Abstract
The oncogene ATPase family AAA domain-containing protein 2 (ATAD2) has been demonstrated to promote malignancy in a number of different types of tumor; however, its expression and role in ovarian cancer (OC) remain unknown. In the present study, it was demonstrated that ATAD2 acts as both a marker and a driver of cell proliferation in OC. Immunohistochemistry (IHC) and bioinformatics analyses were used to evaluate ATAD2 expression in OC, and multi-omics integrated analyses were used to dissect which factor resulted in its upregulation. Multiplex IHC assay was used to reveal the specific expression of ATAD2 in proliferating OC cells. CRISPR-Cas9-mediated gene editing was performed to investigate the effect of ATAD2 deletion on OC proliferation. The results demonstrated that ATAD2 is elevated in primary OC tissues compared with the adjacent normal tissue and metastases from the stomach. Genetic copy number amplification is a primary cause resulting in upregulation of ATAD2, and this was most frequently observed in OC. High ATAD2 expression was associated with advanced progression and predicted an unfavorable prognosis. ATAD2 could be used to identify cases of OC with a high proliferation signature and could label proliferating cells in OC. CRISPR-Cas9-mediated ATAD2 deletion resulted in a significant decrease in both cell proliferation and colony formation ability. Mechanistically, ATAD2-knockdown resulted in deactivation of the mitogen-activated protein kinase (MAPK) pathways, particularly the JNK-MAPK pathway, resulting in suppression of proliferation. Collectively, the data from the present study demonstrated that the ATD2 gene was frequently amplified and protein expression levels were upregulated in OC. Therefore, ATAD2 may serve as an attractive diagnostic and prognostic OC marker, which may be used to identify patients with primary OC, whom are most likely to benefit from ATAD2 gene-targeted proliferation intervention therapies.
Keywords: ATPase family AAA domain-containing protein 2, upregulation, ovarian cancer, proliferation, progression and prognosis, mitogen-activated protein kinase pathway
Introduction
Ovarian cancer (OC) is the most lethal type of tumor of the female reproductive system and has the third highest incidence rate amongst gynecological malignancies worldwide (1). It accounted for 2.5% of all malignancies in women and 5% of all cancer-associated mortalities in females in the USA in 2018, due to a poor overall survival rate (2). There were an estimated 22,240 new cases of OC and 14,070 OC-associated mortalities in the USA in 2018 (3). Despite advances in surgical techniques and adjunct therapies, the survival rate for patients with OC has remained relatively unchanged. Therefore, determining the precise oncogenic mechanisms and developing novel effective therapies may improve the survival rates for patients with OC.
ATPase family AAA domain-containing protein 2 (ATAD2), a member of the ATPase family, is a relatively recently identified oncogene (4), which contains two conserved domains; a bromodomain responsible for histone binding and an ATPase domain that regulates ATPase activity and protein multimerization (5). The ATAD2 gene is located on chromosome 8q24.13, a region that is frequently amplified in different types of tumor (6). ATAD2 expression is upregulated in several different types of cancer, including breast (7), colorectal (8), endometrial (9), gastric (10) and liver cancer (11). Studies have demonstrated that ATAD2 promotes cell proliferation (7), inhibits apoptosis (11) and enhances metastasis (12,13) during tumorigenesis and progression. Overexpression of ATAD2 is typically associated with poor clinical outcomes and predicts unfavorable survival outcomes (8,10,14). ATAD2 expression has also recently been reported to be upregulated in male germ cells and embryonic stem cells, and facilitates chromatin remodeling under normal physiological conditions (15). Although ATAD2 drives cell proliferation and cancer progression in several types of cancer, its expression and significance in OC remains unclear.
In the present study, ATAD2 was identified to be frequently amplified in OC and that upregulation of ATAD2 was associated with OC tissues and sub-populations of cells within tissues with increased proliferative capacity, and was correlated with poorer clinical outcomes. Furthermore, CRISPR-Cas9-mediated genomic disruption of the ATAD2 gene resulted in deactivation of the mitogen-activated protein kinase (MAPK) pathway and reduced proliferation of OC.
Materials and methods
Cell lines and culture conditions
The human OC cell lines SKOV3 and A2780 were obtained from the National Infrastructure of Cell Line Resources. The human embryonic kidney cell line 293T was obtained from American Type Culture Collection. All cell lines were cultured in DMEM (HyClone; GE Healthcare Life Sciences) with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) at 37°C with 5% CO2 in a humidified incubator.
Immunohistochemistry (IHC) analysis
A standard IHC protocol was performed on a commercially obtained OC tissue microarray (OV802a) containing 60 primary OC tissues, 10 tissues of stomach metastases to the ovary and 10 adjacent normal tissues from Alenabio to assess the protein expression levels of ATAD2 in OC. Following IHC staining with a specific antibody against ATAD2 (1:500; catalog no. HPA029424; Sigma-Aldrich; Merck KGaA) overnight at 4°C, the samples were stained with a horseradish peroxidase-conjugated secondary goat anti-rabbit antibody (1:2,500; catalog no. ZB-2301; OriGene Technologies, Inc.) for 1 h at room temperature. The immunohistochemical signals were then observed under a light microscope (magnification, ×200). The staining intensity was scored as follows: 0, Negative; 1, weak; 2, moderate; and 3, strong. The percentage of ATAD2-positive cells was also scored as follows: 0, 0-10%; 1, >10-25%; 2, >25-50%; 3, >50-75%; and 4, >75-100%. The final immunoreactive score was determined by multiplying the intensity and percentage scores, with a minimum score of 0 and a maximum score of 12.
CRISPR-Cas9-mediated ATAD2 gene disruption
CRISPR-Cas9 gene editing technology was used to disrupt ATAD2 expression at the genome level. Two pairs of single guide RNA (sgRNA) specifically targeting ATAD2 exon 6 and exon 2 were designed using the E-CRISP online tool (http://www.e-crisp.org/E-CRISP/designcrispr.html) and cloned into the lentiCRISPRv2 vector (Addgene Inc.; catalog no. 52961). The lentiviruses for the ATAD2 sgRNAs or the vector control were generated in 293T cells by co-transfection of the two recombined lentiCRISPRv2 plasmids or the empty vector with the packaging vectors pMD2G and psPAX2 (Addgene, Inc.) using Lipofectamine 3000® (Invitrogen; Thermo Fisher Scientific, Inc.). The cells were treated with 2 µg/ml puromycin 72 h after lentiviral infection. The deletion of ATAD2 expression was determined by western blotting. The target sgRNA sequences were as follows: sgRNA-ATAD2-1 forward, 5′-CACCGGAAACAACTGATAATCAAGA-3′ and reverse, 3′-CCTTTGTTGACTATTAGTTCTCAAA-5′; and sgRNA-ATAD2-2 forward, 5′-CACCGTAGGCAGTTGGCCAGACAGC-3′ and reverse, 3′-CATCCGTCAACCGGTCTGTCGCAAA-5′.
Real-time cell analyzer (RTCA)
An RTCA (xCELLigence; ACEA Biosciences, Inc.) was used to monitor cell proliferation in real time as described previously (16). The RTCA system measures electrical impedance variations and presents the results in the form of the cell index which reflects cellular proliferation. For each cell line, 3×103 cells/well were seeded in an E-plate, which was placed in a RTCA device at 37°C with 5% CO2. The cell index was recorded automatically in 15-min intervals. The recorded curve is presented as the mean of cell index ± standard error.
Colony formation assay
Cells were seeded in a 6-well plate at a density of 500 cells/well and allowed to grow at 37°C with 5% CO2 for 14 days after which, cells were stained with 0.5% crystal violet (Sigma-Aldrich; Merck KGaA) at room temperature for 10 min. The colonies in each well were observed and counted under a light microscope (magnification, ×100). Three fields of view were observed and only colonies containing >50 cells were counted.
