INTRODUCTION
Neuroblastoma, as well defined by Willis,1 is an embryonal tumor of neural crest origin. Tumors of the neuroblastoma group include neuroblastoma, ganglioneuroblastoma, and ganglioneuroma. We believe that all ganglioneuromas were once neuroblastomas in the early stage of tumor development.1,2 They are collectively called peripheral neuroblastic tumors (pNTs) and are known to present with a wide range of clinical behavior, from spontaneous regression and tumor maturation to aggressive progression that is refractory to intensive treatment. Recent advances in research indicate that molecular and/or genomic properties of individual tumors are closely associated with their unique clinical behaviors.3-5 During the past several decades, histopathologic analyses of pNTs have provided invaluable information for predicting prognosis and determining therapy stratification. The origin of International Neuroblastoma Pathology Classification (INPC) dates to the 1984 article by Shimada et al6 that first introduced the age-linked pathology classification system. Since then, the Shimada system has laid the foundation for the current INPC.2,7 The INPC, as a single prognostic parameter, clearly defines clinically aggressive and nonaggressive groups of pNTs.7,8
The INPC, distinguishing favorable histology (FH) and unfavorable histology (UH) groups, defines four categories of pNTs: neuroblastoma (Schwannian stroma poor); ganglioneuroblastoma, intermixed (Schwannian stroma rich); ganglioneuroma (Schwannian stroma dominant); and ganglioneuroblastoma, nodular (composite, Schwannian stroma rich/stroma dominant and stroma poor).2 Cases in the ganglioneuroblastoma, intermixed and ganglioneuroma categories are always classified into the FH group, and the patients enjoy excellent clinical outcomes.9 Patients with disease in the neuroblastoma and ganglioneuroblastoma, nodular categories are classified into either the FH group or UH group on the basis of age-linked evaluation (0 to 18 months, 18 to 60 months, and ≥ 60 months) of neuroblastic differentiation grade (undifferentiated, poorly differentiated, and differentiating) and Mitosis-Karyorrhexis index (low: < 100/5,000 cells; intermediate 100 to 200/5,000 cells; and high: ≥ 200/5,000 cells) of the individual neuroblastoma tumors or neuroblastoma component of ganglioneuroblastoma, nodular tumors.2,7,8,10,11
Whereas survival of patients with FH pNT is excellent (> 90%), survival of patients with UH pNT has only incrementally improved over the years and currently remains approximately 40% to 50%.7,8,10 This seems to be mainly because we have had a limited idea of what molecular mechanism underlies the therapy-resistant behavior of UH tumors. These observations have led us to seek additional immunohistochemical biomarkers that are tightly associated with aggressive behaviors—that is, unresponsiveness or resistance to the current intensive multimodal therapy—of certain UH tumors. To address this problem, we and others have sought and identified the expression of potentially drug-targetable proteins that are responsible for their aggressive progression in the UH group. When we seek such biomarkers, they are preferably actionable/druggable by existing pharmaceutical agents, US Food and Drug Administration approved, or currently tested in human clinical trials. If we could effectively neutralize the activity of such targets with specific drugs, then survival of UH patients as a whole would substantially improve.
