Abstract
Background
With the dissemination of carbapenemase producers, a revival of colistin was observed for the treatment of infections caused by MDR Gram-negatives. Unfortunately, the increasing usage of colistin led to the emergence of resistance. In Klebsiella pneumoniae, colistin resistance arises through addition of 4-amino-l-arabinose (l-Ara4N) or phosphoethanolamine (pEtN) to the native lipid A. The underlying mechanisms involve numerous chromosome-encoded genes or the plasmid-encoded pEtN transferase MCR. Currently, detection of colistin resistance is time-consuming since it still relies on MIC determination by broth microdilution. Recently, a rapid diagnostic test based on MALDI-TOF MS detection of modified lipid A was developed (the MALDIxin test) and tested on Escherichia coli and Acinetobacter baumannii.
Objectives
Optimize the MALDIxin test for the rapid detection of colistin resistance in K. pneumoniae.
Methods
This optimization consists of an additional mild-acid hydrolysis of 15 min in 1% acetic acid. The optimized method was tested on a collection of 81 clinical K. pneumoniae isolates, including 49 colistin-resistant isolates (45 with chromosome-encoded resistance, 3 with MCR-related resistance and 1 with both mechanisms).
Results
The optimized method allowed the rapid (<30 min) identification of l-Ara4N- and pEtN-modified lipid A of K. pneumoniae, which are known to be the real triggers of polymyxin resistance. At the same time, it discriminates between chromosome-encoded and MCR-related polymyxin resistance.
Conclusions
The MALDIxin test has the potential to become an accurate tool for the rapid determination of colistin resistance in clinically relevant Gram-negative bacteria.
Introduction
Currently, antimicrobial resistance is at the top of the agenda for scientists and governments, while XDR organisms, such as carbapenemase-producing Enterobacterales (CPE) are rapidly emerging. The pipeline of new antibiotics is very limited, and colistin is now considered as one of the last-resort therapies for the treatment of infection caused by XDR Gram-negative bacteria.1 In countries that are considered to be endemic for CPE (e.g. Greece, Italy), colistin is often used as empirical treatment for severe infection, such as bacteraemia. Unfortunately, this increased use of colistin in the therapeutic armamentarium has led inexorably to the development of resistance.2–6
In Gram-negative bacteria, acquired resistance to colistin results mostly from modifications of the drug target, i.e. LPS. These modifications correspond to addition(s) of 4-amino-l-arabinose (l-Ara4N) and/or phosphoethanolamine (pEtN) to lipid A, the anchor of LPS. Addition of such cationic components leads to the repulsion of colistin (an old class of cationic antibiotic that targets polyanionic bacterial LPS and disrupts the bacterial outer membranes), resulting in the protection against outer-membrane disruption by the antibiotic.7,8 The mechanisms involved in such modification of lipid A might be chromosome or plasmid encoded. Plasmid-encoded resistance to colistin involves the acquisition of an mcr-like gene encoding a specific pEtN transferase.9 MCR producers have mostly been reported among Escherichia coli and Salmonella spp.9 In contrast, in Klebsiella spp., chromosome-encoded resistance has been reported to be far more prevalent than mcr acquisition. The most prevalent chromosome-encoded mechanisms are mutations in genes encoding the PmrA/PmrB or PhoP/PhoQ two-component systems and (even more prevalent) alterations of the master regulator MgrB.10
Although the epidemiology of acquired colistin resistance varies depending on the bacterial species and geographical area, rapid detection of such resistance is one of the key ways of improving the treatment of patients infected with MDR bacteria for which other alternatives are not available (e.g. MBL producers). Currently, detection of colistin resistance in Enterobacterales relies on MIC determination using broth microdilution, which is the gold standard for colistin susceptibility testing.11 Recently, we developed a novel rapid approach using MALDI-TOF MS that detects colistin resistance directly on intact bacteria in <15 min, the MALDIxin test.12 It has been validated for E. coli and Acinetobacter baumannii, for which it can discriminate between chromosome- and/or plasmid-encoded resistance (i.e. mcr), besides detecting colistin resistance.12,13
Here we report an optimization of the MALDIxin test for the rapid detection of colistin resistance in Klebsiella pneumoniae.
Materials and methods
Bacterial isolates
A collection of 81 K. pneumoniae clinical isolates from Belgian and French national reference centres for antimicrobial resistance were used in this study (Table 1), including 49 colistin-resistant isolates (45 with chromosome-encoded resistance, 3 with MCR-related resistance and 1 with both mechanisms) and 32 colistin-susceptible isolates.
