of cholesterol in cell membranes: from condensed bilayers to lipid rafts, Acc. Chem. Res 47(12) (2014) 3512–21. [DOI] [PubMed] [Google Scholar]
- [2].Barreteau H, Magnet S, El Ghachi M, Touze T, Arthur M, Mengin-Lecreulx D, Blanot D, Quantitative high-performance liquid chromatography analysis of the pool levels of undecaprenyl phosphate and its derivatives in bacterial membranes, J Chromatogr B Analyt Technol Biomed Life Sci 877(3) (2009) 213–20. [DOI] [PubMed] [Google Scholar]
- [3].Kaiden A, Krag SS, Dolichol metabolism in Chinese hamster ovary cells, Biochem. Cell Biol 70(6) (1992) 385–9. [DOI] [PubMed] [Google Scholar]
- [4].Krag SS, The importance of being dolichol, Biochem. Biophys. Res. Commun 243(1) (1998) 1–5. [DOI] [PubMed] [Google Scholar]
- [5].Lechner J, Wieland F, Sumper M, Biosynthesis of sulfated saccharides N-glycosidically linked to the protein via glucose. Purification and identification of sulfated dolichyl monophosphoryl tetrasaccharides from halobacteria, J. Biol. Chem 260(2) (1985) 860–6. [PubMed] [Google Scholar]
- [6].Adair WL Jr., Cafmeyer N, Characterization of the Saccharomyces cerevisiae cis-prenyltransferase required for dolichyl phosphate biosynthesis, Arch. Biochem. Biophys 259(2) (1987) 589–96. [DOI] [PubMed] [Google Scholar]
- [7].Eggens I, Chojnacki T, Kenne L, Dallner G, Separation, quantitation and distribution of dolichol and dolichyl phosphate in rat and human tissues, Biochim. Biophys. Acta 751(3) (1983) 355–68. [DOI] [PubMed] [Google Scholar]
- [8].Guan Z, Meyer BH, Albers SV, Eichler J, The thermoacidophilic archaeon Sulfolobus acidocaldarius contains an unusually short, highly reduced dolichyl phosphate, Biochim. Biophys. Acta 1811(10) (2011) 607–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Swiezewska E, Chojnacki T, The occurrence of unique, long-chain polyprenols in the leaves of Potentilla species, Acta Biochim. Pol 36(2) (1989) 143–58. [PubMed] [Google Scholar]
- [10].Guan Z, Naparstek S, Kaminski L, Konrad Z, Eichler J, Distinct glycan-charged phosphodolichol carriers are required for the assembly of the pentasaccharide N-linked to the Haloferax volcanii S-layer glycoprotein, Mol. Microbiol 78(5) (2010) 1294–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kuntz C, Sonnenbichler J, Sonnenbichler I, Sumper M, Zeitler R, Isolation and characterization of dolichol-linked oligosaccharides from Haloferax volcanii, Glycobiology 7(7) (1997) 897–904. [DOI] [PubMed] [Google Scholar]
- [12].Higashi Y, Strominger JL, Sweeley CC, Structure of a lipid intermediate in cell wall peptidoglycan synthesis: a derivative of a C55 isoprenoid alcohol, Proc Natl Acad Sci U S A 57(6) (1967) 1878–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wolucka BA, de Hoffmann E, Isolation and characterization of the major form of polyprenyl-phospho-mannose from Mycobacterium smegmatis, Glycobiology 8(10) (1998) 955–62. [DOI] [PubMed] [Google Scholar]
- [14].Kaur D, Brennan PJ, Crick DC, Decaprenyl diphosphate synthesis in Mycobacterium tuberculosis, J. Bacteriol 186(22) (2004) 7564–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ishii K, Sagami H, Ogura K, A novel prenyltransferase from Paracoccus denitrificans, Biochem. J 233(3) (1986) 773–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yan N, Liu Y, Zhang H, Du Y, Liu X, Zhang Z, Solanesol biosynthesis in plants, Molecules 22(4) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chen MM, Weerapana E, Ciepichal E, Stupak J, Reid CW, Swiezewska E, Imperiali B, Polyisoprenol specificity in the Campylobacter jejuni N-linked glycosylation pathway, Biochemistry 46(50) (2007) 14342–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hartley MD, Imperiali B, At the membrane frontier: a prospectus on the remarkable evolutionary conservation of polyprenols and polyprenyl-phosphates, Arch. Biochem. Biophys 517(2) (2012) 83–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhou GP, Troy FA 2nd, Characterization by NMR and molecular modeling of the binding of polyisoprenols and polyisoprenyl recognition sequence peptides: 3D structure of the complexes reveals sites of specific interactions, Glycobiology 13(2) (2003) 51–71. [DOI] [PubMed] [Google Scholar]