POSSIBLE DRIVERS OF AGGRESSIVE UH NEUROBLASTOMA
Among UH neuroblastomas, approximately 50% of them carry MYCN amplification.12 Approximately 80% to 90% of MYCN-amplified neuroblastomas show augmented expression of the MYCN protein.12-14 This is associated with adverse clinical outcomes and considered a driver of clinically aggressive neuroblastoma.15,16 What other mechanisms could account for driving aggressive neuroblastomas without MYCN amplification? The majority of neuroblastoma cell lines without MYCN amplification express high levels of MYC proteins, but the prognostic significance of MYC protein overexpression in patients with neuroblastoma was defined only recently.12 We have found that a considerable number of primary neuroblastomas without MYCN amplification—approximately 10% of the entire cohort or approximately 20% of UH cases—actually express high levels of MYC protein and their survival was similar to that of neuroblastoma with high MYCN protein expression (3-year event-free survival, 46.2 ± 12.0% for MYCN overexpressing neuroblastoma and 43.4 ± 23.1% for MYC overexpressing neuroblastoma, respectively).12 On the basis of this finding, we immunohistochemically defined neuroblastomas with MYCN or MYC overexpression as MYC-driven neuroblastoma.12
Recent next-generation sequencing analyses of high-stage neuroblastomas have provided additional clues as to the possible drivers of non–MYC-driven aggressive neuroblastoma.17-19 Those include ATRX (α-thalassemia/mental retardation syndrome X linked) mutations, leading to ATRX loss and the ALT (alternative lengthening of telomere) phenotype, and TERT overexpression as a result of TERT gene rearrangements. Both mechanisms prevent tumor cells from replication senescence/cell death as a result of telomere erosion and provide them with infinite proliferating capability. Of note, these three types of highly aggressive neuroblastomas—that is, MYC-driven neuroblastoma, ALT phenotype neuroblastoma because of ATRX loss, and TERT-overexpressing neuroblastoma with gene rearrangement—are mutually exclusive and seem to comprise the majority of therapy-resistant or refractory UH neuroblastomas. It should be noted that these three markers—that is, MYC family proteins, TERT, and ATRX—can be the subjects of immunohistochemical assay and potentially incorporated in future INPC.
Other than the above gene and expression alterations found in aggressive UH neuroblastomas, ALK mutations and amplification are found in approximately 10% of sporadic neuroblastoma cases.20,21 It was also found that ALK gene amplification and F1174 mutations are associated with MYCN amplification.22 Moreover, ALK overexpression as a result of ALK amplification/mutations seems to be responsible for its oncogenic activity23; however, the prognostic significance of ALK mutations/overexpression has been controversial, as some reports suggest an association of ALK mutations/overexpression with fatal outcomes of the disease, whereas others suggest otherwise.22,23 In particular, Regairaz et al24 showed immunohistochemically that expression of ALK and its active form pALK was observed in many neuroblastomas independent of ALK mutation/amplification. On the basis of these observations, future INPC will not likely incorporate the immunohistochemical detection of ALK and/or pALK expression.
PROPOSED SUBGROUPING OF UH NEUROBLASTOMAS
As indicated above and also detailed in our article,25 we are gaining additional insight into the potential molecular targets that underlie the therapy-resistant and aggressive behavior of UH neuroblastomas. Moreover, therapy resistance is often attributed to a protein functional level but it is difficult to defeat it because of the inherent plasticity/stemness of tumor cells. In this regard, MYC/MYCN and TERT overexpression26-29 has been linked to the stemness of tumor cells, and direct and indirect therapeutic targeting of these alterations suppresses the stemness of aggressive and therapy-resistant UH neuroblastomas. Similarly, ALT is linked to tumor stem cells.30,31 These observations allow us to incorporate the information into the INPC toward the goal of precision prognosis and therapy stratification.