Table 1.
Results of the MALDIxin test on K. pneumoniae clinical isolates
| Isolate name | Colistin MIC (mg/L) | Resistance mechanism to polymyxins | β-Lactam resistance mechanisms (ESBLs, carbapenemases, …) | Percentage modified lipid Aa | Percentage pEtNa | Percentage l-Ara4Na | Reference |
|---|---|---|---|---|---|---|---|
| Colistin-susceptible isolates | |||||||
| 1609079984 | 0.5 | WT | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | this study | |
| 1609056413 | 0.5 | WT | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | 17 | |
| 1609078951 | 0.5 | WT | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | this study | |
| 1609078870 | 0.5 | WT | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | this study | |
| 1609068884 | 0.5 | WT | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | this study | |
| 1609059262 | 0.5 | WT | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | this study | |
| 1609061149 | 1 | WT | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | 17 | |
| 1609072327 | 0.5 | WT | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | this study | |
| 1609075598 | 0.5 | WT | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | this study | |
| 1609071256 | 0.5 | WT | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | this study | |
| 2 E8 | 0.5 | CTX-M-3 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | 17 | |
| 2 F1 | 0.5 | CTX-M-14 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | 17 | |
| 2 F4 | 0.5 | CTX-M-15 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | 18 | |
| 2 F5 | 0.5 | CTX-M-15 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | 18 | |
| 2 I5 | 0.5 | CTX-M-15 + TEM-1 + SHV-11 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | 17 | |
| 3 C4 | 0.5 | CTX-M-15 + TEM-1B + SHV-11 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | this study | |
| 3 D6 | 0.5 | CTX-M-15 + TEM-1B + SHV-28 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | this study | |
| 3 D7 | 0.5 | CTX-M-15 + TEM-1B + SHV-83 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | this study | |
| 2 I3 | 0.5 | CTX-M-15 + TEM-1 + SHV-11 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | 18 | |
| 2 I4 | 0.5 | CTX-M-15 + TEM-1 + SHV-11 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | 17 | |
| 1 F7 | 0.5 | KPC-2 + SHV-11 + CTX-M-15 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | this study | |
| 3 B4 | 0.5 | KPC-3 + TEM-1A, OXA-9, SHV-11 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | 17 | |
| 3 B7 | 0.5 | GES-5 + SHV-12 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | 17 | |
| 1 B6 | 0.5 | NDM-1 + TEM-1 + CTX-M-15 + SHV-12 + OXA-9 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | 17 | |
| 1 C9 | 0.5 | VIM-1 + SHV-5 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | 17 | |
| 1 E3 | 0.5 | IMP-1 + TEM-15 + CTX-M-15 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | this study | |
| 2 B1 | 0.5 | OXA-48 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | 18 | |
| 2 C6 | 0.5 | NDM-1 + OXA-181 + CTX-M-15 + TEM-1 + OXA-1 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | 17 | |
