Summary
Phage defense systems are often found on mobile genetic elements (MGEs), where they constitutively defend against invaders or are induced to respond to new assaults. Phage satellites, one type of MGE, are induced during phage infection to promote their own transmission, reducing phage production and protecting their hosts in the process. One such satellite in Vibrio cholerae, PLE, sabotages the lytic phage ICP1 which triggers PLE excision from the bacterial chromosome, replication, and transduction to neighboring cells. Analysis of patient stool samples from different geographic regions revealed that ICP1 has evolved to possess one of two syntenic loci encoding an SF1B-type helicase, either of which PLE exploits to drive replication. Further, loss of PLE mobilization limits anti-phage activity due to phage-mediated degradation of the bacterial genome. Our work provides insight into the unique challenges facing parasites of lytic phages and underscores the adaptions of satellites to their ever-evolving target phage.
Keywords: phage, mobile genetic element, defense, replication, parasitism, satellite, helicase
Graphical Abstract:
eTOC blurb:
Phage defense systems are often found on mobile genetic elements (MGEs). McKitterick et al. discover that viral satellites in Vibrio cholerae hijack phage-encoded helicases to replicate and escape the host chromosome, which elicits robust phage defense. Mobilization to escape host takeover may explain why some resistance mechanisms cluster on MGEs.
Introduction
Viruses and mobile genetic elements (MGEs) are associated with organisms from all branches of the tree of life (Koonin and Krupovic, 2015). To successfully infect their hosts, viruses employ various host-takeover programs that inhibit host activities and promote viral processes. Bacterial viruses, or phages, have profound effects on bacterial fitness, as well as on human health and disease (Brüssow, Canchaya and Hardt, 2004; Bondy-Denomy and Davidson, 2014). Of interest, lytic phages, which infect and kill their host in a single round of infection, have impactful roles in shaping the composition of bacterial populations, such as the human gut microbiome (Manrique, Dills and Young, 2017), and are potential biocontrol agents for antibiotic resistant infections (Pires et al., 2016). Lytic phages are particularly insidious to their hosts—upon infection, phages like the Escherichia coli phage T4 can shut down and redirect host transcriptional machinery to favor transcription of phage genes, and deploy nucleases that degrade the host chromosome inhibiting host gene expression and freeing up nucelosides to be incorporated into the rapidly replicating phage genome (Warner et al., 1970; Hercules et al., 1971; Hinton et al., 2005).
Paradoxically, phages also contribute to bacterial population diversity and complexity by facilitating horizontal gene transfer (HGT) (Brüssow, Canchaya and Hardt, 2004; Koskella and Brockhurst, 2014). In addition to well characterized mechanisms by which phages can spread bacterial genetic material to neighboring cells, such as generalized and specialized transduction (Penadés et al., 2015), recent work uncovered a means to package large regions of the bacterial chromosome into phage virions in a process termed lateral transduction (Chen et al., 2018). Independent of packaging, phages also facilitate the spread of bacterial plasmids from lysed cells to neighbors, increasing the range of genetic material that can be shared (Keen et al., 2017). In sharp contrast to these forms of “passive” phage-mediated HGT, certain parasitic mobile genetic elements referred to as phage satellites, such as phage inducible chromosomal islands (PICIs), have evolved to explicitly manipulate the phage replication and packaging programs for their own horizontal spread (Penadés and Christie, 2015).
Typically, phage defense in bacteria is attributed to widely characterized systems, including restriction-modification and CRISPR-Cas systems that target and cleave the infecting phage genome, or toxin/antitoxin and abortive infection systems that function by killing the infected host cell (Samson et al., 2013; Dy et al., 2014; Hille et al., 2018). However, phage parasites, which are being discovered with increasing frequency (Martínez-Rubio et al., 2017; O’Hara et al., 2017; Fillol-Salom et al., 2018), can also provide robust phage defense for their bacterial hosts. One type of PICI, the well characterized Staphylococcus aureus pathogenicity islands (SaPIs), are induced by infection with a helper phage, compete with that helper over the bacterial host’s replication machinery, and steal phage packaging proteins to selfishly package the SaPI genome for horizontal transfer. This parasitic interference negatively impacts the helper phage’s ability to complete its lifecycle, thus blocking plaque formation (Ubeda et al., 2009; Tormo-Más et al., 2010; Ram et al., 2012). Despite diverse mechanisms, phage defense systems must overcome phage-mediated host takeover to prevent rampant phage propagation through the community. Genomic analyses to localize anti-phage mechanisms in bacterial genomes reveal that they tend to cluster together on MGEs known as defense islands (DIs) (Makarova et al., 2011), and this observation has enabled discovery of new phage defense systems (Doron et al., 2018). While hypothesized to have roles in HGT, the prevalence of phage defense systems on MGEs has yet to be explained. Likewise, it remains to be seen if such DIs have evolved to parasitize phages for their own dissemination.
Vibrio cholerae, the etiological agent of the diarrheal disease cholera, is constantly under assault by phages in aquatic environments and in human hosts (Faruque et al., 2005; Seed et al., 2011, 2014). The dominant phage that preys on epidemic V. cholerae is ICP1, a lytic myovirus consistently isolated from cholera patient stool samples in Dhaka, Bangladesh where cholera is endemic (Seed et al., 2011; Angermeyer et al., 2018; McKitterick et al., 2019). In response to attack by ICP1, V. cholerae has acquired the phage-inducible chromosomal island-like element (PLE), a highly specific phage satellite that blocks plaque formation by ICP1 while exploiting phage resources to further its lifecycle (O’Hara et al., 2017). PLE excises from the host chromosome during ICP1 infection, replicates to high copy, and is transduced to neighboring cells. PLE encodes a large serine recombinase, Int, that catalyzes PLE excision and circularization by interacting with ICP1-encoded PexA, a protein of unknown function that is specific to ICP1 and is hijacked by PLE to act as a recombination directionality factor (McKitterick and Seed, 2018). Once excised, PLE replicates, decreasing ICP1’s capacity to replicate its genome (Barth et al., 2019), and is hypothesized to steal structural proteins from ICP1 to facilitate its own transmission. PLE then triggers accelerated lysis of the infected cell allowing for release of PLE transducing particles, ultimately killing the infected V. cholerae host but protecting the population as no infectious ICP1 progeny are produced (O’Hara et al., 2017). Five PLEs with a shared genomic architecture have been identified in epidemic V. cholerae isolates, all of which block plaque formation by ICP1 and undergo the aforementioned PLE lifecycle during infection (O’Hara et al., 2017).
Recent work uncovered a PLE-encoded factor that is necessary for PLE replication: the replication initiation factor, RepA (Barth et al., 2019). RepA binds to the PLE origin of replication (ori) to recruit replisome proteins that have yet to be identified. In the absence of ICP1 infection, however, RepA is not sufficient to drive PLE replication. Further, PLE is not predicted to encode additional replication machinery, suggesting that phage-encoded gene products are recruited for PLE amplification. As all PLEs replicate following ICP1 infection (O’Hara et al., 2017), it stands to reason that PLE evolved to exploit components of ICP1’s replication machinery. Similar to PLE excision (O’Hara et al., 2017), PLE replication is essential for PLE transduction, thus further underscoring the role of ICP1 in driving PLE HGT; however, the relatively low rate of transduction suggests that robust PLE replication may have other roles in PLE conflict with ICP1 (Barth et al., 2019).
To exploit ICP1, PLE must escape ICP1-mediated host takeover during infection. While the precise mechanisms that ICP1 uses to rapidly overtake V. cholerae have not been characterized, ICP1 quickly replicates its genome following infection (Barth et al., 2019) and produces virions within 20 minutes of infection (O’Hara et al., 2017). Here, we identify ICP1 ΔpexA mutants that escape PLE by acquiring mutations in the ICP1-encoded SF1B accessory helicase helA. We show that while this helicase is not necessary for ICP1 replication, it is essential for PLE to hijack for its own replication during ICP1 infection. We show that the excision- and replication-deficient PLE is susceptible to ICP1-mediated host takeover, whereby PLE is degraded while it remains integrated in the V. cholerae chromosome. Analysis of natural isolates of ICP1 from cholera patient stool samples in the megacity of Dhaka compared to a rural site in Bangladesh revealed an alternative SF1B helicase allele in phages from the rural site. Functional comparisons between the two alleles revealed that both, though unrelated, can be hijacked by all PLEs for replication. Though neither helicase is essential for ICP1, ICP1 faces impaired fitness in the absence of either accessory helicase, explaining their prevalence in ICP1 and other Vibrio phages. PLE’s capacity to use a variety of phage-encoded helicases to drive PLE replication stresses the critical role that replication plays in the PLE lifecycle to avoid phage-mediated host takeover and to facilitate continued gene expression. The common trend of phage defense islands clustered on MGEs suggests that mobilization of these phage defense islands, such as PLE, is a common mechanism to escape phage-mediated host takeover.
