Abstract
Peanut (Arachis hypogaea L.) is an important nutrient-rich food legume and valued for its good quality cooking oil. The fatty acid content is the major determinant of the quality of the edible oil. The oils containing higher monounsaturated fatty acid are preferred for improved shelf life and potential health benefits. Therefore, a high oleic/linoleic fatty acid ratio is the target trait in an advanced breeding program. The two mutant alleles, ahFAD2A (on linkage group a09) and ahFAD2B (on linkage group b09) control fatty acid composition for higher oleic/linoleic ratio in peanut. In the present study, marker-assisted backcrossing was employed for the introgression of two FAD2 mutant alleles from SunOleic95R into the chromosome of ICGV06100, a high oil content peanut breeding line. In the marker-assisted backcrossing-introgression lines, a 97% increase in oleic acid, and a 92% reduction in linoleic acid content was observed in comparison to the recurrent parent. Besides, the oleic/linoleic ratio was increased to 25 with respect to the recurrent parent, which was only 1.2. The most significant outcome was the stable expression of oil-content, oleic acid, linoleic acid, and palmitic acid in the marker-assisted backcrossing-introgression lines over the locations. No significant difference was observed between high oleic and normal oleic in peanuts for seedling traits except germination percentage. In addition, marker-assisted backcrossing-introgression lines exhibited higher yield and resistance to foliar fungal diseases, i.e., late leaf spot and rust.
Introduction
Peanut or groundnut (Arachis hypogaea L.) is one of the world’s most important legumes for its valuable edible oil and protein content. It is a major cash crop and plays an essential role in the livelihood of millions, especially in semi-arid tropics. It is cultivated globally in around 27.94 million ha with a total production of 47.09 million tons [1]. China, India, Nigeria, and the United States of America are the leading groundnut producers that account for ~70% of the global peanut production. Peanut is traditionally used for the extraction of oil for edible as well as industrial purposes but the quality attributes vary with geographical region. In China, India, and other Asian countries, half of the produce is crushed for oil extraction and the rest is being used for confectionary and food purposes. While in the USA and other European countries more than two-thirds of peanut production are used for confectionary and food purposes and remaining one-third is used in the extraction of oil. Low oil content peanuts are preferred for table purposes and other food preparations of low caloric value.
Different proportions of saturated fatty acids (SFAs), monounsaturated fatty acids (MUFA) and polyunsaturated fatty acids (PUFA) determine the nutritional quality, shelf life, and flavor of peanut oil as well as its products. The peanut oil contains 80% unsaturated fatty acids (UFAs), mainly oleic (MUFA), and linoleic (PUFA) acids, whereas the remaining 20% SFAs comprises of palmitic, stearic, arachidic, behenic and lignoceric acid. Palmitic acid alone contributes half of the total SFAs while the rest five make up the remaining 50% [2]. SFAs are considered to increase serum low-density lipoproteins cholesterol level in the blood [3]. An elevated level of palmitic acid in the oil also increases the risk of cardiovascular diseases (CVD) [4]. A higher proportion of linoleic acid results in off flavors, rancidity, the short shelf life of oil and its derived products, which makes it undesirable for cooking purpose [5]. From a nutritional point of view, MUFA have been desirable in lowering plasma cholesterol levels and reduced risk of CVD [6, 7]. Therefore, a diet with high oleic (HO) acid can reduce the risk of heart diseases, inflammatory diseases tumorigenesis, and slow down atherosclerosis [8, 9]. In addition, oleic acid has ten-fold higher auto-oxidative stability than linoleic acid [10]. Therefore, there is a greater demand for the improved lines with higher oleic/linoleic (O/L) ratio in the peanut oil.
In peanut, fatty acid desaturase enzyme catalyzes desaturation of oleic to linoleic acid. [11, 12]. It is controlled by two homeologous genes ahFAD2A and ahFAD2B, located on A-genome (linkage group a09) and in B-genome (linkage group b09), respectively [13,14]. Mutations in ahFAD2A and ahFAD2B genes results in reduced fatty acid desaturase enzyme activity that leads to higher accumulation of oleic acid [13,15]. A single base pair (bp) substitution mutation (G:C to A:T) in ahFAD2A gene at 448 bp position results in a missense amino acid from aspartic acid to asparagine (D150N). While, an insertion mutation in A:T of ahFAD2B gene at 442 bp position generates premature stop codon [11, 12]. Thus the two mutant fatty acid desaturase alleles stop the conversion of oleic acid to linoleic acid in peanut [16, 17, 18]. Improved breeding lines with HO and lower linoleic and palmitic acids in peanut oil are essential to make peanut of superior quality. Norden et al., [19] first identified F435 as a natural peanut mutant line with approximately 80% oleic acid and 2% linoleic acid. Later on, the first ever HO peanut breeding line, SunOleic95R, was produced with the help of conventional breeding method in the USA [16]. Chen et al., [20] and Chu et al., [13] developed linked allele specific-polymerase chain reaction (AS-PCR) and cleaved amplified polymorphic sequence (CAPS) markers, respectively for both of the ahFAD2 alleles. The development of the associated markers in peanut helped in the improvement of ‘Tifguard High O/L’ variety in the USA through marker-assisted backcrossing (MABC) [21]. Recently, Janila et al., [22] introgressed ahFAD2 alleles from SunOleic95R into the elite breeding lines using MABC and marker-assisted selection (MAS) at ICRISAT, Patancheru, India. Further, Bera et al., [23] developed HO peanut lines through MAS at ICAR-Directorate of Groundnut Research, Junagadh, India. Most of these molecular breeding lines are under examination in All India Coordinated Research Project on Groundnut (AICRP-G) and, recently, Girnar 4 and Girnar 5 genotypes have been identified for release in India.
Peanut is grown in both rainy and post-rainy (as winter and summer crop) seasons across different states of India, varying largely in climatic and edaphic conditions. The chemical composition of peanut oil is influenced by several factors like genotype, geographic location, season, soil humidity, temperature and growing conditions [24, 25,26]. In general, lower temperature (22˗29°C) is associated with more linoleic acid synthesis due to increased activity of oleate desaturase enzyme [27, 28] and high temperature (30˗33°C) during pod filling to harvesting stage reduces the linoleic acid content in peanut oil [29, 30, 31]. Li et al., [32] also reported that season and temperature had a significant influence on fatty acid content in Brassica crops. Flagella et al., [33] reported a reduction in oleic and stearic acid while an increase in linoleic and palmitic acid in sunflower under irrigated cultivation. Furthermore, healthy and vigorous seedlings are one of the important criteria for making HO peanut cultivation profitable. The chemical composition of seed reserve might affect its germination and seedling vigor as seed reserve content is correlated with germination percentage [34]. In oilseeds, the major storage reservoir is lipid that provides essential energy to the growing embryo and thus affects seed germination. The alterations in seed lipid affect membrane lipid composition in respect to membrane function and permeability, which affects germination, vigor, and tolerance to environmental stress [35]. In peanut, germination percentage decreases with increase in O/L and unsaturated/saturated fatty acid ratios especially at lower (16°C and 14°C) temperatures [36]. Sun et al., [35] found that seed vigor of high oleate lines was lower as compared with the lines with normal oleic content in peanut. Upadhyaya et al., [37] reported a poor yield of ICG-2381, a groundnut accession with high O/L ratio.
Considering the demand of peanut with HO both in domestic and international markets, the present study was undertaken with three objectives: i) introgression of ahFAD2 alleles into the higher oil content peanut variety through MABC; ii) multi-location testing of MABC derived HO peanut lines over the two seasons for yield and impact of locations and seasons on the oil quality and oil content iii) determining the effect of HO trait on seed germination and other seedling traits.
Materials and methods
Plant material
For improving the oil quality, ICGV06100 was used as female/recurrent and SunOleic95R as male/donor parents for MABC breeding program. ICGV06100 is a high yielding and high oil containing (~55%) peanut line but with lower oleic acid (~39.3%), developed by ICRISAT, Patancheru, India (ICRISAT, 2012; unpublished). It is a Virginia bunch (semi-spreading) cultivar derived from the cross [(ICGV92069 × ICGV93184) × (NCAc-343 × ICGV86187) × S23]. SunOleic95R, having both ahFAD2 mutant homozygous alleles with HO (~80%) but lower yield and oil contents (~45%) was used as a male/donor parent. It was developed by Florida Experimental Agriculture Station, USA, from the mutant line F435 [16].
Under the second objective, MABC lines were tested for pod yield in multiple seasons and locations. Initial yield evaluation of MABC lines along with elite cultivars (Abhaya, CO-6, GG-20, ICGS-1043, GPBD-4, JL-24, TMV-2, VRI-6, K-6, TAG-24, GJG-31, and TG-37A) was done at a single location over two seasons. Subsequently, advanced yield evaluation of MABC lines together with other breeding lines and elite cultivars was done at three different locations. Besides fatty acids profile, oil and protein contents of MABC lines, SunOleic95R and ICGV06100 were also estimated at three locations.
