Abstract
Objectives
To verify dissemination of daptomycin-non-susceptible Enterococcus faecium in a hospital where daptomycin was not in use and to understand the evolutionary pathways connecting daptomycin hypersusceptibility to non-susceptibility.
Methods
Clonality of 26 E. faecium was assessed by PFGE and the STs of these isolates were determined. The most daptomycin-susceptible isolate was evolved in vitro by stepwise daptomycin selection, generating isolates for genome comparisons.
Results
The spread of a high-risk daptomycin-non-susceptible VRE clone was detected, as was the occurrence of an unusual daptomycin-hypersusceptible strain (HBSJRP18). To determine the basis for daptomycin hypersusceptibility, we evolved HBSJRP18 in vitro and identified candidate genetic alterations potentially related to daptomycin susceptibility. Both lafB, encoding glycosyltransferase, which is putatively involved in lipoteichoic acid (LTA) biosynthesis, and dak, encoding a dihydroxyacetone kinase likely involved in fatty acid metabolism, were mutated in multiple independent experiments. Trans-complementation showed that the lafB polymorphism naturally occurring in HBSJRP18 caused its daptomycin hypersusceptibility. Fourier-transform infrared spectroscopy identified differences between the extracted LTA spectra from the hypersusceptible isolate and its revertant, as well as other non-susceptible variants, supporting a role for LafB in E. faecium LTA biosynthesis. Zeta potential difference was detected in one evolved dak mutant derivative. While much more susceptible to daptomycin, HBSJRP18 showed enhanced growth in the presence of piperacillin, suggesting that this, or another cell wall-targeting antibiotic, may have selected for the daptomycin-hypersusceptible phenotype.
Conclusions
Our findings provide new information on the basis for daptomycin susceptibility in E. faecium, with implications for limiting the development and spread of daptomycin resistance.
Introduction
MDR Enterococcus faecium have spread worldwide and the majority of isolates are now resistant to bactericidal therapy involving ampicillin, aminoglycosides and vancomycin.1,2 As a result, treatment now often includes second-line agents such as daptomycin, a cyclic lipopeptide with potent bactericidal activity against many resistant Gram-positive organisms, including Enterococcus faecalis and E. faecium.3,4 The daptomycin mechanism of action involves calcium-dependent insertion into the cell membrane, where the drug binds with high affinity to phosphatidylglycerol (PG),5–8 leads to membrane integrity disruption by rapid depolarization and efflux of potassium ions9 and causes mislocalization of essential cell division proteins leading to cell wall defects.10,11 In Brazil, daptomycin was approved to treat infections caused by Gram-positive bacteria in 2008 and since then has been a last-line alternative for treating VRE infections. CLSI currently considers enterococcal isolates with daptomycin MIC >4 mg/L as non-susceptible to the drug.12 However, the increasing occurrence of daptomycin-non-susceptible enterococci in hospitals and their emergence during therapy is compromising the use of daptomycin.4
Polymorphisms known to be related to daptomycin non-susceptibility in enterococci include those in: (i) LiaFSR, a regulatory system associated with the cell envelope stress response; (ii) YycFGHIJ, involved in the regulation of cell wall homeostasis; and (iii) Cls and GdpD, both of which are transmembrane proteins involved in phospholipid metabolism.13,14 Here we describe the nature of an unusual daptomycin-hypersusceptible E. faecium clinical isolate (HBSJRP18), as well as four daptomycin-non-susceptible E. faecium strains, isolated at the Hospital de Base de São José do Rio Preto in Brazil. Interestingly, although these strains were isolated after daptomycin was approved for use in Brazil, all strains were obtained before daptomycin was used at this hospital. To our knowledge, this is the first description of a daptomycin-hypersusceptible E. faecium isolate obtained from a clinical specimen. Here we identify the genetic determinant of daptomycin hypersusceptibility in this isolate, as well as other pathways implicated in daptomycin non-susceptibility in E. faecium.
Materials and methods
Source of strains
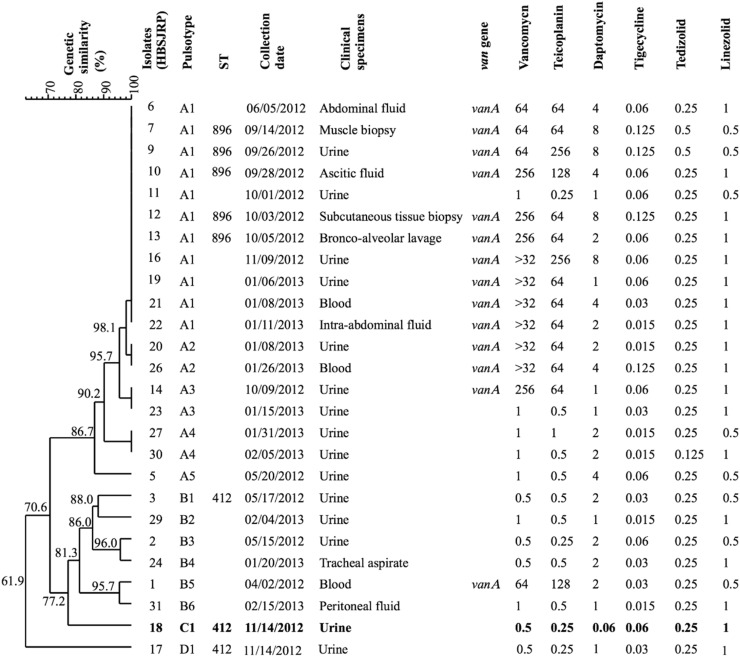
The Hospital de Base de São José do Rio Preto is located in the state of São Paulo and provides care for an area encompassing 1.8 million inhabitants. It includes 889 beds and serves 10 500 patients/month in its first-aid clinic and 560 patients/month in the ICU. During the period from April 2012 to February 2013, 26 E. faecium isolates were obtained from infections in various body sites, with each being isolated from a different patient (Figure 1).
Figure 1.
Dendrogram of E. faecium isolates from infections of patients in a Brazilian hospital. Strains are ordered by genetic similarity (%) determined by PFGE. Date of collection, clinical specimens, vancomycin-resistance gene (van) and MICs (mg/L) of tested antibiotics are indicated. HBSJRP18, the daptomycin-hypersusceptible strain, is shown in bold.
