Abstract
Long non-coding RNAs (lncRNAs) have been reported to be involved in the physiological and pathological processes of tumor pathogenesis, including epithelial-mesenchymal transition (EMT). However, epidermal growth factor receptor (EGFR)-tyrosine kinase inhibitor (TKI) resistance is a major challenge in the treatment of advanced and recurrent EGFR-mutant lung adenocarcinoma. An increased understanding of the underlying mechanisms would aid in the development of effective therapeutic strategies against EGFR-TKI resistance, strategies which are urgently required for clinical therapy. In this study, long non-coding RNA LINC00460 was identified as a novel marker of a poor response to EGFR-TKI and prognosis. In lung cancer cells, LINC00460 promoted EGFR-TKI resistance as a competitive decoy for miR-149-5p, thereby facilitating interleukin (IL)-6 expression and inducing EMT-like phenotypes. The knockdown or knockout of LINC00460 in gefitinib-resistant non-small cell lung cancer cells restored the response to EGFR-TKI. In addition, as compared with patients with a low LINC00460 expression in tumors, those with a high LINC00460 expression had a significantly shorter progression-free survival following gefitinib therapy, and a shorter overall survival. Therefore, LINC00460 may be a predictor of and potential therapeutic target for EGFR-TKI resistance.
Keywords: long non-coding RNAs, LINC00460, non-small cell lung cancer, epidermal growth factor receptor tyrosine kinase inhibitor, interleukin-6
Introduction
The discovery of epidermal growth factor receptor (EGFR)-activating mutations in lung adenocarcinoma and the efficacy of EGFR-tyrosine kinases inhibitors (EGFR-TKIs) has shifted the paradigm for lung cancer treatment from cytotoxic chemotherapies to molecularly targeted therapies (1-3). The majority of patients with lung adenocarcinoma harboring EGFR-activating mutations, such as exon 19 deletion and L858R, have significant positive responses to EGFR-TKIs, including gefitinib, erlotinib, afatinib and osimertinib (4-7). Therefore, EGFR-TKIs are currently the first choice of treatment for lung adenocarcinoma with EGFR-activating mutations (6,8-11). However, despite favorable initial responses to EGFR-TKI treatment, the majority of patients eventually acquire EGFR-TKI resistance and develop recurrence within 1 year. The T790M mutation (12), MET amplification (13,14), hepatocyte growth factor overexpression (15), transformation to small-cell lung cancer (16), epithelial-mesenchymal transition (EMT) (17-20) and the excess secretion of cytokines such as interleukin-6 (IL)-6 (21) may contribute to the acquisition of resistance to EGFR-TKIs. As primary tolerance or acquired resistance is almost inevitable, the effectiveness of EGFR-TKIs appears to be limited (22). Therefore, in order to obtain a better understanding of the molecular events that result in malignancy and drug resistance in lung adenocarcinoma, the identification of novel diagnostic biomarkers is required, which may also lead to the development of more effective evidence-based therapeutics, and improved outcomes.
Recent large-scale transcriptome analyses with next-generation sequencers have reported that the majority of the genome is transcribed to non-coding RNAs that do not code for protein, including small and long non-coding RNAs (lncRNAs) (23). Although small non-coding RNAs, such as microRNAs (miRNAs or miRs) negatively regulate gene expression, few lncRNAs are well characterized (24,25). lncRNAs are arbitrarily defined as non-coding RNAs >200 nucleotides in length (26). Although lncRNAs were initially considered transcriptional noise, recent studies have reported that they play critical roles in cell growth, stem cell pluripotency and metabolism, as well as functioning as regulatory molecules in multiple diseases (27-29). Furthermore, lncRNAs have been shown to be associated with drug resistance (29-32).
Recent studies have reported that the cellular roles of lncRNAs are involved in regulating gene expression at the transcriptional and post-transcriptional levels in various cellular milieu and biological processes (33). As is the case for proteins, the functions of lncRNAs depend on correct subcellular localization. Typically, lncRNAs are responsible for the structural integrity of the nucleus and can modulate expression of nearby genes, when acting in cis in the nucleus, or distant genes, by interacting with various intracellular biomolecules, such as proteins, RNAs and DNAs, when acting in trans in the nucleus or cytoplasm (34,35). The functions of lncRNAs are mediated through distinct modes, such as signaling, scaffolds for protein-protein interactions, guides to target elements and molecular decoys (36,37). lncRNAs function as a molecular signal when they exhibit cell type-specific expression and respond to various stimuli under tight transcriptional controls. Their expression and presence can be used as potential markers of important cellular functions (38). When lncRNAs function as scaffolds, they provide a central platform for the assembly of multicomponent molecular complexes, including complexes of ribonucleoproteins (39). This function is important for the precise control of the specificity and dynamics of molecular interactions (40). Guide lncRNAs interact with molecules, including ribonucleoprotein complexes, to direct them to specific target genes (41). Finally, when lncRNAs function as molecular decoys they limit the activity of regulatory factors by binding to target molecules without exerting other functions. Molecular decoy lncRNAs have been reported to regulate targets by sequestering regulatory molecules, including transcription factors, catalytic proteins and chromatin-modifying complexes, as well as miRNAs (42,43). These types of decoy RNAs, which bind competitively to miRNAs as substitutes of targeted mRNAs, thus inhibiting their activity in the cytoplasm, are referred to as competing endogenous RNAs (ceRNAs) (44).
Although an increasing number of lncRNAs have been identified, the role of lncRNAs in EGFR-mutant lung adenocarcinoma is poorly understood. It was thus hypothesized that lncRNAs aberrantly expressed in EGFR-mutant lung adeno-carcinoma are involved in cancer malignancy and EGFR-TKI resistance. Indeed, recent experimental evidence suggests that lncRNA UCA1 is upregulated in EGFR-mutant non-small cell lung cancer (NSCLC) and induces acquired resistance to EGFR-TKIs by activating the AKT/mTOR pathway and promoting EMT (32).
In this study, we investigated the clinical importance of lncRNA expression in patients with EGFR-mutant lung adenocarcinoma by using the expression profile of lncRNAs in RNA-seq lung cancer datasets to identify a lncRNA associated with malignancy and resistance to EGFR-TKIs. To determine the clinical utility of detecting lncRNA expression in patients with EGFR-mutant lung adenocarcinoma, the lncRNA expression level in EGFR-mutant lung adenocarcinoma and the effects of its expression on lung cancer cells were examined.
Materials and methods
Bioinformatics analysis of RNA-seq and clinical data
RNA-seq and clinical datasets of The Cancer Genome Atlas (TCGA) were downloaded from the Genomic Data Commons (GDC) website (https://portal.gdc.cancer.gov) (45). RNA-seq datasets of the Cancer Cell Line Encyclopedia (CCLE) were also obtained from the GDC website (46). The R software package was used to detect the differential expression of lncRNAs and to construct the figures. The interactions of lncRNA-miRNA and mRNA-miRNA were predicted using the miRDB website (http://www.mirdb.org) and TargetScan website (http://www.targetscan.org/vert_72/).
Cell lines and reagents
A549 (CCL-185), H1299 (CRL-5803) and H1975 (CRL-5908) cells were obtained from the American Type Culture Collection (ATCC®). 293 and PC9 cells were a kind gift from Dr Yuichiro Kanno, Faculty of Pharmaceutical Sciences at Toho University (Chiba, Japan). All non-small cell lung cancer (NSCLC) cell lines were cultured in DMEM (Wako) supplemented with 10% fetal bovine serum and antibiotics. To generate gefitinib-resistant PC9 cells (PC9-GR cells), PC9 cells were continuously exposed to increasing concentrations of gefitinib (0.01-1 µM) for 6 months. The resistant cells were cultured in a gefitinib-free medium for at least 5 days before all experiments. The cell lines used in this study are listed in Table I. Gefitinib, erlotinib, recombinant human EGF and IL-6 were obtained from Wako. Osimertinib was purchased from MedChemExpress. All drugs were dissolved in DMSO and stored at −80°C. Proteins were stored at −20°C in PBS containing 30% glycerol.
Table I.
Summary of parental and engineered cell lines.
| NSCLC cell lines | EGFR mutation | Expression construct | Notes |
|---|---|---|---|
| Parental | |||
| H1299 | Wild-type | NA | - |
| A549 | Wild-type | NA | - |
| PC9 | Del E746-A750 | NA | EMT status: Epithelial |
| Gefitinib-sensitive | |||
| H1975 | L858R/T790M | NA | - |
| HCC827 | Del E746-A750 | NA | Gefitinib-sensitive |
| The lncRNA expression | |||
| levels were obtained from CCLE | |||
| Gene Expression profile dataset (46) | |||
| HCC827 GR5 | Del E746-A750 | NA | Gefitinib-resistant |
| The lncRNA expression | |||
| levels were obtained from CCLE | |||
| Gene Expression profile dataset (46) | |||
| Engineered | |||
| H1299 EGFR WT | Wild-type | pIRESpuro: EGFR WT | - |
| H1299 EGFR 19del | Del E746-A750 | pIRESpuro: EGFR Del E746-A750 | - |
| H1299 EGFR L858R | L858R | pIRESpuro: EGFR L858R | EMT status: Mesenchymal |
| PC9-GR | Del E746-A750 | NA | EMT status: Mesenchymal |
| Gefitinib-resistant | |||
| H1299 EGFR L858R | L858R | pIRESpuro: EGFR L858R | Endogenous LINC00460 |
| LINC00460 KO | knockdown (Cas9) | ||
| H1299 EGFR L858R | L858R | pIRESpuro: EGFR L858R | Endogenous |
| LINC00460 KO EV | pcDNA3-Hyg (Empty Vector) | LINC00460 | |
| knockdown (Cas9) | |||
| H1299 EGFR L858R | L858R | pIRESpuro: | Endogenous |
| LINC00460 KO OE | EGFR L858R pcDNA3-Hyg: | LINC00460 knockdown (Cas9) | |
| LINC00460 | LINC00460 overexpression |
NSCLC, non-small cell lung cancer; EMT, epithelial-mesenchymal transition; KO, knockdown; OE, overexpression.