Western blot analysis
Whole-cell protein was extracted using RIPA lysis buffer (Beijing Solarbio Science & Technology Co., Ltd.) and quantified using a bicinchoninic acid assay protein assay kit (Thermo Fischer Scientific, Inc.). Proteins (40 µg/lane) were denatured, loaded on a 10% gel and resolved using SDS-PAGE. Proteins were transferred to PVDF membranes using semi-dry transfer at 15 V for 2 h (EMD Millipore). The membranes were probed with primary antibodies against ATAD2 (1:500; catalog no. HPA029424l Sigma-Aldrich; Merck KGaA), JNK (1:1,000; catalog no. 9252T; Cell Signaling Technology, Inc.), phosphorylated (p)-JNK (1:1,000; catalog no. 4668T; Cell Signaling Technology, Inc.), ERK (1:1,000; catalog no. 4695T; Cell Signaling Technology, Inc.), p-ERK (1:1,000; catalog no. 4370T; Cell Signaling Technology, Inc.), P38 (1:1,000; catalog no. 8690T; Cell Signaling Technology, Inc.), p-P38 (1:1,000; catalog no. 4511T; Cell Signaling Technology, Inc.) or GAPDH (1:3,000; catalog no. 5174S; Cell Signaling Technology, Inc.) overnight at 4°C, followed by incubation with a horseradish peroxidase-conjugated secondary goat anti-rabbit (catalog no. sc-2004) or anti-mouse (catalog no. sc-2005) poly-clonal immunoglobulin G antibody (both 1:8,000; Santa Cruz Biotechnology, Inc.) at room temperature for 1 h. Signals were visualized using an enhanced chemiluminescence kit (Applygen Technologies Inc.). The results were analyzed using Image J 1.46r software (National Institutes of Health).
Multiplex IHC staining
Multiplex IHC staining was performed to examine the co-expression of ATAD2 and Ki-67 in OC tissue sections on a commercially obtained OC tissue microarray (OV241c; containing 12 primary OC tissues) from Alenabio using an Opal™ 4-Color Manual IHC kit (PerkinElmer, Inc.). Briefly, the sections were first incubated with a specific ATAD2 antibody (1:1,000; catalog no. HPA029424; Sigma-Aldrich; Merck KGaA) for 1 h at room temperature, followed by incubation with a goat anti-rabbit immunoglobulin G antibody (1:5,000; catalog no. NEL840001KT, PerkinElmer, Inc.) for 1 h at room temperature and subsequently labeled with Opal 520 fluorophore (PerkinElmer, Inc.). For Ki-67 staining, an anti-Ki67 antibody (1:1,000; catalog no. ZM-0166; OriGene Technologies, Inc.) was used at room temperature for 1 h and Opal 690 fluorophore (PerkinElmer, Inc.) was used for labeling. Finally, nuclei were stained with DAPI (PerkinElmer, Inc.) for 30 min at room temperature and mounted with anti-fade medium. The inForm 2.4.2 software (PerkinElmer, Inc.) was used to analyze the multispectral images.
Public databases and dataset analyses
Data from public databases were used as provided, without additional processing or normalization of the data, unless otherwise indicated. To assess the expression of ATAD2 mRNA in OC, the CSIOVDB database (version 1.0) (17) was used, which includes transcriptomic profiles for 3,431 OC and normal ovary tissues. To validate the upregulation of ATAD2 mRNA in OC compared with the normal samples, expression of ATAD2 in four independent profiles [GSE12470 (18), GSE14407 (19), GSE18520 (20) and GSE26712 (21)] from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo) were analyzed using the GEO2R online tool (ncbi.nlm.nih.gov/geo/geo2r/).
Putative copy-number calls determined using GISTIC version 2.0 for The Cancer Genome Atlas (TCGA) datasets were obtained for copy-number variation (CNV) analysis (22). Copy-number values were defined as follows: −2, homozygous deletion; −1, hemizygous deletion; 0, neutral / no change; 1, gain; and 2, high level amplification. Comparison of the copy numbers in 607 OC tissues, 130 normal ovary tissues and 431 paired normal blood specimens from TCGA dataset was performed using Oncomine analysis (23). cBioPortal for Cancer Genomics (cbioportal.org/) (24) was used for calculating the association between ATAD2 mRNA expression and copy number values, and also for plotting the CNV status of ATAD2 in cancer tissue samples from TCGA and cell line samples from the Cancer Cell Line Encyclopedia (CCLE; https://portals.broadinstitute.org/ccle).
The association between ATAD2 expression and OC International Federation of Gynecology and Obstetrics (FIGO) grade (25) or FIGO stage (25) was analyzed using the expression data of OC cohorts from the CSIOVDB database and further validated in the OC cohort of the GSE9811 dataset (26).
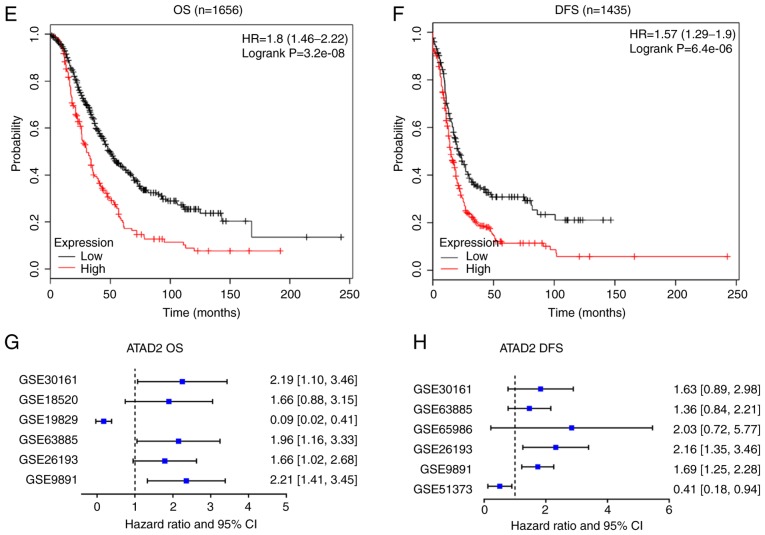
The prognostic value of ATAD2 for patients with OC was analyzed using the Kaplan-Meier plotter online tool (27), which integrates gene expression with disease-free survival (DFS) and overall survival (OS) data of 1,816 OC samples. The survival curves were calculated using the Kaplan-Meier method, and statistical significance was assessed using the log-rank test. The hazard ratio (HR) with 95% confidence interval (CI) was calculated. For further validation, a meta-analysis on ATAD2 HRs for OS and DFS in randomly selected multiple OC cohorts with clinical OS and/or DFS data from the GEO database, GSE30161 (28), GSE18520 (29), GSE 19829 (30), GSE63885 (31), GSE26193 (32), GSE9891 (33), GSE65986 (34) and GSE51373 (35), was performed.
For ATAD2 co-expressed genes analyses, the OC cohort from TCGA (n=307) and another large OC cohort from GSE9891 (33) (n=285) were used to perform co-expression data mining analysis using R2: Genomics Analysis and Visualization Platform (r2.amc.nl). For Gene Ontology (GO) analysis of the enriched biological processes, the Database for Annotation, Visualization and Integrated Discovery (DAVID; version 6.8) bioinformatics resource (36) was used.
Statistical analysis
The data were analyzed by Mann-Whitney U test for comparison of two groups and by Kruskal-Wallis test for multiple groups followed by an uncorrected Dunn's multiple comparisons test in GraphPad Prism version 7.0 (GraphPad Software, Inc.). The expressional correlation analyses between ATAD2 and its correlated genes in TCGA and GSE9891 (33) datasets were conducted by Pearson's correlation analysis and a false discovery rate test using resources within R2, and all data statistical analyses on CSIOVDB samples were calculated automatically by CSIOVDB (http://csibio.nus.edu.sg/CSIOVDB/CSIOVDB.html). P<0.05 was considered to indicate a statistically significant difference.
