Abstract
Using siRNA therapeutics to treat hematologic malignancies has been unsuccessful because blood cancer cells exhibit remarkable resistance to standard transfection methods. Herein we report the successful delivery of siRNA therapeutics with a dual-targeted, layer-by-layer nanoparticle (LbL-NP). The LbL-NP protects siRNA from nucleases in the bloodstream by embedding it within polyelectrolyte layers that coat a polymeric core. The outermost layer consists of hyaluronic acid (a CD44-ligand) covalently conjugated to CD20 antibodies. The CD20/CD44 dual-targeting outer layer provides precise binding to blood cancer cells, followed by receptor-mediated endocytosis of the LbL-NP. We use this siRNA delivery platform to silence B-cell lymphoma 2 (BCL-2), a pro-survival protein, in vitro and in vivo. The dual-targeting approach significantly enhanced internalization of BCL-2 siRNA in lymphoma and leukemia cells, which led to significant downregulation of BCL-2 expression. Systemic administration of the dual-targeted, siRNA-loaded nanoparticle induced apoptosis and hampered proliferation of blood cancer cells both in cell culture and in orthotopic non-Hodgkin’s lymphoma animal models. These results provide the basis for approaches to targeting blood-borne cancers and other diseases, and suggest that LbL nanoassemblies are a promising approach for delivering therapeutic siRNA to hematopoetic cell types that are known to evade transfection by other means.
Keywords: siRNA, hematologic cancer, lymphoma, layer-by-layer nanoparticles, BCL-2
Graphical Abstract
Binary Targeting of siRNA to Hematologic Cancer Cells In Vivo using Layer-by-Layer Nanoparticles. This work demonstrates BCL-2 targeted precision siRNA therapeutics based on a CD20/CD44-dual-targeted layer-by-layer nanoparticle to regulate BCL-2 expression in hematologic malignancies. The binary targeting RNAi approach led to considerable downregulation of BCL-2 expression, and induced apoptosis of the blood cancer cells in culture and in orthotopic blood cancer animal models.
1. Introduction
Each year, over 170,000 people are diagnosed with hematologic malignancies like leukemia, lymphoma and myeloma, and more than 1,200,000 people currently suffer from such diseases in the United States. Blood cancers kill approximately 160 patients every day, and in 2016 alone, they were responsible for nearly 10% of cancer-related deaths in the United States.[1] The considerable mortality from blood cancers is attributed to malignancies that relapse after first line therapy and become resistant to subsequent treatment.
Hematological cancers, particularly chronic lymphocytic leukemia (CLL) and non-Hodgkin lymphoma (NHL), can become resistant to treatment through upregulation of the anti-apoptotic (pro-survival) BCL-2 proteins. Next generation approaches have therefore aimed to target the BCL-2 oncogene, either at the genetic or the protein level, to treat BCL-2-dependent hematologic malignancies.[2] One approach has been to use small molecule BCL-2 inhibitors, such as ABT-737, ABT-263 and ABT-199 (Abbott Laboratories, Abbott Park, IL, USA), inhibitors that control the balance between pro-survival and pro-apoptotic proteins in BCL-2 related cancer.[2a, 3] However, ABT-737 and ABT-263 induce unacceptable levels of thrombocytopenia because they also impact a related off-target protein, BCL-XL. In contrast, ABT-199 (Venclaxta™, Abbvie, North Chicago, IL, USA; Genentech, South San Francisco, CA, USA) has a higher affinity to BCL-2 than to BCL-XL, and it provides anticancer efficacy without major platelet loss in preclinical and clinical studies.[3] It has been approved for the treatment of high-risk relapsed/refractory CLL patients with depletion of 17p, but complete response rates remain below 10%.
In contrast to small molecules, another option for disabling the B-cell lymphoma 2 (BCL-2) oncogene involves specific gene silencing through the delivery of siRNA.[2a, 2c] Because siRNA silences the expression of genes by degrading specific mRNAs, it is unlikely to have the off-target side effects that often accompany small molecule inhibitors. However, the therapeutic efficacy of BCL-2 siRNA (siBCL-2) has not been verified yet in hematologic cancer animal models, mainly due to the lack of effective in vivo RNA delivery systems for hematologic tumors.
Delivery of siRNA therapeutics to specific cell types and tissues can be limited by several barriers, which include the compromised blood serum stability (particularly of unmodified siRNA), efficiency of delivery, and the ability to ensure targeting and selective uptake of siRNA by desired cells. Another important aspect of these barriers is the difficulty in transporting siRNA, an anionic rigid molecule, across the negatively charged lipid bilayer.[4] Additional challenges stem from the intrinsic nature of hematologic cancer. Unlike solid tumors, hematologic malignancies include both continuously circulating blood cancer cells and metastatic lesions in solid tissues,[2a, 2c, 5] so an effective drug delivery strategy must be able to address both free and anchored states. For any RNA delivery system to be successful, it must (i) protect nucleic acids from enzymes in the bloodstream, (ii) selectively bind to the intended cells, and (iii) enable access to the cytoplasm to release its therapeutic payload.[4a, 6]
Herein we report dual-targeted, layer-by-layer nanoparticles (LbL-NPs) that can deliver siRNA to inhibit the progression of hematologic cancers. To shield siRNAs from enzymes in the bloodstream, we embedded siRNA molecules within oppositely charged macromolecular nanolayers on LbL-NPs—a method we have shown to be effective in targeting and silencing oncogenes within solid tumors.[7] The use of alternating electrostatic assembly enables complexation of siRNA in a highly compact form without using a large excess of positively charged polymer, thus enabling an efficiency of packaging difficult to achieve with more typical siRNA polymeric complexes. Within this LbL film, siRNA is shielded from RNA-degrading enzymes that would otherwise prematurely degrade it in the bloodstream, thus greatly boosting serum stability. Finally, of particular note, the final outer layer adsorbed is a negatively charged polyelectrolyte that has the dual role of preventing opsonization of serum proteins and non-specific cell uptake, and presenting specific ligands that can target cells of interest.