Results
ICP1 escapes excision-deficient PLE through mutations in the predicted helicase HelA
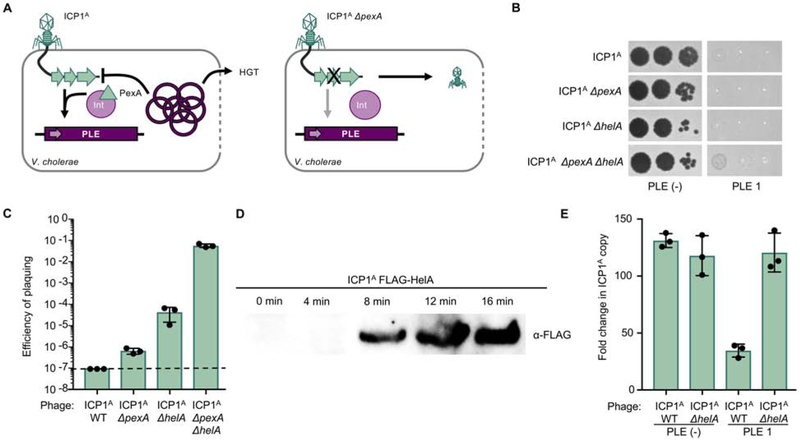
Previous work demonstrated the role for phage-encoded PexA in directing PLE 1 excision during infection with an ICP1 isolate from 2006, referred to as ICP1A (McKitterick and Seed, 2018) (Figure 1A). PLE 1 mediated inhibition of ICP1 does not require excision, so ICP1 ΔpexA is still blocked by PLE 1 (Figure 1B); however, ICP1A ΔpexA forms rare plaques on V. cholerae harboring PLE 1 at a frequency of 1 per 106 phage (Figures 1A and 1C). Due to the low efficiency of plaquing, we consider these phage to be “escape phage” that acquired a mutation in the genome allowing them to overcome PLE 1. To identify the mutated phage gene(s) enabling escape, we collected and sequenced three escape phage. Analysis revealed that all escape phage had mutations in ICP1A gp147, a predicted SF1B-type helicase we have since named helicase A (helA) (Table S1).
Figure 1. ICP1 overcomes excision-deficient PLE through loss of accessory helicase HelA.
A, Schematic of PLE 1 response to ICP1 infection. Left, ICP1 infects PLE (+) V. cholerae and expresses PexA, which interacts with PLE 1-encoded integrase (Int) to direct PLE circularization and excision. Excised PLE 1 replicates to high copy number, inhibits ICP1 replication, and horizontally transduces to neighboring cells when V. cholerae lyses. Right, when ICP1 ΔpexA infects PLE (+) V. cholerae, PLE 1 remains integrated in the host chromosome and rare mutant phage form plaques. B, Tenfold ICP1 dilutions spotted on PLE 1 and PLE (−) V. cholerae lawns (grey). Zones of killing are shown in black. C, Efficiency of plaquing of wild-type (WT) ICP1A or derivatives on PLE 1 relative to a PLE (−) V. cholerae host. Dashed line indicates limit of detection. D, Western blot of endogenously FLAG-tagged HelA during infection of PLE (−) V. cholerae. E, Quantification of change in ICP1 genome copy following infection of the listed V. cholerae host as detected by qPCR.
SF1B-type helicases are found broadly across all domains of life and include the well-studied Pif1 and RecD (Saikrishnan et al., 2009). In eukaryotes such as Saccharomyces cerevisiae, Pif1 is implicated in telomere maintenance, Okazaki fragment processing, and G-quadruplex motif resolution (Byrd and Raney, 2017), while RecD is a core component of the E. coli RecBCD complex involved in DNA processing and repair (Singleton et al., 2004). Another prototypical SF1B-type helicase is phage T4’s Dda, which is a non-essential accessory helicase implicated in origin melting, translocating proteins off DNA, and a wide variety of other functions in vitro, although its exact role in vivo is unknown (Byrd and Raney, 2006; Brister, 2008; He et al., 2012).
To validate the role of helA in ICP1A escape from PLE 1, we constructed a helA deletion in a wild-type ICP1A background and probed the mutant for the ability to overcome PLE 1. ICP1A-encoded helA is not necessary for plaque formation on PLE (−) V. cholerae, and ICP1A ΔhelA is still blocked by PLE 1 (Figure 1B). Similar to ICP1A ΔpexA, ICP1A ΔhelA has an advantage on PLE (+) V. cholerae, allowing rare plaques to form at a frequency two orders of magnitude higher than ICP1A ΔpexA on PLE 1 (Figure 1C). Conversely, the double mutant ICP1A ΔpexA ΔhelA forms small plaques on PLE 1 at a relatively high efficiency (Figures 1B and 1C). ICP1A ΔpexA ΔhelA plaques on PLE (+) V. cholerae were picked and the plaquing efficiency was re-tested to determine if those phage were subsequently able to escape PLE 1 (Figure S1). As these progeny phage re-plaqued at the same efficiency as ICP1A ΔpexA ΔhelA, we conclude that they are not genetic escape phage but instead overcome some aspects of PLE 1 activity through the loss of both ICP1A-encoded pexA and helA.
We next wanted to characterize the role of helA for ICP1A function. HelA is detectable in infected cells via Western blot within eight minutes of ICP1A infection (Figure 1D), consistent with the onset of ICP1A replication (Barth et al., 2019), suggesting that HelA may have a role in ICP1A replication. As PLE 1 reduces ICP1A replication (O’Hara et al., 2017; Barth et al., 2019), we hypothesized that PLE 1 hijacks HelA during infection as a mechanism to interfere with ICP1A replication. To test this hypothesis, we evaluated ICP1A ΔhelA replication in the presence and absence of PLE 1 by qPCR. Consistent with the ability to form plaques on PLE (−) V. cholerae, there were no deficiencies in ICP1A ΔhelA replication relative to a wild-type phage over the course of the infection cycle (Figure 1E), indicating that HelA is not essential for ICP1A replication. Conversely, infection of PLE (+) V. cholerae with ICP1A ΔhelA rescues ICP1 replication to the level that is observed in a PLE (−) host (Figure 1E), suggesting that PLE 1 exploits HelA to interfere with ICP1 during infection. However, because ICP1A ΔhelA is not deficient for replication in the absence PLE 1, the ICP1A replication defect in the presence of PLE 1 is not likely directly due to PLE 1-mediated hijacking of HelA.
ICP1-encoded HelA is necessary for PLE replication
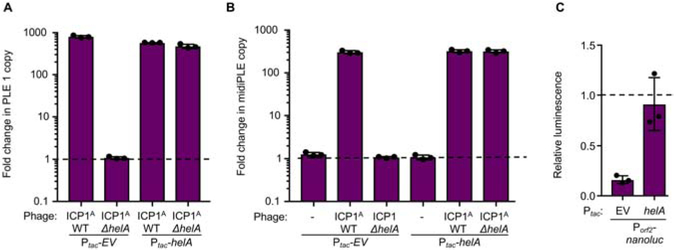
ICP1 and PLE 1 replication are inversely related, wherein ICP1 copy number is restored when PLE 1 replication is abolished via deletion of either the PLE 1 ori or repA (Barth et al., 2019). Therefore, the observed restoration in ICP1A ΔhelA copy number during infection of a PLE (+) host implicates phage-encoded HelA in promoting PLE 1 replication. To test the role of HelA in PLE replication, we infected PLE (+) V. cholerae with ICP1A ΔhelA and monitored the change in PLE 1 copy during infection. While PLE 1 replicates to high copy when infected with wild-type phage, strikingly, PLE 1 does not replicate in the absence of helA (Figure 2A). This phenotype can be complemented by ectopic expression of helA during ICP1A infection, demonstrating that HelA is necessary for PLE 1 replication.
Figure 2. ICP1-encoded HelA is necessary for PLE replication.
A, Quantification of change in PLE 1 copy number following infection by the ICP1 listed as measured by qPCR. Empty vector (Ptac-EV) and helA (Ptac-helA) expression plasmids were induced prior to phage infection. The dashed line indicates no change in copy number. B, Quantification of change in midiPLE copy number following infection of midiPLE (+) V. cholerae ΔlacZ::Ptac-repA with the listed expression plasmid by the ICP1 variant listed as measured by qPCR. C, Change in luminescence of Porf2-nanoluc reporter with the listed expression plasmid following infection by ICP1A ΔhelA relative to the change in luminescence following infection by ICP1A. The dashed line indicates luminescence levels equivalent to WT ICP1A.
SF1B-type helicases are implicated in activities ranging from replication and genome maintenance to transcriptional regulation (Byrd and Raney, 2017). Additionally, the S. aureus phage parasites, SaPIs, use phage-encoded dUTPases as anti-repressors to initiate their transcriptional program, suggesting that these genomic islands can evolve to respond to phage-encoded proteins independent of their biological function for the phage (Tormo-Más et al., 2010; Bowring et al., 2017). As such, we next wanted to determine if HelA has a direct role in PLE 1 replication or if it is necessary to transcriptionally activate the island to allow for production of PLE 1-encoded proteins, such as RepA, that are essential for PLE 1 replication. To test the involvement of HelA in PLE 1 replication, we made use of a minimal PLE replication system referred to as the midiPLE (Barth et al., 2019). The midiPLE contains only the endogenous PLE 1 integrase and ori integrated in the same location as PLE 1 in the V. cholerae chromosome. MidiPLE excises from the chromosome following pexA expression during ICP1A infection but does not replicate without ectopic expression of the PLE 1-encoded replication initiator, RepA. When repA is expressed in trans, midiPLE replicates during ICP1A infection (Figure 2B). In comparison to infection with wild-type phage, midiPLE fails to replicate during infection with ICP1A ΔhelA. This phenotype can be complemented by expressing helA in trans, showing that helA is necessary for PLE 1 replication independent of other PLE 1-encoded genes and supporting the conclusion that HelA is directly involved in PLE 1 replication. Interestingly, helA is not sufficient to stimulate PLE 1 replication in the absence of ICP1A infection (Figure 2B), indicating that other phage components are additionally required to facilitate PLE 1 replication.