| 2 D2 | 0.5 | OXA-204 + CMY-4 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | 17 | |
| 2 D8 | 0.5 | OXA-232 + TEM-1 + CTX-M-15 + OXA-1 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | 18 | |
| S15 | 0.25 | mgrB truncated in ORF by ISKpn25 + PhoP (I201N) | KPC-2 + OXA-9 + TEM-1 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | 19 |
| S17 | 0.5 | mgrB truncated in ORF by ISKpn25 + ArnC (C161Y) | KPC-2 + OXA-9 + TEM-1 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | 19 |
| Chromosome-encoded resistance | |||||||
| TUN-ST-15 | >128 | frameshift mgrB | CTX-M-15 + OXA-1 + SHV-28 | 10.6±1.5 | 0.0±0.0 | 100±0.0 | 20 |
| TUN-ST-101 | 8 | frameshift mgrB | OXA-48 + CTX-M-15 + OXA-1 | 19.5±6.6 | 0.0±0.0 | 100±0.0 | 20 |
| CNR 111 C2 | 16 | frameshift mgrB | OXA-48 + BLSE | 17.0±5.0 | 0.0±0.0 | 100±0.0 | this study |
| CNR 20140042 | 16 | MgrB N42Y and K43I | OXA-48 type + CTX-M grp1 | 18.9±3.4 | 0.0±0.0 | 100±0.0 | 17 |
| CNR 20140423 | 32 | MgrB N42Y and K43I | OXA-48 type + CTX-M grp1 | 26.5±7.6 | 0.0±0.0 | 100±0.0 | this study |
| CNR 20140661 | 64 | MgrB Q30 stop | KPC type | 29.7±4.6 | 0.0±0.0 | 100±0.0 | 17 |
| CNR 20150586 | 64 | MgrB Q30 stop | ESBL | 15.0±3.7 | 0.0±0.0 | 100±0.0 | this study |
| CNR 20150324 | 32 | MgrB Q30 stop | ESBL | 11.8±1.8 | 0.0±0.0 | 100±0.0 | this study |
| CNR 20150655 | 64 | MgrB Q30 stop | CTX-M grp1 | 15.8±7.5 | 0.0±0.0 | 100±0.0 | this study |
| CNR 20150542 | 32 | MgrB L4 stop | OXA-48 type + CTX-M grp1 | 15.2±5.7 | 0.0±0.0 | 100±0.0 | this study |
| CNR 20151119 | 64 | MgrB L4 stop | OXA-427 | 16.7±3.8 | 0.0±0.0 | 100±0.0 | 17 |
| CNR 20150622 | 64 | MgrB Y41 stop | KPC type + ESBL | 27.6±11.4 | 0.0±0.0 | 100±0.0 | 17 |
| CNR 20150777 | 128 | MgrB Y41 stop | CTX-M grp1 | 27.2±9.2 | 0.0±0.0 | 100±0.0 | 17 |
| CNR 20140698 | 32 | MgrB modified from AA 42 (I) | KPC type | 31.1±9.9 | 0.0±0.0 | 100±0.0 | this study |
| CNR 20150944 | 64 | MgrB modified from AA 42 (I) | KPC type | 42.6±8.4 | 0.0±0.0 | 100±0.0 | 17 |
| CNR 20150309 | 64 | MgrB modified from AA 37 (V) | KPC type + CTX-M grp1 | 18.7±3.9 | 0.0±0.0 | 100±0.0 | 17 |
| CNR 20150675 | 64 | mgrB truncated in ORF by IS10 | OXA-427 + CTX-M grp1 | 30.6±8.6 | 0.0±0.0 | 100±0.0 | 17 |
| CNR 20150276 | 32 | mgrB truncated in ORF by IS10 | OXA-427 + CTX-M grp1 | 34.8±12.0 | 0.0±0.0 | 100±0.0 | this study |
| CNR 20140483 | 32 | mgrB truncated in ORF by IS1R-like | OXA-48 type + CTX-M grp1 | 28.3±7.5 | 0.0±0.0 | 100±0.0 | 17 |
| CNR 20140563 | 64 | mgrB truncated in ORF by IS1R | OXA-48 type + DHA type | 31.7±9.3 | 0.0±0.0 | 100±0.0 | 17 |
| CNR 20150295 | 8 | mgrB truncated in ORF by IS1R | OXA-48 type + CTX-M grp1 | 16.7±4.5 | 0.0±0.0 | 100±0.0 | this study |
| CNR 20150713 | 64 | mgrB truncated in ORF by IS1R | OXA-48 type + CTX-M grp1 | 20.9±3.4 | 0.0±0.0 | 100±0.0 | this study |
| S20-003 | 64 | mgrB truncated in ORF by IS1R | OXA-48 type | 17.3±4.3 | 0.0±0.0 | 100±0.0 | this study |
| CNR 20150736 | 32 | mgrB truncated in ORF by IS1R | OXA-48 type + CTX-M grp1 | 34.0±4.3 | 0.0±0.0 | 100±0.0 | this study |