Antimicrobial susceptibility testing
MICs of vancomycin, teicoplanin, daptomycin, linezolid, polymyxin B, tigecycline and tedizolid were determined by broth microdilution, following CLSI 2018 guidelines.12 CLSI breakpoints12 were used for data analysis, with the tedizolid breakpoint established for E. faecalis being adopted. The tigecycline MIC breakpoint (0.25 mg/L) used was that defined by EUCAST.15
Growth inhibition of HBSJRP18 and its evolved derivative HBSJRP18-2.7 was measured in the presence of antibiotics previously administered to the patient for treatment of pneumonia (piperacillin and cefepime) and urinary tract infection (colistin, ceftriaxone and ciprofloxacin). Details are provided in the Supplementary Methods (available as Supplementary data at JAC Online).
Molecular typing
PFGE was performed after SmaI digestion, according to Tenover et al.,16 and data were analysed using BioNumerics v.7.1 (Applied Maths NV, Belgium) by the unweighted pair-group method with arithmetic mean based on Dice coefficients, where optimization and tolerance were set to 0.5% and 1.25%, respectively. Isolates with a genetic similarity coefficient of 80% or greater were considered to be the same pulsotype, but only those with 100% similarity were considered to be the same subtype. The ST of each isolate was determined using the MLSTFinder tool (http://www.genomicepidemiology.org/).17
In vitro evolution of daptomycin-susceptibility variants
Three isolated colonies of the hypersusceptible strain E. faecium HBSJRP18 were used in parallel in vitro evolution experiments to generate increasingly resistant variants. Details are provided in the Supplementary Methods. Susceptibility to daptomycin, ampicillin, vancomycin, polymyxin B, colistin, piperacillin, cefepime, ceftriaxone and ciprofloxacin was assessed for in vitro-evolved variants.
Genome sequencing and comparative analysis
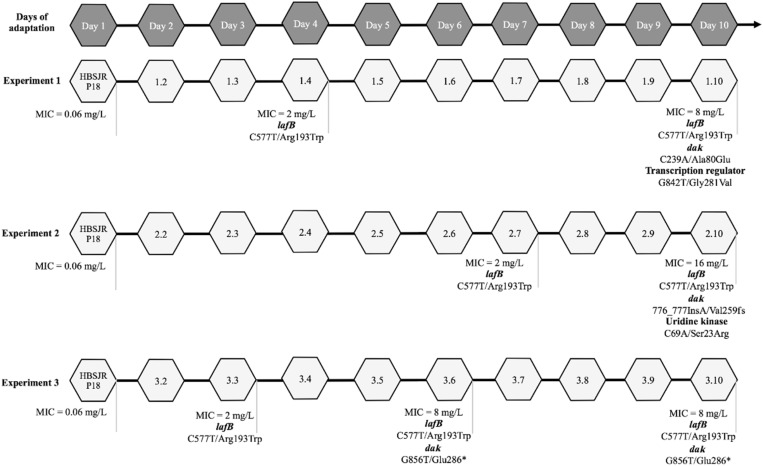
Three daptomycin-resistant E. faecium strains (HBSJRP7, HBSJRP9 and HBSJRP12) and two daptomycin-susceptible strains (HBSJRP10 and HBSJRP13) were selected for genome sequencing. Additionally, the genomes of in vitro-selected daptomycin-susceptibility variants of HBSJRP18 (1.1, 1.4, 1.10, 2.1, 2.7, 2.10, 3.1, 3.3, 3.6, 3.10) were also sequenced. For the latter strains, the first number (1, 2 or 3) identifies which of the triplicate evolution experiments the isolate belonged to. The second number (1–10) corresponds to the day of passage for the isolate (Figure 2). Details are provided in the Supplementary Methods.
Figure 2.
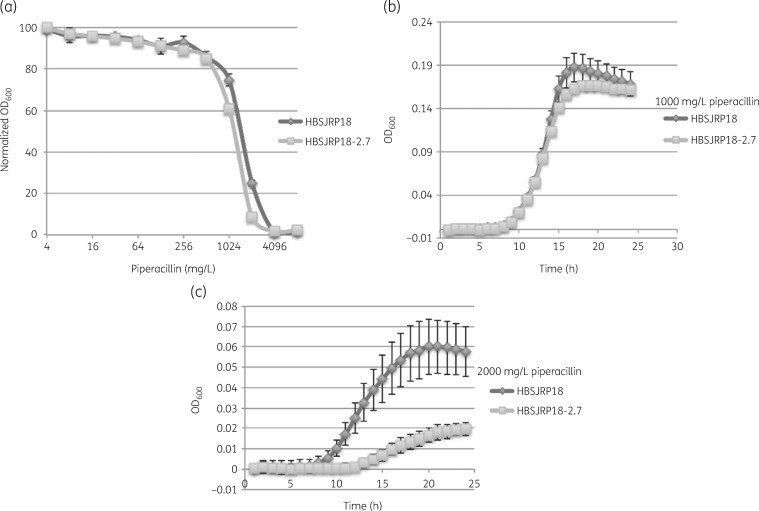
Evaluation of growth in the presence of piperacillin. (a) Dose–response curves of HBSJRP18 and HBSJRP18-2.7 for varying concentrations of piperacillin. (b and c) Growth curves for 1000 mg/L and 2000 mg/L piperacillin, respectively.
Contigs were annotated through the NCBI Prokaryotic Genome Annotation Pipeline and the genomes were deposited at DDBJ/EMBL/GenBank (Table S1, available as Supplementary data at JAC Online). SNPs were confirmed by PCR amplification of the involved region and subsequent Sanger sequencing (Table 1). ResFinder (http://www.genomicepidemiology.org/) was used to identify known antimicrobial resistance genes within the genome sequence data, using a threshold of 90% identity.
Table 1.
