Abstract
Background
Loss-of-function variants in RAD51C are associated with familial ovarian cancer, but its role in hereditary breast cancer remains unclear. The aim of this study was to couple breast tumor sequencing with case-control data to clarify the contribution of RAD51C to hereditary breast cancer.
Methods
RAD51C was sequenced in 3080 breast cancer index cases that were negative in BRCA1/2 clinical tests and 4840 population-matched cancer-free controls. Pedigree and pathology data were analyzed. Nine breast cancers and one ovarian cancer from RAD51C variant carriers were sequenced to identify biallelic inactivation of RAD51C, copy number variation, mutational signatures, and the spectrum of somatic mutations in breast cancer driver genes. The promoter of RAD51C was analyzed for DNA methylation.
Results
A statistically significant excess of loss-of-function variants was identified in 3080 cases (0.4%) compared with 2 among 4840 controls (0.04%; odds ratio = 8.67, 95% confidence interval = 1.89 to 80.52, P< .001), with more than half of the carriers having no personal or family history of ovarian cancer. In addition, the association was highly statistically significant among cases with estrogen-negative (P <. 001) or triple-negative cancer (P < .001), but not in estrogen-positive cases. Tumor sequencing from carriers confirmed bi-allelic inactivation in all the triple-negative cases and was associated with high homologous recombination deficiency scores and mutational signature 3 indicating homologous recombination repair deficiency.
Conclusions
This study provides evidence that germline loss-of-function variants of RAD51C are associated with hereditary breast cancer, particularly triple-negative type. RAD51C-null breast cancers possess similar genomic and clinical features to BRCA1-null cancers and may also be vulnerable to DNA double-strand break inducing chemotherapies and poly ADP-ribose polymerase inhibitors.
RAD51C is one of the five paralogs of RAD51 and is essential for DNA double-strand break repair by homologous recombination (HR). Bi-allelic loss-of-function (LoF) germline variants in RAD51C are responsible for Fanconi anemia-type complementation group O (1), whereas mono-allelic variants have been reported at a low frequency (1.3%) in families with a history of both breast and ovarian cancer (2), but rarely among families with a history of breast cancer only. Although studies have confirmed RAD51C as an ovarian cancer susceptibility gene, evidence for a role in breast cancer remains equivocal (3–9). Resolving the spectrum of cancers associated with pathogenic RAD51C germline variants is important for managing cancer risks in such families. However, given the rarity of RAD51C variants in most populations, current case-control studies remain substantially underpowered to establish a clear role for RAD51C in breast cancer predisposition in isolation. Data from genomic analysis of tumors from carriers of germline variants in candidate genes can provide powerful additional evidence for involvement of a gene in cancer predisposition. Characteristic somatic inactivation events and “mutational signatures” have recently been demonstrated for tumors from carriers of mutations in ATM and PALB2 (10,11). In this study, sequencing data from breast cancer-affected cases in hereditary breast and ovarian cancer (HBOC) families and population-matched cancer-free controls was combined with tumor sequencing data to investigate the role of RAD51C in breast cancer.
Methods
Study Subjects and Sequencing
Case subjects were female index patients diagnosed with breast cancer from 3080 HBOC families that were negative for BRCA1 and BRCA2 pathogenic variants, and were ascertained from the Variants in Practice (ViP) Study from the combined Victorian and Tasmanian Familial Cancer Centres, Australia. Control subjects were 4840 women from the Lifepool study that were cancer free as of September 2017. The average age at diagnosis of the cases and the average age of controls were 45.8 years (range = 17–85 years) and 64.4 years (range = 40–97 years), respectively. This study was approved by the human research ethics committees at each participating ViP study recruitment center and the Peter MacCallum Cancer Centre (approval no. 09/29). All participants provided informed consent for genetic analysis of their germline and tumor DNA.
Germline DNA were sequenced for the coding region and exon-intron boundaries (≥10 bp) of RAD51C using a custom-designed HaloPlex Targeted Enrichment Assay panel (Agilent Technologies, Santa Clara, CA) as described previously (12–15). Tumor DNA was extracted from cancer cells in formalin-fixed, paraffin-embedded slides by needle microdissection and sequenced using an Agilent SureSelect XT Custom Panel that targeted all exons of RAD51C and an additional 487 genes (1.337 Mb total targeted region) including 27 breast cancer driver genes (16).
Statistical Analysis
To analyze data from the case and control study, the conditional maximum likelihood estimate was used to calculate odds ratios (ORs) with 95% confidence intervals (CIs), and the Fisher exact test was used to calculate P values [R 3.3.2 was used (17)]. The Mann–Whitney U test was performed for homologous recombination deficiency (HRD) score comparisons between groups of tumors in GraphPad Prism version 7.00 (California). A P value of less than .05 was considered statistically significant, and all tests were 2-sided.