| CNR 20150960 | 32 | mgrB truncated in ORF by IS1R | ND | 16.8±2.9 | 0.0±0.0 | 100±0.0 | this study |
| CNR 20151181 | 32 | mgrB truncated in ORF by IS1R | OXA-48 type + CTX-M grp1 | 247.6±8.9 | 0.0±0.0 | 100±0.0 | this study |
| CNR 20150050 | 32 | mgrB truncated in promoter by IS1R | DHA type | 22.1±5.9 | 0.0±0.0 | 100±0.0 | 17 |
| CNR 20150943 | 32 | mgrB truncated in promoter by IS1R | DHA type | 26.8±11.7 | 0.0±0.0 | 100±0.0 | 17 |
| CNR 20140591 | 64 | mgrB truncated in ORF by IS5-like | KPC type + ESBL | 31.3±11.0 | 0.0±0.0 | 100±0.0 | 17 |
| CNR 20140862 | 16 | mgrB truncated in ORF by IS5-like | ND | 23.9±6.5 | 0.0±0.0 | 100±0.0 | this study |
| CNR 20150573 | 32 | mgrB truncated in ORF by IS5-like | NDM type + OXA-48 type + CTX-M grp1 | 38.4±25.4 | 0.0±0.0 | 100±0.0 | this study |
| CNR 20140550 | 32 | mgrB truncated in promoter by IS903D | KPC type + CTX-M grp9 | 31.9±8.6 | 0.0±0.0 | 100±0.0 | this study |
| CNR 20151285 | 32 | mgrB truncated in ORF by IS903-like | CTX-M grp1 | 21.3±10.9 | 0.0±0.0 | 100±0.0 | 17 |
| S12-172 | 32 | mgrB truncated in ORF by IS903-like | NDM type | 29.6±4.1 | 0.0±0.0 | 100±0.0 | this study |
| S14-002 | 64 | mgrB truncated in promoter by ISKpn14 | KPC type | 44.2±19.2 | 0.0±0.0 | 100±0.0 | 17 |
| CNR 20140101 | 32 | ΔmgrB | OXA-48 type + CTX-M grp1 | 23.4±5.3 | 0.0±0.0 | 100±0.0 | 17 |
| CNR 20150078 | 32 | ΔmgrB | OXA-48 type + CTX-M grp9 | 20.1±4.7 | 0.0±0.0 | 100±0.0 | 17 |
| CNR 20150066 | 16 | ΔmgrB | OXA-48 type + BLSE | 23.4±3.8 | 0.0±0.0 | 100±0.0 | 17 |
| CNR 20151223 | 32 | ΔmgrB | carbapenem resistant via impermeability | 51.3±8.6 | 0.0±0.0 | 100±0.0 | 17 |
| S1 | 128 | mgrB truncated in ORF by ISKpn25 | KPC-2 + OXA-9 + TEM-1 | 40.9±13.6 | 0.0±0.0 | 100±0.0 | 19 |
| S12 | 64 | mgrB truncated in ORF by ISKpn25 | KPC-2 + OXA-9 + TEM-1 | 26.6±3.8 | 0.0±0.0 | 100±0.0 | 19 |
| CNR 1630 | 32 | mgrB truncated in ORF by IS5 | ND | 27.2±9.1 | 0.0±0.0 | 100±0.0 | 17 |
| CNR 1631 | 8 | mgrB truncated in ORF by IS5 | ND | 42.5±24.7 | 0.0±0.0 | 100±0.0 | this study |
| CNR 1795 | 128 | mgrB truncated in ORF by IS5 | ND | 34.0±4.3 | 0.0±0.0 | 100±0.0 | this study |
| CNR 1861 | 16 | mutated PmrB (T157P) | ND | 28.8±5.1 | 0.0±0.0 | 100±0.0 | 17 |
| MCR-related resistance to colistin | |||||||
| CNR 1732 | 4 | mcr-1 | ND | 32.1±7.7 | 100±0.0 | 0.0±0.0 | 17 |
| CNR 1853 | 4 | mcr-1 | ND | 34.4±11.8 | 95.3±6.2 | 4.7±6.2 | 17 |
| CNR 186 G1 | 8 | mcr-8 | SHV-27 + TEM-1B + SCO-1 | 24.4±5.6 | 100±0.0 | 0.0±0.0 | this study |
| MCR + chromosome-encoded resistance to colistin | |||||||
| CNR 1601 | 32 | mcr-1 + mgrB truncated in ORF by IS5 | ND | 33.6±19.6 | 46.1±13.1 | 53.9±13.1 | 17 |
ND, not determined.
Carbapenemases are shown in bold and ESBLs are underlined.
Mean±standard error of the mean.
Susceptibility testing
Colistin MIC was determined by broth microdilution according to CLSI and EUCAST guidelines.11 Results were interpreted using EUCAST breakpoints as updated in 2019 (http://www.eucast.org/clinical_breakpoints/).