Primers used for sequencing and cloning assays
| Target gene/primers | Sequence (5′–3′) | Size (bp) |
|---|---|---|
| lafB-like gene, E. faecium | ||
| LafB2_F | GTATCCGTGTGCCGATTCAA | 337 |
| LafB2_R | GCACCTGAATACGCACCTT | |
| dak_SNP239, E. faecium | ||
| Dak239_F | GAAATGGTCCAGGCAGGTG | 362 |
| Dak239_2_R | AATCGTTCCTTCTACTGGTTTCA | |
| dak _SNP 777delA, E. faecium | ||
| Dak777_2F | TCATCGCAGTGTCCAAGGTC | 386 |
| Dak777_2R | GGACGCCTTTACCAGCAG | |
| lafB-like gene (cloning and sequencing) | ||
| LafB_F (KpnI) | GGTACCGTGAAGGGGGACGTAAGGTG | 1084 |
| LafB_R (XbaI) | TCTAGACTCCAAACCTAGTCCTTGACCT | |
| dak (cloning and sequencing) | ||
| Dak_F (KpnI) | GGTACCAGCAGTCGATCTGAAGGAGGA | 1814 |
| Dak_R (XbaI) | TCTAGAAAAAGGACTTGGTCAGCGAT | |
| To verify gene in the plasmid vector | ||
| pAT28_F | TAATGCAGCTGGCACGACAGG | 552 |
| pAT28_R | GCGTGATTGCCAAGCACGTCC | |
Cloning and complementation
The lafB and dak genes were amplified from HBSJRP18-2.7 and HBSJRP18, respectively, using primers listed in Table 1. PCR products and pAT2818 were digested with KpnI/XbaI (New England Biolabs) for ligation using T4 DNA ligase (New England Biolabs). Ligation products were first transformed into chemically competent Escherichia coli DH5α cells.19 The desired clones were identified by PCR amplification using primers targeting the pAT28 vector backbone (Table 1) and plasmids were extracted for sequencing of the lafB or dak inserts. Verified pAT28-based clones of lafB from HBSJRP18-2.7, or dak from HBSJRP18, were then transformed into E. faecium HBSJRP18 or HBSJRP18-3.6, respectively, by electroporation.
Sample preparation for infrared spectroscopy analysis
Lipoteichoic acid (LTA) was extracted from HBSJRP18 (daptomycin hypersusceptible), HBSJRP18-2.7 (daptomycin susceptible) and HBSJRP18-3.6 (daptomycin non-susceptible) to obtain Fourier-transform infrared (FTIR) spectra of the LTA. Spectral differences were quantified by multidimensional projection using statistical techniques that reduce the data dimension and help with sample differentiation. For this we used Projection Explorer Sensor (PEx-Sensors)20,21 software, which transforms each FTIR spectrum into a single point, projected onto a visualization map that evaluates the samples by Euclidean distance between the points.
Zeta potential analysis
Changes in cell surface charge were evaluated by assessing zeta potential. Cells were grown overnight in 5 mL of CAMHB, adjusted to a turbidity equivalent to that of a 0.5 McFarland standard and incubated at 37°C for an additional hour. Cells were harvested by centrifugation, washed twice with 1.5 mM NaCl solution, resuspended in 1 mL of the same solution and adjusted to a turbidity equivalent to that of a 1.0 McFarland standard with deionized water. Zeta potential was measured in a Zetasizer Nano ZS (Malvern Panalytical Ltd, Malvern, Worcestershire, UK), using disposable folded capillary cell cuvettes (DTS1070; Malvern Panalytical Ltd) at 25°C. Experiments were repeated using three separate cultures of each strain tested. Results were averaged over three measurements per replicate per strain. Student’s t-test was used to compare zeta potential values and P≤0.01 was considered significant.
Results
Antimicrobial susceptibilities
All 26 E. faecium strains were susceptible to linezolid, tigecycline and tedizolid (Figure 1). One isolate (HBSJRP18) was noted as unusual due to its daptomycin hypersusceptibility (MIC=0.06 mg/L). Half of the 26 isolates were VRE, among which four were daptomycin non-susceptible (Figure 1).
PFGE and MLST analysis
The 26 E. faecium isolates were groupable into four pulsotypes (A–D) (Figure 1). Pulsotype A was predominant and included five subtypes (A1–A5). Daptomycin-non-susceptible isolates HBSJRP7, HBSJRP9, HBSJRP12 and HBSJRP16, together with seven susceptible isolates, clustered in pulsotype A1. Except for HBSJRP16, which was not typed, the other three daptomycin-non-susceptible isolates from pulsotype A1 belonged to ST896 and pulsotypes B1, C1 and D1 belonged to ST412. ST896 is a single-locus variant of ST412 and both belong to clonal complex 17 (CC17)22/clade A.23
Genome sequencing of clinical isolates
The DNA sequence of the genomes of six clinical isolates was determined (Table 2). All ST896 isolates possessed identical resistance gene profiles. HBSJRP18, the daptomycin-hypersusceptible isolate, shared most of these, but lacked the genes responsible for vancomycin and tetracycline resistance. PlasmidFinder identified plasmids belonging to four rep families, three of which are believed to be unique to E. faecium.24 A single rep2 gene was detected in the hypersusceptible E. faecium isolate.
Table 2.
Genome information summary for sequenced E. faecium clinical strains
| Strain | Daptomycin MIC (mg/L) | Genome size (Mb) | ST | Antibiotic resistance genes | Plasmid rep genes |
|---|---|---|---|---|---|
| HBSJRP7 | 8 | 2.874 | 896 | aph(3')-III, ant(6)-Ia, erm(B), msr(C), tet(L), tet(M), vanA | repUS15, rep14, rep17 |
| HBSJRP9 | 8 | 2.907 | 896 | aph(3')-III, ant(6)-Ia, erm(B), msr(C), tet(L), tet(M), vanA | repUS15, rep14, rep17 |
| HBSJRP10 | 4 | 2.892 | 896 | aph(3')-III, ant(6)-Ia, erm(B), msr(C), tet(L), tet(M), vanA | repUS15, rep14, rep17 |
| HBSJRP12 | 8 | 2.898 | 896 | aph(3')-III, ant(6)-Ia, erm(B), msr(C), tet(L), tet(M), vanA | repUS15, rep17 |
| HBSJRP13 | 2 | 2.892 | 896 | aph(3')-III, ant(6)-Ia, erm(B), msr(C), tet(L), tet(M), vanA | repUS15, rep14, rep17 |
| HBSJRP18 | 0.06 | 2.951 | 412 | aph(3')-III, ant(6)-Ia, erm(B), msr(C) | rep17, rep2, repUS15 |
The ST896 strains were highly genetically related to one another, but showed differences in susceptibility to daptomycin (Table 2). Genome comparison using the daptomycin-non-susceptible HBSJRP7 strain as a reference identified a Lys233Ile substitution in a multimodular transpeptidase-transglycosylase (a PBP) in isolate HBSJRP10 and a Thr316Met substitution in the d-alanine-d-alanine ligase in isolate HBSJRP13 (Table 3). These mutations were not found in the other non-susceptible strains that were sequenced in this study. A BLAST search of the mutated, susceptible-associated alleles of both genes in all E. faecium genomes available in NCBI suggests that the mutated alleles of both genes are unique to these isolates and have not been observed before. Because both enzymes are involved in cell envelope assembly and integrity, we hypothesize that these mutations may relate to the lower-level daptomycin resistance observed in these strains.