Results
ATAD2 protein expression is frequently upregulated in OC compared with the non-cancerous normal tissue
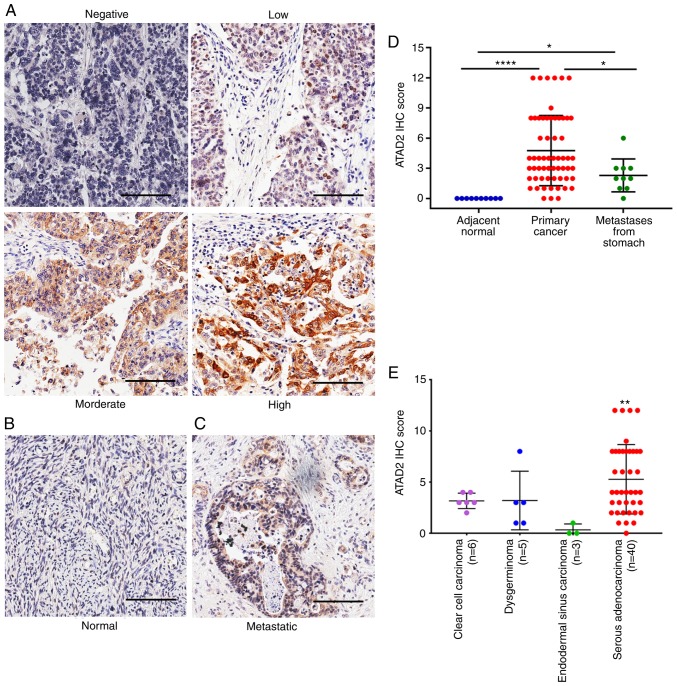
To assess the expression of ATAD2 in OC, IHC analysis was performed on an OC tissue microarray containing 60 primary tissues, 10 metastatic tissues from the stomach and 10 adjacent normal ovary tissues. ATAD2 expression was positive in 83.3% (50/60) of primary OC tissues, including 46.67% of tissues with low expression (28/60), 25.00% of tissues with moderate expression (15/60) and 11.67% of tissues with high expression (7/60) (Fig. 1A). By contrast, none of the adjacent normal tissues (0/10) had detectable levels of ATAD2 expression (Fig. 1B). Metastatic OC from other primary malignancies accounted for 5-30% of overall ovarian malignancies, and stomach cancer is one of the most common primary origins (37). ATAD2 was detectable in 70% (7/10) of the metastatic ovarian malignancies from the stomach (Fig. 1C), with 60% (6/10) of cases exhibiting low expression and 10% (1/10) of cases exhibiting moderate expression.
Figure 1.
Upregulation of ATAD2 protein expression in OC compared with non-cancerous normal tissues and ovarian metastases from the stomach. Representative IHC images showing the expression levels of ATAD2 in (A) primary OC tissues, (B) adjacent normal ovarian tissues (C) and metastatic ovarian cancer from the stomach. (D) Comparison of the ATAD2 IHC scores in primary OC, adjacent normal and in gastric metastases to the ovaries. *P<0.05 and ****P<0.0001 (E) Expression of ATAD2 in different types of OC tissues. Comparison was performed between serous adenocarcinoma samples and non-serous samples of clear cell carcinoma, dysgerminoma and endodermal sinus carcinoma. **P<0.01 vs. endodermal sinus carcinoma. Scale bar, 100 µm. OC, ovarian cancer; IHC, immunohistochemistry; ATAD2, ATPase family AAA domain-containing protein 2.
By comparing IHC scores, ATAD2 expression level in primary OC was significantly higher compared with the adjacent normal tissue (P<0.0001; Fig. 1D). ATAD2 expression in the metastatic ovarian malignancies from stomach was also significantly higher compared with the adjacent ovarian normal tissue (P<0.05), but significantly lower compared with the primary OC tissues (P<0.05; Fig. 1D), suggesting that ATAD2 expression may be used as a diagnostic marker to distinguish secondary ovarian malignancies and primary OC. Regarding different OC subtypes, ATAD2 expression was significantly higher in serous adenocarcinoma compared with endodemermal sinus carcinoma (P<0.01; Fig. 1E).
ATAD2 expression is upregulated in OC based on multiple public datasets
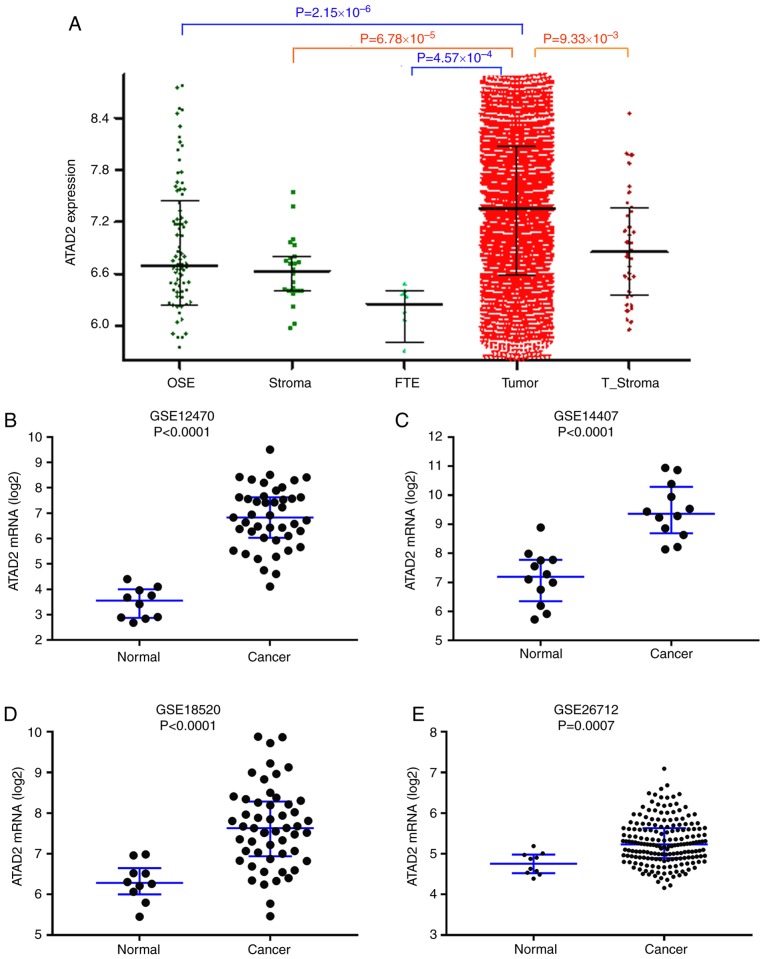
To validate upregulation of ATAD2 in OC, ATAD2 mRNA expression levels were assessed in the CSIOVDB dataset (17), which includes transcriptomic profiles for 3,431 OC and normal ovary tissues. As presented in Fig. 2A, ATAD2 mRNA expression levels were significantly higher in OC compared with the normal tissues. Significant upregulation of ATAD2 mRNA levels in OC compared with normal tissues was further validated in four independent GEO profiles, GSE12470 (Fig. 2B), GSE14407 (Fig. 2C), GSE18520 (Fig. 2D) and GSE26712 (Fig. 2E). Physiological expression levels of ATAD2 in ovarian normal tissues and upregulation of expression in cancerous tissues support the notion that ATAD2 may serve as a biomarker in OC.
Figure 2.
ATAD2 mRNA expression levels are upregulated in OC. (A) ATAD2 expression levels in a series of ovarian tissues including normal OSE, FTE, normal stroma, T_Stroma and paired primary tumor tissues from patients with high grade OC in the CSIOVDB database. mRNA expression levels of ATAD2 in OC and non-cancerous normal tissues were further analyzed in four independent mRNA microarray profiles, (B) GSE12470, (C) GSE14407, (D) GSE18520 and (E) GSE26712. ATAD2 mRNA expression levels were significantly upregulated in OC tissues. Data are presented as the median ± interquartile range. OC, ovarian cancer; ATAD2, ATPase family AAA domain-containing protein 2; OSE, ovarian surface epithelium; FTE, normal fallopian tubes; T_Stroma, tumor stroma.
