Abstract
Objectives
To identify the molecular mechanism of colistin resistance in an MDR Acinetobacter baumannii clinical strain isolated in 2008 from a meningitis case in Brazil.
Methods
Long- and short-read WGS was used to identify colistin resistance genes in A. baumannii strain 597A with a colistin MIC of 64 mg/L. MS was used to analyse lipid A content. mcr was cloned into pET-26b (+) and transformed into Escherichia coli BL21(λDE3)pLysS for analysis.
Results
A novel plasmid (pAb-MCR4.3) harbouring mcr-4.3 within a Tn3-like transposon was identified. The A. baumannii 597A lipid A MS spectra showed a main molecular ion peak at m/z=2034, which indicated the addition of phosphoethanolamine to the lipid A structure. E. coli BL21 transformed with pET-26b-mcr-4.3 gained colistin resistance with a colistin MIC of 8 mg/L.
Conclusions
Colistin resistance in A. baumannii 597A was correlated with the presence of a novel plasmid-encoded mcr-4.3 gene.
Introduction
Over the past 15 years, carbapenem resistance in Gram-negative bacteria has resulted in greater reliance on colistin for treatment. Acquired polymyxin resistance is primarily mediated by chromosomal mutations that modify the bacterial outer membrane (OM).1 However, plasmid-mediated polymyxin resistance (encoded by mcr-1) was first reported in China in 2015.2 The mcr genes encode phosphoethanolamine transferases (MCR enzymes) that modify lipid A of LPS through the addition of phosphoethanolamine. Since the 2015 report, >40 countries have identified mcr genes in Enterobacteriaceae from environmental, animal or human origin.3
To date (June 2019), nine different mcr-like genes have been identified (mcr-1 to mcr-9), which are frequently carried on transmissible plasmids in members of the Enterobacteriaceae.3–5 Notably, mcr-1 has been reported in Acinetobacter lwoffii from hospital surfaces in Italy and mcr-4.3 has been recently identified in Acinetobacter baumannii isolated from pig faeces in China.6,7 In Brazil, mcr-1- and mcr-3.12-harbouring Escherichia coli strains were obtained from clinical, animal and environmental samples.8–10 The aim of this study was to investigate the mechanism of colistin resistance in an MDR A. baumannii recovered in Brazil.
Materials and methods
Bacterial isolates
In 2008, a colistin-resistant A. baumannii strain (termed 597A) with a colistin MIC of 64 mg/L was recovered from the CSF of a patient with meningitis in Brazil.11 The strain was previously assigned to the international clonal complex CC113/79 based on the two MLST schemes, Oxford and Pasteur (https://pubmlst.org/abaumannii/).
WGS analysis
Short-read (550 bp) and long-read sequencing was performed with an Illumina MiSeq desktop sequencer (Illumina Inc., CA, USA) and a PacBio RS II (Pacific Biosciences, CA, USA), respectively. Newbler version 2.7 (454 Life Sciences, CT, USA) and HGAP 2.0 in the SMRT Analysis Portal were used to assemble Miseq and PacBio sequencing reads into de novo contigs. Antimicrobial resistance genes were annotated by ResFinder 2.1; ISs and the transposon were annotated after BLAST search. Plasmid maps were generated with Easyfig 2.1. Identification of plasmid genes, the replicon and the toxin–antitoxin system was based on comparison with sequences archived in the NCBI database (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The plasmid replicon was characterized based on the PCR-based replicon system in A. baumannii.12
Cloning the mcr-4.3 gene into the expression vector pET-26b (+) and transformation of the E. coli BL21(λDE3)pLysS strain
The full coding sequence of mcr-4.3 was amplified by PCR with forward eptA3-Fc (with an NdeI site) and reverse eptA3-Rp (with an XhoI site) primers (Table S1, available as Supplementary data at JAC Online). The purified mcr-4.3 PCR product was inserted into the vector pET-26b (+) (Novagen, WI, USA) digested with the restriction enzymes NdeI and XhoI. Ligation was performed with the Rapid DNA ligation kit (Sigma, St Louis, MO, USA). The recombinant pET-26b-mcr-4.3 was transformed into the chemically competent E. coli DH5α strain (Invitrogen, CA, USA). Transformed cells were grown in LB agar supplemented with 30 mg/L kanamycin (Sigma) overnight at 37°C. The pET-26b-mcr-4.3 in DH5α was sequenced to confirm its integrity.