However, siRNAs are known to silence not only their complementary target genes but also unwanted off-target genes, a phenomena caused by similarity between the seed regions of siRNAs with thousands of mRNAs. Thus using a single siRNA sequence can make it difficult to achieve high on-target silencing without also observing elevated off-target effects.[8] To avoid this potential pitfall, we exploited cocktails of 40 different siRNAs that have different sequences but target the same BCL-2 mRNA. While each sequence contributes to the same on-target effect, the different sequences do not reinforce the same off-target effect. Given the low concentration of each unique siRNA sequence, their off-target effects are significantly reduced. Previous studies show this type of siRNA cocktail (also called siPool) significantly increases on-target silencing and reduces off-target effects.[9]
We hypothesized that LbL-NPs would target blood cancer cells more efficiently if they could simultaneously bind two different surface receptors overexpressed by these malignancies. To investigate this “binary-targeting” approach, we conjugated CD20 antibodies[2b] to the negatively charged polysaccharide hyaluronic acid (HA). In addition to being a native biopolymer, HA is a ligand of the CD44 receptor upregulated on various cancer cells.[10] Thanks to its anionic charge, HA is also readily incorporated as the terminal layer of an LbL-NP. CD20, a pan-B cell epitope, is a well-known blood cancer marker and appears to play an important role in the progression of hematologic cancers. These observations led to the development of a chimeric monoclonal anti-CD20 antibody (Rituximab) to treat CD20-positive malignancies like CLL and NHL.[11] Since CD20 is both a good marker and a potential therapeutic target, we incorporated anti-CD20 antibodies onto the HA modified surface of the LbL nanoparticle platform. However, the CD20-targeted delivery strategy poses a potential pitfall: although precision targeting of CD20 against hematologic cancer has been proven in the clinic over the past 20 years, the actual internalization of CD20 by blood cancer cells remains unclear.[12] Because successful siRNA delivery requires internalization into the cytoplasm, it was unclear whether CD20-targeting alone would provide efficacy.
To provide reliable internalization of our nanoparticles in blood cancer cells, an additional ligand-receptor interaction was exploited. This approach targeted CD44, another cell surface antigen known to be upregulated in various types of cancers, including hematologic malignancies.[13] The CD44 receptor avidly binds to native hyaluronic acid in the body, an interaction that can be exploited by LbL-nanoparticles that incorporate HA.[10] Upon binding with CD44, HA-coated nanoparticles are internalized by cells via CD44-mediated endocytosis. These nanoparticles eventually escape the endosome to deliver their siRNA payload to the cytoplasm. This escape is mediated by a positively charged polypeptide within the LbL-film, poly-L-arginine (PLA), which disrupts endosomes and allows escape.[14]
The resulting dual-targeted LbL-NPs successfully delivered siRNA and downregulated BCL-2 levels in non-Hodgkin’s lymphoma (NHL) and chronic lymphocytic leukemia (CCL) cells. These LbL-NPs allowed for prolonged circulation times of siRNA in mice, with biodistributions typical of nanoparticle systems. Furthermore, these nanoparticles demonstrated in vivo BCL-2 silencing and anticancer activity in an orthotopic NHL mouse model. Overall this work indicates that LbL-NPs may be a promising avenue for advancing siRNA therapeutics for hematologic cancer.
2. Results and Discussion
2.1. In Vitro Transfection of siRNA into Blood Cancer Cells
In this study, we sought to regulate gene expression in two types of blood cancer cells—RS4(11) (human acute lymphoblastic leukemia) and Toledo (human non-Hodgkin’s B cell lymphoma) cell lines using BCL-2 target siRNA cocktails (siBCL2). First of all, to examine the general efficacy of silencing lymphoma cell types, we tried to downregulate a common housekeeping gene, glyceraldehyde 3-phosphate dehydrogenase (GAPDH) in three types of cancer cells—a human breast cancer cell line (MCF 7) and the two blood cancer cell lines (Toledo and RS4(11))—using conventional transfection reagents Lipofectamine®RNAiMAX (Lipo) and HiPerFect (Hiper).
Excellent gene silencing effects were detected in MCF 7 breast cancer cells treated with 1 nM of GAPDH siRNA cocktails (siGAPDH) formulated with Lipo or Hiper. Expression levels of GAPDH dropped to 3.06 ± 0.14% and 11.71 ± 0.90% of the original expression for Lipo and Hiper, respectively (Figure 1a). On the other hand, even after treatment with siGAPDH at a 100-times higher siRNA concentration (100 nM) compared to that used for MCF 7 cells (1 nM), GAPDH expression levels remained at 96.89 ± 10.94% (Lipo) and 100.5 ± 9.744% (Hiper) in RS4(11) cells and at 97.10 ± 13.59% (Lipo) and 86.35 ± 9.713% (Hiper) in Toledo cells (Figure 1b,c). These results clearly signify difficulties in transfecting blood cancer cells and the need for tools to address this more challenging cell type.
Figure 1. In vitro RNA transfection.