Increased copy numbers of the ATAD2 gene is a primary cause of upregulation in OC
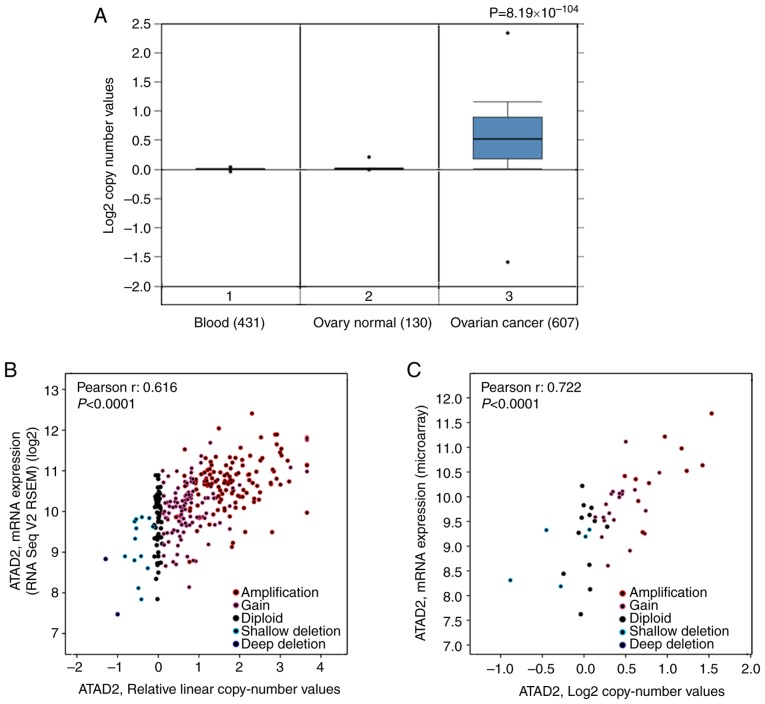
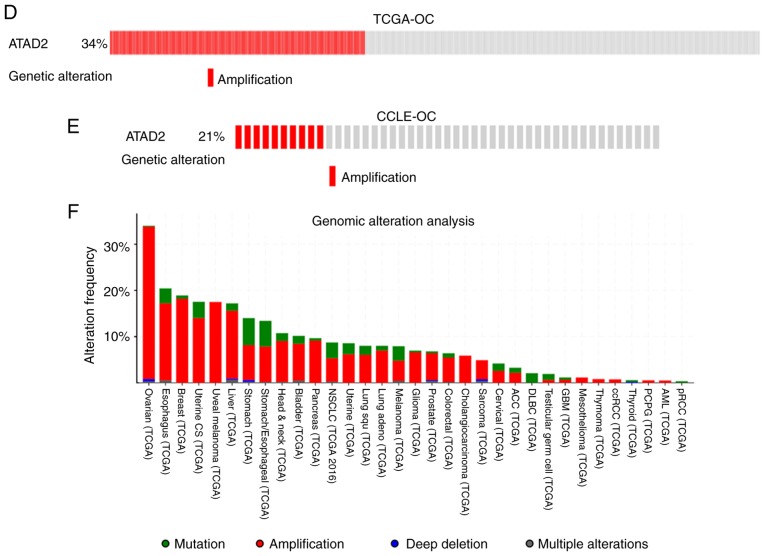
To address the cause resulting in upregulation of ATAD2 in OC, the copy number of the ATAD2 gene in OC was determined compared with the normal ovarian tissues based on data obtained from TCGA. The results revealed that the copy number of ATAD2 in OC tissues was significantly higher compared with the normal ovary tissues and blood samples (P=8.19×10−104; Fig. 3A). To evaluate the contribution of CNV to upregulation of ATAD2 during OC tumorigenesis, correlative analysis of ATAD2 transcriptional levels and its copy number values in OC was performed. ATAD2 copy number increases (gain or amplification) was determined to be high in 74% (429/579) of patients with OC based on the data from TCGA according to the putative copy-number alterations from GISTIC. Additionally, ATAD2 mRNA levels were significantly correlated with the linear copy number value (r=0.616, P<0.0001; Fig. 3B). ATAD2 copy number increase was also found in 64% (30/47) of OC cell lines from the CCLE database, and a significant correlation between ATAD2 mRNA expression levels and copy number value (r=0.722, P<0.0001; Fig. 3C). These results indicate that CNV is a primary cause responsible for upregulation of ATAD2 in OC. Detailed analysis demonstrated that ATAD2 amplification (putative copy-number alteration=2) was present in 34% of OC tissues (Fig. 3D), as well as in 21% of OC cell lines (Fig. 3E). Furthermore, pan-cancer genomic alteration analysis in all types of cancer based on data obtained from TCGA demonstrated that the highest frequency of ATAD2 amplification was in OC (Fig. 3F), suggesting that ATAD2 amplification may be a genomic signature of OC.
Figure 3.
Copy number variation of ATAD2 gene results in its upregulation in OC. (A) Copy number variation analysis between OC and normal ovary tissues and blood samples based on TCGA. (B) Correlation analysis between ATAD2 mRNA expression levels and relative linear copy number values in the TCGA OC dataset. (C) Correlation analysis between ATAD2 mRNA expression levels and copy number values in OC cell lines. High level ATAD2 amplification was observed in (D) 34% of OC tissues and also in (E) 21% of OC cell lines. (F) Genomic alteration analysis of the ATAD2 gene across all types of cancer in TCGA. OC, ovarian cancer; ATAD2, ATPase family AAA domain-containing protein 2; TCGA, The Cancer Genome Atlas; CCLE, Cancer Cell Line Encyclopedia.
ATAD2 upregulation is associated with advanced tumor progression and unfavorable outcomes in patients with OC
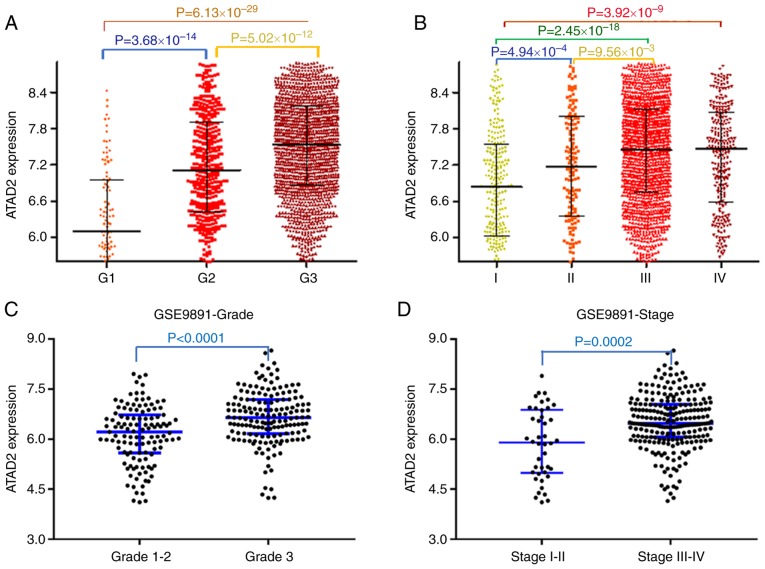
To examine the contribution of upregulation of ATAD2 in the progression of OC, the association between ATAD2 and clinicopathological features of patients with OC was evaluated. Data from the CSIOVDB database, which is comprised of transcriptional data of 3,431 ovarian cancer cases, was first analyzed. The results showed that upregulated ATAD2 expression was associated with increasing tumor FIGO grade (Fig. 4A) and FIGO stage (Fig. 4B). The significant relevance of ATAD2 expression with tumor grade and stage were further validated in the OC cohorts from the GSE9891 GEO dataset, which contains microarray transcription profiles of 285 patients with OC (FIGO grade, P<0.0001; FIGO stage, P=0.0002; Fig. 4C and D, respectively).
Figure 4.