Results
Frequency of Germline RAD51C Variants in HBOC Families and Controls
Breast cancer-affected index cases from 3080 HBOC families and 4840 controls (cancer free as of September 2017) from the Australian population were sequenced for all exons of RAD51C, at average sequencing depths of 147X and 170X, respectively. Overall, 98.7% of targeted bases in the cases and 99.4% in the controls were sequenced to a depth of more than 10-fold. LoF variants were identified in 11 cases (0.4%) and 2 controls (0.04%), suggesting a statistically significant enrichment in the familial cases (OR = 8.67, 95% CI = 1.89 to 80.52, P < .001) (Table 1). Seven of the 10 unique variants identified in this study were previously reported as pathogenic or likely pathogenic, associated with a hereditary cancer syndrome, in the ClinVar database.
Table 1.
RAD51C variants identified in cases and controls study
| CDS change* | Protein change† | Consequence | Case | Control | Exon (of 9) | Intron (of 8) | MAF in gnomAD‡ | Clinical significance§ |
|---|---|---|---|---|---|---|---|---|
| c.68_72dup | p.Val25CysfsTer3 | Frameshift | 1 | 0 | 1 | N/A | 0 | N/A |
| c.146-4_146-2delTCA | N/A | Splice acceptor | 1 | 0 | N/A | 1 | 0 | LP |
| c.394dupA | p.Thr132AsnfsTer23 | Frameshift | 2 | 0 | 2 | N/A | 3.77 × 10−05 | N/A |
| c.397C>T | p.Gln133Ter | Stop gained | 0 | 1 | 3 | N/A | 0 | P |
| c.572-1G>T | N/A | Splice acceptor | 1 | 0 | N/A | 3 | 4.22 × 10−06 | N/A |
| c.577C>T | p.Arg193Ter (rs200293302) | Stop gained | 1 | 1 | 4 | N/A | 3.38 × 10−05 | P |
| c.705 + 1G>A | N/A | Splice donor | 1 | 0 | N/A | 4 | 0 | LP |
| c.706-2A>G | N/A | Splice acceptor | 2 | 0 | N/A | 4 | 0 | P/LP |
| c.904 + 5G>T | N/A | Splice donor | 1 | 0 | N/A | 6 | 2.24 × 10−05 | LP |
| c.905-2_905-1delAG | N/A | Splice acceptor | 1 | 0 | N/A | 6 | 0 | P/LP |
| Total | N/A | N/A | 11 | 2 | N/A | N/A | N/A | N/A |
ENST00000337432 (NM 058216.1). CDS = coding sequence; LP = likely pathogenic; MAF = minor allele frequency; N/A = not applicable; P = pathogenic.
ENSP00000336701(NP 478123.1).
Minor allele frequency in noncancer cohorts, GnomAD v2.1.
Clinical significance reviewed by ClinVar (online database downloaded on July 18, 2018).
The average age at the first breast cancer diagnosis in RAD51C carriers was 44.0 years (range = 26–60) and all were grade 2 or 3 invasive ductal carcinoma, with a proportion (7 of 11) lacking expression of estrogen (ER), progesterone (PR), and HER2 receptors (triple-negative [TN]) as summarized in Table 2.
Table 2.
Cancer diagnosis, pathology, and family history of RAD51C case carriers
| Case | RAD51C variants* | Breast cancer diagnosis |
Other cancer diagnosis |
Family history† |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age, y | Histology | ER | PR | HER2 | Age, y | Site | Histology | Total no. of relatives | Breast cancer | Ovarian cancer | ||
| 3 | p.Thr132AsnfsTer23 | 29, 48 | IDC, G3‡ | ER− | PR− | HER2− | 37 | Thyroid | N/A | 23 | 1 | 0 |
| 4 | c.905-2_905-1delAG | 43 | IDC, G3 | ER− | PR− | HER2− | 26 | Cervix | N/A | 34 | 3 | 0 |
| 5 | c.572-1G>T | 46 | IDC, G3 | ER− | PR− | HER2− | N/A | N/A | N/A | 26 | 2 | 0 |
| 7 | c.706-2A>G | 50 | IDC, G3 | ER− | PR− | HER2− | N/A | N/A | N/A | 33 | 5 | 1 |
| 8 | c.706-2A>G | 40 | IDC, G3 | ER− | PR− | HER2− | N/A | N/A | N/A | 16 | 0 | 0 |
| 9 | p.Arg193Ter | 32 | IDC, G3 | ER− | PR− | HER2− | N/A | N/A | N/A | 20 | 1 | 0 |
| 11 | c.904 + 5G>T | 55 | IDC, G3 | ER− | PR− | HER2− | N/A | N/A | N/A | 13 | 1 | 1 |
| 1 | p.Thr132AsnfsTer23 | 26 | IDC, G2 | ER− | PR− | HER2+ | N/A | N/A | N/A | 46 | 0 | 0 |
| 10 | p.Val25CysfsTer3 | 53 | IDC, G2 | ER+ | PR− | HER2+ | 52 | Ovary | HGS | 29 | 0 | 0 |
| 2 | c.705 + 1G>A | 50 | IDC, G2 | ER+ | PR+ | HER2− | N/A | N/A | N/A | 61 | 2 | 1 |
| 6 | c.146-4_146-2delTCA | 60 | IDC, G2 | ER+ | PR+ | HER2− | 45 | Ovary | HGS | 54 | 2 | 0 |
ENST00000337432 (NM 058216.1), ENSP00000336701(NP 478123.1). ER = estrogen receptor; G = grade; HER2 = human epidermal growth factor receptor 2; HGS = high-grade serous ovarian carcinoma; IDC = invasive ductal carcinoma; N/A = not applicable; PR = progesterone receptor.