Clinical samples
A total of 62 patients with recurrent post-operative EGFR-mutant lung adenocarcinoma treated with gefitinib during the period from January, 2008 to January, 2019 were enrolled in this study. LINC00460 expression was analyzed in 62 samples (FFPE slides of surgical specimens in 23 cases and cell-free RNA in 39 cases). Written informed consent was obtained from all patients prior to their participation in this study. RNA was extracted from the FFPE slides using an RNeasy FFPE kit (cat. no. 73504, Qiagen) and separated serum was stored at −80°C until use. Cell-free RNA in peripheral blood was extracted and purified using a QIAamp Circulating Nucleic Acid kit (cat. no. 55114, Qiagen) in accordance with the manufacturer's protocol and stored at −20°C until use. This single-center study was conducted at the Toho University Omori Medical Center and was approved by the Human Genome/Gene Analysis Research Ethics Committee (authorization no. A17117).
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted using an RNeasy Mini kit, a miRNeasy Mini kit or an RNeasy FFPE kit (Qiagen). To synthesize cDNA, total RNA (500 ng) was reacted with PrimeScript RT Master Mix (Takara Bio Inc.). Real-time PCR with specific primers (10 µM) and SYBR Premix Ex Taq (Takara Bio Inc.) was used to measure all RNA expression levels. All reactions were performed in a total volume of 25 µl. The PCR amplification program consisted of initial denaturation at 95°C for 30 sec, followed by 40 cycles of PCR at 95°C for 5 sec and 60°C for 30 sec. The comparative ΔΔCq method was used to determine the relative expression levels of RNAs (47). The relative amount of target mRNA and lncRNA was normalized to that of GAPDH mRNA. Melting curve analysis validated the specificity of the primers. The sequences of the primers used for qPCR were as follows: LINC00460 forward, 5′-GTGGATGAGAACGAAGGTTACG-3′ and reverse, 5′-CTTTCCCACGCTCAGTCTTTC-3′; human GAPDH forward, 5′-GCACCGTCAAGGCTGAGAAC-3′ and reverse, 5′-TGGTGAAGACGCCAGTGGA-3′; human IL-6 forward, 5′-GGCACTGGCAGAAAACAACC-3′ and reverse, 5′-GCAAGTCTCCTCATTGAATCC-3′; human zinc finger E-box-binding homeobox 1 (ZEB1) forward, 5′-TT CAAACCCATAGTGGTTGCT-3′ and reverse, 5′-TGGGAG ACACCAAACCAACTG-3′; human vimentin (VIM) forward, 5′-AGGCGAGGAGAGCAGGATTTC-3′ and reverse, 5′-AGT GGGTATCAACCAGAGGGAG-3′); human E-cadherin (CDH1) forward, 5′-TCGCTTACACCATCCTCAGC-3′ and reverse, 5′-GGAAACTCTCTCGGTCCAGC-3′; and human MALAT1 forward, 5′-AAAGCAAGGTCTCCCCACAAG-3′ and reverse, 5′-GGTCTGTGCTAGATCAAAAGGC-3′.
Western blot analysis
The cells were lysed with RIPA buffer (Santa Cruz Biotechnology). The total protein concentration was determined by BCA Protein Assay (Takara Bio Inc.). Gels for SDS-PAGE were cast using a TGX Fast Cast Acrylamide kit 10% (cat. no. 1610173, Bio-Rad). The lysates (30 µg of protein in total volume of 20 µl per lane) were run at a constant voltage of 200 V in Tris-Glycine SDS running buffer. The gels were then directly transferred to PVDF membranes (0.2 nm) using a Trans-Blot Turbo Blotting System (Bio-Rad). Following transfer, the membranes were incubated in blocking buffer [Tris-buffered saline with 0.1% Tween-20 (TBS-T)] containing 10% skim milk) at room temperature for 1 h. Each protein of interest was stored with a specific primary antibody in a Dilution Buffer (TBS-T with 2% BSA) at 4°C overnight, washed 3 times with TBS-T, and then reacted with an HRP secondary antibody in a Dilution Buffer at room temperature for 1 h. Chemiluminescence was developed using the Clarity Western ECL substrate, and the blots were scanned with an Amersham Imager 600 device (GE Healthcare Biosciences). The antibodies used in this experiment are listed in Table II.
Table II.
List of antibodies used in this study.
| Antibody | Manufacturer | Species | Dilution |
|---|---|---|---|
| p-EGFR (Y1068) | Abcam (ab5644) | Rabbit monoclonal | 1:1,000 for WB |
| EGFR | Abcam (ab52894) | Rabbit monoclonal | 1:1,000 for WB |
| p-STAT3 (Y705) | Abcam (ab76315) | Rabbit monoclonal | 1:2,000 for WB |
| STAT3 | Abcam (ab68153) | Rabbit monoclonal | 1:2,000 for WB |
| p-AKT (S473) | Cell Signaling Technology (#4060) | Rabbit monoclonal | 1:1,000 for WB |
| AKT | Cell Signaling Technology (#9272) | Rabbit monoclonal | 1:1,000 for WB |
| p-ERK1/2 (T202/Y204) | Cell Signaling Technology (#4370) | Rabbit monoclonal | 1:2,000 for WB |
| ERK1/2 | Cell Signaling Technology (#9102) | Rabbit monoclonal | 1:1,000 for WB |
| GAPDH | Abcam (ab9485) | Rabbit monoclonal | 1:1,000 for WB |
| Lamin B1 | Abcam (ab65986) | Rabbit monoclonal | 1:1,000 for WB |
| Normal Rabbit IgG | Sigma-Aldrich (12-370) | Rabbit | 1:50 for RIP |
| FLAG (M2) WB |
Sigma-Aldrich (F1804) | Mouse monoclonal | 1:50 for RIP, 1:1,000 for |
| Vimentin (V9) | Santa Cruz Biotechnology (sc-6260) | Mouse monoclonal | 1:1,000 for WB, 1:100 for IF |
| E-cadherin (G-10) | Santa Cruz Biotechnology (sc-8426) | Mouse monoclonal | 1:1,000 for WB, 1:100 for IF |
| Cleaved Caspase-3 (Asp175) (5A1E) | Cell Signaling Technology (#9664) | Rabbit monoclonal | 1:1,000 for WB |
| Cleaved PARP (Asp214) | Cell Signaling Technology (#9541) | Rabbit monoclonal | 1:2,000 for WB |
| B-actin | Proteintech (60008-1-Ig) | Mouse monoclonal | 1:5,000 for WB |
| HRP-linked anti-rabbit IgG | Cell Signaling Technology (#7074) | 1:3,000 for WB | |
| HRP-linked anti-mouse IgG | Cell Signaling Technology (#7076) | 1:3,000 for WB |
WB, western blot analysis; IF; immunofluorescence; RIP, RNA immunoprecipitation.
Plasmid construction
The pIRESpuro-EGFR (wild-type, 19del, L858R) expression vector was a gift from Dr Y. Kanno, Faculty of Pharmaceutical Sciences, Toho University. The KOD plus neo (Toyobo) was used for PCR to amplify the full-length LINC00460 sequence from a cDNA library derived from the total RNA from the H1299 cells. The PCR products were then used to clone and express LINC00460 in a pcDNA 3.1/Hygro (+) mammalian expression vector (Invitrogen; Thermo Fisher Scientific). All PCR products were verified by DNA sequencing.
Subcellular fractionation
The PC9 cell pellet was suspended in hypotonic buffer [10 mM Hepes (pH 7.5), 10 mM KCl, and 1.5 mM MgCl2]. Following incubation at 4°C for 10 min, the cells were centrifuged for 20 min at 1,000 x g to collect the cytoplasmic and nuclear fractions. Western blot analysis was used to validate the separation of each fraction. GAPDH was used as a cytoplasm marker. Lamin B1 were used as a nucleus marker. RNA enrichment in each fraction was performed by RT-qPCR.
RNA immunoprecipitation (RIP) assays based on the miTRAP method
miTRAP (an MS2-based RIP assay) was used to pull down endogenous miRNAs that bound to LINC00460. Cell lysates were suspended in RIP buffer containing 20 mM Tris (pH 7.5), 150 mM NaCl, 1 mM MgCl2, 5% glycerol, 0.5% NP-40 and RNase inhibitor. Subsequently, anti-FLAG antibody was added followed by incubation for 2 h at 4°C. Complexes of the MS2 stem-loop RNAs and FLAG-MS2 proteins were purified by using protein G-conjugated magnetic beads (Dynabeads Protein G, Life Technologies; Thermo Fisher Scientific). Precipitated RNA was used for cDNA synthesis and measured by RT-qPCR. The sequences of the primers were as follows: miR-149-5p forward, 5′-TCTGGCTCCGTGTCTTCACTCCC-3′. The reverse primer was supplied with the Mir-X™ miRNA RT-qPCR TB Green® kit (cat. no. 638314, Takara Bio Inc.). The antibodies used in this experiment are listed in Table II.
Luciferase reporter assays
The siCHECK™ vector (Promega) was used to determine the quantity of the RNA interference (RNAi) of miR-149-5p in accordance with the manufacturer's instructions. The cells were co-transfected with a psiCHECK2 dual-luciferase reporter (containing LINC00460, LINC00460 Mut, IL-6, or IL-6 Mut) and miRNA mimics using Lipofectamine™ 2000 (Invitrogen; Thermo Fisher Scientific). Each group was plated in triplicate using 48-well plates. Renilla luciferase activity was normalized to Firefly luciferase activity.
Small interfering RNA (siRNA)-mediated silencing
PC9-GR cells (1×105) were cultured in medium containing 10% FBS (antibiotic-free). siRNAs (100 nM; si-LINC00460#1 and≈#2) were used for transfection using TransIT-siQUEST Transfection Reagent (Mirus Bio) for 24 h after seeding the cells. Following 48 h of incubation, the cells were used to carry out subsequent experimentations (RT-qPCR and WST-8 assay). The target sequences for LINC00460 were as follows: si-LINC00460#1, 5′-UAGCAAUUGCUGGAAUC-3′; and si-LINC00460#2, 5′-CACACUUCTCGGCUAAG-3′. Luciferase siRNA and scrambled siRNA were used as negative controls.