For the accomplishment of the third objective, two separate panels of peanut genotypes were studied for seed and seedling traits. The first panel consisted of normal oleic acid peanut genotypes (GG-20, ICGV06100, ICGV05141, and ICGV06110), while the second panel had HO peanut genotypes (NRCGCS-587, HOP-IL_MAS-191, HOP-IL_MAS-145and HOP-IL_MAS-130) [22, 23, 38].
Molecular markers
Two types of DNA-markers linked to ahFAD2 mutant alleles were used for genotyping. The allele specific-polymerase chain reaction (AS-PCR) markers [20] were used to identify heterozygous plants for the mutant alleles. The cleaved amplified polymorphic sequence (CAPS) markers [13] were deployed to select homozygous plants for both the ahFAD2 alleles.
DNA extraction and marker genotyping
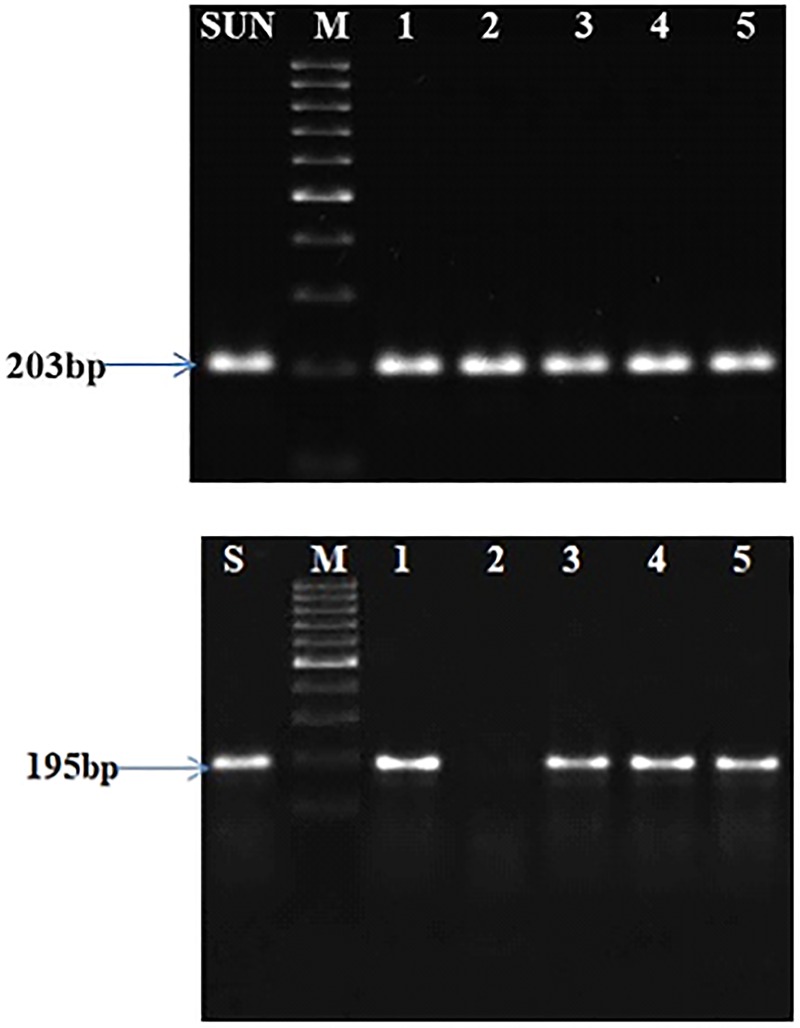
The DNA was extracted from tender fresh leaves of 10 to 15 days old field-grown seedlings using modified cetyltrimethylammonium bromide (CTAB) extraction method [39]. The quality and quantity of DNA were checked [25] and genotyping of the target population was done using AS-PCR and CAPS markers. The primer combination, F435-F and F435SUB-R, amplified 203bp fragment for the mutant allele (substitution from G:C→A:T, ahFAD2A) in the A-genome, while the primer combination, F435-F and F435INS-R amplified 195bp fragment for the mutant allele (A:T insertion, ahFAD2B) in the B-genome (Fig 1). In case of wild type ahFAD2A allele, the 826bp fragment was digested to 598bp and 228bp, while the mutant genotypes had the 826bp fragment intact. For B-genome, 2.0U of restriction enzyme Hpy188I (New England Biolabs, UK) was used for digestion of 10μl of PCR amplicon for about 16 hours at 37°C. The wild type ahFAD2B allele of 1214bp with five restriction sites cleaved into five fragments i.e., 736, 263, 171, 32 and 12bp.While the mutant allele had one additional restriction site in the 736bp fragment which was further cleaved into 550 and 213bp (all together six restriction sites in mutant instead of five in wild type) [23, 25].
Fig 1.
AS-PCR assay, (a) Amplification of ahFAD2A mutant allele-specific 203 bp amplification in 1 to 5 F1 plants; (b) ahFAD2B mutant allele-specific 195 bp in3 to 4 while absent in 1 and 2 F1 plants; where SUN: SunOleic95R, M:100bp DNA ladder.
Estimation of background genome recovery and linkage drag
Eighty polymorphic single sequence repeats (SSRs) from 20 linkage groups (preferably two from each arm of a linkage group) were deployed to determine recurrent parent genome recovery in MABC lines [40, 41]. Furthermore, recurrent parent and MABC lines were assessed based on the passport data. Subsequently, the desirable recombinant plants possessing the smallest size of introgressed segments with minimum linkage drag among MABC lines were identified. For the analysis, additional 10 SSRs, selected from the ~20cM genomic region on either side of ahFAD2 loci from both a09 and b09 linkage groups, were used (S1 Table).
Hybridization and development of MABC lines
Hybridization was done at ICRISAT, Patancheru, India in 2011 during the rainy season. The crossed seeds were planted at ICAR-DGR, Junagadh in post-rainy season in the same year. F1s were genotyped with linked allele-specific markers to identify true F1 plants and plants heterozygous for ahFAD2 alleles were used for backcrossing. The BC1F1 plants were planted in 2012 rainy season and were genotyped with allele-specific markers to identify heterozygous plants at both the loci. Backcrossing and genotyping with AS-PCR markers were continued until the development of BC3F1 generation. The BC3F1 seeds were planted in 2013 rainy season and plants having ahFAD2 alleles were advanced to BC3F2 generation. The BC3F2 seeds were planted in 2013 post-rainy season and plants were genotyped with CAPS marker to identify plants with both the homozygous mutant loci. The BC3F2–3 plants homozygous for ahFAD2 alleles were advanced to BC3F3–4 in 2014 rainy season. Phenotyping for oil content and fatty acid composition was done in BC3F3–4progeny. Finally, introgression lines (ILs) were selected based on oleic acid content and was coded as MABC introgression lines (MABC-ILs).
Yield evaluation of MABC-ILs lines
The initial yield evaluation of MABC lines along with elite peanut cultivars was done in 2014 post rainy and 2015 rainy seasons. In both the seasons, genotypes were planted in a randomized block design (RBD) with three replications. The advanced yield evaluation of MABC-ILs along with other breeding lines and elite cultivars was carried out at three different states, namely Gujarat, Telangana, and Andhra Pradesh in both 2016 rainy and 2016 post-rainy seasons. The crops were sown in RBD with two replications. Each genotype was planted on four-meter beds in four lines. Recommended crop management practices were followed for raising a healthy crop. Pod yield per plot (7.2 m2) was recorded during the harvest on maturity of crop (111–115) days after sowing.
Biochemical analysis for oil content and fatty acid profile
The harvested mature kernels were subjected to oil and fatty acid analysis using Gas chromatograph (model number GC-700, Thermo Fisher, USA) [42] with flame ionization detector (FID) [23].
Seed and seedling traits
The matured kernels harvested from the plants of rainy season 2018 were subjected to the analysis. The pods harvested in the first week of October 2018 were sown in the third week of February 2019. The experiment followed RBD and was conducted in a BOD incubator (San-134, Sanco) under controlled temperature (32 ±2°C), humidity (70 ±5%), and cooled LED lights for 24 h. Each genotype was sown in five replications with 20 kernels per replication in randomized complete block design (RCBD). Ten kernels were sown in a UV protected 7×8 inch black-color plastic plant nursery bags, filed with normal soil (~2.3kg). Thus, two plastic plant nursery bags constituted single replication. The kernels were treated with Bavistin® (2 g per kg of kernels) prior to sowing. After sowing, watering was done until saturation of the polythene bags and kept in BOD for 15 days. Regular watering was maintained on every alternate day. Plastic bags were removed carefully after 15 days so that there was no damage to the root system. The individual plant was collected replication wise from each genotype after thorough washing (Fig 2). Observations on the rate of germination, shoot length, root length, shoot fresh weight, root fresh weight, plant dry weight, root dry weight, and vigor index were recorded. The rate of germination was calculated using the formula: Germination (%) = (number of seeds germinated/total number of seeds sown) × 100. Vigor index was calculated using the formula: Vigor Index = (Seedling dry weight× germination %) /100 [43].