- [20].Kern NR, Lee HS, Wu EL, Park S, Vanommeslaeghe K, MacKerell AD Jr., Klauda JB, Jo S, Im W, Lipid-linked oligosaccharides in membranes sample conformations that facilitate binding to oligosaccharyltransferase, Biophys. J 107(8) (2014) 1885–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Valtersson C, van Duyn G, Verkleij AJ, Chojnacki T, de Kruijff B, Dallner G, The influence of dolichol, dolichol esters, and dolichyl phosphate on phospholipid polymorphism and fluidity in model membranes, J. Biol. Chem 260(5) (1985) 2742–51. [PubMed] [Google Scholar]
- [22].Wang X, Mansourian AR, Quinn PJ, The effect of dolichol on the structure and phase behaviour of phospholipid model membranes, Mol. Membr. Biol 25(6-7) (2008) 547–56. [DOI] [PubMed] [Google Scholar]
- [23].Janas T, Chojnacki T, Swiezewska E, Janas T, The effect of undecaprenol on bilayer lipid membranes, Acta Biochim. Pol 41(3) (1994) 351–8. [PubMed] [Google Scholar]
- [24].Albright CF, Orlean P, Robbins PW, A 13-amino acid peptide in three yeast glycosyltransferases may be involved in dolichol recognition, Proc Natl Acad Sci U S A 86(19) (1989) 7366–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gin P, Clarke CF, Genetic evidence for a multi-subunit complex in coenzyme Q biosynthesis in yeast and the role of the Coq1 hexaprenyl diphosphate synthase, J. Biol. Chem 280(4) (2005) 2676–81. [DOI] [PubMed] [Google Scholar]
- [26].Anderson MS, Eveland SS, Price NP, Conserved cytoplasmic motifs that distinguish sub-groups of the polyprenol phosphate:N-acetylhexosamine-1-phosphate transferase family, FEMS Microbiol. Lett 191(2) (2000) 169–75. [DOI] [PubMed] [Google Scholar]
- [27].Lukose V, Walvoort MTC, Imperiali B, Bacterial phosphoglycosyl transferases: initiators of glycan biosynthesis at the membrane interface, Glycobiology 27(9) (2017) 820–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gandini R, Reichenbach T, Tan TC, Divne C, Structural basis for dolichylphosphate mannose biosynthesis, Nat Commun 8(1) (2017) 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Maeda Y, Kinoshita T, Dolichol-phosphate mannose synthase: structure, function and regulation, Biochim. Biophys. Acta 1780(6) (2008) 861–8. [DOI] [PubMed] [Google Scholar]
- [30].Eichler J, Imperiali B, Stereochemical Divergence of Polyprenol Phosphate Glycosyltransferases, Trends Biochem. Sci 43(1) (2018) 10–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Workman SD, Worrall LJ, Strynadka NCJ, Crystal structure of an intramembranal phosphatase central to bacterial cell-wall peptidoglycan biosynthesis and lipid recycling, Nat Commun 9(1) (2018) 1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Napiorkowska M, Boilevin J, Sovdat T, Darbre T, Reymond JL, Aebi M, Locher KP, Molecular basis of lipid-linked oligosaccharide recognition and processing by bacterial oligosaccharyltransferase, Nat. Struct. Mol. Biol 24(12) (2017) 1100–1106. [DOI] [PubMed] [Google Scholar]
- [33].Sun Lee H, Im W, Transmembrane Motions of PglB Induced by LLO are Coupled with EL5 Loop Conformational Changes Necessary for OST Activity, Glycobiology (2017). [DOI] [PubMed] [Google Scholar]