Optimized MALDIxin test
The MALDIxin procedure was performed as previously described,12 with the addition of a short mild-acid hydrolysis step, which was crucial for K. pneumoniae (Figure S1, available as Supplementary data at JAC Online). Briefly, a single colony cultured on Mueller–Hinton agar (bioMérieux, La Balme-les-Grottes, France) was resuspended in 200 μL of distilled water, washed three times with double-distilled water and resuspended in 100 μL of double-distilled water. A 50 μL aliquot was then submitted to mild-acid hydrolysis by the addition of 50 μL of 2% acetic acid in double-distilled water and heating for 15 min at 100°C. The hydrolysed cells were spun, the supernatant was discarded and the pellet was suspended in 25 μL of double-distilled water. An aliquot of 0.4 μL of the bacterial solution was loaded onto the target and immediately overlaid with 0.8 μL of a super-2,5-dihydroxybenzoic acid matrix (Sigma–Aldrich, Gillingham, UK) used at a final concentration of 10 mg/mL in chloroform/methanol (90:10, v/v). Bacterial solution and matrix were mixed directly on the target by pipetting and the mix was dried gently under a stream of air (<1 min). MALDI-TOF MS analysis was performed on a 4800 Proteomics Analyzer (Applied Biosystems, Foster City, CA, USA) using the reflectron mode. Samples were analysed by operating at 20 kV in the negative ion mode using an extraction delay time set at 20 ns. MS data were analysed using Data Explorer version 4.9 (Applied Biosystems).
Statistical analysis
All experiments were carried out on three independent bacterial cultures. Data were compared two-by-two using the unpaired Welch’s t-test. P values <0.05 were considered statistically different.
Results
Detection of colistin resistance markers in K. pneumoniae using the MALDIxin test
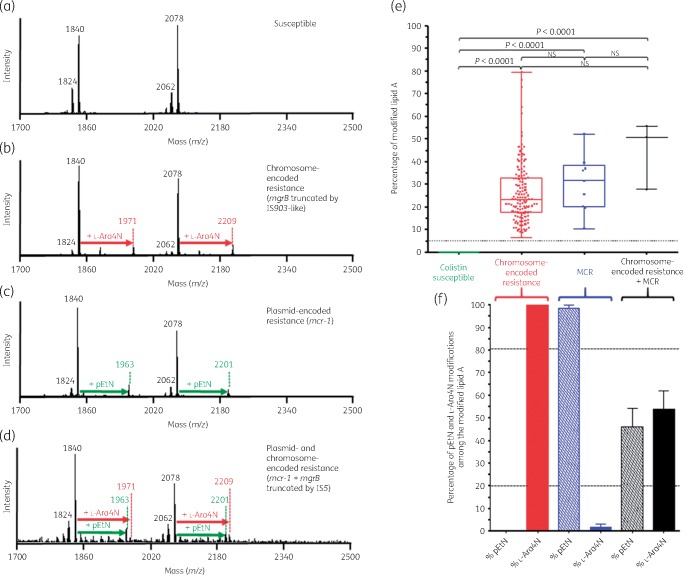
In polymyxin-susceptible K. pneumoniae isolates, the mass spectrum is dominated by two sets of peaks centred at m/z 1840 and m/z 2078 (Figure 1a). The ions at m/z 1824 and m/z 1840 are assigned to a bis-phosphorylated, hexa-acylated lipid A molecule containing or not containing a hydroxylation on the C′-2 fatty acyl chain. The ions at m/z 2062 and m/z 2078 are assigned to a bis-phosphorylated, hepta-acylated lipid A molecule either containing or not containing a hydroxyl group, respectively, on the C′-2 fatty acyl chain and resulting from a palmitoylation at the C-1 acyl-oxo-acyl position of the molecule at m/z 1824 and m/z 1840, respectively.
Figure 1.
Results of the optimized MALDIxin test on K. pneumoniae. Representative spectra of: (a) a polymyxin-susceptible K. pneumoniae isolate; (b) a chromosome-encoded (mgrB disruption) polymyxin-resistant K. pneumoniae isolate; (c) a polymyxin-resistant K. pneumoniae isolate producing MCR-1; and (d) a polymyxin-resistant K. pneumoniae harbouring both chromosome-encoded resistance (mgrB disruption) and plasmid-encoded MCR-1. The peaks at m/z 1824, 1840, 2062 and 2078 (black) correspond to the peaks of native K. pneumoniae lipid A, the peaks at m/z 1971 and 2209 (red) correspond to the addition of l-Ara4N to the native lipid A and the peaks at m/z 1963 and 2201 (green) correspond to the addition of one pEtN to the native lipid A. (e and f) Representation of the percentage of the modified lipid A for colistin-susceptible and colistin-resistant K. pneumoniae isolates. (e) The global percentage of modified lipid A (l-Ara4N+pEtN-modified lipid A/native lipid A) is represented for colistin-susceptible isolates (n=32), chromosome-encoded colistin-resistant K. pneumoniae isolates (n=45), MCR-producing colistin-resistant K. pneumoniae isolates (n=3) and the K. pneumoniae isolate harbouring both mechanisms (n=1). All experiments were performed in triplicate. The error bars represent the standard error of the mean and the dotted horizontal line corresponds to the suggested cut-off for colistin susceptibility related to no modification of lipid A. NS, not significant. (f) Representation of the percentage of l-Ara4N- and pEtN-modified lipid A among the global modified lipid A for colistin-resistant K. pneumoniae isolates. The dotted horizontal lines correspond to the proposed cut-offs for discriminating between chromosome-encoded resistance, MCR production and both mechanisms.