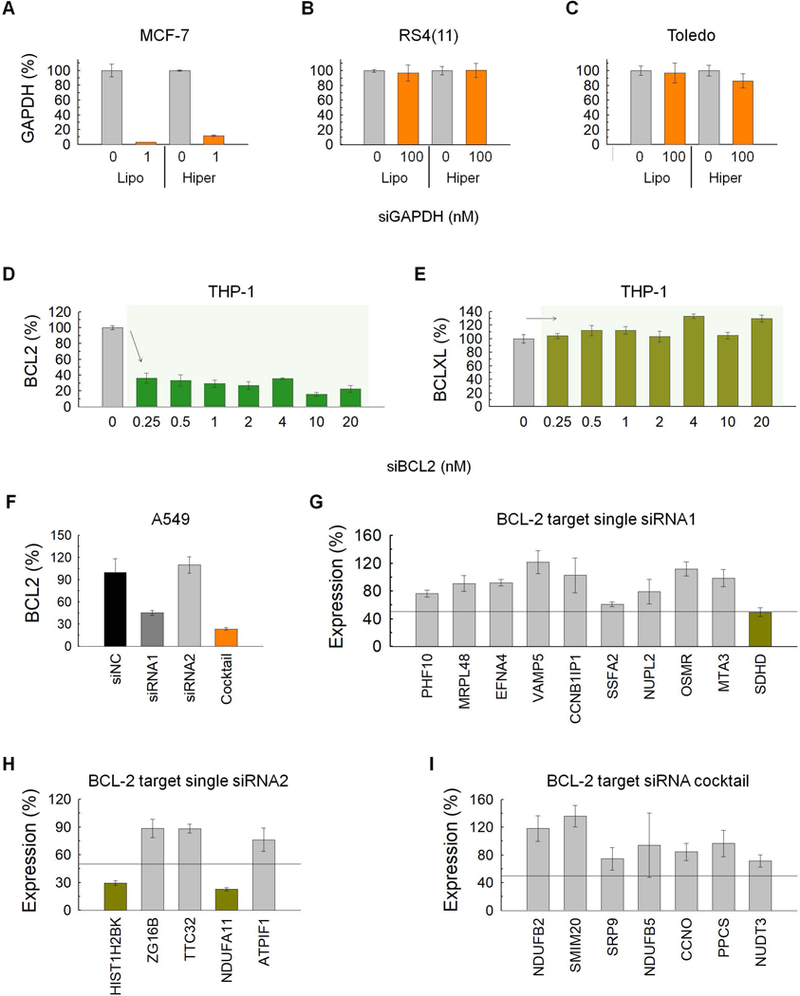
GAPDH expression on (A) MCF-7, (B) Toledo and (C) RS4(11) cells after treatment with GAPDH target siRNA cocktails formulated with Lipofectamine®RNAiMAX (Lipo) and HiPerFect (Hiper). Expression of (D) BCL-2 and (E) BCL-XL on THP-1 cells transfected with BCL-2 target siRNA cocktails formulated with Lipo. (F) Expression of BCL-2 on A549 cells transfected with BCL-2 target single siRNAs and siRNA cocktails formulated with Lipo. Expression of off-target candidate genes on A549 cells transfected with (G) BCL-2 target single siRNA1, (H) single siRNA2 and (I) siRNA cocktails formulated with Lipo.
We next evaluated the BCL-2 silencing activity and off-target effects of siBCL2. Since transfection of siBCL2 into blood cancer cells by conventional transfection agents was unsuccessful, we used a human monocyte cell line, THP-1 instead. We incubated THP-1 with siBCL2 formulated with Lipo; then, we evaluated expression levels of BCL-2 (on-target) and BCL-XL (off-target) genes. Treatment of siBCL2 significantly lowered the expression of BCL-2 to 22.68 ± 0.04% relative to controls at a concentration of 20 nM (Figure 1d); whereas, it did not significantly regulate the expression level of BCL-XL (Figure 1e).
We also evaluated on-target and off-target effects of BCL-2 siRNA cocktails in comparison with single BCL-2 siRNAs. First, to test BCL2 silencing efficiency (on-target effects) of single BCL-2 siRNA and BCL-2 siRNA cocktail, A549 cells were transfected with two types of BCL-2 target single siRNAs (siRNA1 and siRNA2) from commercial sources, the BCL-2 target siRNA cocktail (cocktail) and the negative control siRNA cocktail (siNC) at 3nM total siRNA concentration using Lipo as transfection reagent. BCL2 expression was measured by RT-PCR 24h after transfection. As shown in Figure 1f, siRNA cocktail gave the best knockdown with about 25% remaining mRNA. siRNA1 showed a 50% knock-down, siRNA2 gave no knock-down at all. Possibly, siRNA 2 would require higher concentrations for meaningful down-regulation of BCL-2 gene expression.
Second, to evaluate off-target effects of siRNA1, siRNA2 and cocktail, we ran an off-target detection software TargetScan. Based on the TargetScan scores for single siRNA1, siRNA2 and siRNA cocktail, we selected off-target candidates (Supporting Table S5). We then evaluated the silencing of off-target candidate genes by RT-PCR after treatment of single siRNA1, siRNA2 and siRNA cocktail into A549 cells. Based on the RT-PCR results, we found one off-target gene for siRNA1, SDHD, which shows over 50% knock-down, out of 10 tested candidates (Figure 1g). For siRNA 2, we found two off-target genes, HIST1H2BK and NDUFA11, with > 50% downregulation, out of 5 tested candidates (Figure 1h). For the siRNA cocktail, however, we did not find any single gene down-regulated over 50% out of 7 gene candidates (Figure 1i). These results imply that the BCL-2 siRNA cocktails used in these studies exert substantial gene silencing effects without evidence of significant off-target gene silencing.
2.2. Synthesis and Characterization of Layer-By-Layer siRNA Nanoparticles
To surmount the obstacles to successful siRNA transfection of blood cancer cells, we devised a targeted RNA formulation that can deliver siBCL2 intracellularly while securely protecting the RNA from enzymes present in the bloodstream. The RNA formulation is composed of a negatively charged poly(lactic-co-glycolic acid) (PLGA) nanoparticle (NP) core coated with alternating polyelectrolyte nanolayers using layer-by-layer technology (Figure 2a).