- [34].Lukose V, Luo L, Kozakov D, Vajda S, Allen KN, Imperiali B, Conservation and Covariance in Small Bacterial Phosphoglycosyltransferases Identify the Functional Catalytic Core, Biochemistry 54(50) (2015) 7326–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ray LC, Das D, Entova S, Lukose V, Lynch AJ, Imperiali B, Allen KN, Membrane association of monotopic phosphoglycosyl transferase underpins function, Nat. Chem. Biol 14(6) (2018) 538–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Das D, Kuzmic P, Imperiali B, Analysis of a dual domain phosphoglycosyl transferase reveals a ping-pong mechanism with a covalent enzyme intermediate, Proc Natl Acad Sci U S A 114(27) (2017) 7019–7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Entova S, Billod JM, Swiecicki JM, Martin-Santamaria S, Imperiali B, Insights into the key determinants of membrane protein topology enable the identification of new monotopic folds, Elife 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Datta AK, Lehrman MA, Both potential dolichol recognition sequences of hamster GlcNAc-1-phosphate transferase are necessary for normal enzyme function, J. Biol. Chem 268(17) (1993) 12663–8. [PubMed] [Google Scholar]
- [39].Bligh EG, Dyer WJ, A rapid method of total lipid extraction and purification, Can J Biochem Physiol 37(8) (1959) 911–7. [DOI] [PubMed] [Google Scholar]
- [40].Chen PST, T.Y.; Warner H, Microetermination of phosphorus, Anal. Chem 28 (1956) 1756–1758. [Google Scholar]
- [41].Wang X, Ribeiro AA, Guan Z, Raetz CR, Identification of undecaprenyl phosphate-beta-D-galactosamine in Francisella novicida and its function in lipid A modification, Biochemistry 48(6) (2009) 1162–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Hartley MD, Schneggenburger PE, Imperiali B, Lipid bilayer nanodisc platform for investigating polyprenol-dependent enzyme interactions and activities, Proc Natl Acad Sci U S A 110(52) (2013) 20863–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kozlov VV, Danilov LL, Reversed-phase ion-pair high-performance liquid chromatography assay of polyprenyl diphosphate oligomer homologues, J. Sep. Sci 39(3) (2016) 525–7. [DOI] [PubMed] [Google Scholar]
- [44].Studier FW, Protein production by auto-induction in high density shaking cultures, Protein Expr Purif 41(1) (2005) 207–34. [DOI] [PubMed] [Google Scholar]
- [45].Dorr JM, Koorengevel MC, Schafer M, Prokofyev AV, Scheidelaar S, van der Cruijsen EA, Dafforn TR, Baldus M, Killian JA, Detergent-free isolation, characterization, and functional reconstitution of a tetrameric K+ channel: the power of native nanodiscs, Proc Natl Acad Sci U S A 111(52) (2014) 18607–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Kelly J, Jarrell H, Millar L, Tessier L, Fiori LM, Lau PC, Allan B, Szymanski CM, Biosynthesis of the N-linked glycan in Campylobacter jejuni and addition onto protein through block transfer, J. Bacteriol 188(7) (2006) 2427–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Glover KJ, Weerapana E, Imperiali B, In vitro assembly of the undecaprenylpyrophosphate-linked heptasaccharide for prokaryotic N-linked glycosylation, Proc Natl Acad Sci U S A 102(40) (2005) 14255–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Knowles TJ, Finka R, Smith C, Lin YP, Dafforn T, Overduin M, Membrane proteins solubilized intact in lipid containing nanoparticles bounded by styrene maleic acid copolymer, J. Am. Chem. Soc 131(22) (2009) 7484–5. [DOI] [PubMed] [Google Scholar]
- [49].Long AR, O'Brien CC, Malhotra K, Schwall CT, Albert AD, Watts A, Alder NN, A detergent-free strategy for the reconstitution of active enzyme complexes from native biological membranes into nanoscale discs, BMC Biotechnol. 13 (2013) 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Dorr JM, Scheidelaar S, Koorengevel MC, Dominguez JJ, Schafer M, van Walree CA, Killian JA, The styrene-maleic acid copolymer: a versatile tool in membrane research, Eur. Biophys. J 45(1) (2016) 3–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Prabudiansyah I, Kusters I, Caforio A, Driessen AJ, Characterization of the annular lipid shell of the Sec translocon, Biochim. Biophys. Acta 1848(10 Pt A) (2015) 2050–6. [DOI] [PubMed] [Google Scholar]