Breast cancer and ovarian cancer affected cases in first-, second-, and third-degree relatives of the cases.
Histology and hormone receptor status data in the table are for the second breast cancer diagnosis. The first cancer was high-grade ER-negative breast cancer with no pathology report available.
Subgroup Analysis by Ovarian Cancer Family History and Hormone Receptor Status
To assess if these results could be explained by the known association with an increased risk of ovarian cancer, we considered the distribution of RAD51C variants according to personal and family cancer history of the case cohort. Of the cohort, 21% (638 of 3080 cases) had either a personal and/or a family history of one or more ovarian cancer diagnoses in any first- to third-degree relative. Five RAD51C carriers were identified among the 638 breast and ovarian cancer families (0.8%) compared with 6 among the 2442 breast cancer-only families (0.2%), indicating ovarian cancer family history explained at least part of the association of RAD51C with breast cancer (Table 3). Nevertheless, more than half (6 of 11) of the RAD51C carrier families did not have any history of ovarian cancer, which remained statistically significantly different to the control carrier frequency of 0.04% (OR = 5.96, 95% CI = 1.06 to 60.42, P = .02).
Table 3.
Subgroup analysis of the association between RAD51C and breast cancer by ovarian cancer family history and hormone receptor status
| Personal/family history | Familial breast cancer cases |
Cancer-free controls |
OR (95% CI)* | P† | ||||
|---|---|---|---|---|---|---|---|---|
| RAD51C | Frequency, % | Total | RAD51C | Frequency, % | Total | |||
| All subjects | 11 | 0.4 | 3080 | 2 | 0.04 | 4840 | 8.67 (1.89 to 80.52) | <.001 |
| Have OvCa history‡ | 5 | 0.8 | 638 | 2 | 0.04 | 4840 | 19.09 (3.12 to 200.42) | <.001 |
| Excluding OvCa history§ | 6 | 0.2 | 2442 | 2‖ | 0.04 | 4840 | 5.96 (1.06 to 60.42) | .02 |
| ER+ breast cancer | 3 | 0.2 | 1726 | 2 | 0.04 | 4840 | 4.21(0.48 to 50.44) | .12 |
| ER− breast cancer | 8 | 0.8 | 939 | 2 | 0.04 | 4840 | 20.77 (4.14 to 200.58) | <.001 |
| Triple-negative breast cancer | 7 | 1.1 | 626 | 2 | 0.04 | 4840 | 27.33 (5.19 to 268.54) | <.001 |
The ORs for each subgroup analysis were calculated using the reference group of cancer-free controls. CI = confidence interval; ER = estrogen; OR = odds ratio; OvCa = ovarian cancer.
Fisher exact test, 2-sided.
Ovarian cancer diagnosis in the index case of one or more first- to third-degree relatives.
No ovarian cancer diagnosis in the index case or any first- to third-degree relative.
Both of these control carriers reported having first- and/or second-degree relatives diagnosed with breast and/or ovarian cancer: One control carrier reported a sister diagnosed with breast cancer (age 45 years) and another sister diagnosed with ovarian cancer (age unknown). The other control carrier reported her mother was diagnosed with both breast cancer (age 30 years) and ovarian cancer (age 49) and that “multiple” second-degree relatives were diagnosed with breast or ovarian cancer (age unknown).
We examined the association between RAD51C and hereditary breast cancer by hormone receptor status. Among the cases where ER status was available (2699 of 3080), 1726 (63.9%) were ER-positive, 939 (34.8%) were ER-negative, and 34 were removed from the analysis because they were diagnosed with multiple primary breast cancers with both ER-positive and ER-negative tumors. In the ER-positive group, 0.2% were RAD51C variant carriers, which was not statistically significant (OR = 4.21, 95% CI = 0.48 to 50.44, P = .12) (Table 3). In contrast, 0.8% of the ER-negative group were RAD51C carriers, which was highly statistically significant (OR = 20.77, 95% CI = 4.14 to 200.58, P<.001), and this association was even stronger when considering only the subgroup of TN breast cancer cases, with 1.1% being variant carriers (OR = 27.33, 95% CI = 5.19 to 268.54, P < .001). However, the confidence intervals are broad and the estimates of the odds ratios need to be interpreted with caution.
A similar trend for an excess of missense variants was observed in the cases, particularly for TN cancers (Supplementary Tables 1 and 2, available online). The strength of the association varied with the variant classification tool used, but none of the comparisons would be considered statistically significant when accounting for multiple testing.