PLE replication contributes to anti-phage gene dosage
In the course of replication sampling during ICP1A infection, we observed a defect in PLE 1-mediated accelerated lysis that correlated with a loss of PLE 1 replication. A culture of PLE (+) V. cholerae infected with ICP1 typically lyses 20 minutes post-infection, while an infected PLE (−) culture takes upwards of 90 minutes to lyse (O’Hara et al., 2017). However, cultures infected with ICP1A ΔhelA consistently had delays in lysis, suggesting impaired PLE 1 activity, and ectopic expression of helA led to intermediate lysis phenotypes (Figure S2A). Though the basis for PLE 1-mediated accelerated lysis is not yet known, we reasoned that robust PLE 1 replication enhances expression of PLE 1-encoded genes merely through increasing the template copy number. To test this hypothesis, we created a nanoluciferase transcriptional reporter cloned downstream of PLE 1 orf2 (Porf2nanoluc, Figure S2B) to quantify defects in transcription when PLE 1 is unable to replicate. Relative to infection with wild-type ICP1A, Porf2nanoluc produced 0.16 times as much luminescence during infection with ICP1A ΔhelA (Figure 2C). When PLE 1 replication was restored through ectopic expression of helA, reporter activity during infection with ICP1A ΔhelA was restored to wild-type levels, indicating that PLE 1 copy number contributes to the level of PLE 1 transcription. As such, inhibition of PLE 1 replication leads to phenotypes such as delayed lysis during ICP1A infection and potentially contributes to the ability of ICP1A ΔhelA to escape PLE 1.
ICP1 overcomes replication and excision-deficient PLE through degradation of the V. cholerae chromosome
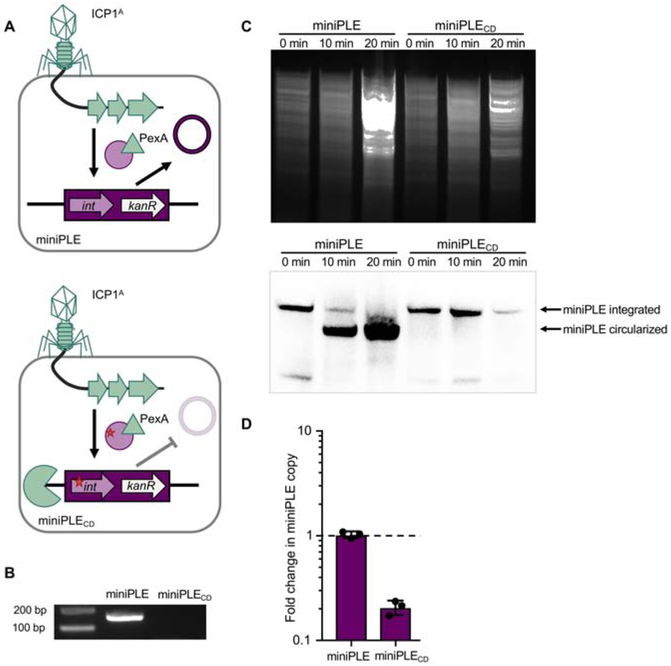
As ICP1-encoded pexA is necessary for PLE 1 excision and helA is necessary for PLE 1 replication during ICP1 infection (Figure 2A), we next wanted to understand how ICP1A ΔpexA ΔhelA overcomes PLE 1 (Figure 1B). Even when PLE 1 is challenged by ICP1A ΔhelA and cannot replicate leading to transcriptional deficiencies, PLE 1 still excises from the V. cholerae chromosome and is more inhibitory than when it is maintained in the chromosome, leading us to speculate that the position of PLE 1 in the cell, either intra- or extrachromosomal, is important for its activity. Phages are known to encode nucleases that attack the bacterial chromosome, freeing up nucleosides to be incorporated into newly synthesized phage genomes (Warner et al., 1970). Additionally, sequencing of total DNA in ICP1 infected V. cholerae cells shows that the proportion of reads mapping to the V. cholerae chromosomes decreases over the course of infection (Barth et al., 2019), leading us to hypothesize that nucleolytic activity deployed by ICP1A to degrade the V. cholerae chromosome during infection is able to degrade PLE 1 when PLE 1 is stuck in the chromosome unable to replicate, allowing for ICP1A to form small plaques on PLE (+) V. cholerae. To test this hypothesis, we made use of a minimal PLE excision system, the miniPLE, that has the PLE 1-encoded integrase but lacks an ori (Figure 3A). Thus during infection, the miniPLE excises from the host chromosome and circularizes, but does not replicate (McKitterick and Seed, 2018). To simulate an excision-deficient miniPLE, we created miniPLECD, which has a point mutation in the catalytic serine residue in the miniPLE-encoded integrase, rendering the construct unable to excise from the chromosome (Figure 3B). Total DNA from ICP1A miniPLE and miniPLECD infected cells was digested and subjected to Southern blot to monitor the stability of the miniPLE (Figure 3C). During the course of ICP1A infection, the miniPLE excises from the V. cholerae chromosome and is maintained as an episome. Conversely, the amount of excision-deficient miniPLECD decreases by 20 minutes following ICP1A infection relative to the amount of total DNA prepped from the cells (Figure 3C), suggesting that the copy number of miniPLECD decreases as a result of ICP1A infection. Quantification of miniPLE via qPCR further demonstrated no change in copy number for the excision-competent miniPLE during ICP1A infection (Figure 3D). In comparison, the miniPLECD that is unable to escape the V. cholerae chromosome decreased in copy number during ICP1A infection, indicating susceptibility to ICP1A-mediated chromosomal degradation. Thus, not only is PLE mobilization important for HGT (O’Hara et al., 2017; Barth et al., 2019), but it is also essential for PLE escape from ICP1 takeover of the V. cholerae host.
Figure 3. Excision and replication deficient PLE is susceptible to ICP1-mediated chromosomal degradation.
A, Cartoon of miniPLE during ICP1 infection. Top, miniPLE-encoded Int (circle) is directed to excise miniPLE during ICP1 infection by ICP1-encoded PexA (triangle), leading to a single-copy circularized episome. Bottom, catalytically dead miniPLECD Int (circle with red star) is unable to excise miniPLE during ICP1 infection, potentially rendering the miniPLE susceptible to phage-mediated chromosomal degradation (pac-man). B, Circularization PCR of miniPLE or miniPLECD following ICP1 infection. C, (Top) Total DNA prepped from V. cholerae with miniPLE or miniPLECD infected by ICP1A at the listed timepoints and imaged via Southern blot (bottom) with a probe against the miniPLE kanR cassette. D, Change in copy number of the miniPLE indicated following ICP1A infection as measured by qPCR. The dashed line indicates no change in copy number.
Diverse SF1B helicases are maintained in ICP1 and contribute to ICP1 fitness
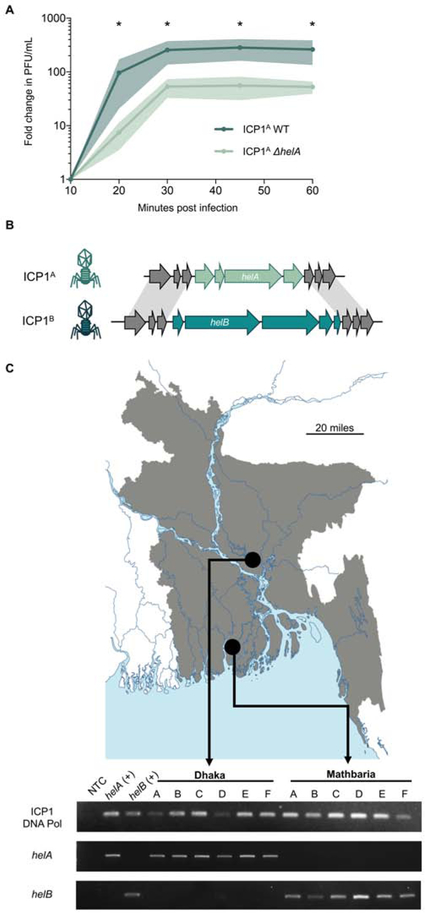
Due to the importance of PLE replication in PLE gene dosage and avoiding ICP1-mediated host takeover, we next hypothesized that ICP1 would evolve to abolish PLE replication by accumulating mutations in helA, indicative of co-evolution between the two entities. To identify signatures of co-evolution, we examined HelA from sequenced isolates of ICP1 that have been recovered from epidemic sampling in Dhaka, Bangladesh. HelA from ICP1 isolated from epidemic sampling from 2001 to 2017 is over 99% identical indicating that there is either little pressure for HelA to evolve over time, or that HelA mutations cannot be tolerated in nature (Table S6). Though there is no change in ICP1 DNA replication in a single round of infection in the absence of helA (Figure 1E), the average burst size of ICP1 A ΔhelA is only one-fifth the size of wild-type ICP1A (Figure 4A). Consistently, ICP1A ΔhelA also forms smaller plaques than wild-type phage (Figure S3A), collectively indicating that ICP1 is less fit in the absence of helA and that functional helA must be maintained by ICP1 in nature.
Figure 4. ICP1 encodes one of two accessory helicase alleles.