ATAD2 upregulation is associated with advanced tumor progression and unfavorable outcomes in patients with OC. ATAD2 expression was stratified by FIGO stage and FIGO grade, and ATAD2 overexpression was closely associated with (A) tumor FIGO stage and (B) FIGO grade in the OC cohort from the CSIOVDB database. Association between ATAD2 and OC (C) stage or (D) grade was further validated in the GSE9891 OC cohort. Data are presented as the median ± interquartile range. Kaplan-Meier survival analysis showed that high ATAD2 expression predicted worse (E) OS and (F) DFS. (G) Meta-analysis diagram showing the general unfavorable effect of ATAD2 on OS of OC patients in multiple GEO cohorts. (H) The meta-analysis diagrams showed the general unfavorable effect of ATAD2 on DFS of patients with OC in multiple GEO cohorts. ATAD2, ATPase family AAA domain-containing protein 2; OC, ovarian cancer; FIDO, International Federation of Gynecology and Obstetrics; OS, overall survival; GEO, Gene Expression Omnibus.
The effects of ATAD2 expression on prognostic outcomes were determined using Kaplan-Meier plotter (27). Based on this, upregulated expression of ATAD2 predicted a significantly worse OS (HR, 1.8; 95% CI, 1.46-2.22; log-rank P=3.2×10−8; Fig. 4E) and DFS (HR,1.57; 95% CI, 1.29-1.90; log-rank P=6.4×10−6; Fig. 4F). The unfavorable effect of ATAD2 on OS (Fig. 4G) and DFS (Fig. 4H) outcomes were further confirmed in the majority of the OC GEO datasets.
ATAD2 identifies a proliferation signature in public OC datasets
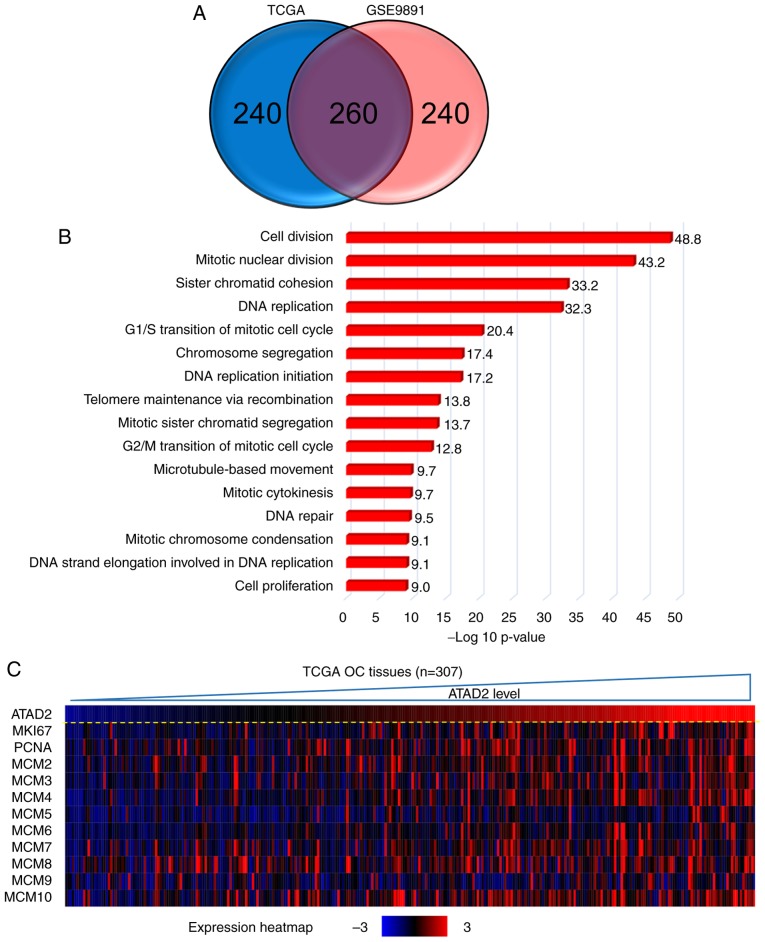
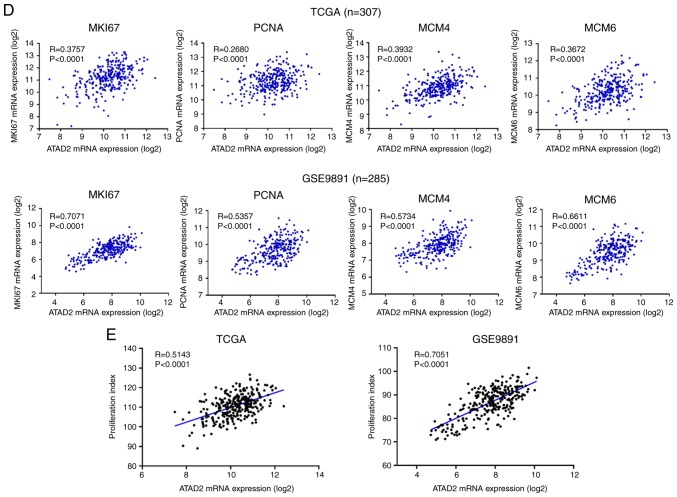
To address the possible role of ATAD2 in OC, gene co-expression network analysis was used. Gene co-expression networks are useful to delineate gene function, identifying a set of genes responding in a coordinated manner to environmental or disease state, and highlighting regulatory associations (38,39). The top 500 positively correlated genes in TCGA and GSE9891 OC datasets were extracted according to the R-value, and they produced a significant overlap in the genes that were co-expressed (260; 52.0%; Fig. 5A), indicating a high reliability and sensitivity of the co-expression analysis. GO analysis was performed on the overlapping 260 ATAD2 co-expressed genes in DAVID, and yielded significant enrichment in fundamental biological processes associated with cell proliferation, such as 'cell division', 'mitotic nuclear division', 'DNA replication', 'G1/S transition of mitotic cell cycle' and 'G2/M transition of mitotic cell cycle' (Fig. 5B), suggesting that ATAD2 may be involved in regulation of proliferation of OC cells. The hypothesized role of ATAD2 on OC proliferation was further supported by a significant association with protein expression levels of classical proliferation makers, including marker of proliferation Ki-67, proliferation cell nuclear antigen and methylmalonyl-CoA mutase in OC tissues in both datasets (Fig. 5C and D; Table SI) and in 47 cell lines from the CCLE database (Table SI). As ATAD2 expression was associated closely with the proliferation index in both datasets (Fig. 5E), this suggested that ATAD2 may be used to label OC tissues with a potentially increased proliferative capacity.
Figure 5.
ATAD2 expression identified OC tissues with increased proliferative capacities. (A) Venn diagram showing the high degree of overlap between the ATAD2 co-expressed genes in the two cohorts. (B) GO analysis of the 260 overlapping ATAD2 co-expressed genes in DAVID. (C) Correlation heatmap showing a significant positive correlation between ATAD2 expression with proliferation markers. (D) Correlation analyses of ATAD2 expression and individual proliferation markers. (E) ATAD2 was significantly positively correlated with the proliferative index in both OC cohorts. Proliferation index is represented by the sum of expression values of proliferation markers including MKI67, PCNA and MCM2-10. ATAD2, ATPase family AAA domain-containing protein 2; OC, ovarian cancer; GO, Gene Ontology; DAVID, Database for Annotation, Visualization and Integrated Discovery; TCGA, The Cancer Genome Atlas; MIK67, marker of proliferation Ki-67; PCNA, proliferation cell nuclear antigen; MCM, methylmalonyl-CoA mutase.