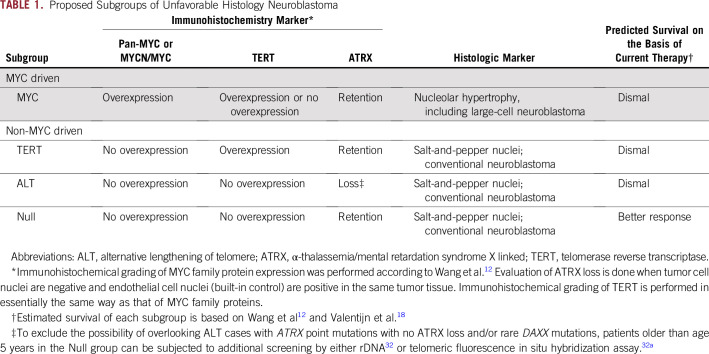
Our plan is to refine the UH neuroblastoma stratification using immunohistopathologic analysis with anti–pan-MYC antibody (or anti-MYCN and anti-MYC), anti-TERT antibody, and anti-ATRX antibody. As shown in Table 1 and Appendix Figure A1 and summarized below, UH neuroblastomas are classified into four subgroups. Taken together, targeting TERT, MYCN/MYC, and ALT via ATRX would contain the stem-cell compartment of the UH neuroblastoma. It is expected that the nonresponder to current high-risk therapy—MYC, TERT, and ATL subgroups—could be separated from the possible responder—Null subgroup—with a high probability before the initiation of therapy:
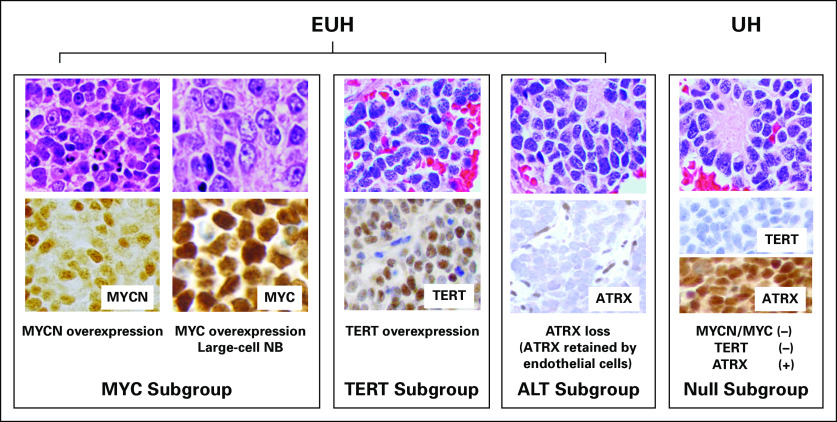
MYC subgroup: MYC-driven neuroblastoma shows augmented expression of MYCN protein and/or MYC protein. As MYC family protein overexpression stimulates rRNA synthesis and protein translation, neuroblastoma cells in this subgroup often exhibit prominent nucleolar formation—nucleolar hypertrophy—and hypertrophic cell morphology.13,14 Because of the presence of one to a few prominent nucleoli, MYC-driven neuroblastoma is cytologically distinguishable from the conventional small blue-cell neuroblastoma with salt-and-pepper nuclei. MYC-driven neuroblastomas, which comprise more than 50% of high-risk neuroblastomas, includes rare and unique tumor, named large-cell neuroblastoma, characterized by the bull’s eye appearance of enlarged and uniquely open euchromatin-rich nuclei that contain highly conspicuous nucleoli.33 Most large-cell neuroblastomas overexpress higher levels of MYCN or MYC protein than other MYC-driven neuroblastomas. Their euchromatin-rich open nuclei suggest the stem cell–like nature of the tumor cells.29 A considerable number of MYC-driven neuroblastomas are also associated with TERT overexpression, as TERT is a direct transcriptional target of MYC family proteins. TERT can also stabilize MYC protein via its noncanonical enzymatic function, creating a positive feedback loop between MYCN/MYC and TERT expression.34,35
TERT subgroup: TERT is the protein component of telomerase, which also includes TERC, the telomerase RNA component. Higher levels of TERT expression are also observed in neuroblastomas without MYCN/MYC protein overexpression. TERT overexpression, in this case, is likely a result of long-range genomic rearrangements but not to promoter mutations in neuroblastoma.36 TERT promoter hypermethylation may also be involved in its activating mechanism.37,38
ALT subgroup: ATRX loss results in the ALT phenotype. ATRX mutation is rarely observed in the MYC and TERT subgroups, and loss of ATRX is exclusively observed in patients with neuroblastoma older than age 5 years.17 Because a normal ATRX function is to insert the variant histone H3, namely H3.3, in concert with DAXX, into chromatin to maintain transcriptionally active euchromatin,39 ATRX loss may result in a more transcriptionally inactive heterochromatic state. Thus, the histologic/cytologic appearance of the ALT subgroup could be the conventional type with salt-and-pepper nuclei.
Null subgroup: There are still UH neuroblastomas without MYC family protein overexpression, TERT overexpression, and ATRX loss. As long as no other aggravated factors are found,40 patients in the Null subgroup would, we hope, respond to current high-risk treatment regimens.
TABLE 1.