LINC00460 knockout by CRISPR-Cas9
A vector expressing LINC00460 dual guide RNAs (gRNAs) and Cas9 was transfected into the H1299 cells in 6-well plates using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific). After 18 h, the cells were re-seeded in a 10-cm dish. The following day, puromycin (1.5 µg/ml, Thermo Fisher Scientific) was added to the cell culture for selection. Individual surviving colonies were removed and expanded in 35-mm dishes. Positive clones were identified by RT-qPCR and further verified by genomic PCR and DNA sequencing. Genomic DNA was extracted from the cell lines with a PureLink™ Genomic DNA Mini kit (cat. no. 1820-00, Thermo Fisher Scientific). The cells subjected to knockout experiments were rescued by transfection with the LINC00460 expression vector (pcDNA3.1-Hyg: LINC00460) and selected on a medium containing hygromycin B (200 µg/ml, Wako). The empty vector (i.e., pcDNA3.1-Hyg) was used as a control. The target sequences of the gRNAs for the LINC00460 coding region were as follows: gRNA1, 5′-TCCCTGTGCGCGCATAGCAG-3′; and gRNA2, 5′-CCATGCAGGGCCCCCCTCAT-3′. The primer sequences of genomic PCR were as follows: Primer-F (forward), 5′-GTGGCAGGGCAGGCCAGCTTTCCCAAAA TG-3′; and Primer-R (reverse), 5′-GGGGCTAAGCTCACC CTTTTATACCAGCACCAATC-3′.
Enzyme-linked immunosorbent assay (ELISA)
The concentrations of human IL-6 in the cell culture supernatants were measured using the Human IL-6 Quantikine ELISA kit (cat. no. D6050, R&D Systems) in accordance with the manufacturer's instructions. The density of each well in a 96-well plate was determined with a microplate reader set to 450 nm. The concentrations of each sample were determined using a standard curve to calculate the values as pg/ml.
Wound healing assays
Scratch wounds of the cell mono-layer were created using a p200 pipet tip, after which the initial images were captured. Second images were captured following 48 h of incubation at 37°C. This assay was performed in the presence of mitomycin C (0.2 mg/ml) to confirm that the result was not due to cell proliferation. The wounded areas were quantified using ImageJ software (https://imagej.nih.gov/ij/).
Immunofluorescence assays
The cells were fixed with 4% paraformaldehyde for 30 min and permeabilized with 0.2% Triton X-100 in PBS for 10 min. Following 1 h of exposure to blocking buffer, coverslips were incubated at 4°C with primary antibodies a Dilution Buffer (PBS with 0.1% Tween-20 and 2% BSA), washed with PBS-T for 3 times, and incubated with secondary antibodies for 30 min. Finally, coverslips were mounted in SlowFade™ Gold Antifade Mountant with DAPI (Invitrogen; Thermo Fisher Scientific) and imaged with a BX61 microscope (Olympus). The antibodies used in this experiment are shown in Table II.
Cell viability assay
The cells were seeded in a 96-well plate (5×103 cells/well). Cell viability was performed using a Cell Counting Kit-8 containing WST-8 (CCK-8; Dojindo Laboratories) according to the manufacturer's protocol. The number of viable cells was quantified by determining the absorbance at 450 nm with a microplate reader.
Caspase-3/7 assays
Each sample was evaluated with a caspase-3/7 activation assay performed on a Caspase-Glo 3/7® Assay System (cat. no. G8090, Promega), according to the manufacturer's instructions.
Cell treatments
293 cells were pre-treated with gefitinib (1 µM) or the solvent control (DMSO). After 1 h, the cells were treated with EGF (200 ng/ml). Following 24 h of incubation at 37°C, the expression level of LINC00460 was measured by RT-qPCR. PC9 cells were treated with rh IL-6 (50 ng/ml) for 24 h, followed by western blot analysis. PC9-GR cells were treated with gefitinib, erlotinib and osimertinib (1 µM) or the solvent control (DMSO) for 24 h following siRNA transfection. After a further 24 h, the cells were used to perform subsequent experiments (RT-qPCR and WST-8 assay).
Determination of clinical outcomes
The progression-free survival (PFS) and overall survival (OS) of patients with a high or low LINC00460 expression was estimated. The PFS of patients treated with EGFR-TKIs was assessed from the date that EGFR-TKI therapy commenced to the first sign of disease progression, as determined by findings from computed tomography or magnetic resonance imaging and the Response Evaluation Criteria In Solid Tumors (RECIST) criteria. OS was defined as the period from the date of diagnosis until death from any cause.
Statistical analyses
Statistical analyses were conducted using SPSS software for Windows, version 12.0 (SPSS, Inc.). Samples were classified as 'high' or 'low' according to whether LINC00460 expression was higher or lower than the median for all samples. Survival curves were drawn with the Kaplan-Meier method and statistical analysis was performed using the log-rank test. The t-test was used to compare data between 2 groups. One-way ANOVA with Tukey's post-hoc test was used for comparisons among multiple groups.
Results
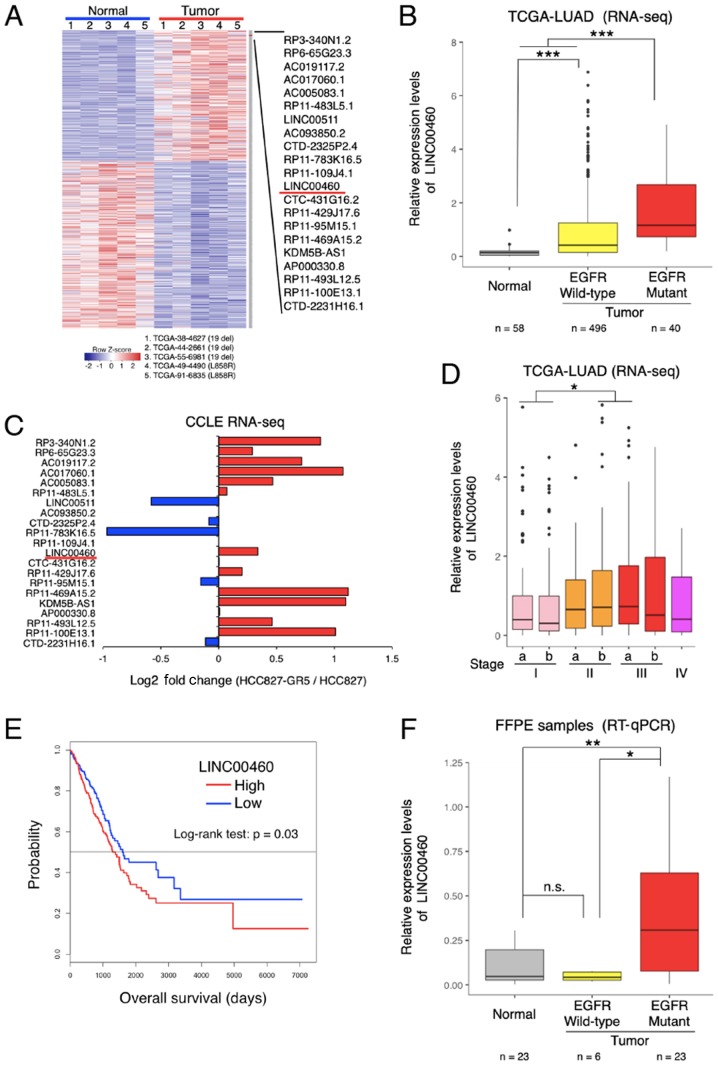
Bioinformatics analysis of RNA-seq and clinical data
The following method was used to screen candidate lncRNAs. Compared with the normal tissues, lncRNA expression was significantly dysregulated in the TCGA database (TCGA-LUAD), indicating the differential expression of lncRNAs in specimens of lung adenocarcinoma with EGFR-activating mutations (exon 19 deletion and L858R) (Fig. 1A). Among the most differentially overexpressed lncRNAs (21 lncRNAs, P<0.01, Z value >2), lncRNA-LINC00460 was selected for further analysis (NCBI Reference Sequence: NR_034119). The LINC00460 expression levels were significantly higher in wild-type EGFR and in specimens with activation mutations (exon 19 deletion and L858R) compared with normal tissues (P<0.0001; Fig. 1B). According to the RNA-seq dataset of the CCLE database, LINC00460 expression was higher in a gefitinib-resistant NSCLC cell line (HCC827 GR5) compared with a gefitinib-sensitive cell line (HCC827) (Fig. 1C). Furthermore, the analysis of the clinical TCGA dataset revealed that LINC00460 expression was significantly associated with an advanced tumor stage (Fig. 1D) and a poor prognosis (Fig. 1E). Consistent with the results of TCGA RNA-seq, LINC00460 expression was found to be significantly higher in lung adenocarcinoma tissues with EGFR-activating mutations than in normal tissues (P=0.0099) by RT-qPCR assay (Fig. 1F).
Figure 1.
Identification of lncRNAs dysregulated in EGFR-activating mutant lung adenocarcinoma. (A) The heatmap shows the expression levels of lncRNAs dysregulated in EGFR-activating mutant lung adenocarcinoma. lncRNAs were identified by RNA-seq analysis (TCGA-LUAD datasets). (B) Relative expression of LINC00460 transcripts in lung adenocarcinoma tissues and normal tissues using TCGA-LUAD RNA-seq datasets. Relative expression levels are presented as log2 (FPKM + 1). (C) Fold change in lncRNA expression in gefitinib-sensitive and -resistant HCC827 cells (HCC827 vs. HCC827-GR5). HCC827 is a gefitinib-sensitive lung adenocarcinoma cell line; HCC827-GR5 is a gefitinib-resistant lung adenocarcinoma cell line. lncRNAs were identified by using the log2 (RPKM + 1) value from RNA-seq datasets (CCLE). (D) LINC00460 expression in relation to clinical stage, based on the T-factor in lung adenocarcinoma, determined using RNA-seq and clinical data of TCGA-LUAD. (E) Kaplan-Meier curves for the overall survival of patients with lung adenocarcinoma. LINC00460 expression levels and clinical data were obtained from TCGA data. (F) LINC00460 expression in lung adenocarcinoma tissues with EGFR-activating mutations or wild-type EGFR, and in normal tissues. The data were determined by using RT-qPCR. GAPDH mRNA was used as an internal control. The level of statistical significance was set at a P-value <0.05 (*P<0.05, **P<0.01, ***P<0.001). n.s., not significant.