A, Relative burst size of ICP1A and ICP1A ΔhelA on PLE (−) V. cholerae as measured by one-step growth curves. *p<0.05. B, Cartoon of ICP1 accessory helicase locus. Grey arrows indicate gene products shared between the two phages, mint arrows indicate gene products unique to the helA locus and turquoise arrows indicate gene products unique to the helB locus. C, Map of distribution of SF1B-type helicases in ICP1 isolates shed by cholera patients in Bangladesh. Top, map (Vecteezy, 2019) of Bangladesh with Dhaka and Mathbaria marked. Bottom, agarose gel showing PCR detection of the conserved DNA polymerase (gp58), helA, and helB in ICP1 isolates from cholera patient stool collected in Dhaka or Mathbaria. Phage isolates are listed in Table S8.
Despite having a high degree of conservation, helA is not considered part of the core ICP1 genome (Angermeyer et al., 2018): two phage isolates recovered from cholera patient stool samples from Dhaka in 2006 do not encode helA, but instead have an alternative SF1B-type helicase in the same locus, which we call helicase B (helB) (Figure 4B). HelB is 24% identical to HelA, with a conserved P-loop ATPase domain, but HelB has an extended C-terminus that contains a domain of unknown function (Figure S3B). In addition to having low sequence identity, helA and helB are flanked by different, unrelated genes each encoding products with no predicted structure or function (Figure 4B), suggesting that while ICP1 is unable to lose its SF1B-type helicase in nature in an attempt to avoid hijacking by PLE for replication, ICP1 may swap helA for a distinct accessory helicase.
Homologs of HelA are commonly found in phages of marine bacteria, particularly, in a group of related myoviruses that infect non-cholera Vibrios (Figure S3C). Of note, two Vibrio phages also encode a homolog of one of the proteins flanking HelA in ICP1A, indicating that the helA locus could have been shared with a common ancestor of these phages. Conversely, HelB is more divergent, with the only identifiable homolog found in a Pseudoalteromonas phage that is also predicted to have the same C-terminus. These HelB proteins cluster on a more distant branch than the HelA homologs (Figure S3C), supporting the hypothesis that helB was horizontally acquired by ICP1. Altogether, SF1B helicases are readily found in marine phages, and ICP1 encoding helA are the dominant ICP1 shed by cholera patients in Dhaka between 2001-2017.
Most epidemic sampling of ICP1 from cholera patients has been done in the urban cholera endemic site in Dhaka; however, we recently began sampling patients at a rural estuarine site in Mathbaria, Bangladesh. In contrast to ICP1 isolates from Dhaka in the 2017 epidemic period, all ICP1 isolates recovered from cholera patients in Mathbaria encoded the helB allele (Figure 4C). One representative isolate from Mathbaria in 2017, referred to here as ICP1B is over 99.8% identical to ICP1A across 90% of the genome, with 205 of 227 ICP1B predicted open reading frames being shared with ICP1A. The resurgence and dominance of helB in the Mathbaria epidemic sampling suggests that there could be a selective advantage for ICP1 encoding helB rather than helA in this region.
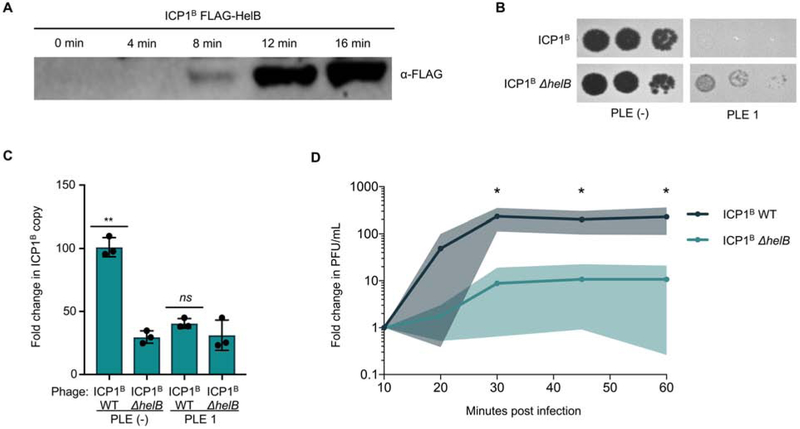
As ICP1B is not isogenic to ICP1A, we first wanted to characterize the role of helB in ICP1B fitness. Similar to HelA, HelB is detectable by Western blot within eight minutes of infection (Figure 5A), again coinciding with ICP1 replication (Barth et al., 2019). Also similar to helA, helB is not essential for ICP1B, and ICP1B ΔhelB forms plaques in the absence and presence of PLE 1 (Figure 5B). Interestingly, ICP1B ΔhelB forms plaques on PLE (+) V. cholerae with a higher efficiency than ICP1A ΔhelA, suggesting that ICP1B has evolved other ways to limit PLE-mediated anti-phage activity.
Figure 5. Loss of helB permits escape from PLE but leads to a defect in ICP1 fitness.
A, Western blot of endogenously FLAG-tagged HelB following infection of PLE (−) V. cholerae. B, Tenfold ICP1 dilutions spotted on the listed V. cholerae lawns. C, Fold change in ICP1 copy number following infection of the listed V. cholerae host as measured by qPCR. D, Relative burst size of ICP1B and ICP1B ΔhelB on PLE (−) V. cholerae as measured by one-step growth curves. *p<0.05, **p<0.001, ns not significant.
We next wanted to see if ICP1B replication was impacted by the helB deletion. In contrast to ΔhelA in ICP1A, ICP1B ΔhelB is significantly impaired for replication during the course of infection compared to wild-type ICP1B (Figure 5C), indicating that although helB is not necessary for ICP1B replication, it does have a more central role in phage fitness. Consistent with the observation that PLE 1 decreases the ability of ICP1A to replicate (Figure 1E), replication of ICP1B, too, is impacted negatively by PLE 1; however, ICP1B ΔhelB does not restore ICP1B replication in the presence of PLE 1 (Figure 5C), demonstrating a more severe fitness cost associated with losing the accessory helicase on ICP1B than on ICP1A independent of the presence of PLE 1.
To confirm the role of helB in ICP1B fitness, we next ectopically expressed helB to complement the mutant phage. However, we could not complement the replication defect for ICP1B ΔhelB, suggesting that the observed decrease in ICP1 fitness may not be due to direct loss of the helB gene product (Figure S4A). To minimize potential polar effects of ΔhelB, we made a targeted deletion of the 25 amino acids encompassing the helicase domain (HD) that contains the Walker A motif necessary for ATP hydrolysis (Blair, Tackett and Raney, 2009). While ICP1B helB ΔHD had increased phage replication relative to the clean helB deletion, there was still a defect in replication that could not be complemented (Figure S4A), suggesting that ectopic expression may not achieve the appropriate timing or level of helB expression, or that the fitness cost is not a result of loss of HelB per se. Due to the complexity of phage genomes and tight regulation of phage gene expression, disrupting even the HD domain of helB could have detrimental effects on uncharacterized in cis sites that could contribute to poor fitness. The fitness cost for ICP1B with ΔhelB or helB ΔHD was observed as a decrease in plaque size (Figure S4B) and ICP1B ΔhelB had a significantly smaller burst, producing one twentieth the number of infectious progeny relative to wild-type (Figure 5D). Altogether, ICP1B is less fit in the absence of helB, consistent with the observation that all natural ICP1 isolates encode one of two SF1B-type helicases, either HelA or HelB.
PLE exploits distinct phage-encoded SF1B-type helicases to drive replication during ICP1 infection
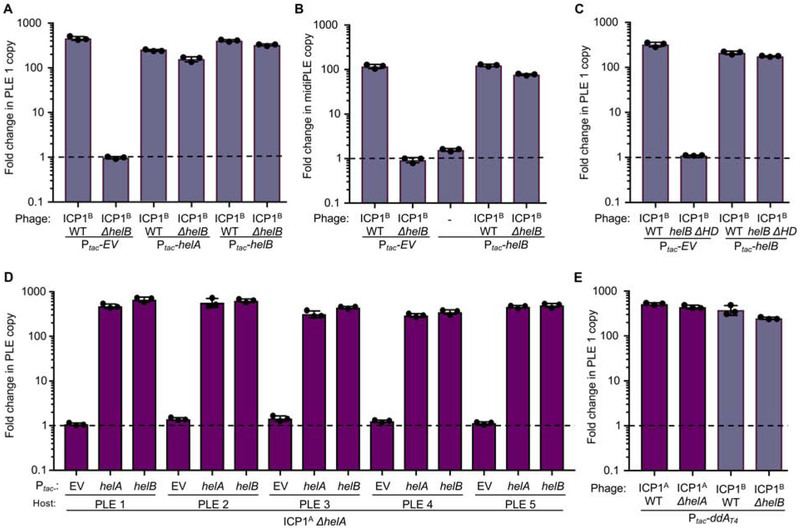
Given that PLE 1 replication requires helA (Figure 2A), and ICP1 with helB are dominant in Mathbaria, we were tempted by the possibility that phage with helB could be selected for as a mechanism to impede PLE 1 replication during infection. Hence, we next assessed if HelB could also support PLE 1 replication. Consistent with the inverse relationship between ICP1 and PLE 1 replication, PLE 1 still replicated upon ICP1B infection and as with ΔhelA, PLE 1 replication was not observed in the absence of helB (Figure 6A), indicating that helB is also necessary for PLE 1 replication. Further, ectopic expression of helB complemented the defect in PLE 1 replication during infection with ICP1B ΔhelB, and ectopic expression of helA was likewise sufficient to restore PLE 1 replication during infection with ICP1B ΔhelB (Figure 6A). These data demonstrate that PLE 1 can harness either ICP1-encoded accessory helicase independent of the infecting ICP1 isolate. Additionally, the shared ability of these non-isogenic ICP1 isolates to drive PLE 1 replication implicates functionally conserved gene products in ICP1 isolates, in addition to helA and helB, that are required for PLE 1 replication.