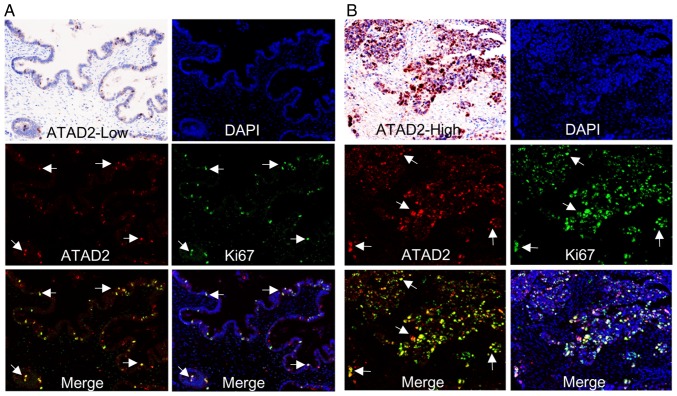
ATAD2 labels proliferating subpopulations in clinical OC samples
To demonstrate a direct association between ATAD2 expression and proliferation of OC, immunostaining of clinical OC samples with specific antibodies against ATAD2 and cell proliferation marker Ki-67 was performed. The staining showed that samples with high levels of ATAD2 expression were also rich in Ki-67 expression. Furthermore, ATAD2 specifically labeled proliferative subpopulations with high levels of Ki-67 expression in the tissues, regardless of whether an OC sample in totality expressed low (Fig. 6A) or high levels of ATAD2 (Fig. 6B). Together, these results demonstrated that ATAD2 is a specific marker for proliferating OC cells and suggest that ATAD2 may increase proliferation of OC.
Figure 6.
ATAD2 labels proliferating cells in clinical OC samples. Co-expression of ATAD2 and Ki-67 in an OC tissue with (A) low or (B) high ATAD2 expression. The arrows indicate representative cells with ATAD2 and/or Ki-67 co-expression. Magnification, ×100. ATAD2, ATPase family AAA domain-containing protein 2; OC, ovarian cancer.
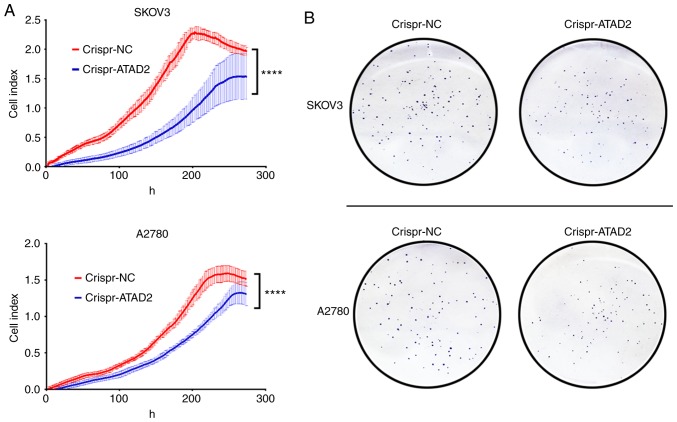
CRISPR-Cas9 mediated ATAD2 gene disruption reduces proliferation and colony formation in OC cell lines
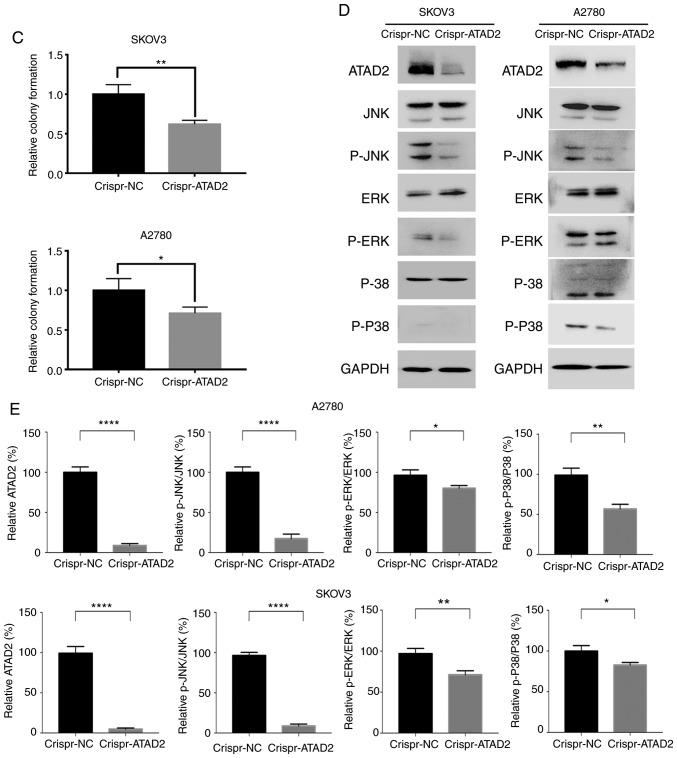
To address whether ATAD2 is necessary for proliferation of OC cells, the ATAD2 gene was disrupted in the SKOV3 and A2780 OC cell lines using CRISPR-Cas9 genome editing technology. RTCA and colony formation experiments were performed to examine the effect of ATAD2 silencing on OC proliferation in vitro. Results from the RTCA analysis showed that ATAD2 deficiency significantly reduced the proliferative ability of both SKOV3 and A2780 cells (both P<0.0001; Fig. 7A). Consistent with the RTCA analysis, colony formation analysis demonstrated that ATAD2 deletion significantly impaired the colony-forming capacity of SKOV3 (P<0.01) and A2780 (P<0.05) cell lines (Fig. 7B and C). Together, these results suggested that ATAD2 was necessary for proliferation of OC cells.
Figure 7.
ATAD2 drives proliferation of OC by targeting MAPK signaling pathways. (A) Real-time cell analyzer analyses of the effect of ATAD2 silencing on the proliferative capacity of SKOV3 and A2780 OC cell lines. ****P<0.0001. (B) Effect of ATAD2 deletion on colony formation abilities. Magnification, ×100. ATAD2 drives proliferation of OC by targeting MAPK signaling pathways. (C) Statistical analysis of colony formation. Data are presented as the mean ± standard deviation, *P<0.05, **P<0.01. (D) Western blots showing the effect of ATAD2 silencing on the MAPK pathway activation in the two OC cell lines. (E) Quantitative analysis of the western blot results. *P<0.05, **P<0.01, ****P<0.0001. ATAD2, ATPase family AAA domain-containing protein 2; OC, ovarian cancer; p-, phosphorylated; NC, negative control; MAPK, mitogen activated protein kinase.
ATAD2 deletion deactivates the JNK-MAPK pathway
To decipher the mechanism by which ATAD2 results in proliferation of OC cells, the effect of ATAD2 deletion on MAPK pathways, which are frequently associated with increased proliferation in various types of cancer (40), was examined. ATAD2 deletion resulted in a marked decrease in phosphorylation levels of all three crucial members of the MAPK sub-pathways, JNK, ERK and p-38 (all P<0.05), of which JNK (P<0.0001) was the most notably affected in both OC cell lines (Fig. 7D). These experiments demonstrate that ATAD2 activates the MAPK pathways, particularly the JNK-MAPK pathway, in OC cells, thus providing a possible explanation for the role of ATAD2 in increasing proliferation of OC.
Discussion
OC is the most lethal type of tumor of the female reproductive system. Despite advances in surgery and chemotherapy in recent years, relatively little progress has been made in reducing the mortality rates of patients with OC (1). Deciphering the oncogenic mechanisms underlying this disease may contribute to the design of more precise and efficient therapeutic strategies.
In the present study, it was demonstrated that the ATPase family member ATAD2 was significantly upregulated in primary OC tissues compared with the adjacent normal tissues and gastric metastases to the ovary, suggesting that ATAD2 may serve as a diagnostic marker to distinguish primary OC from normal and metastatic carcinomas.
Aberrant elevation of ATAD2 expression has also been reported in several other types of cancer, including liver, prostate, lung, colon and cervical cancer (4), emphasizing high oncogenic relevance of ATAD2 expression in cancer.