Bi-Allelic Inactivation Analysis in RAD51C Tumors
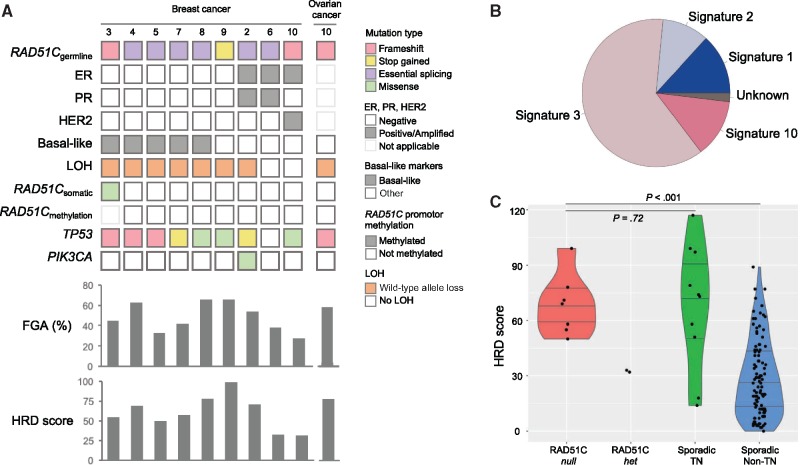
The status of the wild-type RAD51C allele was examined in tumor DNA from nine breast cancers and one ovarian cancer from nine families. Sequencing was performed using a custom gene panel that included all exons of RAD51C to an average depth of 396x. Seven of the nine breast tumors (77.8%) and the high-grade ovarian cancer showed loss of heterozygosity (LOH) across the RAD51C locus and all lost the wild-type allele (RAD51C-null). Six of the seven RAD51C-null breast cancers were TN cancers (Figure 1, A) and five of these were confirmed as basal-like type based on positive immunohistochemical staining for CK5 and EGFR. The two breast tumors that retained heterozygosity across the RAD51C locus were an ER-positive, PR-positive tumor and an ER-positive, HER2-positive tumor. Bisulfite sequencing excluded promoter hypermethylation-mediated silencing of RAD51C in these two tumors, and no somatic RAD51C point mutations were identified, confirming that they only had mono-allelic inactivation of RAD51C (RAD51C-het). As expected, promoter hypermethylation was not detected in any of the RAD51C-null breast cancers. A somatic missense mutation (p.Leu363Met) was identified in one case, but in silico predictions indicate that this is likely to be a benign passenger mutation.
Figure 1.
Genomic characterization of breast and ovarian cancers from carriers of RAD51C germline loss-of-function variants. A) Germline variants, bi-allelic inactivation events, somatic mutations, and genomic alterations of RAD51C-associated tumors. Germline and somatic mutation types are color-coded according to the legend. The phenobar (top) provides information about estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) status, basal-like subtype, loss of heterozygosity (LOH) of the wild-type allele, and somatic mutations in TP53 and PIK3CA. The fraction of the genome altered (FGA) and homologous recombination deficiency (HRD) score are shown for each case below. B) The weighted contribution of mutational signatures from breast cancers of RAD51C germline variant carriers. C) HRD scores of RAD51C-null (n = 7), RAD51C-het (n = 2), sporadic TN breast cancers (n = 10), and sporadic non-TN breast cancers (n = 105). P values were calculated using Mann–Whitney test, 2-tailed. ER = estrogen receptor; FGA = fraction of genome altered; HER2 = human epidermal growth factor receptor 2; HRD = homologous recombination deficiency; LOH = loss of heterozygosity; PR = progesterone receptor; TN = triple-negative.
Overall, 77.8% of the breast tumors from germline RAD51C variant carriers were established to have bi-allelic inactivation of RAD51C, and this were associated with TN breast cancers (100% of the TN breast tumors were RAD51C-null).
Somatic Mutations and Mutational Signature in RAD51C Tumors
Sequencing of the RAD51C-associated cancers identified TP53 as the most common somatically mutated gene found in eight of nine breast tumors (88.9%), including all RAD51C-null tumors and one of the two RAD51C-het tumors. One PIK3CA mutation was detected in an ER-positive RAD51C-null tumor, and one PTEN and one RB1 mutation was observed individually in one TN RAD51C-null tumor.
Somatic mutational signature 3 is a hallmark of tumors with BRCA1 or BRCA2 mutations (18) and is a robust indicator of HR DNA repair deficiency, particularly in tumors that harbor alterations in genes in the same complex of BRCA1/BRCA2-associated DNA repair (eg, PALB2, RAD51C, and BARD1) (19). To evaluate the common mutational processes underlying the RAD51C tumors, we used a pooled sample mutational signature method (10) to overcome the limitation of the small number of somatic variants per sample detected by targeted sequencing. The profile of mutational signatures was generated from 44 single-nucleotide substitution variants identified in nine RAD51C breast tumors and confirmed a predominant mutational signature 3 (Figure 1B) that was consistent with the RAD51C-associated HR deficiency.