Figure 6. PLEs can exploit unrelated phage-encoded SF1B-type helicases for replication.
Replication of PLE 1 (A,C) or midiPLE (B) following infection of V. cholerae with the listed expression vector by the ICP1B variant listed as measured by qPCR. D, Replication of the listed PLE in an isogenic V. cholerae background after infection by ICP1A ΔhelA. E, Replication of PLE 1 following infection by the listed phage as measured by qPCR. Dashed line indicates no change in copy.
We next used ICP1B ΔhelB to probe for midiPLE replication following ectopic expression of repA. As expected, midiPLE replicated when infected with ICP1B but failed to replicate in the absence of helB, indicating that HelB is also directly involved in PLE 1 replication (Figure 6B). Like helA, helB is also not sufficient to stimulate PLE 1 replication in the absence of ICP1B, showing that PLE 1 is dependent on additional replication machinery from ICP1B. We additionally confirmed that ATP hydrolysis is required for HelB to facilitate PLE 1 replication by testing the ICP1B helB ΔHD variant, and, as anticipated, the helicase activity of helB is necessary for PLE 1 replication (Figure 6C).
The first ICP1 isolate identified with helB was from Dhaka in 2006 when PLE 2 V. cholerae was being shed by cholera patients (O’Hara et al., 2017; McKitterick et al., 2019), leading us to evaluate if the two helicase alleles have different capacities to facilitate replication of different PLEs during infection. To test this hypothesis, we first infected isogenic V. cholerae harboring each of the five characterized PLEs with ICP1A and observed that all PLEs replicated equally well (Figure S5). Next, we determined that helA is necessary for replication of all five PLEs during ICP1A infection and that replication can be complemented with ectopic expression of helA (Figure 6D). To evaluate if each PLE can additionally use helB to support replication, we also complemented ICP1A ΔhelA with ectopic expression of helB and found that indeed all five PLEs can use either one of the two ICP1-encoded accessory helicases for replication.
As current data supports the model that PLE responds specifically to ICP1 infection, we next wanted to determine if PLE’s capacity to exploit either HelA or HelB to drive PLE replication is specific to ICP1-encoded proteins or in general to SF1B-type helicases. To address the specificity of the interaction, we ectopically expressed the SF1B-type helicase Dda from E. coli phage T4 during infection with either ICP1A ΔhelA or ICP1B ΔhelB. T4 Dda is only 16% identical to either HelA or HelB and does not group with the marine phage SF1B-type helicases (Figure S3C). Although PLE 1 cannot replicate during infection with either of the Δhel phage alone (Figures 6A and 6D), expression of dda was sufficient to drive PLE 1 replication in the absence of ICP1-encoded accessory helicases (Figure 6E). Despite the apparent specificity between PLE and ICP1, the ability of PLE to exploit a variety of phage-encoded accessory helicases reveals flexibility in at least one requirement for PLE replication, and suggests that swapping of helicase alleles by ICP1 isolates is not a useful strategy to mitigate PLE parasitism.
Discussion
To defend against viral infection, host resistance mechanisms must prevent or bypass virus mediated host takeover. Eukaryotic DNA and RNA viruses broadly use virally encoded ribonucleases to globally degrade host transcripts in the infected cell, which sabotage their hosts through modulation of transcript and protein levels. This decrease in transcript abundance downregulates innate immune responses, processes which are detrimental to the host but are ultimately reversible (Crow et al., 2016; Rivas, Schmaling and Gaglia, 2016). Conversely, degradation of the host chromosome is a host takeover process that is somewhat unique to phages and irreversible. Host chromosome degradation has a twofold benefit for predatory phages: it cleaves and releases nucleosides that can be incorporated into the rapidly replicating phage genome, and it destroys the template needed for expression of host-encoded anti-phage genes. With the imminent shutdown of the host upon phage infection, it is not surprising that many bacterial defense systems, such as restriction-modification and some CRISPR-Cas systems, are expressed constitutively. By discerning between self and non-self, these systems are safely deployed in the absence of infection. Conversely, more self-destructive defense mechanisms, such as toxin/antitoxin and abortive infection systems, cannot be constitutively active and must be induced upon infection. Thus, for an inducible defense system like PLE, and intuitively many phage parasites, mobilization to evade host shutdown is critical. When PLE is unable to replicate or excise from the chromosome it is susceptible to ICP1-mediated chromosomal degradation and can no longer block plaque formation.
As a defense island and phage parasite of ICP1, V. cholerae PLE evolved to use phage-encoded gene products to drive its anti-phage program (McKitterick and Seed, 2018). Here, we characterize an additional ICP1-PLE interaction: PLE hijacks a non-essential ICP1-encoded SF1B-type helicase to drive PLE replication. In comparison, the well-studied SaPIs use their bacterial host’s replication machinery and can autonomously replicate in the absence of helper phage (Úbeda et al., 2008). PLE’s unique requirement for the phage-encoded helicase makes sense given the differences between the helper phages inducing these islands, with PLE induced by a lytic phage that encodes its own replication machinery and SaPIs induced by an activated temperate phage that also exploits bacterial replication machinery (Úbeda et al., 2008). The fact that helA/helB expression is not sufficient to drive midiPLE replication in the absence of ICP1 infection implicates other ICP1-encoded replication proteins in facilitating PLE replication. Aside from the SF1B-type helicases, the potential role for ICP1’s replication machinery in PLE replication remains to be elucidated. As T4 Dda has been observed to have a role in T4 origin initiation during origin-dependent replication (Brister, 2008), we speculate that HelA/HelB has a similar role in facilitating origin firing in PLE by interacting with PLE protein(s) and/or recruiting conserved ICP1 replication proteins. ICP1 is predicted to encode a DNA polymerase and primase/helicase reminiscent of the replication machinery in E. coli phage T7 (Barth et al., 2019). Further work remains to identify what roles, if any, these ICP1 replisome proteins have in PLE mobilization.
Despite not being essential, all ICP1 isolates encode an accessory SF1B-type helicase, as do several marine phages (Figure S3C). Of note, one of these Vibrio phages also encodes a complete Type 1-F CRISPR-Cas system, which is the same type that is encoded by some ICP1 isolates to target and overcome PLE activity (Seed et al., 2013), suggesting that ICP1 may exchange genetic material with or be related to these marine phages infecting non-cholera Vibrios. The fitness cost of losing the accessory SF1B-type helicase, as measured by burst and plaque size, implicate both helA and helB in maintaining optimal phage fitness, though the precise role for these accessory helicases in the phage lifecycle remains to be determined. The ease with which PLE is able to make use of ectopically expressed helB compared to the inability of ectopically expressed helB to complement the ICP1B ΔhelB replication deficiency suggests that these helicases play a specialized role in the phage lifecycle that is more complex than for PLE.
PLE’s capacity to replicate using dissimilar SF1B-type helicases implicates strong evolutionary pressures to maintain PLE replication in response to ICP1 evolution. The ICP1-encoded helicases are only one of the phage-encoded inputs that contribute to PLE activity, similarly, SaPIs have evolved to overcome variability in helper phage induction cues (Bowring et al., 2017). The apparent promiscuity of the SaPI master repressor allows for recognition of structurally dissimilar but functionally conserved phage proteins to ensure SaPI excision, replication and spread, despite their helper phage’s attempts to avoid SaPI induction. It is likewise imperative for PLE to use either of the ICP1-encoded helicases to ensure PLE propagation and host defense despite ICP1 swapping one helicase for another.
The striking spatial separation between the ICP1A and ICP1B populations shed by cholera patients in Bangladesh during the same epidemic period suggests that slight variations in the phage strain, such as the difference between helA and helB, can impact the makeup of phage populations. Indeed, the ability of ICP1B ΔhelB to form plaques in the presence of PLE (Figure 5B) suggests that ICP1B should dominate in the presence of PLE (+) V. cholerae; however, the significant fitness cost for ICP1B ΔhelB suggests that the loss of helB even in the presence of PLE (+) V. cholerae is ultimately detrimental to the phage population.
The necessity of PLE excision and replication during ICP1 infection highlights a crucial role for mobilization of inducible phage defense systems during phage infection. In order for inducible defenses to functionally protect a host cell from phage infection, they must overcome the infecting phage’s destruction of the host chromosome. Elements independent of the host chromosome, such as plasmids, seem to be somewhat protected from degradation by lytic phages (Keen et al., 2017). It thus stands to reason that the observed high prevalence of phage defense systems on genomic islands (Makarova et al., 2011) may be in part due to genomic islands mobilizing during infection and escaping phage-mediated host takeover, with the potential of horizontal transfer and ability to escape from a dying host as an added benefit. Through experimental and in silico validation, more phage defense islands have been identified and characterized, albeit often in a context lacking infection by a native phage. Given the propensity of some phages to degrade their host chromosome during infection and the need for protective MGEs to escape host takeover, it will be interesting to further explore if other inducible defense islands mobilize in response to phage infection and are in fact unrecognized phage satellites.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact Kimberley Seed (kseed@berkeley.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Bacterial Growth Conditions
The bacterial strains and plasmids used in this study are listed in Tables S2 and S5. All bacterial strains were grown at 37°C in LB with aeration or on LB agar plates. The following antibiotics were used as necessary: streptomycin (100 μg/mL), spectinomycin (100 μg/mL), kanamycin (75 μg/mL), ampicillin, (V. cholerae 50 μg/mL, E. coli 100 μg/mL), chloramphenicol (V. cholerae 1.25 μg/mL, E. coli 25 μg/mL). Ectopic expression constructs in V. cholerae were induced 20 minutes prior to ICP1 infection with 1 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) and 1.5 mM theophylline.