In chromosome-encoded colistin-resistant K. pneumoniae isolates, the mass spectrum exhibits two sets of peaks centred at m/z 1971 and m/z 2209, corresponding to the previously observed m/z +131 shifts of mass unit caused by the addition of l-Ara4N to the hexa-and hepta-acylated lipid A structures at m/z 1840 and m/z 2078, respectively (Figure 1b).
In MCR producers, the mass spectrum exhibits two sets of peaks centred at m/z 1963 and m/z 2201, corresponding to the previously observed m/z +123 shifts of mass unit caused by the addition of pEtN to the hexa-and hepta-acylated lipid A structures at m/z 1840 and m/z 2078, respectively (Figure 1c).
In colistin-resistant isolates that exhibit both plasmid (mcr)- and chromosome-encoded resistance, the mass spectrum exhibits three sets of peaks centred at m/z 1963, m/z 2201 and m/z 2209, corresponding to the previously observed m/z +123 shifts of mass unit caused by the addition of pEtN to the hexa-and hepta-acylated lipid A structures at m/z 1840 and m/z 2078, respectively, and m/z +131 shifts of mass unit caused by the addition of l-Ara4N to the hepta-acylated lipid A structures at m/z 2078 (Figure 1d).
To further support this observation, we validated the MALDIxin test on 81 K. pneumoniae clinical isolates, including 45 colistin-resistant and 36 colistin-susceptible isolates. The percentage of modified lipid A corresponding to the sum of the intensities of the peaks associated with pEtN modification (m/z 1963 and m/z 2201) and l-Ara4N modification (m/z 1971 and m/z 2209) divided by the intensities of the peaks assigned to the native lipid A (m/z 1824, m/z 1840 and m/z 2062) allows accurate distinction between colistin-susceptible and colistin-resistant isolates (Figure 1e). Of note, the peak at m/z 2078 was not taken into account in the calculation of the percentage of modified lipid A since this native peak observed in colistin-susceptible isolate spectra might potentially correspond to a peak of modified lipid A resulting from the addition of pEtN plus l-Ara4N to the native bis-phosphorylated, hexa-acylated lipid A (m/z 2078=m/z 1824 + m/z 123 + m/z 131). The percentage of modified lipid A was found to be 0 for all colistin-susceptible K. pneumoniae isolates, while it was >5 for all colistin-resistant isolates (Table 1 and Figure 1e).
Discrimination between chromosome-encoded resistance, MCR-related resistance and both mechanisms
In K. pneumoniae chromosome-encoded colistin resistance results mostly from addition of l-Ara4N to the native lipid A. In contrast, MCR is known to be a pEtN transferase resulting exclusively in addition of pEtN to the native lipid A. Thus, to distinguish between chromosome-encoded resistance and MCR production, the percentages of pEtN and l-Ara4N modifications (% pEtN and % l-Ara4N) were calculated. The % pEtN and % l-Ara4N correspond to the sum of the intensities of the peaks associated with pEtN modifications (m/z 1963 and m/z 2201) or l-Ara-4N modifications (m/z 1971 and m/z 2209) divided by intensities of the peaks associated with both modifications (m/z 1963 and m/z 2201 for pEtN modifications plus m/z 1971 and m/z 2209 for l-Ara4N modifications). As shown in Figure 1(f), for all colistin-resistant K. pneumoniae isolates with chromosome-encoded mechanisms, lipid A was exclusively modified by addition of l-Ara4N. In contrast, MCR-producing isolates had an average % l-Ara4N of 1.6 (0–14.0) and an average % pEtN of 98.4 (86.0–100). When both colistin resistance mechanisms were expressed (only one isolate), % l-Ara4N was 53.9 (36.4–69.5) and % pEtN was 46.1 (30.5–63.6) (Figure 1f). Accordingly, using our isolate collection, arbitrary cut-off values at 20% and 80% for % l-Ara4N and % pEtN, respectively, might be suggested to easily discriminate chromosome-encoded resistance from MCR production and co-expression of both mechanisms.