Table 3.
Mutations identified in ST896 strains compared with the HBSJRP7 reference (Ref) genome
| Strain/Ref contig | Ref position (bp) | Variant type | Ref allele | Mutant allele | Amino acid change | Annotation |
|---|---|---|---|---|---|---|
| HBSJRP9 (daptomycin MIC 8 mg/L) | ||||||
| 25 | 866 | insertion | — | CTTGGTCTTCT | frame shift | hypothetical protein |
| 32 | 5333 | deletion | T | — | NA | intergenic (between sensor histidine kinase VanS and contig break) |
| 41 | 19814 | SNP | T | A | NA | intergenic (between mobile element protein and contig break) |
| 75 | 8628 | NSY SNP | C | T | Gly106Glu | PTS system, sucrose-specific |
| 106 | 26313 | SNP | T | C | NA | intergenic (between S-adenosylmethionine:tRNA ribosyltransferase-isomerase and contig break) |
| 123 | 30223 | NSY SNP | C | T | Thr112Ile | cell division protein FtsI |
| 144 | 5948 | NSY SNP | G | A | Arg208His | SSU ribosomal protein S2p |
| 145 | 9634 | insertion | — | ATCTGTCTCGCAG | NA | intergenic (between methionine ABC transporter ATP-binding protein and CsbD-like protein) |
| HBSJRP10 (daptomycin MIC 4 mg/L) | ||||||
| 59 | 22691 | NSY SNP | A | T | Lys233Ile | multimodular transpeptidase-transglycosylase |
| 66 | 29793 | deletion | G | — | NA | intergenic (between GCN5-related N-acetyltransferase and PTS system, cellobiose-specific IIC component) |
| 75 | 13399 | SYN SNP | G | A | NA | mobile element protein |
| 130 | 23788 | SYN SNP | T | C | NA | ribosomal protein S18 |
| HBSJRP12 (daptomycin MIC 8 mg/L) | ||||||
| 17 | 7425 | SYN SNP | G | A | NA | hypothetical protein |
| 17 | 19639 | NSY SNP | T | A | Asp518Glu | metal-transporting ATPase |
| 32 | 5333 | deletion | T | — | NA | intergenic (downstream of sensor histidine kinase VanS, upstream of contig break) |
| 106 | 26313 | SNP | T | C | NA | intergenic (between S-adenosylmethionine:tRNA ribosyltransferase-isomerase and contig break) |
| HBSJRP13 (daptomycin MIC 2 mg/L) | ||||||
| 106 | 6463 | NSY SNP | C | T | Thr316Met | d-alanine-d-alanine ligase |
NSY SNP, non-synonymous SNP; SYN SNP, synonymous SNP; NA, not applicable (no change in coding sequence; shared between strains).
Bold text indicates that this change is hypothesized to cause the lower daptomycin MIC.
Susceptibility to antibiotics previously administered
The identification of a daptomycin-hypersusceptible strain from a clinical infection seemed paradoxical, especially given that daptomycin acts similarly to antimicrobial peptides produced by mammalian hosts. We wondered whether a particular selective pressure might have given rise to this unusual phenotype. Therefore, we tested the antibiotics administered to the patient from whom HBSJRP18 was isolated. HBSJRP18 was isolated from the urine of a patient who developed a nosocomial urinary tract infection. This patient was initially hospitalized because of pneumonia and had previously received piperacillin/tazobactam and cefepime to treat that condition. The hospital then attempted to resolve the urinary tract infection in this complex patient using polymyxin E (colistin), ceftriaxone and ciprofloxacin. HBSJRP18 had MICs identical to those for HBSJRP18-2.7 of piperacillin, cefepime, ceftriaxone and ciprofloxacin (Table 4). Consistent with its increased susceptibility to daptomycin, HBSJRP18 was also more susceptible to colistin than HBSJRP18-2.7.
Table 4.
MICs of antibiotics used during the treatment of the patient for HBSJRP18 and its in vitro-selected lafB revertant, HBSJRP18-2.7
| MIC (mg/L) |
|||||
|---|---|---|---|---|---|
| Isolate | piperacillin | cefepime | colistin | ceftriaxone | ciprofloxacin |
| HBSJRP18 | 4096 | >2000 | 64 | >10 000 | 128 |
| HBSJRP18-2.7 | 4096 | >2000 | 256 | >10 000 | 128 |
We then examined growth at subMIC levels of the tested drugs and plotted dose–response curves for HBSJRP18 and HBSJRP18-2.7. We noticed that while both isolates showed the same MIC of piperacillin (4096 mg/L), the daptomycin-hypersusceptible strain HBSJRP18 consistently grew to higher ODs at ½ and ¼ of the piperacillin MIC (Figure 2a). To investigate further, we collected growth curves for both strains in the presence of 1000 mg/L or 2000 mg/L piperacillin. At the lower concentration, both isolates grew similarly, with isolate HBSJRP18 showing slightly higher overall growth (Figure 2b). However, HBSJRP18 grew better in the presence of 2000 mg/L piperacillin than HBSJRP18-2.7, showing that HBSJRP18 had higher piperacillin tolerance (Figure 2c). Although these drug concentrations exceed those concentrations usually found in the serum, concentrations >1000 mg/L are found in human urine.25
Mutations found in in vitro-evolved daptomycin-non-susceptible isolates
We sequenced and compared the genomes of independently evolved variants of HBSJRP18 that displayed decreasing levels of daptomycin susceptibility (Figure 3). In all variants, an initial C577T transition (Arg193Trp) in ORF HMPREF0351_10963 of the E. faecium DO genome (NC_017960.1) was observed to precede any other change. ORF HMPREF0351_10963 is annotated as the glycosyltransferase gene lafB and the Arg193Trp mutation restored the coding sequence to that naturally occurring in all other reported E. faecium clinical isolates. Therefore, we classified this transition as a reversion to a WT genotype. The closest homologue of E. faecium lafB in E. faecalis is the biofilm-associated glycolipid synthesis A gene (bgsA) (E. faecalis V583 gene EF2891), which belongs to a two-enzyme system responsible for LTA anchor formation. E. faecalis BgsA is homologous to LafB in Listeria monocytogenes26–28 (Figure S1).
Figure 3.
Schematic representation of the in vitro evolution of daptomycin-susceptibility variant results. We performed this assay in triplicate (Experiments 1, 2 and 3). Each hexagon represents 1 day of the experiment and the number inside is the name of the variant obtained on that day. Daptomycin MIC is shown in mg/L. In the case that a mutation was detected in one of the variants, the following information is below its hexagon: the mutated gene in bold and, on the next line, the changed nucleotide/amino acid and their location. An asterisk denotes a stop codon; fs, frameshift.