Genetic alterations, particularly CNVs, are frequently observed in dysregulation of cancer genes (41). Therefore, it was speculated that CNV may underlie ATAD2 upregulation in OC. To address this possibility, the copy numbers of the ATAD2 gene in OC were compared with matched normal tissues, and a significant increase of ATAD2 copy numbers was shown in OC tissues. Further analysis revealed that 74% of patients with OC had increased ATAD2 CNVs, 34% of which were amplifications and 40% were gains. The association between ATAD2 linear copy number values and expression levels in both patients with OC tissues and cell lines suggested that CNV was the primary cause for elevation of ATAD2 in OC. ATAD2 copy number amplification was also demonstrated in a number of different types of cancer in TCGA, such as esophageal and breast cancer. However, ATAD2 amplification events were most frequently observed in OC, accounting for ~34% of patients with OC, whereas in the next most common cancer, ATAD2 amplification events were only seen in <20% of cases, suggesting that ATAD2 amplification is a characteristic genetic alteration in OC, and that patients with OC are most likely to benefit from ATAD2-targeted genetic disruption therapeutic strategies.
To dissect the role of ATAD2 in OC, the association between ATAD2 expression with clinical features of patients with OC was determined. ATAD2 upregulation was associated with advanced tumor progression and unfavorable prognostic outcomes in patients with OC patients in the majority of cohorts analyzed. Co-expression network analysis suggested that ATAD2 was closely associated with proliferation-related biological processes, including cell division, DNA replication and mitotic cell cycle. ATAD2 specifically identified the patients with OC with evidence of increased proliferation, and ATAD2 directly labeled the proliferating cancer cells in clinical OC tissues, irrespective of the level of proliferation seen overall in each tissue, suggesting that ATAD2 is a specific proliferation marker in OC. To assess the significance of ATAD2 deletion on proliferation of OC, the ATAD2 gene was ablated using CRISPR-Cas9 technology, which is considered the most efficient means of gene editing. It was demonstrated that ATAD2 gene targeting and silencing reduced the proliferation of OC cells. Together, these results collectively indicate that ATAD2 acts as both a specific marker and a potent regulator of cell proliferation in OC.
Although considerable advances in our understanding of OC development and progression have been made in recent years, relatively less is known about the underlying mechanisms of ATAD2 in tumorigenesis. ATAD2 is a direct target of the pRB/E2F pathway and acts as a cofactor of MYC to regulate the Hedgehog pathway (42,43). ATAD2 also acts as a transcriptional co-regulator of estrogen and androgen receptors to induce the expression of genes controlling proliferation and survival of cancer cells (44,45). In the present study, to improve our understanding of the mechanistic role of ATAD2 in proliferation, the effect of ATAD2 on the MAPK pathway was assessed, a pivotal signaling pathway in the control of proliferation. ATAD2 increased phosphorylation of JNK, p38 and ERK, of which JNK was the most notable target. Therefore, it is hypothesized that the MAPK pathway is an important pathway for mediating ATAD2-driven proliferation of OC. Collectively, the data suggest that the amplified and upregulated expression of ATAD2 may be an attractive diagnostic and prognostic marker for OC, and a potential therapeutic oncological target for clinical intervention of OC progression.
Supplementary Data
Acknowledgments
Not applicable.
Funding
This study was financially supported by grants from the National Natural Science Foundation of China (grant nos. 81502384 and 81672338).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JL and TW designed and conceived the study. QL, HL, LL and JL performed the experiments and generated the majority of the data. XD, XR and XF performed the bioinformatics analyses. QL and JL were major contributors in writing the manuscript. All authors have read and approved the final version of this manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Trabert B, DeSantis CE, Miller KD, Samimi G, Runowicz CD, Gaudet MM, Jemal A, Siegel RL. Ovarian cancer statistics, 2018. CA Cancer J Clin. 2018;68:284–296. doi: 10.3322/caac.21456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 4.Hussain M, Zhou Y, Song Y, Hameed HMA, Jiang H, Tu Y, Zhang J. ATAD2 in cancer: A pharmacologically challenging but tractable target. Expert Opin Ther Targets. 2018;22:85–96. doi: 10.1080/14728222.2018.1406921. [DOI] [PubMed] [Google Scholar]
- 5.Boussouar F, Jamshidikia M, Morozumi Y, Rousseaux S, Khochbin S. Malignant genome reprogramming by ATAD2. Biochim Biophys Acta. 2013;1829:1010–1014. doi: 10.1016/j.bbagrm.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Pollack JR, Sørlie T, Perou CM, Rees CA, Jeffrey SS, Lonning PE, Tibshirani R, Botstein D, Børresen-Dale AL, Brown PO. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci USA. 2002;99:12963–12968. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalashnikova EV, Revenko AS, Gemo AT, Andrews NP, Tepper CG, Zou JX, Cardiff RD, Borowsky AD, Chen HW. ANCCA/ATAD2 overexpression identifies breast cancer patients with poor prognosis, acting to drive proliferation and survival of triple-negative cells through control of B-Myb and EZH2. Cancer Res. 2010;70:9402–9412. doi: 10.1158/0008-5472.CAN-10-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hou M, Huang R, Song Y, Feng D, Jiang Y, Liu M. ATAD2 overexpression is associated with progression and prognosis in colorectal cancer. Jpn J Clin Oncol. 2016;46:222–227. doi: 10.1093/jjco/hyv195. [DOI] [PubMed] [Google Scholar]
- 9.Krakstad C, Tangen IL, Hoivik EA, Halle MK, Berg A, Werner HM, Ræder MB, Kusonmano K, Zou JX, Øyan AM, et al. ATAD2 overexpression links to enrichment of B-MYB-translational signatures and development of aggressive endometrial carcinoma. Oncotarget. 2015;6:28440–28452. doi: 10.18632/oncotarget.4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang M, Zhang C, Du W, Yang X, Chen Z. ATAD2 is overexpressed in gastric cancer and serves as an independent poor prognostic biomarker. Clin Transl Oncol. 2016;18:776–781. doi: 10.1007/s12094-015-1430-8. [DOI] [PubMed] [Google Scholar]
- 11.Lu WJ, Chua MS, So SK. Suppression of ATAD2 inhibits hepatocellular carcinoma progression through activation of p53- and p38-mediated apoptotic signaling. Oncotarget. 2015;6:41722–41735. doi: 10.18632/oncotarget.6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong S, Bi M, Yan Z, Sun D, Ling L, Zhao C. Silencing of ATPase family AAA domain-containing protein 2 inhibits migration and invasion of colorectal cancer cells. Neoplasma. 2016;63:846–855. doi: 10.4149/neo_2016_603. [DOI] [PubMed] [Google Scholar]
- 13.Zheng L, Li T, Zhang Y, Guo Y, Yao J, Dou L, Guo K. Oncogene ATAD2 promotes cell proliferation, invasion and migration in cervical cancer. Oncol Rep. 2015;33:2337–2344. doi: 10.3892/or.2015.3867. [DOI] [PubMed] [Google Scholar]
- 14.Caron C, Lestrat C, Marsal S, Escoffier E, Curtet S, Virolle V, Barbry P, Debernardi A, Brambilla C, Brambilla E, et al. Functional characterization of ATAD2 as a new cancer/testis factor and a predictor of poor prognosis in breast and lung cancers. Oncogene. 2010;29:5171–5181. doi: 10.1038/onc.2010.259. [DOI] [PubMed] [Google Scholar]