Genomic Instability and HRD Score of RAD51C Tumors
Tumors demonstrating HR deficiency (including tumors associated with bi-allelic inactivation of BRCA1, BRCA2, or PALB2 through the combination of a germline pathogenic variant and a somatic inactivation event) have been shown to be associated with an increased level of genomic instability (20) and elevated rates of large-scale chromosomal aberrations (21,22). To further confirm that RAD51C bi-allelic inactivation drives breast cancer tumorigenesis through HR deficiency, we evaluated two HR deficiency measures using the genome-wide copy number data. First, we calculated the fraction of genome altered (FGA) (23,24) to measure the degree of broad-range genomic instability, previously reported as the genomic instability index (GII) (25); and second, we calculated an HRD score to measure the HR-deficiency-specific genomic aberrations (26,27), combined from three HRD score components: NtAI (number of telomeric allelic imbalances) (28), HRD-LOH (29), and large-scale state (LST) transitions (18) (Figure 1, A and C). RAD51C-null tumors all had a high level of genomic instability (FGA median = 53.6%, 33.3%–66.5%) and high HRD scores (median 70, 50–99) that exceeded the threshold for HR deficiency proposed for BRCA1-driven breast cancer (20,27). In contrast, neither of the two RAD51C-het tumors met the criteria for HR deficiency (HRD = 33 and 32, FGA = 37.8% and 27.9%, respectively) (Figure 1, A and C). Compared with a cohort of 115 sporadic breast cancers that were sequenced on the same platform, RAD51C-null breast tumors had HRD scores in the upper range observed for the 10 sporadic TN breast cancers and much higher than the 105 non-TN breast cancers (median 69 vs 24, P < .001, Mann–Whitney test, 2-sided) (Figure 1, C). The fraction of the genome that was altered showed good concordance with the HRD score, matching the previously reported positive association (21). The copy number profiles of the RAD51C-null tumors were similar to those described for BRCA1-driven tumors (30) and BRCA-like PALB2-driven breast cancers we previously reported (10). These tumors were also similar to sporadic TN breast cancers but distinct from non-TN breast cancers, for example, frequent gains of 3q, 5p, and 10p and loss of 5q (Supplementary Figure 1, available online). The two RAD51C-het tumors had few alterations and appeared similar to sporadic non-TN breast cancers (Supplementary Figures 1 and 2, available online).
Discussion
RAD51C was first reported as a predisposition gene to HBOC in 2010 (2), and subsequent studies have confirmed its role in ovarian cancer. The recent large-scale study by Song et al. (9) provided strong evidence that RAD51C is a moderate-risk ovarian cancer susceptibility gene and suggested that it should be included alongside BRCA1 and BRCA2 in clinical genetic tests for this indication. In contrast, the relevance of RAD51C to breast cancer predisposition remains unclear. A few studies have reported supportive evidence (3,7,8), whereas others suggested little or no increased risk for breast cancer (4,6,31). This uncertainty is predominantly due to the rarity of pathogenic variants in the population (we observed two carriers, 0.04%, in 4840 cancer-free Australia women), which severely limits the statistical power of case-control studies. To overcome these limitations, augmentation of case-control and family history data with information about the somatic landscape of the tumors occurring in carriers of rare pathogenic variants has recently been used to provide a detailed understanding of the role of ATM and PALB2 in breast cancer predisposition (10,11). Using a similar approach, the data generated in this study have provided strong evidence to conclude that RAD51C is a genuine breast cancer predisposition gene that typically undergoes bi-allelic inactivation and is specifically highly associated with TN breast cancers. With detailed, validated family cancer histories available for our cohort, we were able to demonstrate that the association of RAD51C with breast cancer predisposition was not a consequence of enrichment of ovarian cancer families, as might be anticipated in a clinical cohort selected for BRCA-like features; 6 of the 11 carriers had no personal or family history of ovarian cancer, and there was a similar excess of RAD51C LoF variants compared with controls among the overall familial cases and the breast cancer-only families (8.67-fold vs 5.96-fold). The data shows that RAD51C conforms to Knudson’s 2-hit model with loss of the wild-type allele in all the TN breast cancers, one ER-positive breast cancer and the high-grade serous ovarian cancer, consistent with previous observations (2,6). Together with the high level of genome alteration and HRD scores, the predominant mutational signature 3, and a pattern of copy number alterations consistent with BRCA-like tumors, we conclude that RAD51C-driven breast cancer tumorigenesis occurs via homologous recombination deficiency. Notably, the high proportion of TN breast cancers among carriers and the fact that all TN breast cancers have bi-allelic inactivation strongly links RAD51C LoF variants with TN breast cancer predisposition.