lncRNA expression in cell lines
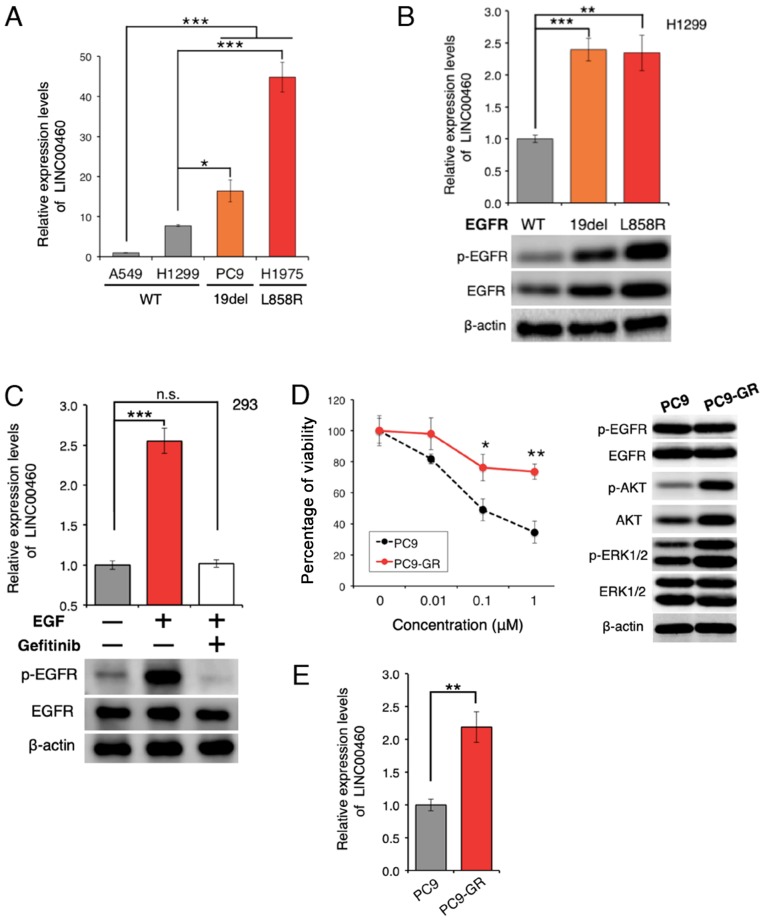
The expression of LINC00460 was analyzed in 4 lung cancer cell lines. LINC00460 expression was found to be significantly higher in lung cancer cell lines with EGFR-activating mutations than in wild-type cell lines (Fig. 2A). Furthermore, transfection with active EGFR mutations resulted in a higher LINC00460 expression, as compared with the expression in H1299 cells expressing wild-type EGFR (Fig. 2B). EGFR activation induced by treatment with EGF yielded similar results; however, EGFR inactivation by pre-treatment with gefitinib significantly attenuated the EGFR-induced increase in LINC00460 expression (Fig. 2C). These results suggest that the abnormal activation of EGFR is associated with the overexpression of LINC00460. To determine whether LINC00460 is overexpressed in gefitinib-resistant cells, gefitinib-resistant PC9 cells (PC9-GR cells) were generated (Fig. 2D). The expression level of LINC00460 was found to be significantly higher in the PC9-GR cells than in the PC9 cells (Fig. 2E), indicating that LINC00460 is involved in resistance against EGFR-TKI.
Figure 2.
Overexpression of LINC00460 in EGFR-mutant lung adenocarcinoma. (A) Relative expression of LINC00460 transcripts in cancer cell lines derived from NSCLC. LINC00460 expression was higher in cell lines harboring EGFR exon 19 deletions and L858R (PC9 and H1975) than in those without EGFR mutations (A549 and H1299) (P<0.001). The data were obtained by RT-qPCR. GAPDH mRNA was used as an internal control. (B) Upregulation of LINC00460 expression triggered by EGFR-activating mutations. LINC00460 expression was significantly upregulated in EGFR-activated mutant-positive cells. H1299 (EGFR wild-type) cells were transfected with pIRES-puro: EGFR-WT, exon 19 deletion, and L858R and selected by puromycin. (C) When EGF (200 ng/ml) was used as an EGFR activator, EGFR activation induced LINC00460 expression. EGFR inactivation was induced by pretreatment with the EGFR-TKI gefitinib (1 µM). (D) Establishment of an acquired gefitinib-resistant cell line. PC9 cells were highly sensitive to gefitinib. PC9-GR refers to gefitinib-resistant PC9 cells. PC9 or PC9-GR cells were treated with gefitinib and cell viability was determined after 72 h by WST-8 assay (left). Western blot analysis was used to observe changes in activation or expression levels of proteins in the indicated cells (right). (E) Aberrant expression of LINC00460 in gefitinib-resistant cells. LINC00460 expression levels were higher in cells with acquired resistance to gefitinib. Expression levels in PC9 and PC9-GR cells were measured by RT-qPCR. GAPDH was used as an internal control. The results of RT-qPCR are presented as the means ± SD of at least 3 independent experiments. The level of statistical significance was set at P-value <0.05 (*P<0.05, **P<0.01, ***P<0.001). n.s., not significant.
Function of LINC00460 as a ceRNA for miR-149-5p facilitating IL-6 production
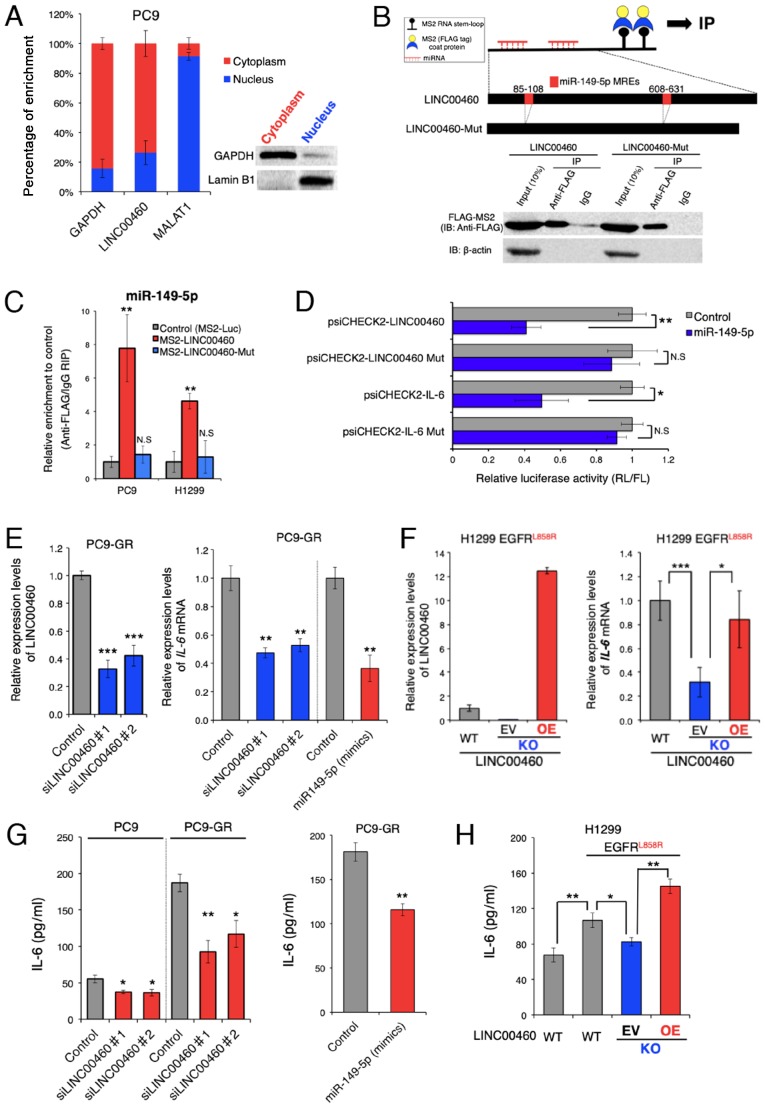
To investigate whether LINC00460 transcripts induce EGFR-TKI resistance, the potential modes of action of LINC00460 were determined in experimental studies. To predict the biological function of LINC00460 transcripts in EGFR-activating mutated lung cancer cells, LINC00460 localization patterns were investigated in PC9 cells. As LINC00460 is mainly localized in the cytoplasm (Fig. 3A), it was hypothesized that it functions as a ceRNA and acts as a decoy for miRNA from the corresponding miRNA-targeted transcripts (43). To examine this hypothesis, we used bioinformatics analysis (miRDB and TargetScan) to identify candidate miRNAs that interact with LINC00460. To pull down endogenous miRNAs associated with LINC00460 in lung cancer cells, an MS2-based RIP was performed based on the previously described miTRAP method (44) (Fig. 3B). Subsequently, the LINC00460-specific enrichment of the candidate miRNAs was validated by RT-qPCR. miR-149-5p enrichment was found to be significantly greater in RNAs retrieved from MS2-LINC00460 than in those from controls (MS2-Luciferase mRNA) and MS2-LINC00460-Mut (MRE-deleted MS2-LINC00460) (Fig. 3C). However, we could not pull down other candidate-targeted miRNAs (data not shown). These data suggest the presence of a specific interaction between LINC00460 and miR-149-5p in NSCLC cells and demonstrate that LINC00460 has two putative miRNA response elements (MREs) of miR-149-5p (Fig. 3B). Of note, bioinformatics analysis revealed that miR-149-5p binds to the MRE in the 3′UTR of IL-6 mRNA. However, in lung adenocarcinoma, the association between LINC00460, miR-149-5p and IL-6 is unknown. Therefore, subsequent experiments focused on the ceRNA function of LINC00460 to target miR-149-5p.
Figure 3.
LINC00460 transcripts function as a ceRNA for miR-149-5p in regulating IL-6 expression. (A) Cell nucleus/cytoplasm fractionation and RT-qPCR indicating the cellular distribution of LINC00460 in PC9 cells. GAPDH was used as a cytoplasm marker. MALAT1 and Lamin B1 were used as nucleus markers. Data are presented as the means ± SD (n=3). (B) Upper panel: Schematic of the binding 2X MS2 stem-loop fused bait RNA to MS2 coat proteins used for the RIP assay based on the miTRAP method and schematic outline of predicted binding sites for miR-149-5p on the LINC00460 sequence. Lower panel: Western blot analysis of FLAG-tag fused MS2 proteins isolated from PC9 input fractions or co-purified with (MS2-)LINC00460 and (MS2-)LINC00460 Mut (deletion mutant of the MREs), respectively. IgG served as a negative control for nonspecific pull downs. β-actin was used as an internal control. (C) MS2-based RIP assay with anti-FLAG antibody in PC9 or H1299 cells. The cells were incubated for 48 h following transfection with a MS2-FLAG plasmid, along with MS2-LINC00460, MS2-LINC00460 Mut, or MS2bs-Luc (control vectors). The results of RT-qPCR are presented as the means ± SD (n=3). (D) Relative luciferase activity of psiCHECK2-LINC00460, LINC00460 Mut, IL-6, and IL-6 Mut upon transfection with miR-149-5p mimics in H1299 cells. psiCHECK2-LINC00460 and psiCHECK2-IL-6 are psiCHECK2 vectors that contain LINC00460 and the 3'UTR of IL-6 mRNA, respectively. Mut stands for deletion mutant of miR-149-5p MREs. Data are presented as the ratio of Renilla luciferase activity (RL) to Firefly luciferase activity (FL). Error bars represent the means ± SD (n=3). (E) Relative expression levels of LINC00460 (left panel) and IL-6 mRNA (right panel) following transfection with control (si-Luc), si-LINC00460#1, and si-LINC00460#2. The results of RT-qPCR are presented as the means ± SD (n=3). (F) Left panel: Detection of LINC00460 in knockout (KO) clones by RT-qPCR. H1299 cells expressing EGFR L858R were transfected with the Cas9 plasmid vector for LINC00460 knockout. The KO cells were rescued by the LINC00460 expression vector. Right panel: Relative expression of IL-6 mRNA in the engineered H1299 cells. The results of RT-qPCR are presented as the means ± SD (n=3). (G and H) Concentrations of IL-6 (pg/ml) in cell culture supernatants were measured by ELISA. (G) Concentration of IL-6 in PC9 or PC9-GR following transfection with control (si-Luc), si-LINC00460#1, si-LINC00460#2 (left), and miR-149-5p mimics (right panel). (H) Concentration of IL-6 in engineered H1299 cells. WT, wild type; KO, knockout; EV, empty vector; OE, overexpression. The results of ELISA are presented as the means ± SD (n=3). The level of statistical significance was P-value set at <0.05 (*P<0.05, **P<0.01, ***P<0.001). n.s., not significant.