Figure 2. Physicochemical characteristics of PLGA NPs and LbL-NPs.
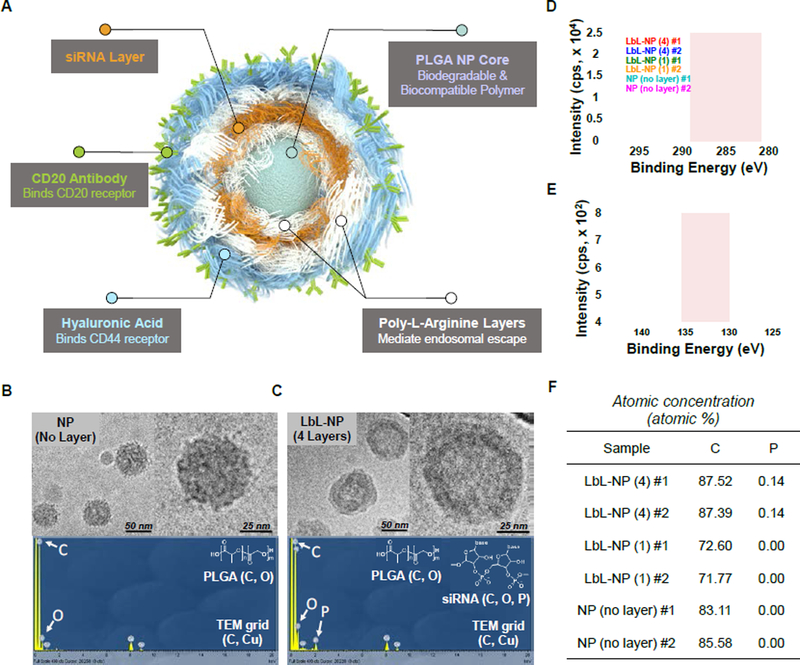
(A) Schematic illustration of a CD20/CD44 dual-targeted LbL-NP for precision siRNA therapeutics. Cryogenic TEM images and Energy-dispersive X-ray spectrum of (B) PLGA NPs and (C) 4-layer LbL-NPs. X-ray photoelectron spectrum indicating (D) carbon and (E) phosphorus contents, and atomic concentration (f) of PLGA NPs, 1-layer and 4-layer NPs.
The PLGA-NP core was prepared via an emulsification-diffusion method previously reported[15] using acid end-functionalized PLGA to achieve a negatively charged colloid. The PLGA-NPs had a hydrodynamic radius of 100.60 ± 0.46 nm and a negative surface charge of – 39.50 ± 1.97 mV. The NPs were evenly dispersed under aqueous conditions; the polydipersity index (PDI) was 0.16 ± 0.02 (Table 1).
Table 1.
Characteristics of PLGA NPs and LbL-NPs
| Sample | Terminal Layer |
Diameter (Z-Avg)a |
Diameter (N-Avg)b |
Zeta potential | PDI |
|---|---|---|---|---|---|
| NP (no layer) | PLGA | 101 ± 0.46 | 63 ± 3.4 | −40 ± 2.0 | 0.16 ± 0.02 |
| LbL−NP (1 layer) | PLA | 104 ± 0.32 | 64 ± 4.2 | 46 ± 1.1 | 0.17 ± 0.019 |
| LbL−NP (2 layers) | siRNA | 106 ± 6.1 | 64 ± 5.0 | −49 ± 2.3 | 0.15 ± 0.032 |
| LbL−NP (3 layers) | PLA | 110 ± 2.1 | 65 ± 3.7 | 40 ± 1.6 | 0.16 ± 0.021 |
| LbL−NP (4 layers) | HA | 114 ± 1.7 | 91 ± 3.6 | −35 ± 2.8 | 0.15 ± 0.0090 |
| LbL−NP (4 layers+Ab) | HA+Ab | 135 ± 1.6 | 92 ± 4.3 | −39 ± 2.1 | 0.13 ± 0.010 |
Z-average is an intensity-based calculated value.
Number average diameter is a number-based calculated value, which can show distributions comparable to those obtained from TEM.
The polyelectrolyte nanolayers were then deposited sequentially onto the PLGA-NP cores via adsorption from aqueous solutions leveraging the electrostatic interactions between oppositely charged polyelectrolytes. Poly-L-arginine (PLA), a positively charged amino acid polymer, was layered onto the negatively charged PLGA-NP substrate first to create a positively charged surface. BCL-2 siRNA molecules were then layered atop the PLA layers as a negative polyelectrolyte layer, onto which an additional PLA layer was added. In turn, hyaluronic acid (HA), a negatively charged natural polysaccharide, was deposited as an outer layer. The HA layer can further act as a ligand of the CD44 receptor, which is overexpressed on various solid and blood-borne cancer cells including many types of hematologic cancers. Moreover, anti-CD20 antibodies (CD20-Ab), which specifically bind with the lymphoma-specific receptor protein CD20[2b], were chemically conjugated on the outer HA layer of the LbL-NP using previously reported conjugation chemistry (Supporting Schemes S1,2). The CD20/CD44-dual-targeting strategy was designed to facilitate targeted nanoparticle binding, endocytosis and ultimately cell uptake and delivery of BCL-2-siRNA into lymphoma cells. The cell-binding and uptake properties gained from the combination of HA and the antibody are meant to assist in targeting cells in solid tissue metastases and in circulating blood flow.