Proposed Subgroups of Unfavorable Histology Neuroblastoma
MOLECULAR TARGETING THERAPIES FOR UH NEUROBLASTOMA RESISTANT TO CURRENT THERAPY
To develop treatment strategies for MYC-driven neuroblastomas, new therapeutic agents should be clearly defined; however, direct targeting of MYC family proteins with small molecules has been a formidable task.41 Hence, many have sought indirect approaches to downregulate MYC family protein expression in cancer cells. Strategies under consideration include, but are not limited to, transcriptional repression of MYC and MYCN genes by G-quadruplex (G4) stabilizers,42,43 bromodomain and extraterminal protein inhibitors,44,45 and inhibitors of a transcriptional cyclin-dependent kinase 746,47; destabilization of MYCN by Aurora kinase A inhibitors48 and of MYC and MYCN proteins by RAS signaling pathway inhibitors49-51; and translational blockade of MYC mRNA by eIF4F inhibition and stabilization of RNA G4 by RNA G4 ligands.52,53 We recognize that UH tumors with high-level MYC family protein expression tend to exhibit hypertrophic nucleoli,13,14 which indicates that they are highly active in rRNA synthesis and protein translation. Small-molecule inhibitors, including CX-5461, an RNA polymerase I inhibitor, and halofuginone, a protein translation inhibitor, could therefore effectively target these pathophysiologic features. As we have shown, these inhibitors in fact downregulate MYC family protein expression in neuroblastoma cells.13
For those patients with TERT-overexpressing neuroblastoma, telomerase inhibitors can be considered. Imetelstat (GRN163L)54 is a potent and specific inhibitor of telomerase that binds with high affinity to TERC (telomerase RNA component). Of interest, sorafenib has been demonstrated to synergize with imetelstat to inhibit the growth of mouse xenografts of human cancer.55 Sorafenib is a US Food and Drug Administration–approved kinase inhibitor,56 but its synergetic effect with imetelstat seems to be a result of p21 attenuating activity.55 On the basis of these observations, a combination of imetelstat and sorafenib may prove to be more efficacious than imetelstat alone to those neuroblastomas with elevated telomerase activity.
ALT inhibition could also be considered for those patients with neuroblastoma with ATRX loss; however, because ATRX loss is a result of structural alterations in the ATRX gene,57 it would be difficult to regain ATRX expression in neuroblastoma. If so, is there any other way by which the ALT phenotype can be suppressed? To address this question, we need to understand the mechanism of how ATRX loss leads to ALT. It has become evident that the acquisition of the ALT phenotype uses the DNA replication stress response,30 which involves a cascade of events, including the obligatory activation of ATR (ataxia telangiectasia and Rad3 related) kinase. AZD6738 is a novel potent and selective inhibitor of ATR kinase with IC50 values of less than 1 µM in cell-based assays58 and would effectively target ALT tumor cells.
CONCLUSION
Prognosis and therapy stratification of neuroblastomas have steadily improved since the introduction of the Evans staging system59 with age used as a prognostic factor60; however, 5-year survival of patients with high-risk neuroblastoma, defined by the International Neuroblastoma Risk Group system (INRG), remains at approximately 40% or lower.61,62 The same is true for the UH neuroblastoma group, which essentially overlaps high-risk neuroblastomas.8,63 Unfortunately, no additional refinement for the INRG has been proposed since 2009.61 The proposed histologic subgrouping of UH neuroblastomas represents a practical and immediately implementable approach with which to create the category leading to the most devastating clinical outcome, namely extremely unfavorable histology (EUH) neuroblastoma. We expect that conventional UH neuroblastoma tumors—that is, the Null subgroup—would, we hope, respond to the current multimodal high-intensity therapy designed for high-risk neuroblastoma, whereas EUH tumors—that is, those of the MYC, TERT, and ALT subgroups—would not. For those EUH tumors, appropriate therapy protocols will have to be designed and tested.
On the basis of our findings,12 MYC family protein immunohistochemistry is already incorporated in the Children’s Oncology Group pathology analysis as an integrated biomarker of neuroblastoma. Similar attempts are now underway for TERT and ATRX immunohistochemistry for precision prognosis and the stratification of neuroblastoma. Furthermore, we are planning to perform correlative studies between TERT protein expression and TERT gene rearrangements and between ATRX protein loss and ATRX gene mutations/deletions. We believe that comprehensive integration of improved INPC with immunohistochemical subgrouping into INRG will contribute significantly to the future management of patients with this devastating disease.
Appendix
FIG A1.