The BL21(λDE3)pLysS strain (Novagen) was used for the expression of the mcr-4.3 gene after induction with 1 mM IPTG. The colistin MIC was determined by broth dilution in a constitutively expressed construct using the recommendations of CLSI. The pET-26b-mcr-4.3 was purified from the E. coli DH5α recombinant and transformed into the chemically competent BL21(λDE3)pLysS strain. The E. coli strain harbouring pET-26b-mcr-4.3 was grown aerobically at 37°C in LB broth (Merck, Darmstadt, Germany) with 30 mg/L kanamycin.
Isolation and characterization of lipid A using MS
A. baumannii 597A, E. coli BL21(λDE3)pLysS and the recombinant pET-26b-mcr-4.3 BL21(λDE3)pLysS were grown with shaking at 37°C in 250 mL of LB broth to an OD600 of 0.05 to 0.8–1.0. Cells were harvested and washed with 1× PBS. Isolation consisted of solvent extractions with chloroform, methanol and water, followed by mild acid hydrolysis.13 For A. baumannii, boiling time was increased to 1 h after mild acid hydrolysis. Dried samples were stored at −20°C until MS analysis.
Lipid extracts were analysed in the negative ion mode with a Synapt G2-Si High Definition Mass Spectrometer equipped with a nanoelectrospray ionization source (Waters, Milford, MA, USA). MS data acquisition and analysis were performed with MassLynx software (version 4.1, Waters).
Results and discussion
mcr-4.3 gene identified in colistin-resistant A. baumannii 597A
We identified the phosphoethanolamine transferase-encoding gene mcr-4.3 in the A. baumannii 597A clinical strain. The gene was located on a novel plasmid, pAb-MCR4.3, and embedded within a Tn3-family transposon (6504 bp) harbouring a resolvase and a Tn3-family transposase (Figure 1). The mcr-4.3 gene (1626 bp), previously annotated as mcr-4.2, shows 100% nucleotide identity to a putative sulphatase present in the chromosome of Shewanella frigidimarina NCIMB 400 (CP000447) (mcr-4).14 In China, the mcr-4.3 allele has been recently identified in A. baumannii recovered from pig faeces (accession number MK016505.1) and in a clinical Enterobacter cloacae (NG_057461) isolate.7,15 Five additional mcr-4 alleles from Salmonella enterica and Shiga toxin-producing E. coli have been reported in swine farms in Italy and Spain.16,17
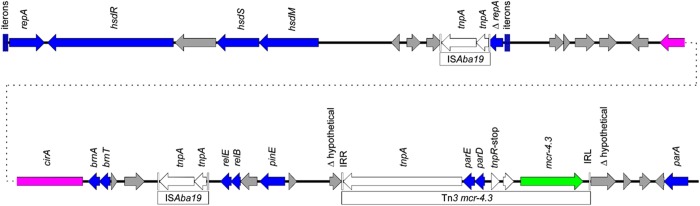
Figure 1.
Schematic representation of the complete sequence of pAb-MCR4.3. Grey arrows indicate confirmed or putative ORFs and their orientations. Arrow size is proportional to predicted ORF length. The Tn3-like transposon carrying mcr-4.3 is highlighted for emphasis, and mcr-4.3 is represented by a green arrow. Transposon genes are indicated with white arrows. Blue arrows indicate genes involved in plasmid replication or maintenance. Pink arrows indicate genes involved in other metabolic functions (cirA from pAb-MCR4.3 is involved in iron acquisition and is a putative virulence factor). TSD, TTTCT; IRL, GGCTAAATTTGCCCAACGCGCCAAAAATGTAAAGTTAGC; and IRR, GGCTAATTTTGCCCAACGCGCCAAAAATGTAAAGTTAGC. Gene nomenclature is based on the closest BLAST match from the NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi). This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Another mcr-4.3 sequence was found in the NCBI database in Acinetobacter nosocomialis (accession number MG948623.2) without related scientific publications.
pAb-MCR4.3 is 35 502 bp in length and harbours two putative type II toxin–antitoxin systems (relBE and parDE); however, the parDE locus was found on the Tn3 mcr4.3 transposon. A GR22 replicase gene of the Rep-3 superfamily is found on pAb-MCR4.3 based on 93% sequence identity of the replicase repB to repAci22. Interestingly, the replicase from GR22 was encoded by a plasmid detected in a carbapenem-resistant A. baumannii assigned to the MLST CC104 isolated in Argentina.18 However, none of the CC104 A. baumannii isolates was colistin resistant or carried mcr.