Fig 2.
Groundnut genotypes grown in BOD; a) Plants grown in polythene bags, b) Plants uprooted for recording observations on seedling traits.
Characterization of genotype
The passport data of MABC-ILs and recurrent parent were recorded on the basis of 16 qualitative and 17 quantitative traits, along with 6 special features, following peanut-descriptor [44] from five plant samples collected from the field at vegetative, reproductive, and harvesting stages.
Statistical analysis
Recurrent parent genome (RPG) recovery was analyzed using the formula: “RPG% = [{2 (R) + (H)}/2N] × 100” [45]; where “R” is the number of loci homozygous for recurrent parent allele; “H” is the number of loci still remaining heterozygous, and “N” is the total number of polymorphic markers used in the background analysis. The stability analysis for the pod yield was performed using AMMI ANOVA and GGE biplot models using R package [46]. A t-test was applied to assess the mean difference between oil, protein, moisture, oleic acid, linoleic acid, and palmitic acid contents among the MABC-ILs and parents. The significant differences between the mean values were determined by Duncan’s multiple range test (DMRT) (Duncan 1955) at a P ≤ 0.05 using CropStat version 7.2 [47]. Significant differences if any, between genotypes were compared using ANOVA.
Results
Development of advanced ILs through MABC
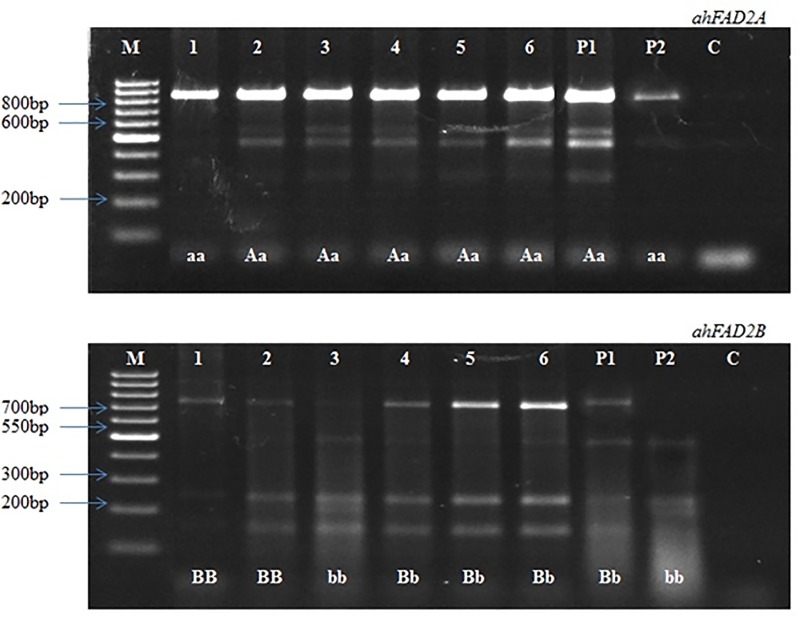
The crossed seeds received from ICRISAT, Patancheru, were planted at ICAR-DGR, Junagadh and resulted in 15 F1 plants. Eight plants were identified as true hybrids carrying both the mutant aFAD2 alleles. These eight F1 plants were used as pollen parents to make the first backcross with the recurrent parent. Out of 28 BC1F1 plants, six plants were found to carry both the ahFAD2 alleles in a heterozygous condition. Second backcrossing resulted in 32 BC2F1 plants and both the mutant alleles were found in nine plants. Third backcrossing resulted in 37 BC3F1 plants, among which six plants carried the ahFAD2 alleles. These six BC3F1 plants were selfed and 67 BC3F2 seeds were harvested and sown in the next season. BC3F2 plants were genotyped with the AS-PCR and CAPS markers, and three plants were finally identified as homozygous for both the ahFAD2 alleles (Fig 3). Subsequently, the fatty acid analysis confirmed single MABC-IL with ~80% oleic acid (which was later coded as NRCGCS-587).
Fig 3.
CAPS assay; (a) Heterozygous and homozygous plants for ahFAD2A mutant allele; (b) Heterozygous and homozygous plants for ahFAD2B mutant allele; where M: 100bp DNA ladder, 1–6: MABC-ILs, P1: ICGV06100, P2: SunOleic95R, C: Control, ‘AA, BB’: homozygous wild alleles, ‘Aa, Bb’: heterozygous alleles and ‘aa, bb’: indicates homozygous mutant alleles.
Recurrent parent genome recovery and linkage drag
Eighty SSRs were polymorphic between the recurrent parents and NRCGCS-587. Homozygosity was found with 73 SSRs in NRCGCS-587 indicating 91.87% recurrent parent genome (RPG) recoveries. However, a genomic segment carrying the ahFAD2 alleles was present in NRCGCS-587. Out of the 10 polymorphic SSRs tested between SunOleic 95R and NRCGCS-587, nine SSRs were amplified only in NRCGCS-587 and not amplified in SunOleic 95R (S1 Table) indicating a linkage drag of additional segments away from the ahFAD2A and ahFAD2B alleles. Therefore, introgression of additional genomic regions in NRCGCS-587 resulted in some linkage drag but it showed no decrease in high oleic content.
Fatty acid profile analysis and estimation of oil content in MABC-IL (NRCGCS-587) and parents
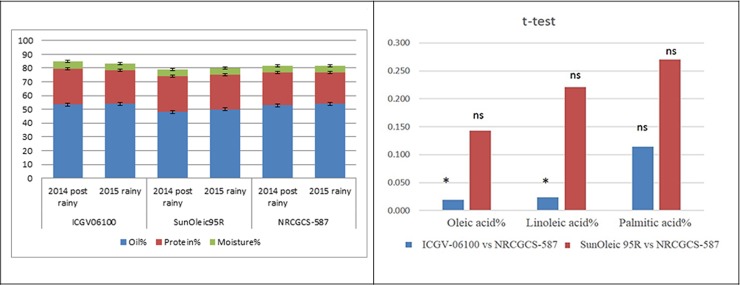
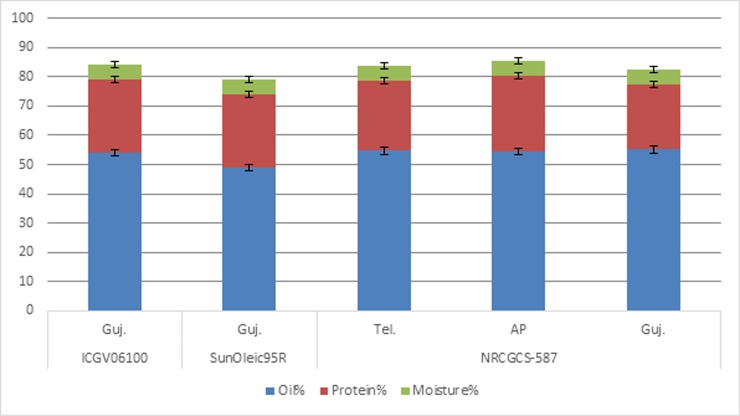
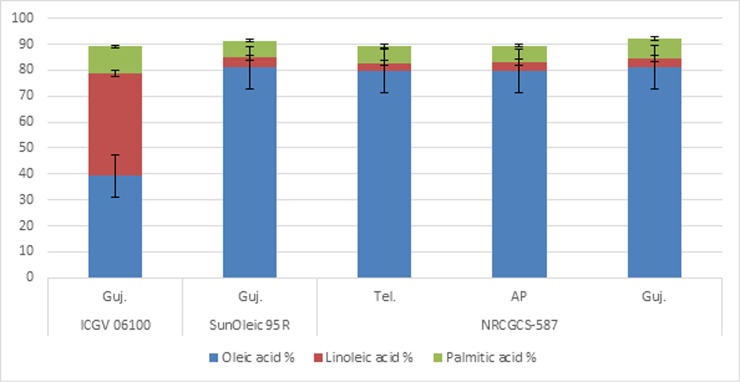
Fatty acid profile analysis of NRCGCS-587 with its parents was done in two seasons (S2 Table). In 2014 post-rainy season plantations, oleic acid and linoleic acid contents in NRCGCS-587 were recorded as 78.8% and 4.0%, respectively. Whereas the same were 42.0% and 35.0% in the recurrent parent, respectively, and as 77.0% and 6.0% in the donor parent, respectively. The O/L ratio in NRCGCS-587 was 19.7, while it was 1.2 in the recurrent parent. The palmitic acid content was 6.8% in NRCGCS-587 as compared to 13.0% and 7.0% in the recurrent and donor parent, respectively (Fig 4). NRCGCS-587 contained 53% oil and 24% protein as compared to 54% oil and 26% protein in the recurrent parent and 48% oil and 26% protein in the donor parent (Fig 5). Further analysis of the oil content and fatty acid composition was done in 2015 rainy season. NRCGCS-587 showed 54% oil and 23% protein content, ICGV-06100 contained 54% oil and 24% protein, and SunOleic95R recorded 50% oil and 25% protein contents. So, there was no significant differences in oil and protein content of NRCGCS-587 with its parents. (Fig 5). In NRCGCS-587, oleic acid, linoleic acid, and palmitic acid contents were 81%, 3%, and 6%, respectively, as compared to 39%, 39%, and 9% in ICGV06100, and 80%, 3.0%, and 6.0%, in SunOleic95R, respectively (Fig 4). The O/L ratio was 27.0 in NRCGCS-587, while it was 1.0 in the recurrent parent and 23.25 in the donor parent.