Histological and immunohistochemical appearances of extremely unfavorable histology and unfavorable histology neuroblastomas. ALT, alternative lengthening of telomere; ATRX, α-thalassemia/mental retardation syndrome X linked; EUH, extremely unfavorable histology; NB, neuroblastoma; TERT, telomerase reverse transcriptase; UH, unfavorable histology.
Footnotes
Presented at the Meeting of the International Neuroblastoma Pathology Committee, Oslo, Norway, June 15-17, 2017.
Supported in part by National Institutes of Health Grant No. U10-CA180886 (H.S.) and P30-CA14089-37 (H.S.).
AUTHOR CONTRIBUTIONS
Conception and design: All authors
Financial support: Hiroyuki Shimada
Administrative support: Hiroyuki Shimada
Provision of study materials or patients: Hiroyuki Shimada
Collection and assembly of data: All authors
Data analysis and interpretation: All authors
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or po.ascopubs.org/site/ifc.
No potential conflicts of interest were reported.
REFERENCES
- 1.Willis R. Neuroblastoma and Ganglioneuroma. Pathology of Tumours. New York, NY: Appleton-Century-Crofts; 1967. [Google Scholar]
- 2.Shimada H, Ambros IM, Dehner LP, et al. Terminology and morphologic criteria of neuroblastic tumors: Recommendations by the International Neuroblastoma Pathology Committee. Cancer. 1999;86:349–363. [PubMed] [Google Scholar]
- 3.Pizzo PA, Poplack DG.eds): Neuroblastoma Principles and Practice of Pediatric Oncology Philadelphia, PA: Lippincott Williams & Wilkins; 2011866–892. [Google Scholar]
- 4.Bosse KR, Maris JM. Advances in the translational genomics of neuroblastoma: From improving risk stratification and revealing novel biology to identifying actionable genomic alterations. Cancer. 2016;122:20–33. doi: 10.1002/cncr.29706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matthay KK, Maris JM, Schleiermacher G, et al. Neuroblastoma. Nat Rev Dis Primers. 2016;2:16078. doi: 10.1038/nrdp.2016.78. [DOI] [PubMed] [Google Scholar]
- 6.Shimada H, Chatten J, Newton WA, Jr, et al. Histopathologic prognostic factors in neuroblastic tumors: Definition of subtypes of ganglioneuroblastoma and an age-linked classification of neuroblastomas. J Natl Cancer Inst. 1984;73:405–416. doi: 10.1093/jnci/73.2.405. [DOI] [PubMed] [Google Scholar]
- 7.Shimada H, Ambros IM, Dehner LP, et al. The International Neuroblastoma Pathology Classification (the Shimada system) Cancer. 1999;86:364–372. [PubMed] [Google Scholar]
- 8.Sano H, Bonadio J, Gerbing RB, et al. International Neuroblastoma Pathology Classification adds independent prognostic information beyond the prognostic contribution of age. Eur J Cancer. 2006;42:1113–1119. doi: 10.1016/j.ejca.2005.11.031. [DOI] [PubMed] [Google Scholar]
- 9.Okamatsu C, London WB, Naranjo A, et al. Clinicopathological characteristics of ganglioneuroma and ganglioneuroblastoma: A report from the CCG and COG. Pediatr Blood Cancer. 2009;53:563–569. doi: 10.1002/pbc.22106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimada H, Umehara S, Monobe Y, et al. International Neuroblastoma Pathology Classification for prognostic evaluation of patients with peripheral neuroblastic tumors: A report from the Children’s Cancer Group. Cancer. 2001;92:2451–2461. doi: 10.1002/1097-0142(20011101)92:9<2451::aid-cncr1595>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 11.Peuchmaur M, d’Amore ES, Joshi VV, et al. Revision of the International Neuroblastoma Pathology Classification: Confirmation of favorable and unfavorable prognostic subsets in ganglioneuroblastoma, nodular. Cancer. 2003;98:2274–2281. doi: 10.1002/cncr.11773. [DOI] [PubMed] [Google Scholar]