To further investigate the RNAi effect, the coding regions of LINC00460 or the 3'UTR of IL-6 were cloned into the psiCHECK2™ vector, which transcribes the fusion of the reporter gene (R. luciferase) and the sequence of interest. Co-transfection of these vectors and a vector expressing miR-149-5p mimics significantly inhibited the luciferase activity of psiCHECK2-LINC00460 and psiCHECK2-IL-6, while each of the MRE deletion mutants (psiCHECK2-LINC00460 Mut or IL-6 Mut) did not exhibit a significant response to the miR-149-5p mimics (Fig. 3D). These results indicate that LINC00460 and IL-6 mRNA contained functional miR-149-5p binding sites.
Subsequently, the effect of LINC00460 expression on IL-6 production was investigated, as IL-6 mRNA expression is regulated by miR-149-5p. The expression of IL-6 mRNA was significantly decreased by the siRNA knockdown of LINC00460 transcripts (Fig. 3E, right panel). This result was consistent with the outcome of the enforced expression of miR-149-5p in PC9-GR cells (Fig. 3E). To accurately determine the potential effect of LINC00460 on the responses to EGFR-TKIs, a Cas9-mediated LINC00460 knockout cell line was generated from H1299 cells, which harbored EGFR L858R rather than EGFR T790M and exhibited intrinsic resistance to EGFR-TKIs (48-50) (Figs. 3F, right panel and S1). The knockdown of LINC00460 significantly deceased IL-6 mRNA expression, whereas the overexpression of LINC00460 in LINC00460 knockout cells (performing a rescue experiment) increased IL-6 mRNA expression (Fig. 3F, left panel). ELISA revealed that IL-6 expression was attenuated at the protein level when LINC00460 was knocked down, and when the expression of miR-149-5p mimics was enforced in PC9-GR cells (Fig. 3G). Although the acquisition of an EGFR-activating mutation (L858R) by the H1299 cells induced the significant production of IL-6 and LINC00460 expression, LINC00460 knockdown decreased the IL-6 protein levels in parental cells (Figs. 2B and 3H). Conversely, the enforced overexpression of LINC00460 significantly promoted IL-6 production in the cells with LINC00460 knockdown (Fig. 3H). Taken together, these findings suggested that LINC00460 expression, at least in part, promoted IL-6 production by directly acting as a decoy for miR-149-5p associated with the degradation of IL-6 mRNA.
Effects of LINC00460 on phenotypes associated with EGFR-TKI resistance in EGFR-mutated lung cancer
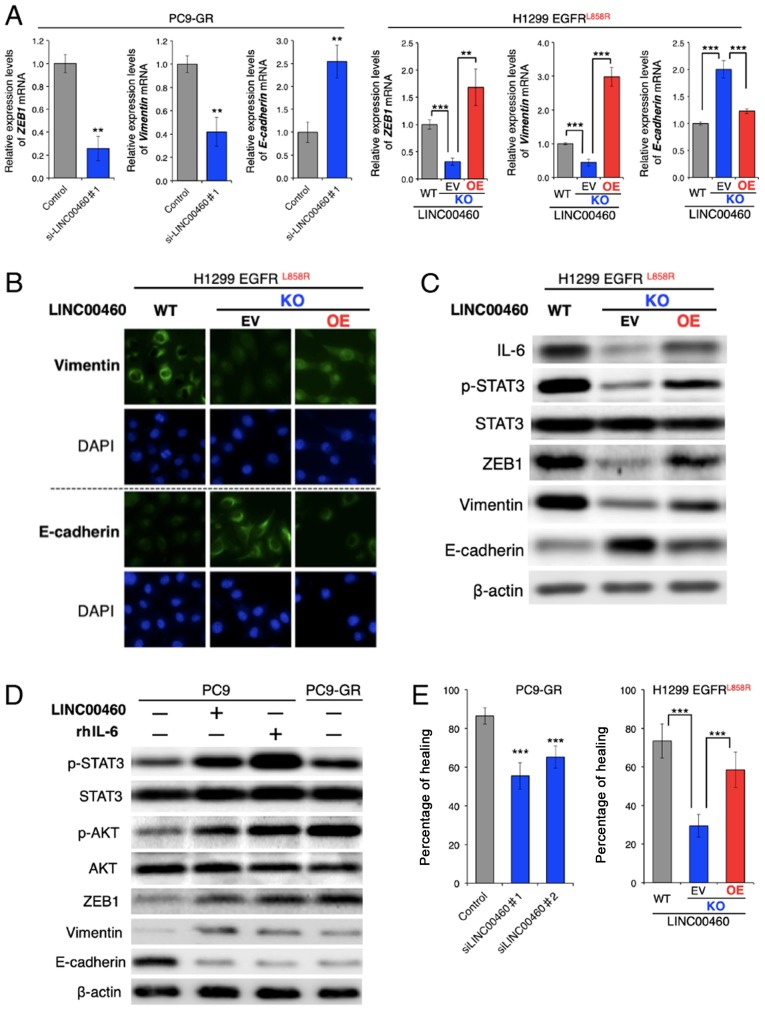
The overexpression of IL-6 in tumors triggers inflammation and primary resistance or a low response to EGFR-TKIs in preclinical models (22). Research on the mechanisms involved in EGFR-TKI resistance has reported that IL-6 induces an EMT phenotype by activating the JAK/STAT3 or AKT signaling pathway (51), which suggests the loss of E-cadherin and increased vimentin and ZEB1 expression (21).
Although the parental PC9-GR and H1299 cells had a mesenchymal phenotype, RT-qPCR analysis of the ZEB1, vimentin (VIM) and E-cadherin (CDH1) mRNA expression levels revealed that LINC00460 knockdown significantly decreased ZEB1 and VIM, and induced CDH1 expression. The overexpression of LINC00460 exerted the opposite effect on the expression levels of these EMT-related mRNAs in the cells in which H1299 LINC00460 had been knocked out (Fig. 4A). In addition, an immunofluorescence assay revealed that LINC00460 knockdown increased the E-cadherin and decreased the vimentin protein levels. Consistently, LINC00460 overexpression exerted the opposite effect (Fig. 4B). Furthermore, western blot analysis clearly indicated that LINC00460 knockdown suppressed the phosphorylation of STAT3 and altered the expression levels of EMT-related proteins, including ZEB1, vimentin and E-cadherin, and that these changes were reversed by LINC00460 overexpression (Fig. 4C). In PC9 cells, which are an epithelial phenotype, transfection with LINC00460 and treatment with human recombinant IL-6 (rhIL-6) induced a similar expression pattern of EMT-related genes to that of gefitinib-resistant PC9-GR cells. These stimuli activated the STAT3 and AKT pathways, promoted the expression levels of ZEB1 and vimentin, and inhibited E-cadherin expression (Fig. 4D). In the context of EMT, silencing LINC00460 expression inhibited the migration of PC9-GR cells. The migration of the cells subjected to LINC00460 knockout was significantly lower than of the parental H1299 cells. Moreover, cells subjected to LINC00460 knockout and subsequently to the overexpression of LINC00460 exhibited a phenotype similar to the original (Fig. 4E). Taken together, these results indicate that LINC00460 overexpression promoted an EMT-like phenotype by upregulating IL-6 production.
Figure 4.
Effects of LINC00460 expression on EMT and cell migration. (A) Changes in gene expression levels of ZEB1, vimentin (VIM) and E-cadherin (CDH1) were measured in PC9-GR and engineered H1299 cells. The results of RT-qPCR are presented as the means ± SD (n=3). (B) Immunofluorescence was used to observe changes in expression levels of vimentin and E-cadherin in engineered H1299 cells. DAPI was used to stain the cell nuclei. (C) Western blot analysis was used to observe changes in activation or expression levels of EMT-related proteins in the indicated cells. β-actin was used as a control. (D) Western blot analysis of the expression of EMT-related proteins and downstream proteins of IL-6 following transient transfection with LINC00460 and treatment of PC9 cells with rh IL-6 (50 ng/ml) or untreated PC9-GR cells. (E) Wound healing assays of PC9-GR cells transfected with siRNAs targeting LINC00460 (left panel) or engineered H1299 cells (right panel). Data for the healing areas are presented as the means ± SD (n=3). WT, wild type; KO, knockout; EV, empty vector; OE, overexpression; rhIL-6, recombinant human interleukin-6. The level of statistical significance was set at P-value <0.05 (**P<0.01, ***P<0.001). n.s., not significant.