This connection is in agreement with the recent finding of germline variants of RAD51C associated with elevated risk in a TN breast cancer cohort (32) and the description of enrichment for epigenetic silencing of RAD51C in basal-like breast cancers in young individuals of African descent (19). Shimelis et al. (32) reported 23 RAD51C pathogenic variant carriers (0.4%) in 6093 Caucasian TN breast cancer patients, and a slightly attenuated frequency was observed after excluding patients with ovarian cancer family history (9 in 3313 patients, 0.3%). Although using slightly different criteria to define pathogenicity (Shimelis et al. included “pathogenic or likely pathogenic” variants with the classification not described in detail, whereas only LoF variants were included in the current study), the frequencies of carriers were similar between Shimelis et al. and our case cohort (0.4% overall, and 0.2% excluding any ovarian cancer family history). However, the overall odds ratio for TN breast cancer reported by Shimelis et al. in their case-only study is lower than reported in our study, reflecting the relatively high number of RAC51C carriers in their population-comparison group of 26 647 ExAC controls, which included 37 (0.1%) reported carriers of pathogenic variants in RAD51C. By comparison, we observe fewer LoF carriers in our control cohort (2 in 4840, 0.04%) compared with the gnomAD database excluding cancer cohorts (142 in 118 423, 0.1%, gnomAD V2.1 noncancer). It is possible that RAD51C variants were underrepresented in our controls, potentially reflecting the healthy, aged nature of our control group (average age 64.4 years). Alternatively, gnomAD or ExAC may overestimate the frequency of RAD51C variants because of the combination of different cohorts, not controlled for age, and lack of cancer diagnosis update. Notably, Song et al. (9) reported 2 carriers identified in 2769 cancer-free individuals aged 35 years or older, which is similar to our controls. However, additional large-scale studies in different populations will be required to determine the frequency of RAD51C LoF variants in the general population.
Deficiencies in the HR pathway are known to sensitize breast tumors to drugs such as platinum compounds that induce DNA damage through DNA double-strand breaks (33). The use of platinum-based chemotherapy regimens in early breast cancer has increased based on recent data suggesting an improved likelihood for pathological complete response in high-risk TN breast cancers regardless of BRCA1 or BRCA2 status (34). In advanced TN breast cancer only, the presence of a germline BRCA1 or BRCA2 variant was associated with improved response to platinum-based chemotherapy (35). With conflicting outcome data together with the additional toxicities associated with the addition of platinum therapies to standard treatments, the ability to better identify the subgroups most likely to benefit is essential. Many TN tumors, particularly of the basal-like subtype, are known to have characteristic features of HR deficiency, and the HRD score has been shown to predict response to platinum-based chemotherapy in the neoadjuvant setting (27). Our results suggest that RAD51C germline variants result in many of the same tumor features previously associated with BRCA germline variants and have the potential to identify a further subgroup of women who may benefit from DNA double-strand break-inducing chemotherapies. Silencing of RAD51C expression in cancer cells has also been reported to be sufficient to induce sensitivity to the synthetic lethality effect of poly ADP-ribose polymerase (PARP) inhibitors (36). Recent clinical trials in ovarian cancer have confirmed that tumors harboring RAD51C variants were responsive to the PARP inhibitor Rucaparib (37,38), suggesting a potential therapeutic opportunity for other HR-deficient cancer types.
This study has two important limitations. First, the broad confidence intervals of the odds ratios indicate that the study is underpowered to give a stable estimate of the odds ratios of RAD51C LoF variants. The high odds ratios may be exaggerated as a result of the “extreme phenotype” cohort design of the study (familial and/or young onset cases vs aged cancer-free controls) and should not be considered as a relative risk, directly transferable to all carriers of RAD51C pathogenic variants in the population. Second, this study did not screen for large genomic rearrangements affecting RAD51C, although a previous study identified none in 141 non-BRCA1 and BRCA2 families, suggesting that this type of variant may be rare in RAD51C (39), in comparison to BRCA1 and BRCA2 (40).
Together, our findings indicate that although pathogenic germline variants in RAD51C are rare, they are associated with a statistically significantly increased risk, particularly of triple-negative breast cancers, and this information has the potential to greatly benefit patients not only in effective risk management but also in improved cancer treatment.
Funding
This work was supported by the National Breast Cancer Foundation, the Victorian Cancer Agency, and the National Health and Medical Research Council.
Notes
Affiliations of authors: Cancer Genetics Laboratory, Peter MacCallum Cancer Centre, Melbourne, Victoria, Australia (NL, MZ, DC, BWXL, SMR, IGC); Sir Peter MacCallum Department of Oncology, University of Melbourne, Melbourne, Victoria, Australia (NL, DC, BWXL, SA, JL, SBF, KLG, PAJ, IGC); Parkville Familial Cancer Centre, Peter MacCallum Cancer Centre and Royal Melbourne Hospital, Melbourne, Victoria, Australia (SM, NG, PAJ); Bioinformatics Consulting Core, Peter MacCallum Cancer Centre, Melbourne, Victoria, Australia (MZ, JL); Lifepool, Peter MacCallum Cancer Centre, Melbourne, Victoria, Australia (LD); Department of Clinical Pathology, University of Melbourne, Melbourne, Victoria, Australia (DB, SBF, KLG, IGC); Molecular Genomics Core Facility, Peter MacCallum Cancer Centre, Melbourne, Victoria, Australia (JEAL); Familial Cancer Centre, Austin Health, Heidelberg, Victoria, Australia (TJ); Cabrini Family Cancer Clinic, Cabrini Hospital, Malvern, Victoria, Australia (YA); Cancer Genomics Program, Peter MacCallum Cancer Centre, Melbourne, Victoria, Australia (KLG).