- 12.Wang LL, Teshiba R, Ikegaki N, et al. Augmented expression of MYC and/or MYCN protein defines highly aggressive MYC-driven neuroblastoma: A Children’s Oncology Group study. Br J Cancer. 2015;113:57–63. doi: 10.1038/bjc.2015.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niemas-Teshiba R, Matsuno R, Wang LL, et al. MYC-family protein overexpression and prominent nucleolar formation represent prognostic indicators and potential therapeutic targets for aggressive high-MKI neuroblastomas: A report from the Children’s Oncology Group. Oncotarget. 2017;9:6416–6432. doi: 10.18632/oncotarget.23740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang LL, Suganuma R, Ikegaki N, et al. Neuroblastoma of undifferentiated subtype, prognostic significance of prominent nucleolar formation, and MYC/MYCN protein expression: A report from the Children’s Oncology Group. Cancer. 2013;119:3718–3726. doi: 10.1002/cncr.28251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brodeur GM, Seeger RC, Schwab M, et al. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science. 1984;224:1121–1124. doi: 10.1126/science.6719137. [DOI] [PubMed] [Google Scholar]
- 16.Seeger RC, Brodeur GM, Sather H, et al. Association of multiple copies of the N-myc oncogene with rapid progression of neuroblastomas. N Engl J Med. 1985;313:1111–1116. doi: 10.1056/NEJM198510313131802. [DOI] [PubMed] [Google Scholar]
- 17.Cheung NK, Zhang J, Lu C, et al. Association of age at diagnosis and genetic mutations in patients with neuroblastoma. JAMA. 2012;307:1062–1071. doi: 10.1001/jama.2012.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valentijn LJ, Koster J, Zwijnenburg DA, et al. TERT rearrangements are frequent in neuroblastoma and identify aggressive tumors. Nat Genet. 2015;47:1411–1414. doi: 10.1038/ng.3438. [DOI] [PubMed] [Google Scholar]
- 19.Peifer M, Hertwig F, Roels F, et al. Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature. 2015;526:700–704. doi: 10.1038/nature14980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chmielecki J, Bailey M, He J, et al. Genomic profiling of a large set of diverse pediatric cancers identifies known and novel mutations across tumor spectra. Cancer Res. 2017;77:509–519. doi: 10.1158/0008-5472.CAN-16-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bellini A, Bernard V, Leroy Q, et al. Deep sequencing reveals occurrence of subclonal ALK mutations in neuroblastoma at diagnosis. Clin Cancer Res. 2015;21:4913–4921. doi: 10.1158/1078-0432.CCR-15-0423. [DOI] [PubMed] [Google Scholar]
- 22.De Brouwer S, De Preter K, Kumps C, et al. Meta-analysis of neuroblastomas reveals a skewed ALK mutation spectrum in tumors with MYCN amplification. Clin Cancer Res. 2010;16:4353–4362. doi: 10.1158/1078-0432.CCR-09-2660. [DOI] [PubMed] [Google Scholar]
- 23.Schulte JH, Bachmann HS, Brockmeyer B, et al. High ALK receptor tyrosine kinase expression supersedes ALK mutation as a determining factor of an unfavorable phenotype in primary neuroblastoma. Clin Cancer Res. 2011;17:5082–5092. doi: 10.1158/1078-0432.CCR-10-2809. [DOI] [PubMed] [Google Scholar]
- 24.Regairaz M, Munier F, Sartelet H, et al. Mutation-independent activation of the anaplastic lymphoma kinase in neuroblastoma Am J Pathol 186435–445.2016[Erratum: Am J Pathol 186:1710, 2016] [DOI] [PubMed] [Google Scholar]
- 25.Shimada H, Ikegaki N. Neuroblastoma Pathology and Classification for Precision Prognosis and Therapy Stratification, Neuroblastoma. New York, NY: Elsevier; 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shay JW, Wright WE. Telomeres and telomerase in normal and cancer stem cells. FEBS Lett. 2010;584:3819–3825. doi: 10.1016/j.febslet.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joseph I, Tressler R, Bassett E, et al. The telomerase inhibitor imetelstat depletes cancer stem cells in breast and pancreatic cancer cell lines. Cancer Res. 2010;70:9494–9504. doi: 10.1158/0008-5472.CAN-10-0233. [DOI] [PubMed] [Google Scholar]