Effect of LINC00460 on responses to EGFR-TKIs
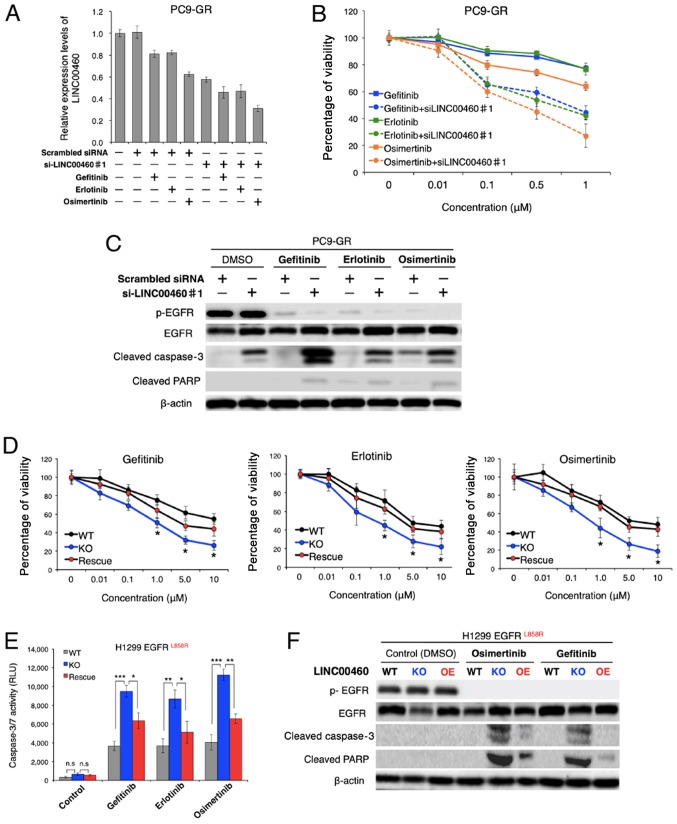
To evaluate the therapeutic potential of LINC00460 for gefitinib-resistant NSCLC, the issue of whether LINC00460 silencing restores responses to EGFR-TKIs (gefitinib, erlotinib and osimertinib) was investigated in NSCLC harboring activating EGFR mutations in vitro. Of note, while treatment with each EGFR-TKIs alone exerted a minimal inhibitory effect on the viability of PC9-GR cells, the combined application of single EGFR-TKIs and LINC00460 siRNA markedly decreased the LINC00460 expression level and cell viability of PC9-GR (Fig. 5A and B). Moreover, the results of western blot analysis revealed that all combinations of a single EGFR-TKI and LINC00460 siRNA promoted the cleavage of caspase-3 and PARP in PC9-GR cells, indicating that apoptosis was induced in these cells (Fig. 5C). Subsequently, the question of whether the expression levels of LINC00460 are associated with cell viability in response to EGFR-TKIs in NSCLC harboring EGFR-activating mutations without T790M was investigated. As shown in Fig. 5D, viability following treatment with various EGFR-TKIs was significantly decreased by LINC00460 knockout in H1299 cells harboring L858R. Conversely, the overexpression of LINC00460 in the cells which had been subjected to LINC00460 knockout cells increased the number of surviving cells in all experiments. In addition, caspase 3/7 activity in these cells following treatment with EGFR-TKIs was investigated. Caspase-Glo 3/7 assay and western blot analysis indicated that the effects of LINC00460 expression on cell viability were, at a minimum, involved in significantly inducing or inhibiting the apoptotic pathway (Fig. 5E and F). Taken together, these findings suggested that the combined use of siRNA targeting LINC00460 and EGFR-TKIs induced the apoptosis of EGFR-mutated, LINC00460-overexpressing, NSCLC cells resistant to EGFR-TKI monotherapy.
Figure 5.
LINC00460 silencing restores responses to EGFR-TKIs in NSCLC cells. (A-C) PC9-GR cells were treated with gefitinib, erlotinib and osimertinib or combinations of a EGFR-TKI (1 µM) and siRNA (100 nM) targeting LINC00460 for 24 h. Scrambled siRNA and DMSO was used as negative control. (A) Relative expression of LINC00460 was measured by RT-qPCR. GAPDH mRNA was used as an internal control. (B) Cell viability was determined by WST-8 assay. (C) Cell apoptosis was detected by western blot analysis. (D-F) Effect of LINC00460 expression on cell viability of engineered H1299 cells treated with EGFR-TKIs. (D) Percentage viability following treatment with various EGFR-TKIs was determined after 72 h by WST-8 assay. (E) Relative activation of caspase-3/7 following treatment with EGFR-TKIs (5 µM) was determined by caspase-Glo 3/7 assay after 72 h. DMSO was used as a control. RLU, relative light units. (F) Western blot analysis of cells treated with each EGFR-TKI (5 µM) for 72 h. WT, wild-type; KO, knockout; EV, empty vector; OE, overexpression. Results are presented as the means ± SD (n=3). The level of statistical significance was P-value set <0.05 (*P<0.05, **P<0.01, ***P<0.001). n.s., not significant.
LINC00460 expression predicts a shorter PFS and OS of patients with EGFR-mutant NSCLC treated with EGFR-TKIs
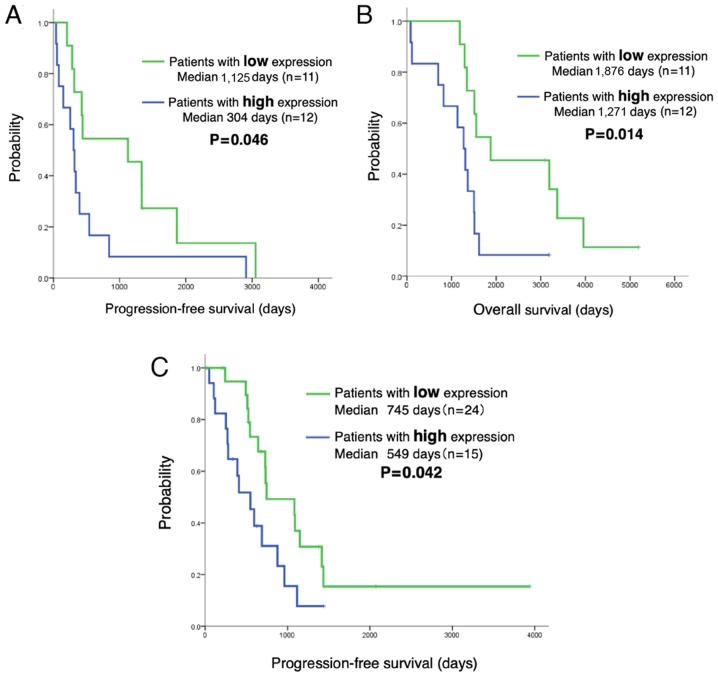
The analysis of the LINC00460 status in 62 patients with EGFR-mutant lung adenocarcinoma treated with EGFR-TKI revealed no significant differences in the clinical characteristics (age, sex, performance status, smoking history, exon 19 deletion or L858R, EGFR-TKI and metastatic site) between those with a high LINC00460 expression (n=27) and those with a low LINC00460 expression (n=35). The PFS and OS of patients in relation to LINC00460 expression was then estimated. Notably, Kaplan-Meier survival analysis indicated that patients with a high LINC00460 expression in their tumors (n=12) had a significantly shorter PFS and OS following gefitinib therapy than those with a low expression (n=11) (median PFS: 304 vs. 1,125 days, P=0.046, respectively; OS: 1,271 vs. 1,876 days, P=0.014; Fig. 6A and B). Moreover, PFS was significantly shorter for patients with a high LINC00460 expression in cell-free RNA (n=15) following EGFR-TKI therapy than for those with a low LINC00460 expression (n=24) (median PFS: 549 vs. 745 days, P=0.042, respectively) (Fig. 6C).
Figure 6.
LINC00460 expression predicts a shorter PFS and OS in patients with EGFR-mutant NSCLC treated with EGFR-TKI therapy. (A) PFS following gefitinib therapy. LINC00460 expression in patients with EGFR-mutant lung adenocarcinoma was classified as low or high by using median LINC00460 expression as the cut-off. PFS after gefitinib therapy was significantly shorter for those with a high LINC00460 expression in tumors compared with those with a low LINC00460 expression in tumors (median PFS: 304 vs. 1,125 days, respectively; P=0.046). (B) OS. OS was significantly shorter for patients with a high LINC00460 expression in tumors compared with those with a low LINC00460 expression in tumors (median OS: 1,271 vs. 1,876 days, respectively; P=0.014). (C) Prediction of PFS using LINC00460 expression levels in plasma cell-free RNA. PFS was significantly shorter for patients with a high LINC00460 expression in cell-free RNA (n=15) following EGFR-TKI therapy than for those with a low LINC00460 expression (n=24) (median PFS, 549 vs. 745 days, respectively; P=0.042).
Discussion
The present study on the role of lncRNAs in lung adenocarcinoma with EGFR mutation revealed that LINC00460 overexpression in lung cancer cells was caused by the abnormal activation of EGFR signaling attributable to the excessive stimulation of EGF and acquisition of a cancer driver mutation in EGFR. EGFR expression further increased when these cells acquired gefitinib resistance. The assessment of the biological significance of LINC00460 in lung cancer cells suggested that this RNA functioned as a competing endogenous RNA decoy for the tumor suppressive miRNA-miR-149-5p, which then significantly promoted IL-6 production and EMT-like traits. In addition, with respect to the novel clinical importance of LINC00460 in EGFR mutation-positive lung cancer, it was found that the silencing LINC00460 in combination with treatment with EGFR-TKIs (gefitinib, erlotinib and osimertinib) exerted a more potent inhibitory effect on the viability of EGFR-mutant NSCLC cells. These findings suggest that a high LINC00460 expression renders EGFR-TKI treatment ineffective and may prevent the development of intrinsic resistance to EGFR-TKIs and the emergence of drug-tolerant cells in EGFR-mutated lung cancers that overexpress LINC00460.
LINC00460 has recently been reported to be an oncogenic lncRNA in types of various cancer (29). These findings suggest that LINC00460 is overexpressed in various types of cancer, including nasopharyngeal, colorectal, ovarian and lung cancers, and that LINC00460 expression promotes cell growth and migration and increases tumor volume and weight in vivo (52-55). These studies also indicate that the binding of LINC00460 to various miRNAs is the molecular mechanism through which LINC00460 affects cell function.