Phage Growth Conditions
The phage isolates used in this study are listed in Table S3. Phage were propagated using the soft agar overly method and high titer stocks were made by polyethylene glycerol precipitation and stored in sodium chloride-tris-EDTA (STE) buffer (Clokie and Kropinski, 2009).
Phage isolation from rice water stool
The collection of cholera patient rice water stool (RWS) was approved by the icddr,b institutional review board. All samples were deidentified and written informed consent was obtained from adult participants and from the guardians of children. Stool samples were mixed with glycerol in cryovials, frozen, until being processed at the University of California, Berkeley. For processing, samples were thawed and grown on thiosulfate-citrate-bile salts-sucrose agar, or used to inoculate alkaline peptone water (APW) for outgrowth. Liquid APW cultures were struck out on agar plates and aliquots were frozen with glycerol. Individual colonies selected from plates were confirmed as V. cholerae by PCR. These isolates of V. cholerae, in addition to the PLE (−) laboratory strain, were used to isolate phages directly from the RWS glycerol stocks and from frozen APW outgrowths. Isolated phages were plaque purified twice after isolation.
METHOD DETAILS
Bacterial and phage cloning conditions
Bacterial mutants were cloned using splicing by overlap extension PCR and introduced by natural transformation (Dalia, Lazinski and Camilli, 2014). Plasmids were constructed using Gibson Assembly or Golden Gate Assembly. Phage mutants were constructed using CRISPR-Cas engineering as previously described (Box et al., 2016; McKitterick and Seed, 2018). Briefly, an editing template with the desired deletion was cloned into a plasmid and V. cholerae harboring this plasmid was infected by the ICP1 strain of interest. Ten plaques of the passaged phage were collected and mutants were selected on V. cholerae engineered to encode an inducible Type 1-E CRISPR-Cas system and a plasmid with a spacer targeting the gene of interest. Mutant phages were verified via Sanger sequencing and purified two times on the targeting host before storing in STE. Total phage gDNA was prepped with a DNeasy Blood & Tissue Kit (Qiagen).
Phage plaquing conditions
Spot plates were performed as before (McKitterick and Seed, 2018). Briefly, mid-log V. cholerae was added to 0.5% molten LB agar poured on a solid agar plate and allowed to solidify. Ten-fold dilutions of phage were applied to the surface in 3 μL spots and allowed to dry. Plates were incubated at 37°C. Images are representative of at least two independent experiments. The efficiency of plaquing (EOP) was calculated by comparing the number of plaques (in an agar overlay with each dilution plated on a single petri dish) that a given phage forms on PLE (−) V. cholerae relative to the number of plaques formed on PLE (+) V. cholerae. Each EOP was calculated in triplicate, and the limit of detection is the point at which the phage is unable to productively infect the PLE (+) host while still forming plaques on a PLE (−) host. Burst size was measured in biological triplicate by one-step growth curves. PLE (−) V. cholerae was infected with ICP1 at MOI ~ 0.1 and allowed to adsorb. Cells were then diluted 1/2500, and samples were taken at the time points listed and mechanically lysed with chloroform, with the 10 minute timepoint representing unadsorbed phage. Lysates were pelleted and titered on PLE (−) V. cholerae in technical duplicate. Burst was calculated as the fold change in PFU/mL from the timepoint of interest relative to the 10 minute timepoint. Significance was determined by unpaired T Test. Plaque size was determined by imaging and quantifying with ImageJ at least 20 plaques each from 3 independent replicates in 0.5% agar overlay (with each dilution plated on a single petri dish) on PLE (−) V. cholerae. Significance was determined through a nonparametric T Test.
qPCR conditions
Fold change in genome copy was performed as before (O’Hara et al., 2017) with a slight modification. Fold change in ICP1 copy number was measured by growing cells to an OD600 of 0.3, infecting with a multiplicity of infection (MOI) of 0.01, and taking a sample that was boiled for 10 minutes as a starting value. Infected cells were returned to the incubator for 20 minutes, at which point another sample was taken and boiled. Boiled samples were diluted 1:50 and used as a template in the qPCR reaction. To measure the fold change in PLE and midiPLE copy number, cells were grown to an OD600 = 0.3 and the initial sample was immediately taken prior to addition of phage at an MOI of 2.5 and boiled for 10 minutes. Samples were taken at 20 minutes after infection, boiled for 10 minutes, and diluted 1:1000. Quantification of the fold change in miniPLE copy was measured during infection with an MOI of 5, with samples taken immediately prior to infection and 30 minutes after infection and boiled for 10 minutes. Boiled samples were diluted 1:100 and used as template. Experiments with ectopic expression constructs were induced at OD600 = 0.2 for 20 minutes and then normalized to OD600 = 0.3 prior to infection. All samples were run in biological triplicates and technical duplicates. The template was mixed with the primers listed in Table S4 and IQ Sybr Green Master Mix (Bio-rad) and run on a CFX Connect Real-Time PCR Detection System (Bio-rad). Fold change was measured as the amount of DNA in the sample at 20 minutes after infection for all experiments except for measuring miniPLE fold change (this sample was taken 30 minutes after infection) relative to the amount of DNA in the sample at T=0. Significance was measured by 2-tailed T Test.
Western Blots
PLE (−) V. cholerae was grown to an OD600 = 0.3 and infected with the endogenously FLAG-tagged ICP1 listed. At the listed timepoints, 1 mL samples were collected and mixed with equal volume ice-cold methanol and centrifuged at 21000 × g for 3 minutes at 4°C. Pellets were washed with ice-cold PBS, resuspended in 1x Laemmli buffer, and boiled for 10 minutes at 99°C. Total protein was run on a 10% Stain Free TGX SDS-PAGE gel (Bio-rad). Primary Rabbit-α-FLAG antibodies (Sigma) were used at a dilution of 1:5000 and detected with goat-α-rabbit-HRP conjugated secondary antibodies at a dilution of 1:5000 (Bio-rad). Clarity Western ECL Substrate (Bio-rad) was used to develop the blots and a Chemidoc XRS Imaging System (Bio-rad) was used to image.
Lysis kinetics and Nanoluciferase assay
PLE (+) V. cholerae cells were grown to an OD600 = 0.2 and the listed ectopic expression constructs were induced for 20 minutes. Cells were then normalized to an OD600 = 0.3 and infected at an MOI of 2.5. For lysis kinetics, OD600 was monitored for 30 minutes. For nanoluciferase, 100 μL cells were sampled at T=0 and T=20 minutes after infection and added to 100 μL ice cold methanol. Luminescence was measured in a Spectra Max i3x plate reader (Molecular Devices) using the Nano-Glo Luciferase Assay System (Promega). Relative luminescence was calculated by dividing the luminescence detected after infection with the knockout phage relative to the luminescence detected after infection with the wild-type phage.
PCR conditions
PLE circularization PCRs were performed as described (McKitterick and Seed, 2018). Briefly, plaques on the miniPLE or miniPLECD hosts were picked into 50 μL of water and boiled for 10 minutes. Boiled template (2 μL) was used with the primers listed in Table S4 to detect miniPLE circularization. Detection of ICP1 gp58, helA, and helB from ICP1 isolates were performed on 5 – 30 ng prepped gDNA from isolated phage with the primers listed in Table S4. PCRs were run on 2% agarose gels and visualized with GelGreen.
Southern Blots
A probe against miniPLE was created using the DIG-High Prime DNA Labeling and Detection Started Kit I (Sigma). Cells were grown up to OD600 = 0.3 with kanamycin and infected with ICP1A at an MOI of 5. At the timepoints indicated, 5 mL of cells were harvested and mixed with 5 mL ice cold methanol. Samples were spun at 7000 × g at 4°C for 5 minutes. Pellets were washed with ice cold PBS and spun again. Total DNA was extracted from the pellets with the DNeasy Blood & Tissue Kit (Qiagen). Equal volumes of samples (between 1.5 and 4.1 μg DNA) were digested overnight with EcoRV-HF and SalI-HF (NEB) and run on a 0.7% agarose gel and visualized with GelRed. The agarose gel was washed briefly and incubated with 0.25 N HCl for 15 minutes, washed again, denatured in 0.4 M NaOH for 20 minutes, and transferred overnight. DNA was fixed by baking the blot at 120ΰC for 30 minutes, and hybridized with 17 ng/mL miniPLE probe overnight at 42ΰC. The blot was detected with the DIG-High Prime DNA Labeling and Detection Started Kit I (Sigma) and CSPD™ Substrate (ThermoScientific) and visualized on a Chemidoc XRS Imaging System (Bio-rad).