- 28.Cotterman R, Knoepfler PS. N-Myc regulates expression of pluripotency genes in neuroblastoma including lif, klf2, klf4, and lin28b. PLoS One. 2009;4:e5799. doi: 10.1371/journal.pone.0005799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ikegaki N, Shimada H, Fox AM, et al. Transient treatment with epigenetic modifiers yields stable neuroblastoma stem cells resembling aggressive large-cell neuroblastomas. Proc Natl Acad Sci USA. 2013;110:6097–6102. doi: 10.1073/pnas.1118262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flynn RL, Cox KE, Jeitany M, et al. Alternative lengthening of telomeres renders cancer cells hypersensitive to ATR inhibitors. Science. 2015;347:273–277. doi: 10.1126/science.1257216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wakimoto H, Tanaka S, Curry WT, et al. Targetable signaling pathway mutations are associated with malignant phenotype in IDH-mutant gliomas. Clin Cancer Res. 2014;20:2898–2909. doi: 10.1158/1078-0432.CCR-13-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Udugama M, Sanij E, Voon HP, et al. Ribosomal DNA copy loss and repeat instability in ATRX-mutated cancers. Proc Nat Acad Sci. 2018;115:4737–4742. doi: 10.1073/pnas.1720391115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32a.Koschmann C, Calinescu A-A, Nunez FJ, et al. ATRX loss promotes tumor growth and impairs nonhomologous end joining DNA repair in glioma. Sci Trans Med. 2016;8:328ra28–328ra28. doi: 10.1126/scitranslmed.aac8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tornóczky T, Kálmán E, Kajtár PG, et al. Large cell neuroblastoma: A distinct phenotype of neuroblastoma with aggressive clinical behavior. Cancer. 2004;100:390–397. doi: 10.1002/cncr.20005. [DOI] [PubMed] [Google Scholar]
- 34.Koh CM, Khattar E, Leow SC, et al. Telomerase regulates MYC-driven oncogenesis independent of its reverse transcriptase activity. J Clin Invest. 2015;125:2109–2122. doi: 10.1172/JCI79134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu KJ, Grandori C, Amacker M, et al. Direct activation of TERT transcription by c-MYC. Nat Genet. 1999;21:220–224. doi: 10.1038/6010. [DOI] [PubMed] [Google Scholar]
- 36.Lindner S, Bachmann HS, Odersky A, et al. Absence of telomerase reverse transcriptase promoter mutations in neuroblastoma. Biomed Rep. 2015;3:443–446. doi: 10.3892/br.2015.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lindsey JC, Schwalbe EC, Potluri S, et al. TERT promoter mutation and aberrant hypermethylation are associated with elevated expression in medulloblastoma and characterise the majority of non-infant SHH subgroup tumours. Acta Neuropathol. 2014;127:307–309. doi: 10.1007/s00401-013-1225-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee S, Borah S, Bahrami A. Detection of aberrant TERT promoter methylation by combined bisulfite restriction enzyme analysis for cancer diagnosis. J Mol Diagn. 2017;19:378–386. doi: 10.1016/j.jmoldx.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salomoni P. The PML-interacting protein DAXX: Histone loading gets into the picture. Front Oncol. 2013;3:152. doi: 10.3389/fonc.2013.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dagg RA, Pickett HA, Neumann AA, et al. Extensive proliferation of human cancer cells with ever-shorter telomeres. Cell Reports. 2017;19:2544–2556. doi: 10.1016/j.celrep.2017.05.087. [DOI] [PubMed] [Google Scholar]
- 41.Darnell JE., Jr Transcription factors as targets for cancer therapy. Nat Rev Cancer. 2002;2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 42.Brooks TA, Hurley LH. Targeting MYC expression through G-quadruplexes. Genes Cancer. 2010;1:641–649. doi: 10.1177/1947601910377493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benabou S, Ferreira R, Aviñó A, et al. Solution equilibria of cytosine- and guanine-rich sequences near the promoter region of the n-myc gene that contain stable hairpins within lateral loops. Biochim Biophys Acta. 2014;1840:41–52. doi: 10.1016/j.bbagen.2013.08.028. [DOI] [PubMed] [Google Scholar]