The authors have no conflict of interest to declare.
PAJ and IGC contributed equally to this work.
The funders had no role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; and the decision to submit the manuscript for publication.
The authors thank Lyon Mascarenhas, Rebecca Driessen, the ViP study site principal investigators Geoffrey Lindeman, Marion Harris, and Ingrid Winship, and the staff at the Victorian and Tasmanian Familial Cancer Centres who enrolled participants and provided clinical data. We also thank all the participants of the ViP and Lifepool studies for donating their DNA samples and clinical information. We thank the following staff from Peter MacCallum Cancer Centre: Tim Semple, Gisela Mir Arnau from Molecular Genomics core facility for sequencing the tumor DNA; Kaushalya Amarasinghe, Niko Thio, and Richard Lupat from Bioinformatics core facility for helping with the bioinformatic analysis; Peter Mac Tissue Bank for preparing sections of tumor blocks; and Heather Thorne, Genna Glavich, and other staff from kConFab for assisting with obtaining the tumor blocks.
Supplementary Material
References
- 1. Vaz F, Hanenberg H, Schuster B, et al. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat Gen. 2010;42(5):406–409. [DOI] [PubMed] [Google Scholar]
- 2. Meindl A, Hellebrand H, Wiek C, et al. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet. 2010;42(5):410–414. [DOI] [PubMed] [Google Scholar]
- 3. Blanco A, Gutierrez ES, Santamarina M, et al. RAD51C germline mutations found in Spanish site-specific breast cancer and breast-ovarian cancer families. Breast Cancer Res Treat. 2014;147(1):133–143. [DOI] [PubMed] [Google Scholar]
- 4. Loveday C, Turnbull C, Ruark E, et al. Germline RAD51C mutations confer susceptibility to ovarian cancer. Nat Genet. 2012;44(5):475–476; author reply 476. [DOI] [PubMed] [Google Scholar]
- 5. Thompson ER, Boyle SE, Johnson J, et al. Analysis of RAD51C germline mutations in high-risk breast and ovarian cancer families and ovarian cancer patients. Hum Mutat. 2012;33(1):95–99. [DOI] [PubMed] [Google Scholar]
- 6. Pelttari LM, Heikkinen T, Thompson D, et al. RAD51C is a susceptibility gene for ovarian cancer. Hum Mol Genet. 2011;20(16):3278–3288. [DOI] [PubMed] [Google Scholar]
- 7. Clague J, Wilhoite G, Adamson A, et al. RAD51C germline mutations in breast and ovarian cancer cases from high-risk families. PLoS One. 2011;6(9):e25632.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jonson L, Ahlborn LB, Steffensen AY, et al. Identification of six pathogenic RAD51C mutations via mutational screening of 1228 Danish individuals with increased risk of hereditary breast and/or ovarian cancer. Breast Cancer Res Treat. 2016;155(2):215–222. [DOI] [PubMed] [Google Scholar]
- 9. Song H, Dicks E, Ramus SJ, et al. Contribution of germline mutations in the RAD51B, RAD51C, and RAD51D genes to ovarian cancer in the population. J Clin Oncol. 2015;33(26):2901–2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee JEA, Li N, Rowley SM, et al. Molecular analysis of PALB2-associated breast cancers. J Pathol. 2018;245(1):53–60. [DOI] [PubMed] [Google Scholar]
- 11. Weigelt B, Bi R, Kumar R, et al. The landscape of somatic genetic alterations in breast cancers from ATM germline mutation carriers. J Natl Cancer Inst. 2018;110(9):1030–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li N, Rowley SM, Thompson ER, et al. Evaluating the breast cancer predisposition role of rare variants in genes associated with low-penetrance breast cancer risk SNPs. Breast Cancer Res. 2018;20(1):3.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li N, Rowley SM, Goode DL, et al. Mutations in RECQL are not associated with breast cancer risk in an Australian population. Nat Genet. 2018;50(10):1346–1348. [DOI] [PubMed] [Google Scholar]
- 14. Thompson ER, Rowley SM, Li N, et al. Panel testing for familial breast cancer: calibrating the tension between research and clinical care. J Clin Oncol. 2016;34(13):1455–1459. [DOI] [PubMed] [Google Scholar]
- 15. Li N, Thompson ER, Rowley SM, et al. Reevaluation of RINT1 as a breast cancer predisposition gene. Breast Cancer Res Treat. 2016;159(2):385–392. [DOI] [PubMed] [Google Scholar]
- 16. Pereira B, Chin SF, Rueda OM, et al. The somatic mutation profiles of 2 433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. 2016;7:11479.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2016. [Google Scholar]
- 18. Popova T, Manie E, Rieunier G, et al. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res. 2012;72(21):5454–5462. [DOI] [PubMed] [Google Scholar]