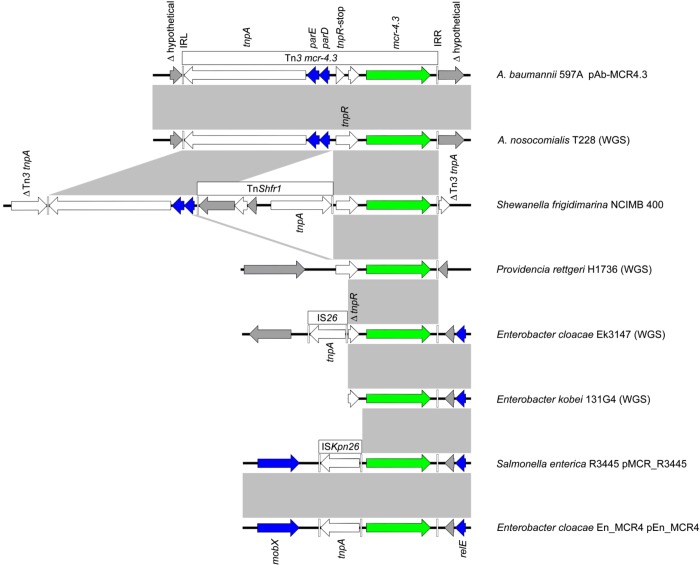
The Tn3-family transposon harbouring mcr-4.3 is characterized by inverted repeats and a 5 bp target site duplication (TSD) (TTTCT). Similar TSDs were found in the mcr-4.3 region from the S. frigidimarina chromosome NCIMB 400 (accession number CP000447). mcr-4 variants are found closely related to the Tn3-family transposons or truncated sequences (CP000447, MF543359, MH061380, CVLT01000089, JRUA01000041, QKNF01000006 and QMCU01000033). Other mcr genes including mcr-1 and mcr-2 are found in composite transposons, while mcr-3.5 and mcr-5 are embedded within Tn3-family and Tn21-subfamily transposons, respectively.19–23 Subsequent genetic events have resulted in gene loss or gain due to the movement of other mobile genetic elements including IS26 and ISKpn26 (Figure 2).
Figure 2.
Alignment of mcr-4.3. Comparison of the Tn3-family structure carrying mcr-4.3 in pAb-MCR4.3 from A. baumannii 597A and seven other sequences obtained from the NCBI. Subsequent genetic events in and around the Tn3-family structure are evidenced, including deletions, insertions and rearrangements. Arrow size is proportional to predicted ORF length and the colour scheme is the same as that used in Figure 1. The grey-shaded rectangles indicate areas of identity >95% between each sequence. WGS results of A. baumannii 597A and pAb-MCR4.3 were deposited in NCBI GenBank with accession numbers CP033869 and CP033872, respectively. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Cloning and lipid A structure experiments confirmed the function of the mcr-4 gene
As a phosphoethanolamine transferase, MCR catalyses the attachment of phosphoethanolamine to lipid A of LPS. A. baumannii 597A lipid A MS spectra had the main ion peak at m/z=2034 consisting of the addition of phosphoethanolamine (+124 Da) to the heptacylated bisphosphorylated form of lipid A (m/z=1910) (Figure S1).