Deposition of the four nanolayers that comprise the final LbL-NP was confirmed by monitoring zeta potential values and observing slight increases in particle diameter. Surface charge reversal following each adsorption step suggests successful layering of the polyelectrolytes including BCL-2-siRNAs. After deposition of 4 polyelectrolyte layers (PLA, siRNA, PLA, HA polyelectrolytes) and conjugation of anti-CD20 antibodies, the hydrodynamic radius increased. The final LbL-NPs were evenly distributed, indicating an average hydrodynamic radius of 135 ± 1.6 nm, surface charge of −39 ± 2.1 mV and PDI of 0.13 ± 0.010 (Table 1).
The nanolayers on the particles were also examined with cryogenic transmission electron microscopy (cryo-TEM). Cryo-TEM images clearly displayed the uniform nanolayer coating on the spherical PLGA nanoparticle core (Figure 2b,c). The presence of siRNA molecules in the nanolayers was further verified via Energy-dispersive X-ray spectroscopy (EDX) and X-ray photoelectron spectroscopy (XPS) techniques. Distinct peaks for phosphate atoms obtained within the nucleic acids were observed in 4 layer LbL-NPs indicated the presence of siRNA molecules in the tetralayered LbL-NPs. Phosphate was not detected in the uncoated PLGA-NPs or single-layered NPs (Figure 2d–f).
2.3. Targeting Blood Cancer Cells via CD20 and CD44
After studying the physicochemical properties of the LbL-NPs, we monitored in vitro cellular uptake of LbL-NPs into blood cancer cells—RS4(11) and Toledo cells. Their surface receptor proteins were first monitored; particularly, expression levels of CD20 and CD44 receptors were evaluated by flow cytometry. Characterized CD20 is known as a lymphoma-specific receptor and CD44 is overexpressed on various cancer cells including blood cancer cells. As presented in Figure 3a and b, RS4(11) cells were upregulated in CD44, but downregulated in CD20 (CD44+ / CD20-). On the other hand, Toledo cells were upregulated in both CD44 and CD20 receptor proteins (CD44+ / CD20+).
Figure 3. Flow cytometry analyses.
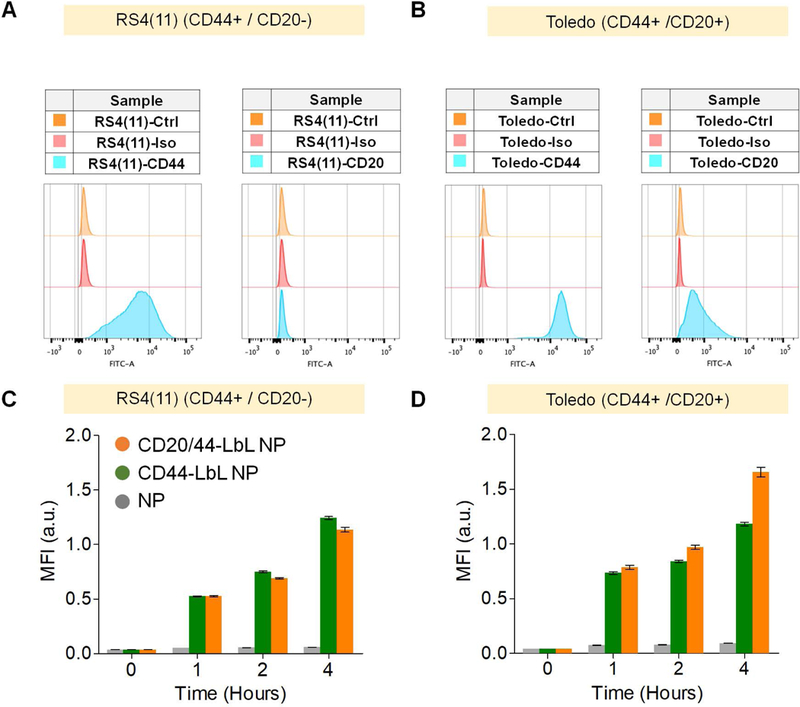
Flow cytometry analysis of (A) RS4(11) and (B) Toledo cells incubated with FITC-labeled anti-CD44 and anti-CD20 antibodies. Relative fluorescence intensities of (C) RS4(11) and (d) Toledo cells treated with fluorescently labeled PLGA NPs, CD44-targeted LbL-NPs and CD20/CD44 dual-targeted LbL-NPs in comparison with the control cells without treatment. At the indicated time point after treatment of NPs, 10,000 cells were collected, and fluorescence intensities were quantified by flow cytometry. Mean fluorescence intensities of the cells were used in this experiment.
We then studied cellular uptake of dye-labeled LbL-NPs including (i) PLGA-NPs, (ii) CD44-targeted LbL-NPs layered with HA (CD44-LbL-NPs), and (iii) CD20/CD44-targeted LbL-NPs (CD20/44-LbL-NPs) covered with HA-CD20-Ab conjugate into the RS4(11) and Toledo cells via flow cytometry. The flow cytometry results demonstrated that the cellular uptake was time-dependent; fluorescence intensities from LbL-NPs and PLGA-NPs increased with time. In comparison with PLGA-NPs, CD44-LbL-NPs and CD20/44-LbL-NPs showed 21-fold and 13-fold stronger fluorescence intensities in RS4(11) and Toledo cells, respectively, which can be attributed to receptor-mediated strong interaction between the LbL-NPs and the cells (Figure 3c,d). Of note, cellular uptake of dual-targeted CD20/44-LbL-NPs by RS4(11) cells (CD44+ / CD20-) was not significantly higher than that of single-targeted CD44-LbL-NPs. The dual-targeted CD20/44-LbL-NPs, however, demonstrated 1.4-fold greater targeting than the single-targeted CD44-LbL-NPs to Toledo cells (CD44+ / CD20+), suggesting that the CD20/CD44-dual-targeting approach is an effective way to target lymphoma cells that upregulate both CD44 and CD20 receptor proteins.