Another gene repeatedly altered in the in vitro selection experiments was dak, which acquired distinct mutations in all three experimental replicates (Figure 3). In one replicate, a C239A transversion in dak was identified that results in a non-conservative Ala80Glu change. Additionally, in this strain, a mutation was also observed in a second related gene encoding a diacylglycerol kinase, dagK (Table S2 and Figure 3). In another evolved strain, that with the highest daptomycin MIC, an adenine insertion in dak (776_777insA) was found that changed the reading frame causing premature protein truncation. Finally, the third evolved strain acquired a G to T transversion at position 856 in dak (G856T), creating a stop codon and a truncated protein (Glu286*).
Antibiotic susceptibility of in vitro-evolved isolates
Isolates HBSJRP18-2.7 and HBSJRP18-3.6, which possessed identical lafB reversions followed by different dak mutations, were screened for susceptibility to daptomycin and other antibiotics by broth microdilution (Table 5). We observed a 2-fold increase in vancomycin MIC and a 2-fold decrease in ampicillin MIC for the selected strains, suggesting that changes in the cell envelope may have accompanied the increase in daptomycin MIC.
Table 5.
Comparison of antibiotic MICs for HBSJRP18 and its variants
| MIC (mg/L) |
||||
|---|---|---|---|---|
| Isolate | daptomycin | vancomycin | ampicillin | polymyxin B |
| HBSJRP18 | 0.06 | 0.5 | 256 | >256 |
| HBSJRP18-2.7 | 2 | 1 | 128 | >256 |
| HBSJRP18-3.6 | 8 | 1 | 128 | >256 |
Trans-complementation analysis of lafB and dak
To test whether the daptomycin-hypersusceptibility phenotype in HBSJRP18 was due to a diffusible product encoded by its lafB (577C) allele, we introduced the WT allele (577T) into the strain in trans cloned into the shuttle vector pAT28.18 Expression of the WT glycosyltransferase (lafB) allele (obtained from revertant HBSJRP18-2.7) raised the daptomycin MIC for the hypersusceptible strain to 2 mg/L, consistent with the phenotype of WT revertants observed during daptomycin selection. Expression of the dak WT allele in the HBSJRP18-3.6 daptomycin-resistant strain background, however, did not restore susceptibility, likely because the resistant form is dominant (Table 6).
Table 6.
Daptomycin MIC for the isolates complemented in trans
| Isolate | Daptomycin MIC (mg/L) |
|---|---|
| HBSJRP18 | 0.06 |
| HBSJRP18 + pAT28 | 0.125 |
| HBSJRP18 + pAT28lafB18-2.7 | 2 |
| HBSJRP18-2.7 | 2 |
| HBSJRP18-3.6 | 8 |
| HBSJRP18-3.6 + pAT28 | 4 |
| HBSJRP18-3.6 + pAT28dak_18 | 4 |
pAT28, empty vector; pAT28lafB18-2.7, lafB gene from HBSJRP18-2.7 was amplified and cloned into pAT28; pAT28dak_18, dak gene from HBSJRP18 was amplified and cloned into pAT28.
FTIR characterization
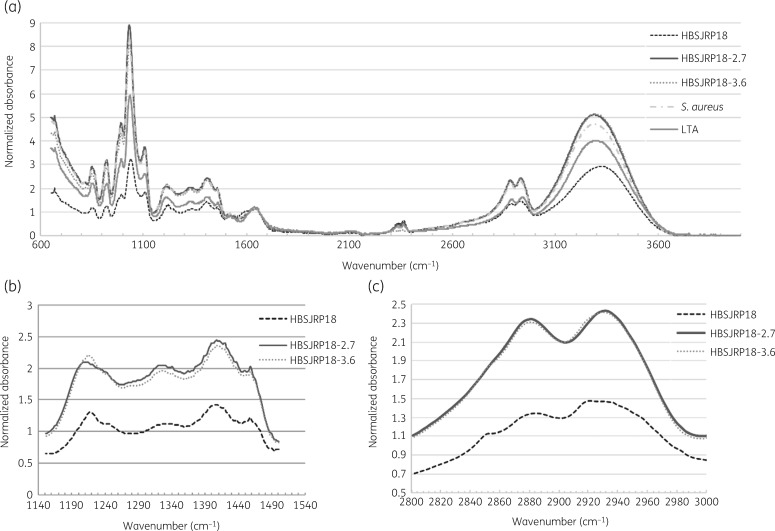
Because LafB is predicted to be involved in LTA biosynthesis, we examined the LTA from HBSJRP18 and its derivatives using FTIR. LTA extracted from HBSJRP18, HBSJRP18-2.7, HBSJRP18-3.6 and Staphylococcus aureus ATCC 25923 were assessed by infrared spectroscopy, using commercial S. aureus-derived LTA (Sigma–Aldrich Inc.) as a positive control. Whole spectra showed differences between the LTA extracted from the different strains (Figure 4a). Cell content that could be detected in the spectral regions from wavenumber 1150 to 1500 cm−1 (Figure 4b) and from wavenumber 2800 to 3000 cm−1 (Figure 4c) were less represented in the LTA extracted from HBSJRP18 when compared with that from its variants. These regions are influenced by the bending modes of >CH2 and –CH3 groups found in proteins, fatty acids and phosphate-bearing compounds29 and by oscillation energy of the C–H dipoles of the CH3 and CH2 radicals, which are abundant in membrane fatty acids. When we analysed FTIR bandwidth, we extracted the amount of dipoles present, since the area is directly proportional to the amount of dipole present in the analysed molecules. We found that the hypersusceptibility in HBSJRP18 was directly linked to the decrease of dipoles CH2 and CH3. CH3 decrease was the most evident, as demonstrated in Figure S2.
Figure 4.
FTIR spectra of LTA extracted from E. faecium HBSJRP18, its variants and S. aureus. (a) FTIR spectra of LTA extracted from E. faecium HBSJRP18, E. faecium HBSJRP18-2.7, E. faecium HBSJRP18-3.6 and S. aureus ATCC 25923 plus a commercial S. aureus-derived LTA (Sigma–Aldrich Inc.). (b) Spectrum region from wavenumber 1150 to 1500cm−1 comparing E. faecium HBSJRP18 and its variants E. faecium HBSJRP18-2.7 and E. faecium HBSJRP18-3.6. (c) Spectrum region from wavenumber 2800 to 3000cm−1 with all samples described before.