Discussion
Here we optimized the MALDIxin test for the detection of colistin resistance in K. pneumoniae. The procedure used included a preliminary short (15 min) mild-acid hydrolysis step, which allowed the rapid identification of l-Ara4N- and pEtN-modified lipid A, which are known to be the real triggers of colistin resistance. In K. pneumoniae, chromosome-encoded resistance is more frequent than MCR plasmid-encoded resistance. It mainly involves alteration of MgrB, leading to activation of the arn operon and subsequent addition of l-Ara4N to the native lipid A.10 This modification results in an m/z +131 shift of the native lipid A-related peaks. In contrast, expression of MCR enzymes results in the addition of pEtN to the native lipid A.14 Accordingly, a shift of m/z +123 is observed. Using the optimized MALDIxin test, we could (i) easily predict colistin resistance in K. pneumoniae by checking whether any modified (l-Ara4N or pEtN) lipid A is present in the bacterial membrane, but also (ii) discriminate between chromosome- and mcr-encoded resistance by looking at the percentage of l-Ara4N or pEtN modification in the modified lipid A. As expected, 100% l-Ara4N modification was observed in the case of chromosome-encoded resistance while close to 100% modified lipid A was related to pEtN addition in the case of MCR expression. Although only one isolate was available, detection of concomitant mechanisms (MCR production + disruption of chromosome-encoded MgrB) repeatedly resulted in a mixture of pEtN- and l-Ara4N-modified lipid A (about 50%/50%). In addition, despite the fact that the MALDIxin test was able to accurately detect colistin-resistant isolates, there was no strong correlation between the modification level of lipid A and the resistance level of colistin in terms of MIC.
In the context of MCR-related colistin resistance, molecular biology is widely used for the detection of MCR-producing isolates.15 However, the increasing number of mcr variants (mcr-1 to mcr-9) that do not share a strong nucleotide identity will inexorably lead to false-negative results. By targeting the pEtN modification of lipid A, which corresponds to the result of all MCR variants, the MALDIxin test might be an accurate screening test for the identification of a new MCR variant.
To the best of our knowledge, this is the first MALDI-TOF MS-based method that allows the rapid detection of colistin resistance and at the same time discrimination between chromosome-encoded and MCR-related polymyxin resistance in K. pneumoniae without necessitating any complex lipid extraction steps. Indeed, Liang et al.16 recently described another MALDI-TOF MS-based method that has the ability to differentiate colistin-susceptible from colistin-resistant K. pneumoniae, but that requires fastidious sample preparation of membrane lipids with incubation in a special buffer, incubation in cooled ice, washes in ethanol and final extraction in chloroform/methanol/water (12:6:1, v/v/v).
We should acknowledge that MALDI-TOF MS analysis was performed on a 4800 Proteomics Analyzer, which is not commonly available in clinical microbiology laboratories. In addition, samples were analysed by operating in the negative ion mode of the mass spectrometer, which is not currently and widely usable on routine mass spectrometers. Accordingly, a few optimization steps are still needed to implement this test in routine use.
Funding
This work was partially funded by the University Paris-Sud, France. Laurent Dortet is a member of the Laboratory of Excellence in Research on Medication and Innovative Therapeutics (LERMIT) supported by a grant from the French National Research Agency (ANR-10-LABX-33). Gerald Larrouy-Maumus was supported by the Department of Life Sciences in the Faculty of Natural Sciences Imperial College London, UK, and MRC-Confidence in Concept grant number 105603/Z/14/Z.
Transparency declarations
Laurent Dortet, Alain Filloux and Gerald Larrouy-Maumus are co-inventors of the MALDIxin test for which a patent has been filed by Imperial Innovations. All other authors: none to declare.
Author contributions
Laurent Dortet and Gerald Larrouy-Maumus had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: Laurent Dortet and Gerald Larrouy-Maumus. Acquisition, analysis or interpretation of data: all authors. Drafting of the manuscript: Laurent Dortet, Gerald Larrouy-Maumus and Alain Filloux. Critical revision of the manuscript for important intellectual content: all authors.