Subsequently, this study whether LINC00460 overexpression disrupts the miRNA regulatory machinery, which may affect EGFR-TKI resistance in lung cancer cells and worsens the outcomes of lung cancer patients. It was found that LINC00460, the oncogenic function of which was attributed to the inhibition of miR-149-5p, promoted its target gene, IL-6, in EGFR-mutated lung cancer with limited responsiveness to EGFR-TKIs. The tumor suppressor, miR-149-5p, has been shown to inhibit fibroblast activation by reducing IL-6 expression (56). In addition, cancer-associated fibroblasts enhance EMT and the stem-like traits of human gastric cancer cells by inducing IL-6 expression by means of an miR-149-5p dependent mechanism (56). Moreover, a previous study on nasopharyngeal cancer indicated that LINC00460 facilitated carcinogenesis by acting as a decoy for miR-149-5p to upregulate IL-6 expression (52). The discovery of the function of this LINC00460 supports the current findings regarding EGFR-mutated lung cancer. IL-6 is a cytokine associated with a number of chronic inflammatory diseases. Furthermore, IL-6 expression is involved in the regulation of tumor growth and metastatic spread, including that of lung cancers. Activated mutant EGFRs activates the gp130/JAK/STAT3 pathway by upregulating IL-6 in primary human lung adenocarcinoma cells (57). The IL-6-inducible activation of JAK1/STAT3 signaling also induces de novo resistance to EGFR-TKIs in NSCLC with a T790M mutation (58). These results do not contradict the current findings that LINC00460 overexpression activates STAT3 by promoting IL-6 production in EGFR-activating mutant lung cancer cells with EGFR-TKI resistance (Fig. 4C and D).
In addition, the acquisition of mesenchymal-like traits by the activation of IL-6 inducible STAT3 and AKT signaling contributes to EGFR-TKI resistance (59). EMT also functions as a potential mechanism of EGFR-TKI resistance in NSCLC (18,23). This property has also been observed in a subset of NSCLC patients who developed EGFR-TKI resistance (59). EMT was originally reported to be an essential process in the developmental stage and cell migration during early embryogenesis. When EMT occurs in cancer cells, they lose their epithelial properties and gain mesenchymal characteristics. The epithelial cell marker, E-cadherin, is a cell adhesion molecule that suppresses cancer cell motility, whereas the mesenchymal cell markers, VIM and ZEB1 enhance cancer cell migration and invasion (23). Consistent with these results, a previous study indicated that LINC00460 promoted EMT, as well as cell migration and invasion (53). Taken together, these previous findings support those of the current study, indicating that the upregulation of LINC00460 is involved in the mechanism of cancer malignancy and EGFR-TKI resistance induced by IL-6-inducible EMT. However, the effects of LINC000460 on cell phenotypes and drug sensitivity suggest that LINC00460 interacts with various other miRNAs and proteins to control cell signaling, in addition to the molecular mechanism described in the present study. Furthermore, LINC00460 may code for a small peptide under certain conditions (60), although we were unable to identify this peptide in the present study (data not shown). Therefore, further studies of the molecular mechanisms underlying LINC00460 efficacy are warranted.
A high LINC00460 expression has recently been reported to be associated with a poor prognosis in certain types of cancer, including nasopharyngeal carcinoma (52) and colorectal carcinoma (55). A similar association was observed in the present analysis of EGFR-mutated lung adenocarcinoma. In this study, patients with a high LINC00460 expression in tumors had a significantly shorter PFS following gefitinib therapy and OS compared with those with a low LINC00460 expression. A recent clinical study on patients harboring EGFR-mutant lung cancer suggested that patients with high IL-6 levels in tumors had a significantly shorter PFS following gefitinib therapy (61), which is in accordance with the current findings that LINC00460 regulated IL-6 levels in lung cancer. Moreover, it was found that the sensitivity to EGFR-TKIs (gefitinib, erlotinib and osimertinib) was reduced by an increased LINC00460 expression. Osimertinib is a third-generation EGFR-TKI and is approved for the treatment of EGFR-T790M-positive NSCLC in patients with acquired resistance to first- or second-generation EGFR-TKIs. Currently, first-line treatment with osimertinib is considered a standard therapy for patients with EGFR-mutant NSCLC. However, resistance to osimertinib in these patients is similar to that observed with other EGFR-TKIs. The current study deepens our understanding of the molecular mechanisms underlying resistance to osimertinib in EGFR-mutated NSCLC. It is expected that the LINC00460 expression level may be a novel predictor of the effects of EGFR-TKI and a prognostic indicator of EGFR mutation-positive NSCLC.
This study had some limitations. First, it was a retrospective single-center study with a small sample size. Although differences in clinical outcome in relation to LINC00460 expression were identified, the number of patients enrolled was too small to consider the association of LINC00460 expression with the PFS and OS. Thus, a large-scale multicenter study is required to confirm the validity of these results.
In conclusion, the findings of this study suggest that LINC00460 overexpression is associated with a poor response to the latest EGFR-TKI (i.e., osimertinib), as well as to first-generation EGFR-TKIs (i.e., gefitinib and erlotinib), and with poor outcomes following gefitinib treatment. A novel therapeutic strategy targeting LINC00460 may overcome EGFR-TKI resistance and improve the prognosis of patients with EGFR-mutant lung adenocarcinoma. Advances in the preclinical and clinical development of RNAi drugs and their drug delivery systems, and in technologies for RNA detection, have increased the sophistication of drug and diagnostic development processes (62,63). The discovery of the importance of LINC00460 may lead to its use as a prognostic predictor, diagnostic indicator of EGFR-TKIs, and as a molecular target of pharmaceuticals. Moreover, the present findings may lead to development of individualized treatments for lung cancer. Therefore, the clinical importance of research on LINC00460 is likely to increase.
Supplementary Data
Acknowledgments
The authors would like to thank Mr. Atsushi Kakimoto of Konica Minolta Inc., as well as Mr. Yuichiro Kanno and Mr. Tomohiro Ariyama, of the Faculty of Pharmaceutical Sciences, Toho University for providing assistance with this study. The authors would also like to thank Mr. David Kipler for his editorial review of the article. The abstract for this article was presented at the 2019 Annual Meeting of the American Society of Clinical Oncology (May, 31 to June 4, 2019) in Chicago, IL, USA and was published as abstract no. e20529 in the Journal of Clinical Oncology 37, 2019.
Glossary
- lncRNAs
long non-coding RNAs
- NSCLC
non-small cell lung cancer
- EGFR
epidermal growth factor receptor
- EGFR-TKI
epidermal growth factor receptor-tyrosine kinase inhibitor
- IL-6
interleukin-6
- PFS
progression-free survival
- OS
overall survival
- PCR
polymerase chain reaction
- HGF
hepatocyte growth factor
- EMT
epithelial-mesenchymal transition
- TCGA
The Cancer Genome Atlas
- GDC
Genomic Data Commons
- CCLE
Cancer Cell Line Encyclopedia
- RECIST
response evaluation criteria in solid tumors
- EGF
epidermal growth factor
- KO
knockout
- OE
overexpression
Funding
This study was supported by JSPS KAKENHI (grant nos. JP15K09195 and JP18K08189).
Availability of data and materials
All data generated or analyzed during this study are included in this published article or are available from the corresponding author on reasonable request.
Authors' contributions
KI, YN, TM and SH conceived and designed the study. KKa, HK, TI and YN performed the experiments. KI and YN wrote the manuscript. SS, YT, NT, AI, SH and KKi were involved in the conception and design of the study and reviewed and edited the manuscript. All authors have read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
This single-center study was conducted at the Toho University Omori Medical Center and was approved by the Human Genome/Gene Analysis Research Ethics Committee (autho-rization no. A17117). Written informed consent was obtained from all patients prior to participation.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Hoffman PC, Mauer AM, Vokes EE. Lung cancer. Lancet. 2000;355:479–485. doi: 10.1016/S0140-6736(99)08003-4. [DOI] [PubMed] [Google Scholar]
- 2.Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 3.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 4.Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I, et al. North-East Japan Study Group Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 5.Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, Palmero R, Garcia-Gomez R, Pallares C, Sanchez JM, et al. Spanish Lung Cancer Group in collaboration with Groupe Français de Pneumo-Cancérologie and Associazione Italiana Oncologia Toracica: Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): A multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 6.Yang JC, Wu YL, Schuler M, Sebastian M, Popat S, Yamamoto N, Zhou C, Hu CP, O'Byrne K, Feng J, et al. Afatinib versus cisplatin-based chemotherapy for EGFR mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6): Analysis of overall survival data from two randomised, phase 3 trials. Lancet Oncol. 2015;16:141–151. doi: 10.1016/S1470-2045(14)71173-8. [DOI] [PubMed] [Google Scholar]
- 7.Mok TS, Wu YL, Ahn MJ, Garassino MC, Kim HR, Ramalingam SS, Shepherd FA, He Y, Akamatsu H, Theelen WS, et al. AURA3 Investigators Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N Engl J Med. 2017;376:629–640. doi: 10.1056/NEJMoa1612674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, Tsurutani J, Seto T, Satouchi M, Tada H, Hirashima T, et al. West Japan Oncology Group Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): An open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 9.Zhou C, Wu YL, Chen G, Feng J, Liu XQ, Wang C, Zhang S, Wang J, Zhou S, Ren S, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): A multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 10.Kuan FC, Kuo LT, Chen MC, Yang CT, Shi CS, Teng D, Lee KD. Overall survival benefits of first-line EGFR tyrosine kinase inhibitors in EGFR-mutated non-small-cell lung cancers: A systematic review and meta-analysis. Br J Cancer. 2015;113:1519–1528. doi: 10.1038/bjc.2015.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soria JC, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, Dechaphunkul A, Imamura F, Nogami N, Kurata T, et al. FLAURA Investigators Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med. 2018;378:113–125. doi: 10.1056/NEJMoa1713137. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi S, Boggon TJ, Dayaram T, Jänne PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG, Halmos B. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 13.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 14.Tong JH, Yeung SF, Chan AW, Chung LY, Chau SL, Lung RW, Tong CY, Chow C, Tin EK, Yu YH, et al. MET Amplification and Exon 14 Splice Site Mutation Define Unique Molecular Subgroups of Non-Small Cell Lung Carcinoma with Poor Prognosis. Clin Cancer Res. 2016;22:3048–3056. doi: 10.1158/1078-0432.CCR-15-2061. [DOI] [PubMed] [Google Scholar]