Computational analyses
Escape ICP1A ΔpexA phage were isolated from PLE 1 V. cholerae and purified twice on the same host. Total gDNA was prepped as above. NEBNext Ultra II DNA Library Preparation Kit for Illuminia (New England Biolabs) was used to prep genomic DNA which was sequenced by paired-end sequencing (2 × 150 bp) on an Illumina HiSeq4000 (University of California, Berkeley QB3 Core Facility). The wild-type phage genome was assembled using SPAdes (Bankevich et al., 2012) with paired-end reads and default settings. This assembly was used as the reference sequence for comparison to escape phage sequence reads with breseq (Deatherage and Barrick, 2014) in ‘consensus’ mode and default settings. Protein alignments were analyzed using Praline (Bawono and Heringa, 2014). HelA conservation was determined by analyzing HelA from 17 phages isolated between 2001 and 2017 (Angermeyer et al., 2018; McKitterick et al., 2019) using a Praline alignment (Bawono and Heringa, 2014). Phages that did not have whole genome information were Sanger sequenced from previously prepped phage gDNA (McKitterick et al., 2019) with the primers listed in Table S4. Phages included in the phylogenetic analysis were selected from a BLASTP search of HelA and HelB. Each hit was included if it had over 30% identity to either protein across 90% of the protein. A multiple alignment of helicase amino acid sequences was generated with MUSCLE v3.8.31 (Edgar, 2004) using default settings. The alignment file was converted to the PHYLIP format with Clustal X v2.0 (Larkin et al., 2007) and a bootstrapped (n=100) maximum-likelihood phylogenetic tree was solved using PhyML v20120412 (Guindon et al., 2005) with the following settings: -d aa -s BEST --rand_start --n_rand_starts 100 -o tlr -b 100).
Quantification and statistical analysis
Statistical tests used for experiments are listed in the Methods section. Data was analyzed using Prism GraphPad. For EOPs, qPCR, lysis kinetics, and nanoluciferase assays, error bars indicate standard deviation of average fold change from three independent biological replicates. For relative burst, the shaded area indicates the standard deviation of the average fold change from three independent biological replicates. Spot plate, agarose gel, and blot images are representative of at least two independent experiments.
Data and code availability
The data supporting the study are found in the manuscript, supplementary information, or from the corresponding author upon request.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-FLAG polyclonal antibody | Sigma | Cat# SAB4301135 RRID# AB_2811010 |
| Goat anti-Rabbit IgG antibody, peroxidase conjugated | Sigma | Cat# AP132P RRID# AB_90264 |
| Bacterial and Virus Strains | ||
| Bacterial Strains, see Table S2 | This paper | N/A |
| Phage Strains, see Table S3 | This paper | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| iQ SYBR Green Supermix | Bio-Rad | Cat# 1708880 |
| CSPD™ Substrate (0.25 mM Ready-To-Use) Southern AP Substrate | ThermoScientific | Cat# T2141 |
| Clarity Western ECL Substrate | Bio-Rad | Cat# 1705060 |
| GelGreen | Biotium | Cat# 41005 |
| GelRed | Biotium | Cat# 41003 |
| BsaI | New England Biolabs | Cat# R0535S |
| BseRI | New England Biolabs | Cat# R0581S |
| T4 Ligase | New England Biolabs | Cat# M0202S |
| EcoRV-HF | New England Biolabs | Cat# R3195S |
| SalI-HF | New England Biolabs | Cat# R3138S |
| Critical Commercial Assays | ||
| DNeasy Blood & Tissue Kit | Qiagen | Cat# 69506 |
| Nano-Glo Luciferase Assay System | Promega | Cat# N1110 |
| DIG-High Prime DNA Labeling and Detection Started Kit I | Sigma | Cat# 11745832910 |
| TGX Stain-Free FastCast Acrylamide Kit | Bio-Rad | Cat# 1610183 |
| Deposited Data | ||
| ICP1 _2017_F_Mathbaria | This paper | Deposited at GenBank: accession number MN419153 |
| Escape phages | This paper | Deposited in the Sequence Read Archive (SRA) under BioProject ID PRJNA563933 |
| Oligonucleotides | ||
| Primers used in this study, see Table S4 | This paper | N/A |
| Recombinant DNA | ||
| Plasmids used in this study, see Table S5 | This paper | N/A |
| Software and Algorithms | ||
| SPAdes V3.11.1 | (Bankevich et al. 2012) | http://cab.spbu.ru/software/spades/ |
| Breseq v0.33.0 | (Deatherage & Barrick 2014) | https://github.com/barricklab/breseq/releases/tag/v0.33.0 |
| MUSCLE v3.8.31 | (Edgar 2004) | https://www.drive5.com/muscle/ |
| Clustal X v2.0 | (Larkin et al. 2007) | http://www.clustal.org/clustal2/ |
| Praline | (Bawono & Heringa 2014) | http://www.ibi.vu.nl/programs/pralinewww/ |
| BLASTP | N/A | https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins |
| Prism GraphPad | N/A | https://www.graphpad.com/scientific-software/prism/ |
| CLC Main Workbench 7 | Qiagen | https://www.qiagenbioinformatics.com/products/clc-main-workbench/ |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| PhyML v20120412 | (Guindon et al. 2005) | https://github.com/stephaneguindon/phyml |
| Other | ||
| Vecteezy | N/A | https://www.vecteezy.com/ |
Highlights:
Phage satellites exploit helicases of a V. cholerae lytic phage to selfishly replicate
Phylogenetically distinct helicases from patient stool samples are sensitive to hijacking
Mobilization of phage satellites is critical to avoid phage takeover of bacterial host
ACKNOWLEDGMENTS
This research was funded by the National Institute of Allergy and Infectious Diseases grant R01AI127652 (K.D.S.). A.C.M. was supported by the Kathleen L. Miller Fellowship from the Henry Wheeler Center for Emerging and Neglected Diseases. K.D.S. is a Chan Zuckerberg Biohub Investigator and holds an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. The authors are thankful to icddr,b hospital and lab staff for support, in particular Shirajum Monira, Marzia Sultana, Kazi Zillur Rahman, and Monika Sultana. M.A., and the governments of Bangladesh, Canada, Sweden, and United Kingdom for providing core/unrestricted support. We thank Angus Angermeyer for help with the annotation/computational analyses.
Footnotes
DECLARATION OF INTERESTS
K.D.S. is a scientific advisor for Nextbiotics, Inc.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Angermeyer A et al. (2018) ‘Analysis of 19 Highly Conserved Vibrio cholerae Bacteriophages Isolated from Environmental and Patient Sources Over a Twelve-Year Period’, Viruses, 10(6), p. 299. doi: 10.3390/v10060299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A et al. (2012) ‘SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing’, Journal of Computational Biology, 19(5), pp. 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth ZK et al. (2019) ‘Genome replication dynamics of a bacteriophage and its satellite reveal strategies for parasitism and viral restriction’, bioRxiv. Available at: https://www.biorxiv.org/content/10.1101/639039v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bawono P and Heringa J (2014) ‘PRALINE: A Versatile Multiple Sequence Alignment Toolkit’, Methods Mol Bio, 1079, pp. 245–62. doi: 10.1007/978-1-62703-646-7. [DOI] [PubMed] [Google Scholar]
- Blair LP, Tackett AJ and Raney KD (2009) ‘Development and evaluation of a structural model for SF1B helicase dda’, Biochemistry, 48(11), pp. 2321–2329. doi: 10.1021/bi801919s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondy-Denomy J and Davidson AR (2014) ‘When a virus is not a parasite: The beneficial effects of prophages on bacterial fitness’, Journal of Microbiology, 52(3), pp. 235–242. doi: 10.1007/s12275-014-4083-3. [DOI] [PubMed] [Google Scholar]
- Bowring J et al. (2017) ‘Pirating conserved phage mechanisms promotes promiscuous staphylococcal pathogenicity island transfer’, eLife, 6, p. e26487. doi: 10.7554/eLife.26487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box AM et al. (2016) ‘Functional analysis of bacteriophage immunity through a Type I-E CRISPR-Cas system in Vibrio cholerae and its application in bacteriophage genome engineering’, Journal of Bacteriology, 198(3), pp. 578–90. doi: 10.1128/JB.00747-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brister JR (2008) ‘Origin Activation Requires both Replicative and Accessory Helicases during T4 Infection’, Journal of Molecular Biology, 377(5), pp. 1304–1313. doi: 10.1021/acsnano.6b07096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brüssow H, Canchaya C and Hardt W-D (2004) ‘Phages and the Evolution of Bacterial Pathogens: from Genomic Rearrangements to Lysogenic Conversion’, Microbiol Mol Biol Rev, 68(3), pp. 560–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd AK and Raney KD (2006) ‘Displacement of a DNA binding protein by Dda helicase’, Nucleic Acids Research, 34(10), pp. 3020–3029. doi: 10.1093/nar/gkl369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd AK and Raney KD (2017) ‘Structure and function of Pif1 helicase’, Biochemical Society Transactions, 45(5), pp. 1159–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J et al. (2018) ‘Genome hypermobility by lateral transduction’, Science, 362(6411), pp. 207–212. [DOI] [PubMed] [Google Scholar]
- Clokie MRJ and Kropinski AM (eds) (2009) Bacteriophages : methods and protocols, Methods in molecular biology. Humanan Press. [Google Scholar]
- Crow MS et al. (2016) ‘Diverse mechanisms evolved by DNA viruses to inhibit early host defenses’, Critical Reviews in Biochemistry and Molecular Biology, 51 (6), pp. 452–481. doi: 10.1080/10409238.2016.1226250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalia AB, Lazinski DW and Camilli A (2014) ‘Identification of a Membrane-Bound Transcriptional Regulator That Links Chitin and Natural Competence in Vibrio cholerae’, mBio, 5(1), pp. e01028–13. doi: 10.1128/mBio.01028-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deatherage DE and Barrick J (2014) Evolved Microbes from Next-Generation Sequencing Data Using breseq, in Engineering and Analyzing Multicellular Systems: Methods and Protocols. Edited by Sun L and Shou W. New York, New York: Springer. [Google Scholar]