Comparison of the IdMap plots for the LTA of the four bacterial strains indicated a clear distinction between E. faecium HBSJRP18/E. faecium HBSJRP18-3.6 and E. faecium HBSJRP18-2.7/E. faecium HBSJRP18-3.6, showing that the loss of susceptibility of HBSJRP18-3.6 substantially changed the molecular characteristics of the cell envelope and, in particular, the content isolated from the LTA extraction (Figure S2). The difference between isolates HBSJRP18 and HBSJRP18-2.7 suggests that the mutation in lafB present in HBSJRP18 altered the content of LTA and/or components of the cell membrane. The LTA from HBSJRP18-2.7 closely resembled LTA from S. aureus ATCC 25923, which is also susceptible to daptomycin30 (Figure S3).
Zeta potential analysis
Earlier studies comparing daptomycin-non-susceptible versus daptomycin-susceptible isolates have shown an increase in positive surface charge.31,32 Mean cell surface zeta potential, described in Table 7, distinguished only the most resistant strain.
Table 7.
Zeta potential of E. faecium HBSJRP18 and its mutants
| Strain | Zeta potential (mV) |
|---|---|
| E. faecium HBSJRP18 | −26.5±0.47 |
| E. faecium HBSJRP18-2.7 | −26.4±0.53 |
| E. faecium HBSJRP18-3.6 | −25.6±0.55a |
The data are expressed as mean value±SD of three replications of three independent experiments.
P<0.01 for E. faecium HBSJRP18-3.6 when compared with the other two isolates.
Discussion
Even though daptomycin was not used in the hospital prior to November 2012, daptomycin non-susceptibility was found in 4 out of 10 E. faecium isolates of pulsotype A, subtype A1/ST896 (CC17). Other cases of daptomycin-non-susceptible E. faecium from patients with no prior exposure to daptomycin have been reported,33 including among VRE strains.34 Vancomycin exposure is known to induce cell wall changes that result in decreased daptomycin susceptibility in S. aureus.35,36 The daptomycin-non-susceptible isolates examined here were all VRE, although the patients had not been treated with vancomycin during residence in this hospital. This suggests that the strains were acquired by nosocomial spread, perhaps from facilities where vancomycin and daptomycin were in broader use. All E. faecium isolates studied here retained susceptibility to tigecycline, linezolid and tedizolid, which are alternatives for treating daptomycin-non-susceptible VRE infection.
Genome comparison of the first daptomycin-non-susceptible isolate of this study (HBSJRP7) with two other daptomycin-non-susceptible isolates, plus two daptomycin-susceptible isolates of the same ST, identified mutations that appeared to be associated with increased susceptibility to daptomycin (Table 3). We hypothesize that these patients were treated with β-lactams or other cell wall-targeting agents, which could have selected for the mutation we observed in a multimodular transpeptidase-transglycosylase (E. faecium HBSJRP10). This protein belongs to a family of enzymes responsible for the polymerization of the peptidoglycan strand and the cross-linking between glycan chains, which is also the target of the β-lactam class of antibiotics.37 Similarly, we identified a mutation in the d-alanine-d-alanine ligase (identified in HBSJRP13), which acts by cross-linking peptidoglycan strands,38 that was also associated with increased daptomycin susceptibility. We believe that these mutations are responsible for the increased susceptibility we observed, but this remains to be validated.
Based on genome comparisons of an unusual daptomycin-hypersusceptible clinical isolate and three independently evolved variants, we discovered a new and unexpected vulnerability to daptomycin that appears to reside within the E. faecium LTA biosynthetic pathway. When we exposed the hypersusceptible strain to increased daptomycin pressure, all independently evolved laboratory lineages first reverted to the naturally occurring base at position 577 in lafB, which is found in all E. faecium genomes so far reported in GenBank. Complementation by providing the WT allele in trans raised the daptomycin MIC for HBSJRP18 to a level of susceptibility typical of E. faecium (MIC of 2 mg/L), indicating that the 577T allele is dominant and represents a gain of function over the 577C allele. The 577C allele encodes a non-conservative Arg in place of a Thr at codon 193 and presumably compromises LafB glycosyltransferase function. Because LafB is implicated in LTA biosynthesis, this observation unveils a previously unaccounted-for role for LTA in the mechanism of action of daptomycin.
Unrelated daptomycin-hypersusceptible phenotypes derived by mutagenesis in vitro have been described.39–42 In E. faecalis, the deletion of the LiaR response regulator reversed resistance to daptomycin and also resulted in hypersusceptibility to the drug.39 In E. faecium, the same experimental deletion of LiaR was also responsible for reversion of daptomycin resistance.40 In another study, hypersusceptibility to daptomycin in E. faecium was found to be due to disruption of the liaFSR operon by various IS elements.41
To understand what type of selection may have given rise to clinical isolate HBSJRP18, we investigated the MICs of other antibiotics for the strain. HBSJRP18 was found to have a growth advantage at very high concentrations of piperacillin. A seesaw effect is already known in S. aureus showing that when daptomycin MIC increases, β-lactam resistance decreases. Moreover, a combination of daptomycin and a β-lactam provides synergy against E. faecium and E. faecalis.43 The higher resistance observed to β-lactams could have served as a selective pressure that offered the daptomycin-hypersusceptible isolate a selective advantage compared with other isolates. The significance of this finding is speculative, but we hypothesize that biological or therapeutic selection of an altered cell wall played a role in selecting for daptomycin hypersusceptibility.
The dak gene, encoding a dihydroxyacetone kinase, was the only other gene in which mutations occurred in all strains evolved for elevated resistance to daptomycin. Recently described by Miller et al.,44 Dak is homologous to FakA in S. aureus, a fatty acid kinase, previously demonstrated to be essential for phosphorylating exogenous fatty acids and incorporating them into the S. aureus phospholipid bilayer.45 The IdMap plots from FTIR showed that the loss of daptomycin susceptibility in HBSJRP18-3.6 substantially changed the molecular characteristics of the cell envelope, in particular the content accessed by the LTA extraction. The precise role of Dak in this modulation, however, remains to be clarified in future work.
FTIR analysis identified qualitative differences in the LTA extracted from the parental hypersusceptible clinical isolate and reverted evolved strains. FTIR showed that the hypersusceptible lafB genotype results in a cell envelope with changed content of fatty acids, phospholipids and glycolipids. Lipidomic analysis will provide a clearer understanding of the effect of this change in cell envelope biogenesis.