Fig 4. Oleic acid, linoleic acid, and palmitic acid in NRCGCS-587 and parents grown in ICAR-DGR during 2014 post rainy and 2015 rainy; “*” indicates significance at 5%; “ns” indicates non-significant.
Fig 5. Oil, protein, and moisture in NRCGCS-587 and parents grown in ICAR-DGR during 2014 post rainy and 2015 rainy seasons; “*” indicates significance at 5%; “ns” indicates non-significant.
Pod yield of MABC-IL
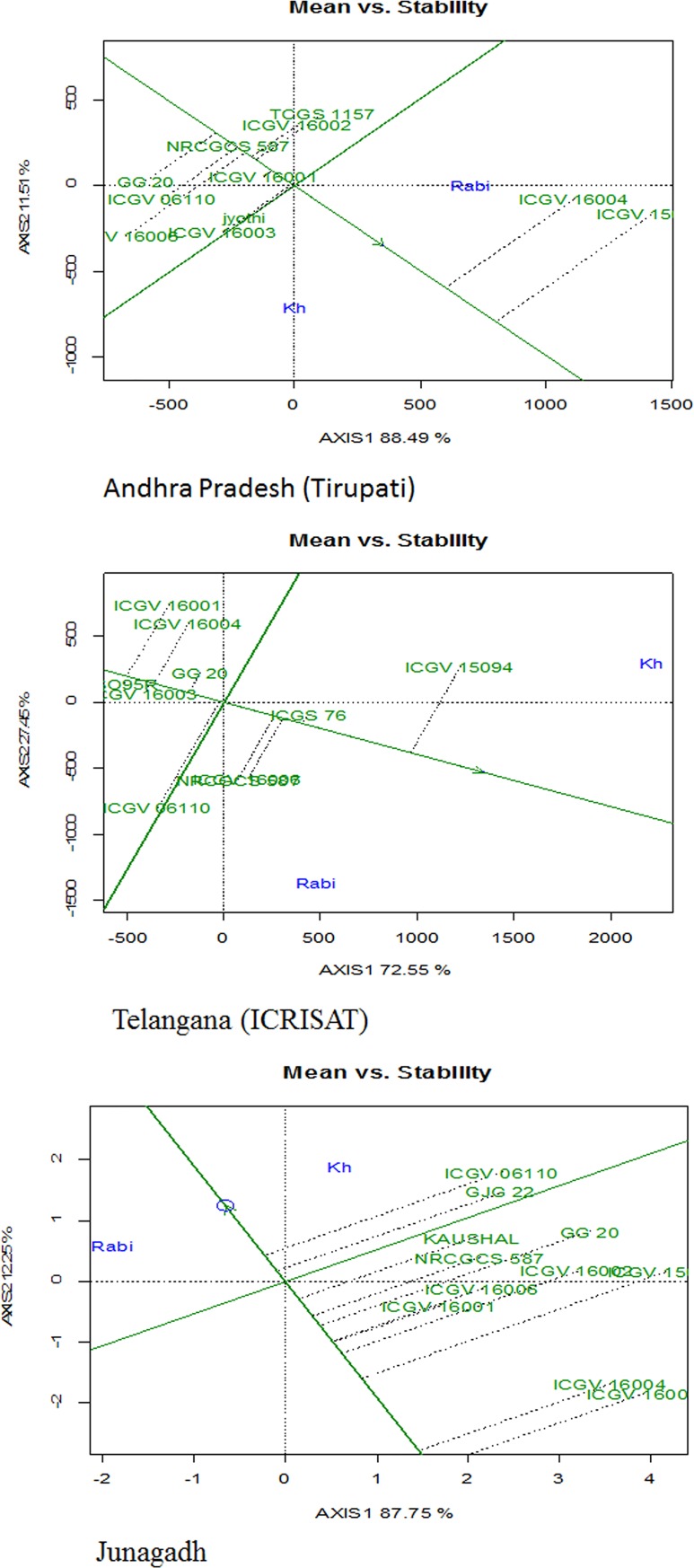
NRCGCS-587, along with 12 elite cultivars, was tested for yield and related traits. The analysis of variance revealed significant differences among the genotypes and genotype × environment interaction for pod yield. In 2014 post-rainy season, pod yield of NRCGCS-587 was 1464 kg/ha that was significantly higher than the check cultivars Abhaya, CO-6, GG-20, ICGS-1043, JL-24, TMV-2 and VRI-6; on par with K-6, TAG-24 and GJG-31; and lower than TG-37A (Table 1). During 2015 rainy season, pod yield of NRCGCS-587 (1714 kg/ha) was significantly higher than check cultivars except for TG-37A and GG-20. The pooled pod yield of NRCGCS-587 (1589 kg/ha) was significantly higher than all the check cultivars except TG-37A. Shelling percentage (73%) and hundred-kernel weight (50g) of NRCGCS-587 were higher with the check cultivars. Besides, NRCGCS-587 was tested at three different states over two seasons. AMMI analysis of variance (Table 2) revealed a significant interaction effect of genotype × location on pod yield followed by location and genotype, individually. Stability analysis in all the three locations by GGE biplot showed that pod yield of NRCGCS-587 was higher (Fig 6) with local check cultivars in Telangana (ICGS76) and Andhra Pradesh (TCGS˗157) and superior to common check cultivar (GG˗20).
Table 1. Yield and the related traits of NRCGCS-587 grown in ICAR-DGR, Gujarat, during 2014 post rainy and 2015 rainy season.
| Genotypes | Pod Yield (kg/ha) | Shelling (%) |
100 kernel weight (g) | ||
|---|---|---|---|---|---|
| 2015 rainy | 2014 post rainy | Mean | |||
| Abhaya | 1418.4 c-d | 1376.3 b-d | 1397.3 b-d | 72.1 a-c | 49.3 b-d |
| Co-6 | 1485.4 c-d | 1062.5 d-e | 1274.0 b-e | 70.7 a-d | 57.3 a-b |
| NRCGCS-587 | 1714.0 b-c | 1463.9 b-c | 1588.9 b | 72.1 a-c | 59.0 a-b |
| GG-20 | 1883.0 a-b | 967.9 e | 1425.4 b-d | 74 a-b | 65.7 a |
| GJG-31 | 1488.2 c-d | 1354.8 b-e | 1421.5 b-d | 66.4 d | 51.3 b-d |
| GPBD-4 | 1485.0 c-d | 1569.5 b | 1527.2 b-c | 74.1 a-b | 52.7 b-d |
| ICGS-1043 | 1450.8 c-d | 1178.5 c-e | 1314.7 b-e | 71.7 a-c | 54.3 b-c |
| JL-24 | 1385.6 c-e | 964.1 e | 1174.9 c-e | 69.7 b-d | 46.0 c-e |
| K-6 | 1336.5 c-e | 1499.4 b-c | 1417.9 b-d | 74.5 a | 53.3 b-d |
| TAG-24 | 1271.6 d-e | 1575.9 b | 1423.8 b-d | 70.9 a-d | 49.3 b-d |
| TG-37A | 2163.8 a | 2105.5 a | 2134.7 a | 70.4 a-d | 45.3 c-e |
| TMV-2 | 866.0 f | 1122.4 c-e | 994.2 e | 72.8 a-c | 44 d-e |
| VRI-6 | 1042.0 e-f | 1038.1 d-e | 1040.1 d-e | 68.7 c-d | 39.0 e |
| CV% | 15.08 | 14.97 | 14.28 | 13.90 | 18.61 |
Means followed by same letter are not significantly different (less than or equal) at P = 0.05.