2.4. Carrier Toxicity and Regulation of BCL-2 Gene Expression In Vitro
After the cellular uptake experiments, we investigated cytotoxicity of CD20/44-LbL-NPs in blood cancer cells in comparison with that of the in vitro transfection agent Lipo. We treated Toledo and RS4(11) cells with siBCL2 or negative control siRNA cocktails (siNC) using Lipo. Notably, we found that over 40% of cells were dead after incubation with 8 pmol siRNA (siBCL2 or siNC) formulated with Lipo; over 60% were dead after treatment of 200 nM siRNA, showing high toxicity of the transfection agent Lipo (Supporting Figure S4a,d). Moreover, incubation of the cells with 200 nM siBCL2 formulated with Lipo did not induce down-regulation of BCL-2 gene expression (Supporting Figure S4c,f). The intrinsic toxicity of Lipo also hindered us from increasing the dose of siBCL2 over 200 nM. On the other hand, treatment of CD20/44-LbL-NPs did not induce significant toxicity to RS4(11) and Toledo cells at a dose of 200 nM siBCL2 or siNC (Supporting Figure S4b,e), and it could slightly suppress BCL-2 expression levels of the cells (Supporting Figure S4c,f).
More importantly, the non-toxic, biocompatible properties of LbL-NPs allowed us to further increase the dose of siRNA up to 2.6 μM—13 times higher than the maximum dose of siRNA formulated with Lipo. Even at the dose of 2.6 μM siNC, siNC-LbL-NPs did not show any significant toxicity, and also did not suppress BCL-2 gene expression. However, treatment of CD44-siBCL2-LbL-NPs markedly suppressed BCL-2 expression level (Figure 4a,d). After treatment of 2.6 μM siBCL2 formulated with CD20/44-LbL-NPs, the BCL-2 expression level dropped dramatically down to 19.34 ± 2.69% and 10.02 ± 0.23% in RS4(11) and Toledo cells, respectively, leading to cell death. Dual-targeted LbL-NPs successfully deliver siBCL2 to blood cancer cells, causing apoptosis of 73.18 ± 2.43% of RS4(11) cells and 74.03 ± 0.23% of Toledo cells at an siRNA concentration of 2.6 μM (Figure 4b,e). The CD20/44-siBCL2-LbL-NPs showed slightly greater BCL-2 gene silencing effects than CD44-siBCL2-LbL-NPs at doses of 0.7 μM and 1.3 μM siBCL2 in Toledo cells (CD44+ / CD20+). As expected, CD20/44-siBCL2-LbL-NPs did not deliver better BCL-2 silencing in RS4(11) cells (CD44+ / CD20-). Despite the minor improvement of CD20/44-siBCL2-LbL-NPs in BCL-2 suppression activities in Toledo cells, there was no added advantage in cell killing effects, which might be expected given that both the CD44-LbL-NPs and CD20/44-LbL-NPs have sufficient time to interact with cells while in culture.
Figure 4. In vitro gene silencing effect of siBCL2 LbL-NPs.
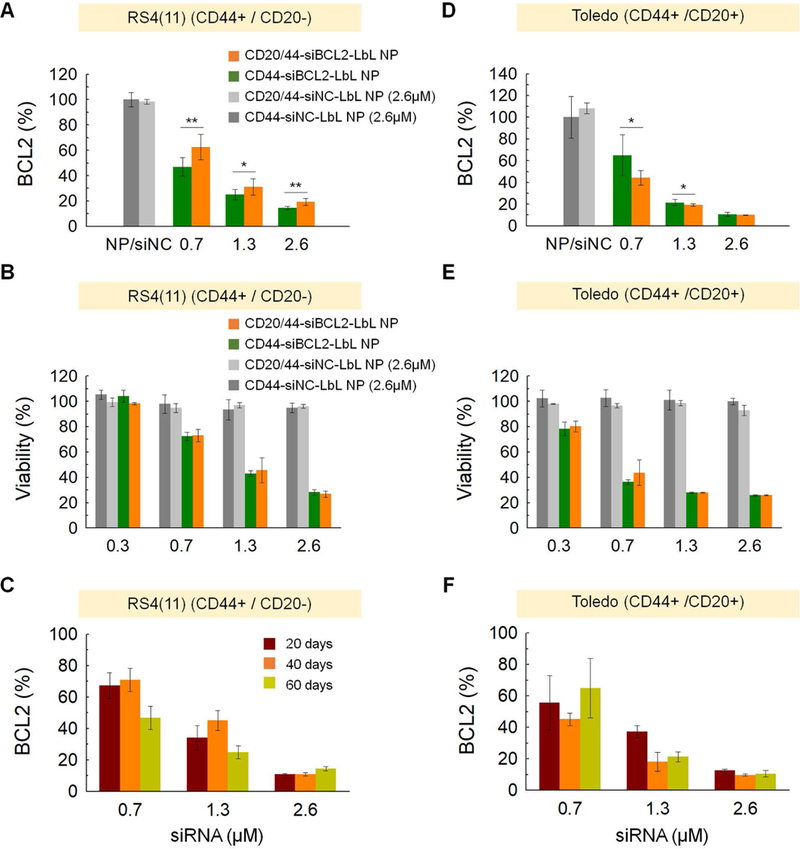
BCL-2 expression and cell viability of (A,B) RS4(11) cells or (D,E) Toledo cells after treatment with negative control siRNA cocktails or BCL-2 target siRNA cocktails formulated with CD44-targeted LbL-NPs or CD20/CD44-dual-targeted LbL-NPs for 72 h. Biological stability of BCL-2 target siRNA cocktails formulated with CD20/CD44-dual-targeted LbL-NPs, which were administered to (C) RS4(11) cells and (F) Toledo cells after storage in distilled water at 4 ˚C for 20, 40 or 60 days. Error represents SD of 3 of biological replicates. P-values were determined by student’s t-test: ***, p ≤ 0.05; **, p ≤ 0.1; *, p ≤ 0.5, ns, p > 0.5.