An alteration of zeta potential, as has been observed by others for some daptomycin-resistant strains,31,32 was only observed for evolved strain HBSJP18-3.6. Interestingly, this strain exhibited the highest level of daptomycin resistance (8 mg/L). Although the zeta potential was statistically significant, whether the 1 mV difference is biologically relevant remains to be demonstrated.
In summary, characterization of a daptomycin-hypersusceptible E. faecium clinical isolate identified a loss-of-function mutation in the glycosyltransferase gene lafB, revealing a new role for LTA in the mechanism of action of daptomycin. Since reversion of the lafB mutation was observed as a first step toward restoring WT daptomycin susceptibility, this mutation might be more dominant than other mechanisms for daptomycin resistance as well. This suggests that small molecule inhibition of lafB may yield daptomycin susceptibility in strains possessing mutations in LiaR44 and CLS46 and represents a new therapeutic opportunity for treatment and prevention of the emergence of daptomycin resistance by these mechanisms.
Our results also implicate a potential role for dak in daptomycin resistance, although its contribution is yet to be understood. This work also documents dissemination of a high-risk vancomycin-resistant clone of E. faecium with reduced susceptibility to daptomycin, despite the fact that daptomycin had not yet been used at the hospital and vancomycin had not been used in these patients. Although vancomycin resistance and daptomycin non-susceptibility removed the most rapidly bactericidal options, these strains retained susceptibility to tigecycline, linezolid and tedizolid, although resistance in enterococci to both of the classes of these drugs is known and spreading.46–50 This work highlights the importance of continued research to salvage the activity of bactericidal antibiotics such as daptomycin and an opportunity within the LTA biosynthetic pathway for potentially doing so.
Supplementary Material
Acknowledgements
We thank Lilian Tan Moriyama for assistance with the spectrophotometer Cary 630 FTIR (Agilent Technologies, Inc.). We also thank Andrey Coatrini for assistance with FTIR analysis. Tedizolid phosphate was kindly provided by Bayer.
Funding
This study was supported by Sao Paulo Research Foundation (FAPESP 2016/23810-6) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (PVE/CAPES no. 88881.030524/2013-01), the NIH/NIAID-sponsored Harvard-wide Program on Antibiotic Resistance AI083214, National Institutes of Health grant AI146715, and the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES)—Finance Code 001.
Transparency declarations
None to declare.
References
- 1. Arias CA, Contreras GA, Murray BE.. Management of multidrug-resistant enterococcal infections. Clin Microbiol Infect 2010; 16: 555–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Osman KM, Ali MN, Radwan I. et al. Dispersion of the vancomycin resistance genes vanA and vanC of Enterococcus isolated from Nile tilapia on retail sale: a public health hazard. Front Microbiol 2016; 7: 1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tran TT, Munita JM, Arias CA.. Mechanisms of drug resistance: daptomycin resistance. Ann N Y Acad Sci 2015; 1354: 32–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Desai H, Wong R, Pasha AK.. A novel way of treating multidrug-resistant enterococci. North Am J Med Sci 2016; 8: 229–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Taylor SD, Palmer M.. The action mechanism of daptomycin. Bioorg Med Chem 2016; 24: 6253–68. [DOI] [PubMed] [Google Scholar]
- 6. Hachmann A-B, Sevim E, Gaballa A. et al. Reduction in membrane phosphatidylglycerol content leads to daptomycin resistance in Bacillus subtilis. Antimicrob Agents Chemother 2011; 55: 4326–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jung D, Rozek A, Okon M. et al. Structural transitions as determinants of the action of the calcium-dependent antibiotic daptomycin. Chem Biol 2004; 11: 949–57. [DOI] [PubMed] [Google Scholar]
- 8. Lee MT, Hung WC, Hsieh MH. et al. Molecular state of the membrane-active antibiotic daptomycin. Biophys J 2017; 113: 82–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Steenbergen JN, Alder J, Thorne GM. et al. Daptomycin: a lipopeptide antibiotic for the treatment of serious Gram-positive infections. J Antimicrob Chemother 2005; 55: 283–8. [DOI] [PubMed] [Google Scholar]
- 10. Pogliano J, Pogliano N, Silverman JA.. Daptomycin-mediated reorganization of membrane architecture causes mislocalization of essential cell division proteins. J Bacteriol 2012; 194: 4494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Muller A, Wenzel M, Strahl H. et al. Daptomycin inhibits cell envelope synthesis by interfering with fluid membrane microdomains. Proc Natl Acad Sci USA 2016; 113: E7077–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.CLSI. Performance Standards for Antimicrobial Susceptibility Testing–Twenty-Eighth Edition: M100 2018.
- 13. Miller WR, Munita JM, Arias CA.. Mechanisms of antibiotic resistance in enterococci. Expert Rev Anti Infect Ther 2014; 12: 1221–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Palmer KL, Daniel A, Hardy C. et al. Genetic basis for daptomycin resistance in enterococci. Antimicrob Agents Chemother 2011; 55: 3345–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.EUCAST. Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 9.0, 2019. http://www.eucast.org/clinical_breakpoints/.