We used the expression vector pET-26b (+) to assess the function of the mcr-4.3 gene. The colistin MIC for the recipient E. coli strain increased from 0.0125 to 8 mg/L, indicating that the gene conferred phenotypic resistance. Furthermore, the mass spectrum of E. coli BL21(λDE3)pLysS carrying the plasmid exhibited an ion peak at m/z=1920, indicating the addition of phosphoethanolamine to lipid A (m/z=1796; Figure S1). Our findings contradict the lack of functionality of the mcr-4.3 gene previously observed in E. cloacae strains carrying the gene, where no phosphoethanolamine addition to lipid A or increased colistin MIC was reported.13–20 In our additional analysis, multiple zinc-binding sites (E240, T278, H377, D452 and H453) were conserved in MCR-4 variants as compared with MCR-1 and MCR-2 that could support the enzymatic function of the protein.24
Mutations in genes related to colistin resistance and other antimicrobial resistance genes
In Acinetobacter species, colistin resistance has primarily been mediated by lipid A modification with the addition of phosphoethanolamine due to mutations in the pmrCAB operon.1 However, no mutations in the pmrCAB operon were identified in A. baumannii strain 597A. An additional pmrC-related gene, defined as a phosphoethanolamine transferase-encoding gene (eptA-1), was found in the chromosome of the 597A strain with 50.5% nucleotide identity to the mcr-4.3 gene. The phosphoethanolamine transferase EptA and MCR catalytic domains are highly conserved in A. baumannii; however, their different functionality related to the mechanism of colistin resistance is unclear.24 No mutations that have been implicated in A. baumannii colistin resistance were found in lpxCAD (lipid A biosynthesis pathway proteins), naxD (deacetylase), lpsB (glycosyltransferase), lolA (OM lipoprotein), lolB (OM lipoprotein), pldA (OM phospholipase A) or ttg2E (putative toluene tolerance protein).25 However, we cannot exclude that additional secondary unknown mechanisms of colistin resistance could be associated with the high-level colistin resistance in A. baumannii 597A.
Multiple antimicrobial resistance-encoding genes, including resistance to β-lactams (blaOXA-23 and blaTEM-1A), aminoglycosides [strB, strA, aph(3')-Vla and aadA1], sulphonamides (sul2) and trimethoprim (dfrA1), were identified in A. baumannii 597A. The antimicrobial resistance genotype correlated with the phenotype.
Conclusions
We provide evidence that colistin resistance in A. baumannii 597A strain CC113/79 is mediated by mcr-4.3. The gene was carried on a novel plasmid within a Tn3-family transposon.
Supplementary Material
Acknowledgements
We would like to thank the staff of the Multidrug-Resistant Organism Repository and Surveillance Network (MRSN) at the Walter Reed Army Institute of Research (Silver Spring, MD, USA) for WGS support and analysis.
Funding
This work was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), INPRA - Instituto Nacional de Pesquisa em Resistência Antimicrobiana—Brazil, CNPq 465718/2014-0, Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES)-PNPD and Finance Code 001, Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ). D.E.X. received financial support from Programa de Excelência em Pesquisa-CNPq/Fundação Oswaldo Cruz (PROEP/FIOCRUZ) (grant # APQ-1549-2.12/15). The QB3/Chemistry Mass Spectrometry Facility at the University of California (Berkeley, CA, USA) receives support from the National Institutes of Health (grant number 1S10OD020062-01).
Transparency declarations
None to declare.
References
- 1. Lima WG, Alves MC, Cruz WS. et al. Chromosomally encoded and plasmid-mediated polymyxins resistance in Acinetobacter baumannii: a huge public health threat. Eur J Clin Microbiol Infect Dis 2018; 37: 1009–19. [DOI] [PubMed] [Google Scholar]
- 2. Liu Y-Y, Wang Y, Walsh TR. et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 2016; 16: 161–8. [DOI] [PubMed] [Google Scholar]