Supplementary Material
References
- 1. Falagas ME, Kasiakou SK.. Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin Infect Dis 2005; 40: 1333–41. [DOI] [PubMed] [Google Scholar]
- 2. Kontopidou F, Giamarellou H, Katerelos P. et al. Infections caused by carbapenem-resistant Klebsiella pneumoniae among patients in intensive care units in Greece: a multi-centre study on clinical outcome and therapeutic options. Clin Microbiol Infect 2014; 20: O117–23. [DOI] [PubMed] [Google Scholar]
- 3. Kontopidou F, Plachouras D, Papadomichelakis E. et al. Colonization and infection by colistin-resistant Gram-negative bacteria in a cohort of critically ill patients. Clin Microbiol Infect 2011; 17: E9–11. [DOI] [PubMed] [Google Scholar]
- 4. Neonakis IK, Samonis G, Messaritakis H. et al. Resistance status and evolution trends of Klebsiella pneumoniae isolates in a university hospital in Greece: ineffectiveness of carbapenems and increasing resistance to colistin. Chemotherapy 2010; 56: 448–52. [DOI] [PubMed] [Google Scholar]
- 5. Rojas LJ, Salim M, Cober E. et al. Colistin resistance in carbapenem-resistant Klebsiella pneumoniae: laboratory detection and impact on mortality. Clin Infect Dis 2017; 64: 711–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Venditti C, Nisii C, D'Arezzo S. et al. Letter to the Editor: surveillance of mcr-1 and mcr-2 genes in carbapenem-resistant Klebsiella pneumoniae strains from an Italian hospital. Euro Surveill 2017; 22: pii=30604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jeannot K, Bolard A, Plesiat P.. Resistance to polymyxins in Gram-negative organisms. Int J Antimicrob Agents 2017; 49: 526–35. [DOI] [PubMed] [Google Scholar]
- 8. Olaitan AO, Morand S, Rolain JM.. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol 2014; 5: 643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu YY, Wang Y, Walsh TR. et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 2016; 16: 161–8. [DOI] [PubMed] [Google Scholar]
- 10. Poirel L, Jayol A, Nordmann P.. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev 2017; 30: 557–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.CLSI and EUCAST Joint Subcommittee. Recommendations for MIC Determination of Colistin (Polymyxin E) as Recommended by the Joint CLSI-EUCAST Polymyxin Breakpoints Working Group. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/General_documents/Recommendations_for_MIC_determination_of_colistin_March_2016.pdf.
- 12. Dortet L, Bonnin RA, Pennisi I. et al. Rapid detection and discrimination of chromosome- and MCR-plasmid-mediated resistance to polymyxins by MALDI-TOF MS in Escherichia coli: the MALDIxin test. J Antimicrob Chemother 2018; 73: 3359–67. [DOI] [PubMed] [Google Scholar]
- 13. Dortet L, Potron A, Bonnin RA. et al. Rapid detection of colistin resistance in Acinetobacter baumannii using MALDI-TOF-based lipidomics on intact bacteria. Sci Rep 2018; 8: 16910.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yu H, Qu F, Shan B. et al. Detection of the mcr-1 colistin resistance gene in carbapenem-resistant Enterobacteriaceae from different hospitals in China. Antimicrob Agents Chemother 2016; 60: 5033–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jousset AB, Bernabeu S, Bonnin RA. et al. Development and validation of a multiplex polymerase chain reaction assay for detection of the five families of plasmid-encoded colistin resistance. Int J Antimicrob Agents 2019; 53: 302–9. [DOI] [PubMed] [Google Scholar]
- 16. Liang T, Leung LM, Opene B. et al. Rapid microbial identification and antibiotic resistance detection by mass spectrometric analysis of membrane lipids. Anal Chem 2019; 91: 1286–94. [DOI] [PubMed] [Google Scholar]
- 17. Girlich D, Naas T, Dortet L.. Comparison of the Superpolymyxin and ChromID Colistin R screening media for the detection of colistin-resistant Enterobacteriaceae from spiked rectal swabs. Antimicrob Agents Chemother 2019; 63: e01618-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dortet L, Fusaro M, Naas T.. Improvement of the Xpert Carba-R kit for the detection of carbapenemase-producing Enterobacteriaceae. Antimicrob Agents Chemother 2016; 60: 3832–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jousset AB, Bonnin RA, Rosinski-Chupin I. et al. A 4.5-year within-patient evolution of a colistin-resistant Klebsiella pneumoniae carbapenemase-producing K. pneumoniae sequence type 258. Clin Infect Dis 2018; 67: 1388–94. [DOI] [PubMed] [Google Scholar]
- 20. Jaidane N, Bonnin RA, Mansour W. et al. Genomic insights into colistin-resistant Klebsiella pneumoniae from a Tunisian teaching hospital. Antimicrob Agents Chemother 2018; 62: e01601-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.