We further tested the physicochemical and functional stability of CD20/44-BCL2-LbL-NPs because the shelf life of RNA formulations is an important characteristic for clinical translation. To investigate the stability of the LbL-NP formulations, we kept CD20/44-BCL2-LbL-NPs in aqueous solution at 4 °C, and characterized their physicochemical properties. The NP formulations remained stable for up to 60 days, showing consistent hydrodynamic diameter, zeta potential and PDI values over time (Table2). Of note, CD20/44-BCL2-LbL-NPs were also fully functioning and could suppress BCL-2 gene expression after storage in a 4 °C refrigerator for up to 60 days (Figure 4c,f).
Table 2.
Physicochemical Stability of PLGA NPs and LbL-NPs
| Sample | Diameter (Z-Avg)a |
Diameter (N-Avg)b |
Zeta potential | PDI |
|---|---|---|---|---|
| LbL-NP (4 layers+Ab) | 135 ± 1.6 | 92 ± 4.3 | −39 ± 2.1 | 0.13 ± 0.010 |
| LbL−NP (4+Ab) 20 days | 129 ± 0.62 | 94 ± 2.4 | −45 ± 0.66 | 0.15 ± 0.0090 |
| LbL−NP (4+Ab) 40 days | 129 ± 4.8 | 90 ± 4.4 | −42 ± 1.1 | 0.10 ± 0.015 |
| LbL−NP (4+Ab) 60 days | 130 ± 2.8 | 92 ± 5.2 | −36 ± 0.87 | 0.096 ± 0.013 |
Z-average is an intensity-based calculated value.
Number average diameter is a number-based calculated value.
2.5. In Vivo Pharmacokinetics and Biodistribution Study
We also studied in vivo pharmacokinetics (PK) and tissue-distributions of siRNAs loaded in LbL-NPs. First, to investigate the PK, we employed Alexa-Fluor 647-labeled negative control siRNA molecules as a negatively-charged polyelectrolyte layer to prepare siNC embedded LbL-NPs. We intravenously injected the CD20/44- Alexa-Fluor 647/siNC-LbL-NPs into healthy balb/C mice and collected blood at pre-determined time points. We then measured the concentration of siRNA in blood samples and calculated the half-lives of Alexa-Fluor 647-siNC molecules by a two-compartment model fitting.
The PK results indicated a fast half-life of 0.2505 h and a slow half-life of 65.86 h (Figure 5a). The half-lives of siRNAs formulated in LbL-NPs were found to be significantly longer than that of unmodified free siRNA (~2 min) reported in a previous study[16]. After collecting blood samples, we sacrificed the mice and harvested major organs, from which fluorescence intensities were examined under an in vivo fluorescence imaging instrument. The fluorescence images show relative tissue distribution of the siRNA, indicating that siRNA molecules mainly accumulated in liver, kidney and heart tissues (Figure 5b,c). Liver uptake can be attributed to the mononuclear phagocyte system to filter out and destroy foreign particles, and liver-accumulation is typical for most systemically-delivered nanoparticles. Accumulation in heart and kidney tissues can be attributed to blood-circulation and renal clearance of siRNAs released from nanoparticles in the bloodstream.
Figure 5. In vivo pharmacokinetics and tissue distribution.
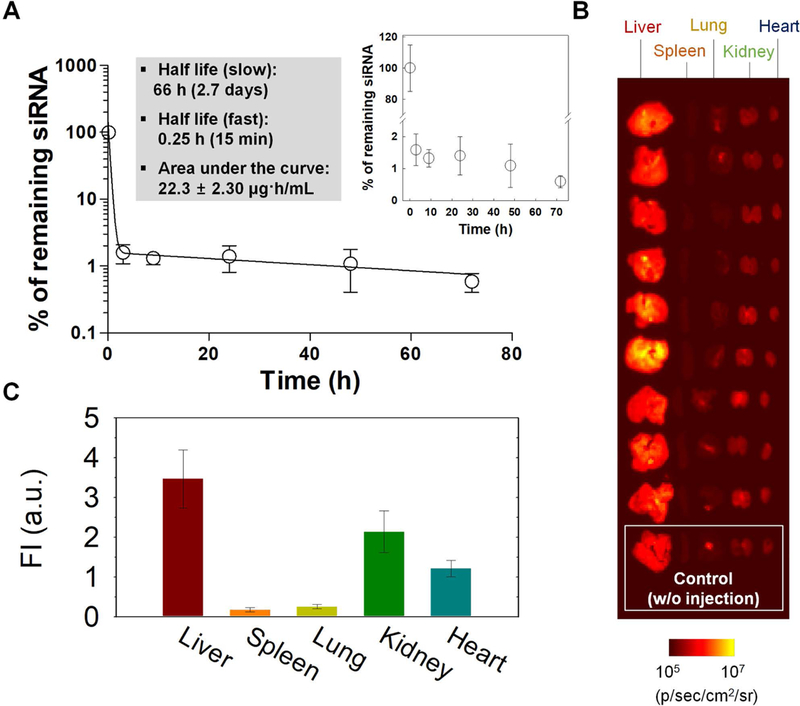
(A) Blood half-life and (B,C) tissue distribution of Alexa-647-labeled negative control siRNA formulated with CD20/CD44-dual-targeted LbL-NPs after systemic administration of the NPs into healthy BALB/c mice at 1 mg/kg of siRNA. The organs were collected at 72 h after injection.