Table 2. AMMI Analysis of variance for pod yield evaluated at the three locations.
| df | MSS | Pr (>F) |
% Sum of Squares | |
|---|---|---|---|---|
| Locations (L) | 5 | 4758501 | <0.001 | 36.8 |
| Rep (L) | 6 | 125004 | 0.22 | 1.2 |
| Genotype (G) | 9 | 968802 | <0.001 | 13.5 |
| G*L | 45 | 594607 | <0.001 | 41.3 |
| PC1 | 13 | 1174680 | 0 | 57.1 |
| PC2 | 11 | 539693.9 | 0 | 22.2 |
| PC3 | 9 | 349655.6 | <0.001 | 11.8 |
| PC4 | 7 | 241216.7 | 0.015 | 6.3 |
| PC5 | 5 | 142883.3 | 0.165 | 2.7 |
| Residuals | 54 | 87212 | 7.3 |
PC1, PC2 …PC5 indicates principal components 1, 2….5 (denotes variation accounted by each components); df–Degrees of freedom; MSS- Mean sum of squares. P- value at 5%.
Fig 6. Average environment coordination (AEC) views of the GGE-biplot based on environment-focused scaling peanut genotypes evaluated for pod yield in Andhra Pradesh, Telangana, and Gujarat, India.
Oil content and fatty acid profile of MABC-IL in three different states
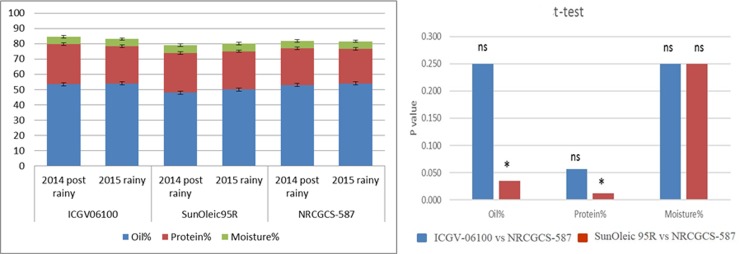
The pod samples of NRCGCS-587 were collected from three different states viz., Andhra Pradesh, Telangana and Gujarat in 2016 post-rainy season and subjected to biochemical analysis (S2 Table). Oil content in NRCGCS-587 did not differ much across the states, i.e., 54.7%, 54.5%, and 55.1% in Telangana, Andhra Pradesh, and Gujarat, respectively. Oleic acid content was almost the same in the pods of the two states, viz., Telangana (79.8%) and Andhra Pradesh (79.6%), while it was slightly higher in Gujarat (81.2%). Furthermore, linoleic acid (Telangana-3.0%, Andhra Pradesh-3.5%, and Gujarat-3.2%) and palmitic acid contents (Telangana-6.5%, Andhra Pradesh-6.4%, and Gujarat-7.8%) across the locations were similar (Figs 7 and 8). Likewise, oleic to the linoleic ratio in NRCGCS-587 also remained almost the same.
Fig 7. Oil, protein, and moisture in NRCGCS-587 and parents grown in Andhra Pradesh, Telangana, and Gujarat, India during 2016 rainy season.
Fig 8. Oleic acid, linoleic acid, and palmitic acid in NRCGCS-587 and parents grown in Andhra Pradesh, Telangana, and Gujarat, India during 2016 rainy season.
Passport data of NRCGCS-587 (MABC-IL) and recurrent parent
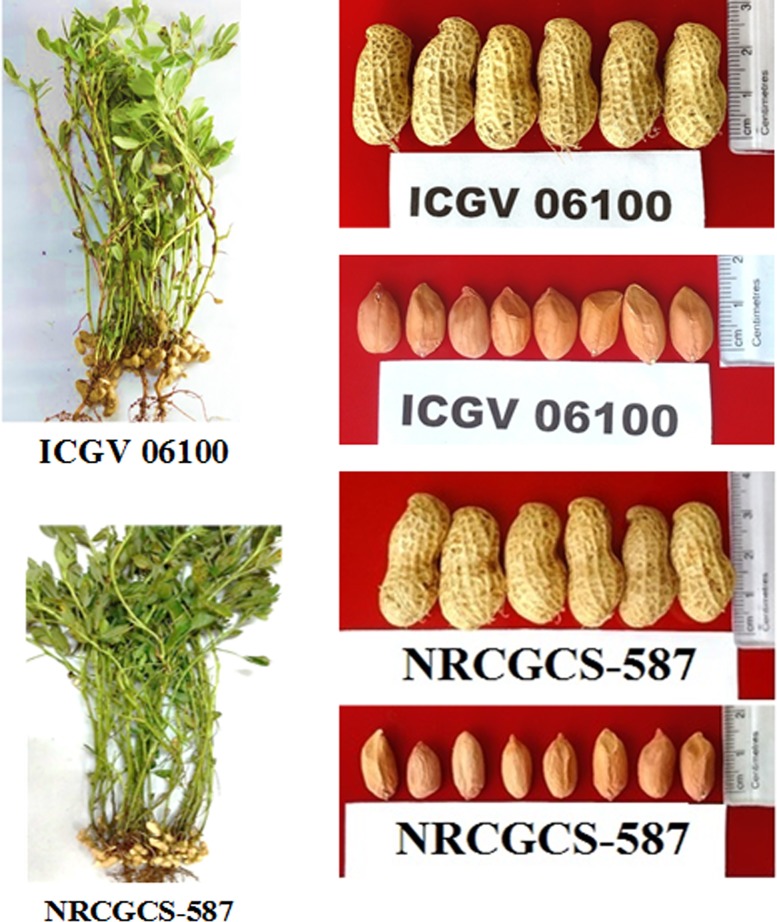
NRCGCS-587 is a Virginia bunch genotype characterized by decumbent-3 growth habit, alternate branching, green color, ovate leaf, and simple inflorescence. It takes about 23 days after germination for 50% flowering and 115 days for maturity. Average plant height, leaf length and leaf width are 42.6 cm, 40.1 mm, and 13.2 mm, respectively. It produces an average of five primary branches per plant and 2–3 flowers per inflorescence. Pods are mostly two seeded and the average length and width of pods are 26.0 mm and 12.4 mm, respectively. The mean length and width of kernels are 13.8 mm and 6.8 mm, respectively and it is rose in color (Fig 9). It yields 108.0 g of pods per square meter with 20% harvest index, 70% shelling-out-turn, ~55% oil content,~80% oleic acid, and ~4% linoleic acid content (S3 Table). Most importantly, NRCGCS-587 has also shown resistance to rust and late leaf spot, i.e., 1 and 3 disease severity scores, respectively in 1–9 modified scale (data not shown).
Fig 9. Plant, pod, and kernels of ICGV06100 and NRCGCS-587.
Seed and seedling traits
Average seed germination of 93.3% was found in normal oleic peanut, while it was 81.7% in HO peanut. A significant difference in germination percentage was recorded between normal and HO peanut (Table 3). There were no significant differences between normal and HO peanut for vigor index, fresh and dry plant weight, shoot and root length, fresh shoot and root weight, dry shoot and root weight, shoot length/root length, fresh shoot weight/fresh root weight, dry shot weight/dry root weight, and plant fresh weight/plant dry weight. However, the genotypic difference was observed within the normal and HO peanut groups. In both, the groups shoot length, fresh shoot biomass, and dry shoot biomass were higher than fresh root length, fresh root biomass, and dry root biomass.
Table 3. Details of seedling traits in normal oleic and high oleic peanut genotypes.