- 16. Tenover FC, Arbeit RD, Goering RV. et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 1995; 33: 2233–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Larsen MV, Cosentino S, Rasmussen S. et al. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 2012; 50: 1355–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Trieu-Cuot P, Carlier C, Poyart-Salmeron C. et al. A pair of mobilizable shuttle vectors conferring resistance to spectinomycin for molecular cloning in Escherichia coli and in gram-positive bacteria. Nucl Acids Res 1990; 18: 4296.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kostylev M, Otwell AE, Richardson RE. et al. Cloning should be simple: Escherichia coli DH5α-mediated assembly of multiple DNA fragments with short end homologies. PLoS One 2015; 10: e0137466.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Paulovich FV, Maki RM, de Oliveira MC. et al. Using multidimensional projection techniques for reaching a high distinguishing ability in biosensing. Anal Bioanal Chem 2011; 400: 1153–9. [DOI] [PubMed] [Google Scholar]
- 21. Paulovich FV, Moraes ML, Maki RM. et al. Information visualization techniques for sensing and biosensing. Analyst 2011; 136: 1344–50. [DOI] [PubMed] [Google Scholar]
- 22. Cattoir V, Leclercq R.. Twenty-five years of shared life with vancomycin-resistant enterococci: is it time to divorce? J Antimicrob Chemother 2013; 68: 731–42. [DOI] [PubMed] [Google Scholar]
- 23. Lebreton F, van Schaik W, McGuire AM. et al. Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. MBio 2013; 4: e00534-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jensen LB, Garcia-Migura L, Valenzuela AJ. et al. A classification system for plasmids from enterococci and other Gram-positive bacteria. J Microbiol Methods 2010; 80: 25–43. [DOI] [PubMed] [Google Scholar]
- 25. Landersdorfer CB, Bulitta JB, Kirkpatrick CMJ. et al. Population pharmacokinetics of piperacillin at two dose levels: influence of nonlinear pharmacokinetics on the pharmacodynamic profile. Antimicrob Agents Chemother 2012; 56: 5715–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Webb AJ, Karatsa-Dodgson M, Grundling A.. Two-enzyme systems for glycolipid and polyglycerolphosphate lipoteichoic acid synthesis in Listeria monocytogenes. Mol Microbiol 2009; 74: 299–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reichmann NT, Gründling A.. Location, synthesis and function of glycolipids and polyglycerolphosphate lipoteichoic acid in Gram-positive bacteria of the phylum Firmicutes. FEMS Microbiol Lett 2011; 319: 97–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Theilacker C, Sanchez-Carballo P, Toma I. et al. Glycolipids are involved in biofilm accumulation and prolonged bacteraemia in Enterococcus faecalis. Mol Microbiol 2009; 71: 1055–69. [DOI] [PubMed] [Google Scholar]
- 29. Alvarez-Ordóñez A. Technical and methodological aspects of Fourier transform infrared spectroscopy in food microbiology research. In: Fourier Transform Infrared Spectroscopy in Food Microbiology: SpringerBriefs in Food, Health, and Nutrition. Springer, 2012; 1–18 [Google Scholar]
- 30. Ma W, Zhang D, Li G. et al. Antibacterial mechanism of daptomycin antibiotic against Staphylococcus aureus based on a quantitative bacterial proteome analysis. J Proteomics 2017; 150: 242–51. [DOI] [PubMed] [Google Scholar]
- 31. Hall Snyder AD, Werth BJ, Nonejuie P. et al. Fosfomycin enhances the activity of daptomycin against vancomycin-resistant enterococci in an in vitro pharmacokinetic-pharmacodynamic model. Antimicrob Agents Chemother 2016; 60: 5716–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cui L, Isii T, Fukuda M. et al. An RpoB mutation confers dual heteroresistance to daptomycin and vancomycin in Staphylococcus aureus. Antimicrob Agents Chemother 2010; 54: 5222–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kelesidis T, Humphries R, Uslan DZ. et al. De novo daptomycin-nonsusceptible enterococcal infections. Emerg Infect Dis 2012; 18: 674–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fraher MH, Corcoran GD, Creagh S. et al. Daptomycin-resistant Enteroccoccus faecium in a patient with no prior exposure to daptomycin. J Hosp Infect 2007; 65: 376–8. [DOI] [PubMed] [Google Scholar]
- 35. Sakoulas G, Alder J, Thauvin-Eliopoulos C. et al. Induction of daptomycin heterogeneous susceptibility in Staphylococcus aureus by exposure to vancomycin. Antimicrob Agents Chemother 2006; 50: 1581–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cui L, Tominaga E, Neoh HM. et al. Correlation between reduced daptomycin susceptibility and vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob Agents Chemother 2006; 50: 1079–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sauvage E, Kerff F, Terrak M. et al. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev 2008; 32: 234–58. [DOI] [PubMed] [Google Scholar]
- 38. Ellsworth BA, Tom NJ, Bartlett PA.. Synthesis and evaluation of inhibitors of bacterial d-alanine: d-alanine ligases. Chem Biol 1996; 3: 37–44. [DOI] [PubMed] [Google Scholar]
- 39. Reyes J, Panesso D, Tran TT. et al. A liaR deletion restores susceptibility to daptomycin and antimicrobial peptides in multidrug-resistant Enterococcus faecalis. J Infect Dis 2015; 211: 1317–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Panesso D, Reyes J, Gaston EP. et al. Deletion of liaR reverses daptomycin resistance in Enterococcus faecium independent of the genetic background. Antimicrob Agents Chemother 2015; 59: 7327–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sinel C, Cosquer T, Auzou M. et al. Sequential steps of daptomycin resistance in Enterococcus faecium and reversion to hypersusceptibility through IS-mediated inactivation of the liaFSR operon. J Antimicrob Chemother 2016; 71: 2793–7. [DOI] [PubMed] [Google Scholar]
- 42. Blake KL, O’Neill AJ.. Transposon library screening for identification of genetic loci participating in intrinsic susceptibility and acquired resistance to antistaphylococcal agents. J Antimicrob Chemother 2013; 68: 12–6. [DOI] [PubMed] [Google Scholar]
- 43. Smith JR, Barber KE, Raut A. et al. β-Lactam combinations with daptomycin provide synergy against vancomycin-resistant Enterococcus faecalis and Enterococcus faecium. J Antimicrob Chemother 2015; 70: 1738–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Miller WR, Tran TT, Diaz L. et al. LiaR-independent pathways to daptomycin resistance in Enterococcus faecalis reveal a multilayer defense against cell envelope antibiotics. Mol Microbiol 2019; 111: 811–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Parsons JB, Broussard TC, Bose JL. et al. Identification of a two-component fatty acid kinase responsible for host fatty acid incorporation by Staphylococcus aureus. Proc Natl Acad Sci USA 2014; 111: 10532–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang Y, Lv Y, Cai J. et al. A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J Antimicrob Chemother 2015; 70: 2182–90. [DOI] [PubMed] [Google Scholar]
- 47. Bourgeois-Nicolaos N, Nguyen TT, Defrance G. et al. The emergence of linezolid resistance among enterococci in intestinal microbiota of treated patients is unrelated to individual pharmacokinetic characteristics. Antimicrob Agents Chemother 2014; 58: 2681–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bi R, Qin T, Fan W. et al. The emerging problem of linezolid-resistant enterococci. J Glob Antimicrob Resist 2018; 13: 11–9. [DOI] [PubMed] [Google Scholar]
- 49. Bender JK, Cattoir V, Hegstad K. et al. Update on prevalence and mechanisms of resistance to linezolid, tigecycline and daptomycin in enterococci in Europe: towards a common nomenclature. Drug Resist Updat 2018; 40: 25–39. [DOI] [PubMed] [Google Scholar]
- 50. Dhand A, Lee L, Lobo S. et al. Characteristics of tedizolid non-susceptible enterococcal clinical isolates. Open Forum Infect Dis 2017; 4 Suppl 1: S646–7. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.