- 3. Wang R, van Dorp L, Shaw LP. et al. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat Commun 2018; 9: 1179.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Partridge SR, Di Pilato V, Doi Y. et al. Proposal for assignment of allele numbers for mobile colistin resistance (mcr) genes. J Antimicrob Chemother 2018; 73: 2625–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carroll LM, Gaballa A, Guldimann C. et al. Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype Typhimurium isolate. mBio 2019; 10: e00853-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Caselli E, D’Accolti M, Soffritti I. et al. Spread of mcr-1-driven colistin resistance on hospital surfaces. Emerg Infect Dis 2018; 24: 1752–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ma F, Shen C, Zheng X. et al. Identification of a novel plasmid carrying mcr-4.3 in an Acinetobacter baumannii strain in China. Antimicrob Agents Chemother 2019; 63: e00133-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rossi F, Girardello R, Morais C. et al. Plasmid-mediated mcr-1 in carbapenem-susceptible Escherichia coli ST156 causing a blood infection: an unnoticeable spread of colistin resistance in Brazil? Clinics 2017; 72: 642–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kieffer N, Nordmann P, Moreno AM. et al. Genetic and functional characterization of an MCR-3-like enzyme-producing Escherichia coli isolate recovered from swine in Brazil. Antimicrob Agents Chemother 2018; 62: e00278-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lentz SA, de Lima-Morales D, Cuppertino VM. et al. Letter to the editor: Escherichia coli harbouring mcr-1 gene isolated from poultry not exposed to polymyxins in Brazil. Euro Surveill 2016; 21: pii=30267. [DOI] [PubMed] [Google Scholar]
- 11. Coelho-Souza T, Reis JN, Martins N. et al. Longitudinal surveillance for meningitis by Acinetobacter in a large urban setting in Brazil. Clin Microb Infect 2013; 19: E241–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bertini A, Poirel L, Mugnier PD, Villa L. et al. Characterization and PCR-based replicon typing of resistance plasmids in Acinetobacter baumannii. Antimicrob Agents Chemother 2010; 54: 4168–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Henderson JC, O’Brien JP, Brodbelt JS. et al. Isolation and chemical characterization of lipid A from Gram-negative bacteria. J Vis Exp 2013; e50623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Teo JWP, Kalisvar M, Venkatachalam I. et al. mcr-3 and mcr-4 variants in carbapenemase-producing clinical Enterobacteriaceae do not confer phenotypic polymyxin resistance. J Clin Microbiol 2018; 56: e01562-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chavda B, Lv J, Hou M. et al. Coidentification of mcr-4.3 and blaNDM-1 in a clinical Enterobacter cloacae isolate from China. Antimicrob Agents Chemother 2018; 62: e00649-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Carattoli A, Villa L, Feudi C. et al. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill 2017; 22: pii=30589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. García V, García-Meniño I, Mora A. et al. Co-occurrence of mcr-1, mcr-4 and mcr-5 genes in multidrug-resistant ST10 enterotoxigenic and Shiga toxin-producing Escherichia coli in Spain (2006-2017). Int J Antimicrob Agents 2018; 52: 104–8. [DOI] [PubMed] [Google Scholar]
- 18. Cameranesi MM, Limansky AS, Morán-Barrio J. et al. Three novel Acinetobacter baumannii plasmid replicase-homology groups inferred from the analysis of a multidrug-resistant clinical strain isolated in Argentina. J Infect Dis Epidemiol 2017; 3: 046. [Google Scholar]
- 19. Snesrud E, McGann P, Chandler M.. The birth and demise of the ISApl1-mcr-1-ISApl1 composite transposon: the vehicle for transferable colistin resistance. mBio 2018; 9: e02381-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Snesrud E, Maybank R, Kwak YI. et al. Chromosomally encoded mcr-5 in colistin-nonsusceptible Pseudomonas aeruginosa. Antimicrob Agents Chemother 2018; 62: e00679-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Partridge SR. mcr-2 in the IncX4 plasmid pKP37-BE is flanked by directly oriented copies of ISEc69. J Antimicrob Chemother 2017; 72: 1533–5. [DOI] [PubMed] [Google Scholar]
- 22. Liu L, Feng Y, Zhang X. et al. New variant of mcr-3 in an extensively drug-resistant Escherichia coli clinical isolate carrying mcr-1 and blaNDM-5. Antimicrob Agents Chemother 2017; 61: e01757-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hammerl JA, Borowiak M, Schmoger S. et al. mcr-5 and a novel mcr-5.2 variant in Escherichia coli isolates from food and food-producing animals, Germany, 2010 to 2017. J Antimicrob Chemother 2018; 73: 1433–5. [DOI] [PubMed] [Google Scholar]
- 24. Huang J, Zhu Y, Han M-L. et al. Comparative analysis of phosphoethanolamine transferases involved in polymyxin resistance across 10 clinically relevant Gram-negative bacteria. Int J Antimicrob Agents 2018; 51: 586–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Henry R, Crane B, Powell D. et al. The transcriptomic response of Acinetobacter baumannii to colistin and doripenem alone and in combination in an in vitro pharmacokinetics/pharmacodynamics model. J Antimicrob Chemother 2015; 70: 1303–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.