| Trait | Name of genotypes | Oil%* | Oleic acid %* | Germination% | Shoot Length (SL) |
Root Length(RL) | SL/RL | Fresh Shoot wt. (FSW) (g) |
Fresh Root wt. (FRW) (g) |
FSW/ FRW |
Dry Shoot wt. (DSW) (g) |
Dry Root wt. (DRW) (g) |
DSW/DRW | Plant Fresh wt. (PFW) (g) |
Plant Dry wt. (PDW) (g) |
PFW/PDW | Vigor index |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
High oleic (~80%) peanuts |
NRCGCS-587 | 55 | 80 | 80.00 | 17.78 | 7.65 | 2.34 | 1.97 | 0.16 | 11.94 | 0.32 | 0.05 | 6.90 | 2.13 | 0.37 | 5.77 | 0.3 |
| HOP-IL_MAS-191 | 53.2 | 79.8 | 73.33 | 21.24 | 10.89 | 1.93 | 2.22 | 0.13 | 17.32 | 0.24 | 0.02 | 14.60 | 2.35 | 0.26 | 8.99 | 0.19 | |
| HOP-IL_MAS-145 | 54.5 | 80.3 | 76.7 | 23.00 | 9.55 | 2.40 | 2.56 | 0.15 | 17.03 | 0.32 | 0.01 | 26.55 | 2.71 | 0.33 | 8.23 | 0.25 | |
| HOP-IL_MAS-130 | 54.7 | 80.5 | 96.7 | 17.17 | 5.80 | 3.01 | 1.35 | 0.06 | 21.26 | 0.12 | 0.02 | 6.77 | 1.41 | 0.14 | 10.00 | 0.14 | |
| Mean | 81.70 | 19.79 | 8.47 | 2.42 | 2.02 | 0.13 | 15.98 | 0.25 | 0.02 | 13.71 | 2.15 | 0.28 | 7.81 | 0.22 | |||
| Normal oleic
(~50–55%) peanuts |
GG-20 | 51 | 64 | 90 | 22.40 | 6.75 | 3.32 | 2.58 | 0.12 | 22.43 | 0.28 | 0.03 | 10.24 | 2.70 | 0.31 | 8.82 | 0.27 |
| ICGV-06100 | 55 | 39 | 83.30 | 15.46 | 5.93 | 2.63 | 2.02 | 0.35 | 5.77 | 0.27 | 0.05 | 6.36 | 2.37 | 0.32 | 7.38 | 0.26 | |
| ICGV-05141 | 54.7 | 55 | 100.00 | 14.68 | 6.63 | 2.28 | 1.24 | 0.08 | 15.48 | 0.14 | 0.01 | 10.37 | 1.32 | 0.16 | 8.32 | 0.16 | |
| ICGV-06110 | 53 | 38.3 | 100.00 | 17.20 | 4.15 | 4.42 | 1.62 | 0.06 | 29.36 | 0.10 | 0.01 | 20.40 | 1.67 | 0.11 | 15.61 | 0.11 | |
| Mean | 93.30 | 17.44 | 5.87 | 3.16 | 1.86 | 0.15 | 12.42 | 0.20 | 0.02 | 11.84 | 2.01 | 0.22 | 9.03 | 0.20 | |||
| CD@5% | 7.55 | 5.16 | 1.42 | 0.9 | 0.73 | 0.04 | 3.65 | 0.1 | 0.01 | 5.51 | 0.76 | 0.1 | 2.57 | 0.08 | |||
| CV% | 4.93 | 15.84 | 11.34 | 18.8 | 21.4 | 14.55 | 11.6 | 24.31 | 22.14 | 24.75 | 20.88 | 23.24 | 15.9 | 22.04 |
Discussion
Peanut with HO is preferred over normal peanut due to its extended shelf life and multiple health benefits. High oil and oleic acid content in the peanuts are necessary for producing superior quality of oil to meet the nutritional needs and for industrial purposes. Moreover, the high oil containing peanuts can be used to combat malnutrition due to its higher caloric value over normal peanut. [48]. Therefore, improvement of oleic acid content in peanut for higher oxidative stability and better dietary properties is one of the important breeding objectives worldwide. Availability of molecular markers linked to the ahFAD2 gene has facilitated marker-assisted breeding for HO. MABC breeding further ensures the transfer of desirable gene together with maximum genome recovery of the recurrent parent [49, 50]. Previously, nematode resistance [51], rust resistance [52], and high oleic acid [22, 23] traits were transferred to elite peanut cultivars using MABC breeding. The use of CAPS and SNP markers has considerably reduced the time and volume of breeding material in different backcross generations [25]. In the first objective, a high oil content peanut genotype, ICGV06100, was targeted to improve oleic acid content using MABC breeding. The studies reported the development of a peanut genotype, NRCGCS-587, with high oil and HO content. The HO trait was introgressed from SunOleic95R into the genetic background of ICGV06100 through MABC approach and developed an improved version of ICGV06100 with 97% increase in oleic acid content over the recurrent parent.
The increase in oleic acid content in NRCGCS-587 led to a reduction in linoleic acid. There was a 90% and 24% reduction in linoleic acid and palmitic acid, respectively, in NRCGCS-587 as compared to the recurrent parent. Moreover, linoleic acid content ranged from 3.0% to 4.0% and palmitic acid ranged from 6.1% to 7.8% over different locations indicating their stable expression. The O/L ratio was increased to 27 in NRCGCS-587 from 1.2 in the recurrent parent. A similar trend of increase in oleic acid and O/L ratio, as well as a reduction in linoleic acid and palmitic acid, has already been reported [22, 23]. Commonly, an alteration in any of the metabolite biosynthesis also has a negative feedback effect on the production of other metabolites in a related pathway. Likewise, a significant reduction in palmitic acid level in NRCGCS-587 was recorded. Several previous studies have also reported a similar effect of ahFAD2 alleles on palmitic acid content [14, 22, 23, 53].
Generally, variation in oil content and fatty acid composition was reported in different environments due to the quantitative nature of these traits that are controlled by complex pathways [25, 26, 54]. However, limited or no variation was observed in NRCGCS-587 regarding oil, oleic, linoleic, and palmitic acid contents over locations indicating the minimal environmental effect on oil and HO traits. It seems that only a few independent genes, with the major effect, control oil and oleic acid production in NRCGCS-587. The selection for improved fatty acid composition would not affect the oil content of seed since there was no significant correlation between percent oil and any of the fatty acids or related variables [55]. Although fatty acid composition showed variation with the growth habit and environment, the oil content remained constant [56, 57, 58]. As a result, NRCGCS-587 with stable oil content across locations would be a better choice for use as a parent in the future breeding program on enhancing oleic acid and oil content in peanut.
NRCGCS-587 had more than 90% background genome recovery as well as precise introgression of ahFAD2 alleles. Moreover, identical passport data of NRCGCS-587 and ICGV06100 except oleic acid content corroborate maximum genome recovery from recurrent parent and precise introgression of ahFAD2 alleles in NRCGCS-587. Thus, NRCGCS-587 is an improved version of ICGV06100 having ~80% oleic acid content. The combined approach of both genotypic and phenotypic selections was found appropriate and effective in selecting improved lines [23, 59]. High oleic acid content did not affect seedling traits except the rate of germination. Significant variation in the rate of germination between HO and normal oleic peanut groups might be due to the alteration in lipid composition of seeds leading to changed membrane function and permeability. The germination decreased as O/L and unsaturated/saturated ratios increased in peanut, especially at lower (16°C and 14°C) temperatures [35]. Jungman and Schubert [36] reported that HO lines had lower seed vigor than their paired lines with normal oleic content. In general, the processes of germination initiates at a temperature below 15°C in peanut. Lower germination rate observed in HO peanut in this research might be due to the change in fatty acid composition since the temperature was maintained constant at 32°C. In sorghum, the α-amylase activity of seeds and subsequent seed germination percentage were affected by long-chain fatty acid composition [60].
In Pinus pinea, an increase in caprylic or oleic acids retarded the seed germination. The inhibition was dependent on fatty acid concentration and chain-length [61]. Short-chain fatty acids could infiltrate membrane lipids and change the physical properties that lower the seed germination [62].
In conclusion, there was a narrow but significant difference in seedling establishment between HO and normal oleic peanut under optimum temperature. Poor seed germination rate in HO peanut than normal peanut could be a cause of concern if a significant difference is more and needs further investigation to overcome it. A perfectly stable genotype having constant yield across geographical locations is a key to a successful variety [63]. The higher pod yield in the post-rainy season than a rainy season in NRCGCS-587 indicated that it might be more remunerative under irrigation than rain-fed conditions. It yielded either significantly higher or on par with all check cultivars except TG-37A indicating the potential to excel the local elite varieties from different peanut-growing states in India. Shelling percent and hundred-kernel weight were also on par with elite cultivars. Furthermore, NRCGCS-587 recorded maximum pod yield (2445 kg/ha) in Telangana and Andhra Pradesh that makes it suitable for these states. Stable pod yield, oil content, and HO content of NRCGCS-587 over the locations make it more rewarding for the peanut growing farmers. NRCGCS-587 is an improved version of ICGV06100 having genotypically 91% RPG and ahFAD2 alleles, and phenotypically high oil and yield. Thus, improved nutritional qualities would fetch premium price to the farmers without compromising the yield and meet the demand of peanut oil for industrial purposes.
Supporting information
(XLSX)
(DOCX)
(DOCX)
Acknowledgments
Authors acknowledge the financial support received from Integrated Scheme of Oilseeds, Pulses, Oilpalm and Maize (ISOPOM), Ministry of Agriculture and Farmers Welfare, the Government of India.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The work presented in this article is a contribution from research projects sponsored by Integrated Scheme of Oilseeds, Pulses, Oilpalm and Maize (ISOPOM), Department of Agriculture and Co-operation (DAC), Ministry of Agriculture, Government of India.
References
- 1.FAOSTAT 2017 Available: http://faostat.fao.org accessed on 22-03-2019.