- 44.Delmore JE, Issa GC, Lemieux ME, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Puissant A, Frumm SM, Alexe G, et al. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov. 2013;3:308–323. doi: 10.1158/2159-8290.CD-12-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chipumuro E, Marco E, Christensen CL, et al. CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell. 2014;159:1126–1139. doi: 10.1016/j.cell.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Christensen CL, Kwiatkowski N, Abraham BJ, et al. Targeting transcriptional addictions in small cell lung cancer with a covalent CDK7 inhibitor Cancer Cell 26909–922.2014[Erratum: Cancer Cell 27:149, 2015] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gustafson WC, Meyerowitz JG, Nekritz EA, et al. Drugging MYCN through an allosteric transition in Aurora kinase A. Cancer Cell. 2014;26:414–427. doi: 10.1016/j.ccr.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burotto M, Chiou VL, Lee JM, et al. The MAPK pathway across different malignancies: A new perspective. Cancer. 2014;120:3446–3456. doi: 10.1002/cncr.28864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Manning BD, Toker A. AKT/PKB signaling: Navigating the network. Cell. 2017;169:381–405. doi: 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Janku F, Yap TA, Meric-Bernstam F. Targeting the PI3K pathway in cancer: Are we making headway? Nat Rev Clin Oncol. 2018;15:273–291. doi: 10.1038/nrclinonc.2018.28. [DOI] [PubMed] [Google Scholar]
- 52.Cammas A, Millevoi S. RNA G-quadruplexes: Emerging mechanisms in disease. Nucleic Acids Res. 2017;45:1584–1595. doi: 10.1093/nar/gkw1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolfe AL, Singh K, Zhong Y, et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature. 2014;513:65–70. doi: 10.1038/nature13485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dikmen ZG, Gellert GC, Jackson S, et al. In vivo inhibition of lung cancer by GRN163L: A novel human telomerase inhibitor. Cancer Res. 2005;65:7866–7873. doi: 10.1158/0008-5472.CAN-05-1215. [DOI] [PubMed] [Google Scholar]
- 55.Gupta R, Dong Y, Solomon PD, et al. Synergistic tumor suppression by combined inhibition of telomerase and CDKN1A. Proc Natl Acad Sci USA. 2014;111:E3062–E3071. doi: 10.1073/pnas.1411370111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilhelm S, Carter C, Lynch M, et al. Discovery and development of sorafenib: A multikinase inhibitor for treating cancer. Nat Rev Drug Discov. 2006;5:835–844. doi: 10.1038/nrd2130. [DOI] [PubMed] [Google Scholar]
- 57.Cheung NK, Dyer MA. Neuroblastoma: Developmental biology, cancer genomics and immunotherapy. Nat Rev Cancer. 2013;13:397–411. doi: 10.1038/nrc3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vendetti FP, Lau A, Schamus S, et al. The orally active and bioavailable ATR kinase inhibitor AZD6738 potentiates the anti-tumor effects of cisplatin to resolve ATM-deficient non-small cell lung cancer in vivo. Oncotarget. 2015;6:44289–44305. doi: 10.18632/oncotarget.6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Evans AE, D’Angio GJ, Randolph J. A proposed staging for children with neuroblastoma. Children’s Cancer Study Group A. Cancer. 1971;27:374–378. doi: 10.1002/1097-0142(197102)27:2<374::aid-cncr2820270221>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 60.Breslow N, McCann B. Statistical estimation of prognosis for children with neuroblastoma. Cancer Res. 1971;31:2098–2103. [PubMed] [Google Scholar]
- 61.Cohn SL, Pearson AD, London WB, et al. The International Neuroblastoma Risk Group (INRG) classification system: An INRG Task Force report. J Clin Oncol. 2009;27:289–297. doi: 10.1200/JCO.2008.16.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park JR, Bagatell R, London WB, et al. Children’s Oncology Group’s 2013 blueprint for research: Neuroblastoma. Pediatr Blood Cancer. 2013;60:985–993. doi: 10.1002/pbc.24433. Erratum: Pediatr Blood Cancer 61:958, 2014. [DOI] [PubMed] [Google Scholar]
- 63.Uryu K, Nishimura R, Kataoka K, et al. Identification of the genetic and clinical characteristics of neuroblastomas using genome-wide analysis. Oncotarget. 2017;8:107513–107529. doi: 10.18632/oncotarget.22495. [DOI] [PMC free article] [PubMed] [Google Scholar]