- 19. Polak P, Kim J, Braunstein LZ, et al. A mutational signature reveals alterations underlying deficient homologous recombination repair in breast cancer. Nat Genet. 2017;49(10):1476–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stefansson OA, Jonasson JG, Johannsson OT, et al. Genomic profiling of breast tumours in relation to BRCA abnormalities and phenotypes. Breast Cancer Res. 2009;11(4):R47.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Knijnenburg TA, Wang L, Zimmermann MT, et al. Genomic and molecular landscape of DNA damage repair deficiency across the Cancer Genome Atlas. Cell Rep. 2018;23(1):239–254.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Watkins JA, Irshad S, Grigoriadis A, et al. Genomic scars as biomarkers of homologous recombination deficiency and drug response in breast and ovarian cancers. Breast Cancer Res. 2014;16(3):211.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Burrell RA, McClelland SE, Endesfelder D, et al. Replication stress links structural and numerical cancer chromosomal instability. Nature. 2013;494(7438):492–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chin SF, Teschendorff AE, Marioni JC, et al. High-resolution aCGH and expression profiling identifies a novel genomic subtype of ER negative breast cancer. Genome Biol. 2007;8(10):R215.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vollan HK, Rueda OM, Chin SF, et al. A tumor DNA complex aberration index is an independent predictor of survival in breast and ovarian cancer. Mol Oncol. 2015;9(1):115–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Marquard AM, Eklund AC, Joshi T, et al. Pan-cancer analysis of genomic scar signatures associated with homologous recombination deficiency suggests novel indications for existing cancer drugs. Biomark Res. 2015;3:9.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Telli ML, Timms KM, Reid J, et al. Homologous recombination deficiency (HRD) score predicts response to platinum-containing neoadjuvant chemotherapy in patients with triple-negative breast cancer. Clin Cancer Res. 2016;22(15):3764–3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Birkbak NJ, Wang ZC, Kim JY, et al. Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer Discov. 2012;2(4):366–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Abkevich V, Timms KM, Hennessy BT, et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br J Cancer. 2012;107(10):1776–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Joosse SA, van Beers EH, Tielen IH, et al. Prediction of BRCA1-association in hereditary non-BRCA1/2 breast carcinomas with array-CGH. Breast Cancer Res Treat. 2009;116(3):479–489. [DOI] [PubMed] [Google Scholar]
- 31. Thompson ER, Boyle SE, Johnson J, et al. Analysis of RAD51C germline mutations in high-risk breast and ovarian cancer families and ovarian cancer patients. Hum Mutat. 2012;33(1):95–99. [DOI] [PubMed] [Google Scholar]
- 32. Shimelis H, LaDuca H, Hu C, et al. Triple-negative breast cancer risk genes identified by multigene hereditary cancer panel testing. J Natl Cancer Inst. 2018;110(8):855–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schouten PC, Marme F, Aulmann S, et al. Breast cancers with a BRCA1-like DNA copy number profile recur less often than expected after high-dose alkylating chemotherapy. Clin Cancer Res. 2015;21(4):763–770. [DOI] [PubMed] [Google Scholar]
- 34. Loibl S, O'Shaughnessy J, Untch M, et al. Addition of the PARP inhibitor veliparib plus carboplatin or carboplatin alone to standard neoadjuvant chemotherapy in triple-negative breast cancer (BrighTNess): a randomised, phase 3 trial. Lancet Oncol. 2018;19(4):497–509. [DOI] [PubMed] [Google Scholar]
- 35. Tutt A, Tovey H, Cheang MCU, et al. Carboplatin in BRCA1/2-mutated and triple-negative breast cancer BRCAness subgroups: the TNT Trial. Nat Med. 2018;24(5):628–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Min A, Im SA, Yoon YK, et al. RAD51C-deficient cancer cells are highly sensitive to the PARP inhibitor olaparib. Mol Cancer Ther. 2013;12(6):865–877. [DOI] [PubMed] [Google Scholar]
- 37. Colombo I, Lheureux S, Oza AM.. Rucaparib: a novel PARP inhibitor for BRCA advanced ovarian cancer. Drug Des Devel Ther. 2018;12:605–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Evans T, Matulonis U.. PARP inhibitors in ovarian cancer: evidence, experience and clinical potential. Ther Adv Med Oncol. 2017;9(4):253–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sanchez-Bermudez AI, Sarabia-Meseguer MD, Garcia-Aliaga A, et al. Mutational analysis of RAD51C and RAD51D genes in hereditary breast and ovarian cancer families from Murcia (southeastern Spain). Eur J Med Genet. 2018;61(6):355–361. [DOI] [PubMed] [Google Scholar]
- 40. James PA, Sawyer S, Boyle S, et al. Large genomic rearrangements in the familial breast and ovarian cancer gene BRCA1 are associated with an increased frequency of high risk features. Fam Cancer. 2015;14(2):287–295. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.