- 2.Kavera B, Nadaf HL, Hanchinal RR. Near infrared reflectance spectroscopy (NIRS) for large scale screening of fatty acid profile in peanut (Arachis hypogaea L.), Legume res. 2014; 37:272–280. 10.5958/j.0976-0571.37.3.041 [DOI] [Google Scholar]
- 3.Johnson S, Saikia N. Fatty acids profile of edible oils and fat in India, Centre for Science and Environment, New Delhi, 2008; pp. 1–48. [Google Scholar]
- 4.Kratz M, Cullen P, Kannenberg F, Kassner A, Fobker M, Abuja PM, et al. Effects of dietary fatty acids on the composition and oxidizability of low density lipoprotein. European Journal of Clinical Nutrition. 2002; 56:72–81. 10.1038/sj.ejcn.1601288 [DOI] [PubMed] [Google Scholar]
- 5.WHO. Diet, nutrition and the prevention of chronic diseases, WHO technical report series 916, Report of a joint WHO/FAO expert consultation, World Health Organization, Geneva: 2003; 88 [PubMed] [Google Scholar]
- 6.Wang CT. Peanut production, trade and utilization Peanut Science and Technology Bulletin, National Peanut Agri-Indus Res Sys. 2009; 1 (5&6), 8–32. [Google Scholar]
- 7.Vassiliou EK, Gonzalez A, Garcia C, Tadros JH, Chakraborty G, Toney JH. Oleic acid and peanut oil high in oleic acid reverse the inhibitory effect of insulin production of the inflammatory cytokine TNF-alpha both in vitro and in vivo systems. Lipids in Health and Disease. 2009; 8:25 10.1186/1476-511X-8-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Byrne DJ, Knauft DA, Shireman RB. Low fat-monounsaturated rich diets containing high-oleic peanuts improve serum lipoprotein profiles. Lipids. 1997; 32(7):687–695. 10.1007/s11745-997-0088-y [DOI] [PubMed] [Google Scholar]
- 9.Yamaki T, Nagamine I, Fukumoto K, Yano T, Miyahara M, Sakurai H. High oleic peanut oil modulates promotion stage in lung tumorigenesis of mice treated with methyl nitrosourea. Food Science and Technology Research. 2005; 11:231–235. 10.3136/fstr.11.231 [DOI] [Google Scholar]
- 10.O’keefe SF, Wiley VA, Knauft DA. Comparison of oxidative stability of high-and normal-oleic peanut oils. J Am Oil Chem Soc. 1993; 70(5):489–492. BF02542581 [Google Scholar]
- 11.Jung S, Powell G, Moore K, Abbott A. The high oleate trait in the cultivated peanut (Arachis hypogaea L.). II. Molecular basis and genetics of the trait. Mol Gen Genet. 2000. b 263:806–811. 10.1007/s004380000243 [DOI] [PubMed] [Google Scholar]
- 12.Lopez Y, Nadaf HL, Smith OD, Connell JP, Reddy AS, Fritz AK. Isolations and characterization of the Δ12 fatty acid desaturase in peanut (Arachis hypogaea L.) and search for polymorphism for the high oleate trait in Spanish market-type lines. Theor Appl Genet. 2000; 101:1131–1138. 10.1007/s001220051589 [DOI] [Google Scholar]
- 13.Chu Y, Holbrook CC, Ozias-Akins P. Two alleles of control the high oleic acid trait in cultivated peanut. Crop sci. 2009; 49:2029–2036. 10.2135/cropsci2009.01.0021 [DOI] [Google Scholar]
- 14.Pandey MK, Wang ML, Qiao L, Feng S, Khera P, Wang H, et al. Identification of QTLs associated with peanut oil contents in RIL populations and mapping FAD2 genes and their relative contribution towards oil quality. BMC Genetics. 2014; 15:133 10.1186/s12863-014-0133-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung S, Swift D, Sengoku E, Patel M, Teule F, Powell G, et al. The high oleate trait in the cultivated peanut (Arachis hypogaea L.) I. Isolation and characterization of two genes encoding microsomal oleoyl-PC desaturases. Mol Gen Genet. 2000. a; 263:796–805. 10.1007/s004380000244 [DOI] [PubMed] [Google Scholar]
- 16.Gorbet DW, Knauft DA. Registration of ‘SunOleic 95R’peanut. Crop sci. 1997; 37:1392. [Google Scholar]
- 17.Wang ML, Chen CY, Tonnis B, Barkley NA, Pinnow DL, Pittman RN, et al. Oil, fatty acid, flavonoid, and resveratrol content variability and FAD2A functional SNP genotypes in the US peanut mini-core collection. J Agric Food Chem. 2013; 61:2875–2882. 10.1021/jf305208e [DOI] [PubMed] [Google Scholar]
- 18.Wang ML, Khera P, Pandey MK, Wang H, Qiao L, Feng S, et al. Genetic mapping of QTLs controlling fatty acids provided insights into the genetic control of fatty acid synthesis pathway in peanut (Arachis hypogaea L.). PLoS One. 2015. a; 10(4):e0119454 10.1371/journal.pone.0119454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Norden AJ, Gorbet DW, Knauft DA, Young CT. Variability in oil quality among peanut genotypes in the Florida breeding program. Peanut Sci. 1987; 14:7–11. 10.3146/i0095-3679-14-1-3 [DOI] [Google Scholar]
- 20.Chen Z, Wang ML, Barkley NA, Pittman RN. A simple allele-specific PCR assay for detecting FAD2 alleles in both A and B genomes of the cultivated peanut for high-oleate trait selection. Plant Mol Biol Rep. 2010; 28:542–548. 10.1007/s11105-010-0181-5 [DOI] [Google Scholar]
- 21.Chu Y, Ramos L, Holbrook CC, Ozias-Akins P. Frequency of a loss-of-function mutation in Oleoyl-PC Desaturase (ahFAD2A) in the mini-core of the US peanut germplasm collection. Crop sci. 2007; 47:2372–2378. 10.2135/cropsci2007.02.0117 [Google Scholar]
- 22.Janila P, Pandey MK, Shasidhar Y, Variatha MT, Sriswathi M, Khera P, et al. Molecular breeding for introgression of fatty acid desaturase mutant alleles (ahFAD2A and ahFAD2B) enhances oil quality in high and low oil containing peanut genotypes. Plant Sci. 2016; 242:203–213. 10.1016/j.plantsci.2015.08.013 [DOI] [PubMed] [Google Scholar]
- 23.Bera SK, Kamdar JH, Kasundra SV, Dash P, Maurya AK, Jasani MD, et al. Improving oil quality by altering levels of fatty acids through marker-assisted selection of ahfad2 alleles in peanut (Arachis hypogaea L.). Euphytica. 2018; 214:162 10.1007/s10681-018-2241-0 [DOI] [Google Scholar]
- 24.Grosso NR, Lamarque A, Maestri D, Zygadlo J, Guzmán C. Fatty acid variation of runner peanut (Arachis hypogaea L.) among geographic localities from Cordoba (Argentina). J Am Oil Chem Soc. 1994; 71:541–542. 10.1007/BF02540669 [DOI] [Google Scholar]
- 25.Grosso N, Guzmán C. Chemical composition of aboriginal peanut (Arachis hypogaea L.) seeds from Peru. J Agric Food Chem. 1995; 43:102–105. 10.1021/jf00049a019 [DOI] [Google Scholar]
- 26.Dwivedi SL, Nigam SN, Jambunathan R, Sahrawat KL, Nagabhushanam GVS, Raghunath K. Effect of genotypes and environments on oil content and oil quality parameters and their association in peanut (Arachis hypogaea L.). Peanut Sci. 1993; 20:84–89. 10.3146/i0095-3679-20-2-5 [DOI] [Google Scholar]
- 27.Andersen PC, Gorbet DW. Influence of year and planting date on fatty acid chemistry of high oleic acid and normal peanut genotypes. Journal of Agricultural and Food Chemistry. 2002; 50:1298–1305. 10.1021/jf0113171 [DOI] [PubMed] [Google Scholar]
- 28.Sogut T, Ozturk F, Kizil S. Effect of sowing time on peanut (Arachis hypogaea L.) cultivars: i. yield, yield components, oil and protein content. Scientific Papers-Series A, Agronomy. 2016; 59: 415–420. 10.1016/j.aaspro.2016.09.018 [DOI] [Google Scholar]
- 29.Chaiyadee S, Jogloy S, Songsri P, Singkham N, Vorasoot N, Sawatsitang P, et al. Soil moisture affects fatty acids and oil quality parameters in peanut. International Journal of Plant Production. 2013; 7:81–96. [Google Scholar]
- 30.Dwivedi SL, Nigam SN, Rao RN, Singh U, Rao KVS. Effect of drought on oil, fatty acids and protein contents of groundnut (Arachis hypogaea L.) seeds. Field crops research. 1996; 48(2–3):125–133. 10.1016/S0378-4290(96)01027-1 [DOI] [Google Scholar]
- 31.Golombek D, Sridhar R, Singh U. Effect of soil temperature on the seed composition of three Spanish cultivars of groundnut (Arachis hypogaea L.). J Agric Food Chem. 1995; 43:2067–2070. 10.1021/jf00056a021 [DOI] [Google Scholar]