- 15.Morozumi Y, Boussouar F, Tan M, Chaikuad A, Jamshidikia M, Colak G, He H, Nie L, Petosa C, de Dieuleveult M, et al. Atad2 is a generalist facilitator of chromatin dynamics in embryonic stem cells. J Mol Cell Biol. 2016;8:349–362. doi: 10.1093/jmcb/mjv060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu J, Liu Z, Liu Q, Li L, Fan X, Wen T, An G. CLEC3B is downregulated and inhibits proliferation in clear cell renal cell carcinoma. Oncol Rep. 2018;40:2023–2035. doi: 10.3892/or.2018.6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan TZ, Yang H, Ye J, Low J, Choolani M, Tan DS, Thiery JP, Huang RY. CSIOVDB: A microarray gene expression database of epithelial ovarian cancer subtype. Oncotarget. 2015;6:43843–43852. doi: 10.18632/oncotarget.5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshihara K, Tajima A, Komata D, Yamamoto T, Kodama S, Fujiwara H, Suzuki M, Onishi Y, Hatae M, Sueyoshi K, et al. Gene expression profiling of advanced-stage serous ovarian cancers distinguishes novel subclasses and implicates ZEB2 in tumor progression and prognosis. Cancer Sci. 2009;100:1421–1428. doi: 10.1111/j.1349-7006.2009.01204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bowen NJ, Walker LD, Matyunina LV, Logani S, Totten KA, Benigno BB, McDonald JF. Gene expression profiling supports the hypothesis that human ovarian surface epithelia are multipotent and capable of serving as ovarian cancer initiating cells. BMC Med Genomics. 2009;2:71. doi: 10.1186/1755-8794-2-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mok SC, Bonome T, Vathipadiekal V, Bell A, Johnson ME, Wong KK, Park DC, Hao K, Yip DK, Donninger H, et al. A gene signature predictive for outcome in advanced ovarian cancer identifies a survival factor: Microfibril-associated glycoprotein 2. Cancer. Cell. 2009;16:521–532. doi: 10.1016/j.ccr.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emanuelsson O, Nagalakshmi U, Zheng D, Rozowsky JS, Urban AE, Du J, Lian Z, Stolc V, Weissman S, Snyder M, Gerstein MB. Assessing the performance of different high-density tiling microarray strategies for mapping transcribed regions of the human genome. Genome Res. 2007;17:886–897. doi: 10.1101/gr.5014606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBio-Portal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prat J, FIGO Committee on Gynecologic Oncology Staging classification for cancer of the ovary, fallopian tube, and peritoneum: Abridged republication of guidelines from the international federation of gynecology and obstetrics (FIGO) Obstet Gynecol. 2015;126:171–174. doi: 10.1097/AOG.0000000000000917. [DOI] [PubMed] [Google Scholar]
- 26.Trimarchi JM, Stadler MB, Cepko CL. Individual retinal progenitor cells display extensive heterogeneity of gene expression. PLoS One. 2008;3:e1588. doi: 10.1371/journal.pone.0001588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gyorffy B, Lánczky A, Szállási Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr Relat Cancer. 2012;19:197–208. doi: 10.1530/ERC-11-0329. [DOI] [PubMed] [Google Scholar]
- 28.Ferriss JS, Kim Y, Duska L, Birrer M, Levine DA, Moskaluk C, Theodorescu D, Lee JK. Multi-gene expression predictors of single drug responses to adjuvant chemotherapy in ovarian carcinoma: Predicting platinum resistance. PLoS One. 2012;7:e30550. doi: 10.1371/journal.pone.0030550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonome T, Levine DA, Shih J, Randonovich M, Pise-Masison CA, Bogomolniy F, Ozbun L, Brady J, Barrett JC, Boyd J, Birrer MJ. A gene signature predicting for survival in suboptimally debulked patients with ovarian cancer. Cancer Res. 2008;68:5478–5486. doi: 10.1158/0008-5472.CAN-07-6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Konstantinopoulos PA, Spentzos D, Karlan BY, Taniguchi T, Fountzilas E, Francoeur N, Levine DA, Cannistra SA. Gene expression profile of BRCAness that correlates with responsiveness to chemotherapy and with outcome in patients with epithelial ovarian cancer. J Clin Oncol. 2010;28:3555–3561. doi: 10.1200/JCO.2009.27.5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lisowska KM, Olbryt M, Dudaladava V, Pamuła-Piłat J, Kujawa K, Grzybowska E, Jarząb M, Student S, Rzepecka IK, Jarząb B, Kupryjańczyk J. Gene expression analysis in ovarian cancer-faults and hints from DNA microarray study. Front Oncol. 2014;4:6. doi: 10.3389/fonc.2014.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mateescu B, Batista L, Cardon M, Gruosso T, de Feraudy Y, Mariani O, Nicolas A, Meyniel JP, Cottu P, Sastre-Garau X, Mechta-Grigoriou F. miR-141 and miR-200a act on ovarian tumorigenesis by controlling oxidative stress response. Nat Med. 2011;17:1627–1635. doi: 10.1038/nm.2512. [DOI] [PubMed] [Google Scholar]
- 33.Tothill RW, Tinker AV, George J, Brown R, Fox SB, Lade S, Johnson DS, Trivett MK, Etemadmoghadam D, Locandro B, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res. 2008;14:5198–5208. doi: 10.1158/1078-0432.CCR-08-0196. [DOI] [PubMed] [Google Scholar]
- 34.Makii C, Oda K, Ikeda Y, Sone K, Hasegawa K, Uehara Y, Nishijima A, Asada K, Koso T, Fukuda T, et al. MDM2 is a potential therapeutic target and prognostic factor for ovarian clear cell carcinomas with wild type TP53. Oncotarget. 2016;7:75328–75338. doi: 10.18632/oncotarget.12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koti M, Gooding RJ, Nuin P, Haslehurst A, Crane C, Weberpals J, Childs T, Bryson P, Dharsee M, Evans K, et al. Identification of the IGF1/PI3K/NF κB/ERK gene signalling networks associated with chemotherapy resistance and treatment response in high-grade serous epithelial ovarian cancer. BMC Cancer. 2013;13:549. doi: 10.1186/1471-2407-13-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 37.Lee SJ, Bae JH, Lee AW, Tong SY, Park YG, Park JS. Clinical characteristics of metastatic tumors to the ovaries. J Korean Med Sci. 2009;24:114–119. doi: 10.3346/jkms.2009.24.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saha A, Kim Y, Gewirtz ADH, Jo B, Gao C, McDowell IC, GTEx Consortium. Engelhardt BE, Battle A. Co-expression networks reveal the tissue-specific regulation of transcription and splicing. Genome Res. 2017;27:1843–1858. doi: 10.1101/gr.216721.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Dam S, Võsa U, van der Graaf A, Franke L, de Magalhães JP. Gene co-expression analysis for functional classification and gene-disease predictions. Brief Bioinform. 2018;19:575–592. doi: 10.1093/bib/bbw139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang W, Liu HT. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 2002;12:9–18. doi: 10.1038/sj.cr.7290105. [DOI] [PubMed] [Google Scholar]
- 41.Shlien A, Malkin D. Copy number variations and cancer. Genome Med. 2009;1:62. doi: 10.1186/gm62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ciró M, Prosperini E, Quarto M, Grazini U, Walfridsson J, McBlane F, Nucifero P, Pacchiana G, Capra M, Christensen J, Helin K. ATAD2 is a novel cofactor for MYC, overexpressed and amplified in aggressive tumors. Cancer Res. 2009;69:8491–8498. doi: 10.1158/0008-5472.CAN-09-2131. [DOI] [PubMed] [Google Scholar]
- 43.Wu G, Lu X, Wang Y, He H, Meng X, Xia S, Zhen K, Liu Y. Epigenetic high regulation of ATAD2 regulates the Hh pathway in human hepatocellular carcinoma. Int J Oncol. 2014;45:351–361. doi: 10.3892/ijo.2014.2416. [DOI] [PubMed] [Google Scholar]
- 44.Zou JX, Guo L, Revenko AS, Tepper CG, Gemo AT, Kung HJ, Chen HW. Androgen-induced coactivator ANCCA mediates specific androgen receptor signaling in prostate cancer. Cancer Res. 2009;69:3339–3346. doi: 10.1158/0008-5472.CAN-08-3440. [DOI] [PubMed] [Google Scholar]
- 45.Zou JX, Revenko AS, Li LB, Gemo AT, Chen HW. ANCCA, an estrogen-regulated AAA+ ATPase coactivator for ERalpha, is required for coregulator occupancy and chromatin modification. Proc Natl Acad Sci USA. 2007;104:18067–18072. doi: 10.1073/pnas.0705814104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.