- [52].Oluwole AO, Danielczak B, Meister A, Babalola JO, Vargas C, Keller S, Solubilization of Membrane Proteins into Functional Lipid-Bilayer Nanodiscs Using a Diisobutylene/Maleic Acid Copolymer, Angew. Chem. Int. Ed. Engl 56(7) (2017) 1919–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Grinberg AV, Gevondyan NM, Grinberg NV, Grinberg VY, The thermal unfolding and domain structure of Na+/K+-exchanging ATPase. A scanning calorimetry study, Eur. J. Biochem 268(19) (2001) 5027–36. [DOI] [PubMed] [Google Scholar]
- [54].Vogel R, Siebert F, Conformation and stability of alpha-helical membrane proteins. 2. Influence of pH and salts on stability and unfolding of rhodopsin, Biochemistry 41(11) (2002) 3536–45. [DOI] [PubMed] [Google Scholar]
- [55].Aricha B, Fishov I, Cohen Z, Sikron N, Pesakhov S, Khozin-Goldberg I, Dagan R, Porat N, Differences in membrane fluidity and fatty acid composition between phenotypic variants of Streptococcus pneumoniae, J. Bacteriol 186(14) (2004) 4638–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Funari SS, Barcelo F, Escriba PV, Effects of oleic acid and its congeners, elaidic and stearic acids, on the structural properties of phosphatidylethanolamine membranes, J. Lipid Res 44(3) (2003) 567–75. [DOI] [PubMed] [Google Scholar]
- [57].Hoff B, Strandberg E, Ulrich AS, Tieleman DP, Posten C, 2H-NMR study and molecular dynamics simulation of the location, alignment, and mobility of pyrene in POPC bilayers, Biophys. J 88(3) (2005) 1818–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Saxena R, Shrivastava S, Chattopadhyay A, Exploring the organization and dynamics of hippocampal membranes utilizing pyrene fluorescence, J. Phys. Chem. B 112(38) (2008) 12134–8. [DOI] [PubMed] [Google Scholar]
- [59].Neidhardt FC, Physiology of the Bacterial Cell: A Molecular Approach, Sinauer Associates Inc.1990. [Google Scholar]
- [60].Jorgenson MA, Kannan S, Laubacher ME, Young KD, Dead-end intermediates in the enterobacterial common antigen pathway induce morphological defects in Escherichia coli by competing for undecaprenyl phosphate, Mol. Microbiol 100(1) (2016) 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Jorgenson MA, Young KD, Interrupting biosynthesis of O antigen or the lipopolysaccharide core produces morphological defects in Escherichia coli by sequestering undecaprenyl phosphate, J. Bacteriol 198(22) (2016) 3070–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Leaver J, Hancock IC, Baddiley J, Fractionation studies of the enzyme complex involved in teichoic acid synthesis, J. Bacteriol 146(3) (1981) 847–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Anderson RG, Hussey H, Baddiley J, The mechanism of wall synthesis in bacteria. The organization of enzymes and isoprenoid phosphates in the membrane, Biochem. J 127(1) (1972) 11–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Guan Z, Breazeale SD, Raetz CR, Extraction and identification by mass spectrometry of undecaprenyl diphosphate-MurNAc-pentapeptide-GlcNAc from Escherichia coli, Anal. Biochem 345(2) (2005) 336–9. [DOI] [PubMed] [Google Scholar]
- [65].Zhou GP, Troy FA 2nd, NMR study of the preferred membrane orientation of polyisoprenols (dolichol) and the impact of their complex with polyisoprenyl recognition sequence peptides on membrane structure, Glycobiology 15(4) (2005) 347–59. [DOI] [PubMed] [Google Scholar]
- [66].Piela L, Nemethy G, Scheraga HA, Proline-induced constraints in alpha-helices, Biopolymers 26(9) (1987) 1587–600. [DOI] [PubMed] [Google Scholar]
- [67].Sezgin E, Levental I, Mayor S, Eggeling C, The mystery of membrane organization: composition, regulation and roles of lipid rafts, Nat. Rev. Mol. Cell Biol 18(6) (2017) 361–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.