- 32.Li X, Wu L, Qiu G, Wang T, Liu C, Yang Y, et al. Effects of sowing season on agronomic traits and fatty acid metabolic profiling in three Brassica napus L. Cultivars. Metabolites. 2019; 9(2):37 10.3390/metabo 9020037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flagella Z, Rotunno T, Tarantino E, Di Caterina R, De Caro A. Changes in seed yield and oil fatty acid composition of high oleic sunflower (Helianthus annuus L.) hybrids in relation to the sowing date and the water regime. European journal of agronomy. 2002; 17(3):221–230. 10.1016/S1161-0301(02)00012-6 [DOI] [Google Scholar]
- 34.Copeland LO, McDonald MB. Seed vigor and vigor testing In Principles of Seed Science and Technology.Springer, Boston, MA: 2001. [Google Scholar]
- 35.Sun M, Spears JF, Isleib TG, Jordan DL, Penny B, Johnson D, et al. Effect of production environment on seed quality of normal and high-oleate large seeded Virginia-type peanut (Arachis hypogaea L.). Peanut Sci. 2014; 41(2):90–99. 10.3146/PS12-16.1 [DOI] [Google Scholar]
- 36.Jungman BS, Schubert AM. The effect of fatty acid profiles on peanut seed germination at low soil temperatures. Proc Amer Peanut Res Educ Soc. 2000; 32:36. [Google Scholar]
- 37.Upadhyaya HD, Dwivedi SL, Vadez V, Hamidou F, Singh S, Varshney RK, et al. Multiple resistant and nutritionally dense germplasm identified from mini core collection in peanut. Crop Sci. 2014; 54(2): 679–693. 10.2135/cropsci2013.07.0493 [DOI] [Google Scholar]
- 38.Nawade B, Bosamia TC, Thankappan R, Rathnakumar AL, Kumar A, Dobaria JR, et al. Insights into the Indian Peanut Genotypes for ahFAD2 Gene Polymorphism Regulating Its Oleic and Linoleic Acid Fluxes. Front. Plant Sci. 2016; 7:1271 10.3389/fpls.2016.01271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mace ES, Buhariwalla KK, Buhariwalla HK, Crouch JH. A high-throughput DNA extraction protocol for tropical molecular breeding programs. Plant Mol Biol Rep. 2003; 21:459–460. 10.1007/BF02772596 [DOI] [Google Scholar]
- 40.Gautami B, Foncéka D, Pandey MK, Moretzsohn MC, Sujay V, Qin H, et al. An international reference consensus genetic map with 897 marker loci based on 11 mapping populations for tetraploid groundnut (Arachis hypogaea L.). PLoS One. 2012; 7:41213 10.1371/journal.pone.0041213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bera SK, Kamdar JH, Kasundra SV, Ajay BC. Identification of a novel QTL governing resistance to Sclerotial stem rot disease in peanut. Australasian Plant Pathology. 2016; 45: 637 10.1007/s133313-016-0448-x [DOI] [Google Scholar]
- 42.Jokić S, Sudar R, Svilovic S, Vidović S, Bilić M, Velić D. Fatty Acid Composition of Oil Obtained from Soybeans by Extraction with Supercritical Carbon Dioxide. Czech J Food Sci. 2013; 31:116–125. [Google Scholar]
- 43.Kharb RPS, Lather BPS, Deswal DP. Prediction of Field Emergence through Heritability and Genetic Advance of Vigour Parameters. Seed Science and Technology. 1994; 22:461–466. [Google Scholar]
- 44.IBPGR & ICRISAT. Descriptors for groundnut, 125 International Board for Plant Genetic Resources, Rome, Italy and International Crops Research Institute for the Semi-Arid Tropics, Andhra Pradesh, India. 1992; ISBN 92-9043-139-3.
- 45.Vishwakarma MK, Mishra VK, Gupta PK, Yadav PS, Kumar H, Joshi AK. Introgression of the high grain protein gene Gpc-B1 in an elite wheat variety of Indo-Gangetic Plains through marker assisted backcross breeding. Current Plant Biology. 2014; 1:60–7. [Google Scholar]
- 46.R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna: 2015. [Google Scholar]
- 47.IRRI. CropStat for Windows, Version 7.2, IRRI 2007.
- 48.Arya SS, Salve AR, Chauhan S. Peanuts as functional food: a review. J Food Sci Technol. 2016; 53:31–41. 10.1007/s13197-015-2007-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pandey MK, Monyo E, Ozias-Akins P, Liang X, Guimarães P, Nigam SN, et al. Advances in Arachis genomics for peanut improvement. Biotechnology Advances. 2012; 30 (3): 639–651. 10.1016/j.biotechadv.2011.11.001 [DOI] [PubMed] [Google Scholar]
- 50.Varshney RK, Mohan SM, Gaur PM, Gangarao NVPR, Pandey MK, Bohra A, et al. Achievements and prospects of genomics-assisted breeding in three legume crops of the semi-arid tropics. Biotechnology Advances. 2013; 31:1120–1134. 10.1016/j.biotechadv.2013.01.001 [DOI] [PubMed] [Google Scholar]
- 51.Chu Y, Wu CL, Holbrook CC, Tillman BL, Person G, Ozias-Akins P. Marker-assisted selection to pyramid nematode resistance and the high oleic trait in peanut. Plant Genome. 2011; 4:110–117. 10.3835/plantgenome2011.01.0001 [DOI] [Google Scholar]
- 52.Varshney RK, Pandey MK, Janila P, Nigam SN, Sudini H, Gowda MVC, et al. Marker-assisted introgression of a QTL region to improve rust resistance in three elite and popular varieties of peanut (Arachis hypogaea L.). Theor Appl Genet. 2014; 127:1771–1781. 10.1007/s00122-014-2338-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang XZ, Wu Q, Tang YY, Sun QX, Wang CT. FAD2B from a peanut mutant with high oleic acid content was not completely dysfunctional. Advances in Applied Biotechnology. 2015. b; 332: 265–271. 10.1007/978-3-662-45657-6_28 [DOI] [Google Scholar]
- 54.Sarvamangala C, Gowda MVC, Varshney RK. Identification of quantitative trait loci for protein content, oil content and oil quality for groundnut (Arachis hypogaea L.). Field Crops Res. 2011; 122:49–59. 10.1016/j.fcr.2011.02.010 [DOI] [Google Scholar]
- 55.Mercer LC, Wynne TC, Young CT. Inheritance of fatty acid content in peanut oil. Peanut Sci. 1990; 17:17–21. 10.3146/i0095-3679-17-1-7 [DOI] [Google Scholar]
- 56.Gulluoglu L, Bakal H, Onat B, El Sabagh A, Arioglu H. Characterization of peanut (Arachis hypogaea L.) seed oil and fatty acids composition under different growing season under Mediterranean environment. Journal of Experimental Biology and Agricultural Sciences. 2016; 4(5S):564–571. 10.18006/2016 [DOI] [Google Scholar]
- 57.Bansal UK, Satija DR, Ahula KL. Oil composition of diverse groundnut (Arachis hypogaea L.) genotypes relation to different environments. J Sci Food Agric. 1993; 63:17–19. 10.1002/jsfa.2740630104 [DOI] [Google Scholar]
- 58.Raheja RK, Battai SK, Ahuja KL, Labana KS, Singh M. Comparison of oil content and fatty acid composition of peanut genotypes differing in growth habit. Plant Food Hum Nutr. 1987; 37:103–108. 10.1007/BF01092045 [DOI] [Google Scholar]
- 59.Hasan MM, Rafii MY, Ismail MR, Mahmood M, Rahim HA, Alam MA, et al. Marker-assisted backcrossing: a useful method for rice improvement. Biotechnology & Biotechnological Equipment. 2015; 29(2):237–254. 10.1080/13102818.2014.995920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marambe B, Nagaoka T, Anso T. Identification and biological activity of germination—inhibiting long-chain fatty acids in animal-waste composts. Plant Cell Physiol. 1993; 34, 605–612. [PubMed] [Google Scholar]
- 61.Vincenzini MT, Vincieri F, Vanni P. The effects of octanoate and oleate on isocitrate lyase activity during the germination of Pinus pinea seeds. Plant Physiol. 1973; 52, 549–553. 10.1104/pp.52.6.549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hendricks SB, Taylorson RB. Variation in germination and amino acid leakage of seeds with temperature related to membrane phase change. Plant Physiol. 1976; 58, 7–11. 10.1104/pp.58.1.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Allard RW. Principles of Plant Breeding. John Willey and Sons Inc, New York: 1960. [Google Scholar]