- Doron S et al. (2018) ‘Systematic discovery of antiphage defense systems in the microbial pangenome’, Science, 359(6379), p. eaar4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dy RL et al. (2014) ‘Remarkable Mechanisms in Microbes to Resist Phage Infections’, Annual Review of Virology, 1(1), pp. 307–331. doi: 10.1146/annurev-virology-031413-085500. [DOI] [PubMed] [Google Scholar]
- Edgar RC (2004) ‘MUSCLE : multiple sequence alignment with high accuracy and high throughput’, Nucleic Acids Res, 32(5), pp. 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faruque SM et al. (2005) ‘Self-limiting nature of seasonal cholera epidemics: Role of host-mediated amplification of phage’, Proceedings of the National Academy of Sciences, 102(17), pp. 6119–6124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillol-Salom A et al. (2018) ‘Phage-inducible chromosomal islands are ubiquitous within the bacterial universe’, ISME Journal. Springer US, 12(9), pp. 2114–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S et al. (2005) ‘PHYML Online — a web server for fast maximum likelihood-based phylogenetic inference’, Nucleic Acids Research, 33(Web Server issue), pp. W557–W559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z et al. (2017) ‘Isolation and Complete Genome Sequence of a Novel Pseudoalteromonas Phage PH357 from the Yangtze River Estuary’, Current Microbiology. Springer US, 74(7), pp. 832–839. [DOI] [PubMed] [Google Scholar]
- He X et al. (2012) ‘The T4 phage SF1B helicase Dda is structurally optimized to perform DNA strand separation’, Structure. Elsevier Ltd, 20(7), pp. 1189–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hercules K et al. (1971) ‘Mutants in a Nonessential Gene of Bacteriophage T4 Which Are Defective in the Degradation of Escherichia coli Deoxyribonucleic Acid’, J Virol, 7(I), pp. 95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille F et al. (2018) ‘The Biology of CRISPR-Cas: Backward and Forward’, Cell, 172(6), pp. 1239–1259. [DOI] [PubMed] [Google Scholar]
- Hinton DM et al. (2005) ‘Transcriptional takeover by σ appropriation: Remodelling of the σ70 subunit of Escherichia coli RNA polymerase by the bacteriophage T4 activator MotA and co-activator AsiA’, Microbiology, 151(6), pp. 1729–1740. doi: 10.1099/mic.0.27972-0. [DOI] [PubMed] [Google Scholar]
- Kauffman KM et al. (2018) ‘Viruses of the Nahant Collection, characterization of 251 marine Vibrionaceae viruses’, Scientific Data, 5, p. 180114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen EC et al. (2017) ‘Novel “Superspreader” Bacteriophages Promote Horizontal Gene Transfer by Transformation’, mBio, 8(1), pp. e02115–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV and Krupovic M (2015) ‘Evolution of adaptive immunity from transposable elements combined with innate immune systems’, Nat Rev Genet. Nature Publishing Group, 16(3), pp. 184–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskella B and Brockhurst MA (2014) ‘Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities’, FEMS Microbiology Reviews, 38(5), pp. 916–931. doi: 10.1111/1574-6976.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA et al. (2007) ‘Clustal W and Clustal X version 2.0’, Bioinformatics, 23(21), pp. 2947–2948. [DOI] [PubMed] [Google Scholar]
- Levine MM et al. (1982) ‘The Pathogenicity of Nonenterotoxigenic Vibrio cholerae Serogroup O1 Biotype El Tor Isolated from Sewage Water in Brazil’, Journal of Infectious Diseases, 145(3), pp. 296–299. [DOI] [PubMed] [Google Scholar]
- Luhtanen A-M et al. (2014) ‘Isolation and characterization of phage–host systems from the Baltic Sea ice’, Extremophiles, 18(1), pp. 121–130. [DOI] [PubMed] [Google Scholar]
- Makarova KS et al. (2011) ‘Defense Islands in Bacterial and Archaeal Genomes and Prediction of Novel Defense Systems’, Journal of Bacteriology, 193(21), pp. 6039–6056. doi: 10.1128/JB.05535-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manrique P, Dills M and Young MJ (2017) ‘The Human Gut Phage Community and Its Implications for Health and Disease’, Viruses, 9(6), p. E141. doi: 10.3390/v9060141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Rubio R et al. (2017) ‘Phage-inducible islands in the Gram-positive cocci’, The ISME Journal. Nature Publishing Group, 11(4), pp. 1029–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKitterick AC et al. (2019) ‘Competition between mobile genetic elements drives optimization of a phage-encoded CRISPR-Cas system: Insights from a natural arms-race’, Phil. Trans. R. Soc. B, 374(1772), p. 20180089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKitterick AC and Seed KD (2018) ‘Anti-phage islands force their target phage to directly mediate island excision and spread’, Nature Communications. Springer US, 9(1), p. 2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naser I. Bin et al. (2017) ‘Analysis of the CRISPR-Cas system in bacteriophages active on epidemic strains of Vibrio cholerae in Bangladesh’, Scientific Reports. Springer US, 7(1), p. 14880. doi: 10.1038/s41598-017-14839-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara BJ et al. (2017) ‘A highly specific phage defense system is a conserved feature of the Vibrio cholerae mobilome’, PLoS Genetics, 13(6), p. e1006838. doi: 10.1371/journal.pgen.1006838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penadés JR et al. (2015) ‘Bacteriophage-mediated spread of bacterial virulence genes’, Current Opinion in Microbiology, 23, pp. 171–8. [DOI] [PubMed] [Google Scholar]
- Penadés JR and Christie GE (2015) ‘The Phage-Inducible Chromosomal Islands: A Family of Highly Evolved Molecular Parasites’, Annual Review of Virology, 2(1), pp. 181–201. [DOI] [PubMed] [Google Scholar]
- Petrov VM et al. (2010) ‘Genomes of the T4-related bacteriophages as windows on microbial genome evolution’, Virol J, 7(292). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires DP et al. (2016) ‘Genetically Engineered Phages: a Review of Advances over the Last Decade’, Microbiology and Molecular Biology Reviews, 80(3), pp. 523–543. doi: 10.1128/MMBR.00069-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram G et al. (2012) ‘Staphylococcal pathogenicity island interference with helper phage reproduction is a paradigm of molecular parasitism’, Proceedings of the National Academy of Sciences, 109(40), pp. 16300–16305. doi: 10.1073/pnas.1204615109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramphul C et al. (2017) ‘Genome analysis of three novel lytic Vibrio coralliilyticus phages isolated from seawater, Okinawa, Japan’, Marine Genomics. Elsevier B.V., 35, pp. 69–75. [DOI] [PubMed] [Google Scholar]
- Rivas HG, Schmaling SK and Gaglia MM (2016) ‘Shutoff of host gene expression in influenza A virus and herpesviruses: Similar mechanisms and common themes’, Viruses, 8(4), p. 102. doi: 10.3390/v8040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikrishnan K et al. (2009) ‘Mechanistic Basis of 5′-3′ Translocation in SF1B Helicases’, Cell, 137(5), pp. 849–859. doi: 10.1016/j.cell.2009.03.036. [DOI] [PubMed] [Google Scholar]
- Samson JE et al. (2013) ‘Revenge of the phages: defeating bacterial defences’, Nat Rev Microbiol. Nature Publishing Group, 11(10), pp. 675–87. doi: 10.1038/nrmicro3096. [DOI] [PubMed] [Google Scholar]
- Seed KD et al. (2011) ‘Evidence of a Dominant Lineage of Vibrio cholerae-Specific Lytic Bacteriophages Shed by Cholera Patients over a 10-Year Period in Dhaka, Bangladesh’, mBio, 2(1), pp. e00334–10. doi: 10.1128/mBio.00334-10.Editor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seed KD et al. (2013) ‘A bacteriophage encodes its own CRISPR/Cas adaptive response to evade host innate immunity’, Nature. Nature Publishing Group, 494(7438), pp. 489–91. doi: 10.1038/nature11927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seed KD et al. (2014) ‘Evolutionary consequences of intra-patient phage predation on microbial populations’, eLife, 3, p. e03497. doi: 10.7554/eLife.03497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton MR et al. (2004) ‘Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks’, Nature, 432(7014), pp. 187–193. [DOI] [PubMed] [Google Scholar]
- Tormo-Más MA et al. (2010) ‘Moonlighting bacteriophage proteins derepress staphylococcal pathogenicity islands.’, Nature, 465(7299), pp. 779–782. doi: 10.1038/nature09065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda C et al. (2009) ‘Specificity of staphylococcal phage and SaPI DNA packaging as revealed by integrase and terminase mutations.’, Mol Microbiol., 72(1), pp. 98–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Úbeda C et al. (2008) ‘SaPI mutations affecting replication and transfer and enabling autonomous replication in the absence of helper phage’, Molecular Microbiology, 67(3), pp. 493–503. doi: 10.1111/j.1365-2958.2007.06027.x. [DOI] [PubMed] [Google Scholar]
- Vecteezy (2019) Vector Bangladesh Map. Available at: https://www.vecteezy.com/.
- Warner HR et al. (1970) ‘Isolation of Bacteriophage T4 Mutants Defective in the Ability to Degrade Host Deoxyribonucleic Acid’, J Virol, 5(6), pp. 700–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the study are found in the manuscript, supplementary information, or from the corresponding author upon request.