- 15.Yano S, Yamada T, Takeuchi S, Tachibana K, Minami Y, Yatabe Y, Mitsudomi T, Tanaka H, Kimura T, Kudoh S, et al. Hepatocyte growth factor expression in EGFR mutant lung cancer with intrinsic and acquired resistance to tyrosine kinase inhibitors in a Japanese cohort. J Thorac Oncol. 2011;6:2011–2017. doi: 10.1097/JTO.0b013e31823ab0dd. [DOI] [PubMed] [Google Scholar]
- 16.Marcoux N, Gettinger SN, O'Kane G, Arbour KC, Neal JW, Husain H, Evans TL, Brahmer JR, Muzikansky A, Bonomi PD, et al. EGFR-Mutant Adenocarcinomas That Transform to Small-Cell Lung Cancer and Other Neuroendocrine Carcinomas: Clinical Outcomes. J Clin Oncol. 2019;37:278–285. doi: 10.1200/JCO.18.01585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suda K, Tomizawa K, Fujii M, Murakami H, Osada H, Maehara Y, Yatabe Y, Sekido Y, Mitsudomi T. Epithelial to mesenchymal transition in an epidermal growth factor receptor-mutant lung cancer cell line with acquired resistance to erlotinib. J Thorac Oncol. 2011;6:1152–1161. doi: 10.1097/JTO.0b013e318216ee52. [DOI] [PubMed] [Google Scholar]
- 18.Weng CH, Chen LY, Lin YC, Shih JY, Lin YC, Tseng RY, Chiu AC, Yeh YH, Liu C, Lin YT, et al. Epithelial-mesenchymal transition (EMT) beyond EGFR mutations per se is a common mechanism for acquired resistance to EGFR TKI. Oncogene. 2019;38:455–468. doi: 10.1038/s41388-018-0454-2. [DOI] [PubMed] [Google Scholar]
- 19.Chung JH, Rho JK, Xu X, Lee JS, Yoon HI, Lee CT, Choi YJ, Kim HR, Kim CH, Lee JC. Clinical and molecular evidences of epithelial to mesenchymal transition in acquired resistance to EGFR-TKIs. Lung Cancer. 2011;73:176–182. doi: 10.1016/j.lungcan.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 20.Li L, Gu X, Yue J, Zhao Q, Lv D, Chen H, Xu L. Acquisition of EGFR TKI resistance and EMT phenotype is linked with activation of IGF1R/NF-κB pathway in EGFR-mutant NSCLC. Oncotarget. 2017;8:92240–92253. doi: 10.18632/oncotarget.21170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yao Z, Fenoglio S, Gao DC, Camiolo M, Stiles B, Lindsted T, Schlederer M, Johns C, Altorki N, Mittal V, et al. TGF-β IL-6 axis mediates selective and adaptive mechanisms of resistance to molecular targeted therapy in lung cancer. Proc Natl Acad Sci USA. 2010;107:15535–15540. doi: 10.1073/pnas.1009472107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: The mechanistic link and clinical implications. Nat Rev Clin Oncol. 2017;14:611–629. doi: 10.1038/nrclinonc.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Takamatsu K, Chuma S, Kojima-Kita K, Shiromoto Y, Asada N, Toyoda A, Fujiyama A, et al. MVH in piRNA processing and gene silencing of retrotransposons. Genes Dev. 2010;24:887–892. doi: 10.1101/gad.1902110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santosh B, Varshney A, Yadava PK. Non-coding RNAs: Biological functions and applications. Cell Biochem Funct. 2015;33:14–22. doi: 10.1002/cbf.3079. [DOI] [PubMed] [Google Scholar]
- 27.Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yarmishyn AA, Kurochkin IV. Long noncoding RNAs: A potential novel class of cancer biomarkers. Front Genet. 2015;6:145. doi: 10.3389/fgene.2015.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tao H, Yang JJ, Zhou X, Deng ZY, Shi KH, Li J. Emerging role of long noncoding RNAs in lung cancer: Current status and future prospects. Respir Med. 2016;110:12–19. doi: 10.1016/j.rmed.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 30.Wang Q, Cheng N, Li X, Pan H, Li C, Ren S, Su C, Cai W, Zhao C, Zhang L, et al. Correlation of long non-coding RNA H19 expression with cisplatin-resistance and clinical outcome in lung adenocarcinoma. Oncotarget. 2017;8:2558–2567. doi: 10.18632/oncotarget.13708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang B, Jiang H, Wang L, Chen X, Wu K, Zhang S, Ma S, Xia B. Increased MIR31HG lncRNA expression increases gefitinib resistance in non-small cell lung cancer cell lines through the EGFR/PI3K/AKT signaling pathway. Oncol Lett. 2017;13:3494–3500. doi: 10.3892/ol.2017.5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng N, Cai W, Ren S, Li X, Wang Q, Pan H, Zhao M, Li J, Zhang Y, Zhao C, et al. Long non-coding RNA UCA1 induces non-T790M acquired resistance to EGFR-TKIs by activating the AKT/mTOR pathway in EGFR-mutant non-small cell lung cancer. Oncotarget. 2015;6:23582–23593. doi: 10.18632/oncotarget.4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen LL. Linking Long Noncoding RNA Localization and Function. Trends Biochem Sci. 2016;41:761–772. doi: 10.1016/j.tibs.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 34.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 35.Goff LA, Rinn JL. Linking RNA biology to lncRNAs. Genome Res. 2015;25:1456–1465. doi: 10.1101/gr.191122.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flynn RA, Chang HY. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell. 2014;14:752–761. doi: 10.1016/j.stem.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 39.Yang L, Froberg JE, Lee JT. Long noncoding RNAs: Fresh perspectives into the RNA world. Trends Biochem Sci. 2014;39:35–43. doi: 10.1016/j.tibs.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Good MC, Zalatan JG, Lim WA. Scaffold proteins: Hubs for controlling the flow of cellular information. Science. 2011;332:680–686. doi: 10.1126/science.1198701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hung T, Chang HY. Long noncoding RNA in genome regulation: Prospects and mechanisms. RNA Biol. 2010;7:582–585. doi: 10.4161/rna.7.5.13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, Lu L, Liu C, Yi JS, Zhang H, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52:101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, Lin L, Yao H, Su F, Li D, et al. A cytoplasmic NF-κB interacting long noncoding RNA blocks IκB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27:370–381. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 44.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu J, Lichtenberg T, Hoadley KA, et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell. 2018;173:400–416.e411. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV, Sonkin D, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 48.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aparicio-Prat E, Arnan C, Sala I, Bosch N, Guigó R, Johnson R. DECKO: Single-oligo, dual-CRISPR deletion of genomic elements including long non-coding RNAs. BMC Genomics. 2015;16:846. doi: 10.1186/s12864-015-2086-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen YR, Fu YN, Lin CH, Yang ST, Hu SF, Chen YT, Tsai SF, Huang SF. Distinctive activation patterns in constitutively active and gefitinib-sensitive EGFR mutants. Oncogene. 2006;25:1205–1215. doi: 10.1038/sj.onc.1209159. [DOI] [PubMed] [Google Scholar]
- 51.Yadav A, Kumar B, Datta J, Teknos TN, Kumar P. IL-6 promotes head and neck tumor metastasis by inducing epithelial-mesenchymal transition via the JAK-STAT3-SNAIL signaling pathway. Mol Cancer Res. 2011;9:1658–1667. doi: 10.1158/1541-7786.MCR-11-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kong YG, Cui M, Chen SM, Xu Y, Xu Y, Tao ZZ. LncRNA-LINC00460 facilitates nasopharyngeal carcinoma tumorigenesis through sponging miR-149-5p to up-regulate IL6. Gene. 2018;639:77–84. doi: 10.1016/j.gene.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 53.Li K, Sun D, Gou Q, Ke X, Gong Y, Zuo Y, Zhou JK, Guo C, Xia Z, Liu L, et al. Long non-coding RNA linc00460 promotes epithelial-mesenchymal transition and cell migration in lung cancer cells. Cancer Lett. 2018;420:80–90. doi: 10.1016/j.canlet.2018.01.060. [DOI] [PubMed] [Google Scholar]
- 54.Lian Y, Yan C, Xu H, Yang J, Yu Y, Zhou J, Shi Y, Ren J, Ji G, Wang K. A Novel lncRNA, LINC00460, Affects Cell Proliferation and Apoptosis by Regulating KLF2 and CUL4A Expression in Colorectal Cancer. Mol Ther Nucleic Acids. 2018;12:684–697. doi: 10.1016/j.omtn.2018.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu X, Wen J, Wang H, Wang Y. Long non-coding RNA LINC00460 promotes epithelial ovarian cancer progression by regulating microRNA-338-3p. Biomed Pharmacother. 2018;108:1022–1028. doi: 10.1016/j.biopha.2018.09.103. [DOI] [PubMed] [Google Scholar]
- 56.Li P, Shan JX, Chen XH, Zhang D, Su LP, Huang XY, Yu BQ, Zhi QM, Li CL, Wang YQ, et al. Epigenetic silencing of microRNA-149 in cancer-associated fibroblasts mediates prostaglandin E2/interleukin-6 signaling in the tumor microenvironment. Cell Res. 2015;25:588–603. doi: 10.1038/cr.2015.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gao SP, Mark KG, Leslie K, Pao W, Motoi N, Gerald WL, Travis WD, Bornmann W, Veach D, Clarkson B, et al. Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. J Clin Invest. 2007;117:3846–3856. doi: 10.1172/JCI31871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim SM, Kwon OJ, Hong YK, Kim JH, Solca F, Ha SJ, Soo RA, Christensen JG, Lee JH, Cho BC. Activation of IL-6R/JAK1/STAT3 signaling induces de novo resistance to irreversible EGFR inhibitors in non-small cell lung cancer with T790M resistance mutation. Mol Cancer Ther. 2012;11:2254–2264. doi: 10.1158/1535-7163.MCT-12-0311. [DOI] [PubMed] [Google Scholar]
- 59.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, Bergethon K, Shaw AT, Gettinger S, Cosper AK, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Matsumoto A, Pasut A, Matsumoto M, Yamashita R, Fung J, Monteleone E, Saghatelian A, Nakayama KI, Clohessy JG, Pandolfi PP. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature. 2017;541:228–232. doi: 10.1038/nature21034. [DOI] [PubMed] [Google Scholar]
- 61.Tamura T, Kato Y, Ohashi K, Ninomiya K, Makimoto G, Gotoda H, Kubo T, Ichihara E, Tanaka T, Ichimura K, et al. Potential influence of interleukin-6 on the therapeutic effect of gefitinib in patients with advanced non-small cell lung cancer harbouring EGFR mutations. Biochem Biophys Res Commun. 2018;495:360–367. doi: 10.1016/j.bbrc.2017.10.175. [DOI] [PubMed] [Google Scholar]
- 62.Matsui M, Corey DR. Non-coding RNAs as drug targets. Nat Rev Drug Discov. 2017;16:167–179. doi: 10.1038/nrd.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Setten RL, Rossi JJ, Han SP. The current state and future directions of RNAi-based therapeutics. Nat Rev Drug Discov. 2019;18:421–446. doi: 10.1038/s41573-019-0017-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article or are available from the corresponding author on reasonable request.