2.6. BCL-2 Targeted Blood Cancer Therapy In Vivo
Given that blood cancer cells are circulating in the bloodstream and randomly accumulate in diverse tissues and lymph nodes, targeted delivery of therapeutic agents into lymphoma cells is essential for treatment of blood cancer cells; however, it remains highly challenging to date[2]. In an effort to tackle the issue, we formulated siBCL2 in the CD20/CD44-dual-targeted LbL-NPs for precision delivery of siBCL2 into circulating and localized blood cancer cells in vivo. For real-time monitoring of blood cancer cells in live animals, Toledo cells were engineered to express firefly luciferase. The cells were injected into the tail vein of the SCID-beige mice for preparation of orthotopic NHL models, which were then intravenously administrated with CD44-siBCL2-LbL-NPs, CD20/44-siBCL2-LbL-NPs or saline twice a week for 5 weeks from a week inoculation of Toledo cells. The injection schedule was determined based on the pharmacokinetic properties of the LbL-NPs. In vivo fate of the cells was monitored at serial time points using an in vivo luminescence imaging instrument.
Overall, luminescence signals for the cells evenly spread over the mice, implying blood-circulation of the cancer cells. The luminescence intensities of all 3 groups increased with time, indicating proliferation of the blood cancer cells in vivo (Figure 6a). Among the three groups, the control group injected with saline showed the most rapid increase in cell proliferation, whereas the cancer cells treated with CD20/44-siBCL2-LbL-NPs proliferated more sluggishly than those of the other groups. Proliferation of the cells treated with CD20/44-siBCL2-LbL-NPs was significantly hindered, with 9-fold and 2-fold lower luminescence intensities respectively than those administrated with saline, and also 1.7-fold and 1.3-fold lower than that of CD44-siBCL2-LbL-NP group at 10 days and 20 days after administration of NPs, respectively (Figure 6b). Notably, the control group without NP treatment and the vehicle control group treated with CD20/44-siNC-LbL-NPs were euthanized at 23 days due to their severe tumor burdens according to ethical guidelines placed by the Division of Comparative Medicine (DCM) at Massachusetts Institute of Technology. In contrast, 100% of the dual-targeted CD20/44-siBCL2-LbL-NP group survived for the entire test period of 46 days, and 80% survival was observed for single-targeted CD44-siBCL2-LbL-NPs group, indicating an improved drug delivery effect thanks to the added CD20 targeting. The results also indicate that the CD20-NPs do not exert antitumor effects such as those observed with Rituximab. This is likely due to the inaccessibility of the antibody Fc domains, which are covalently attached to the nanoparticle surface and therefore unable to bind to Fcy receptors on innate immune cells. Lacking this interaction the CD20 NPs are unlikely to illicit antibody-mediated cell-dependent cytotoxicity (ADCC) or complement mediated cytotoxicity (CMC), two important mechanisms of action for therapeutic CD20 mAbs.[17] Overall these results reveal considerable anticancer effect with minimal toxicity of CD20/44-siBCL2-LbL-NPs.
Figure 6. In vivo anticancer effect of siBCL2 LbL-NPs.
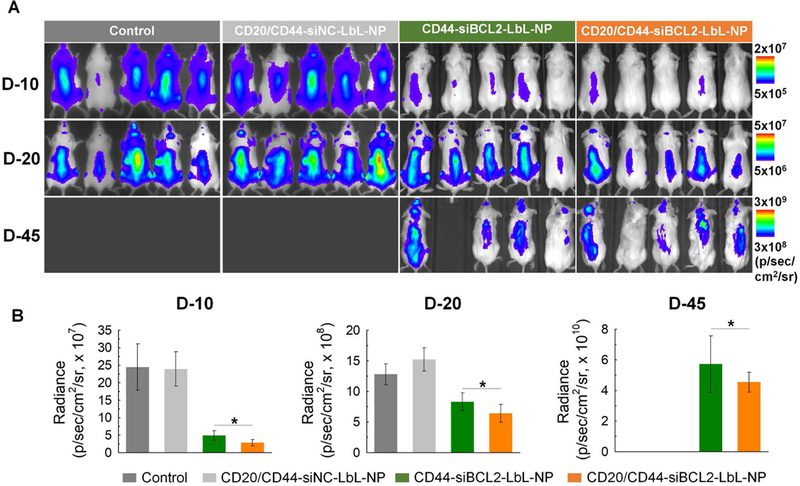
Saline, CD44-targeted siBCL2 LbL-NPs or CD20/CD44-dual-targeted siBCL2 LbL-NPs were intravenously injected into SCID beige mice intravenously inoculated with firefly luciferase expressing Toledo cells. The saline and NPs were given twice in a week for 5 weeks from a week after inoculation of firefly luciferase expressing Toledo cells into the mice. (A) Luminescence images of Toledo cells in SCID beige mice. (B) Quantitative analyses of luminescence intensities from Toledo cells. Error represents SD of 5 biological replicates. P-values were determined by student’s t-test: ***, p ≤ 0.05; **, p ≤ 0.1; *, p ≤ 0.5.
2.7. Tissue Analyses and Blood Chemistry Tests
After monitoring proliferation of cancer cells in vivo for 45 days, we excised tumors and major organs from the mice. The tumors were collected from metastatic solid tumors found throughout the body as shown in luminescence signals that indicate tumor locations (Fig. 6a). In particular, the strongest luminescence signals were detected from the spine, and a substantial portion of tumors were found at the spine, implying that the primary site of metastasis may be the bone marrow. We sectioned the tumor tissues and stained BCL-2 proteins in the tumor sections by an immunohistochemistry (IHC) staining method. IHC results revealed that treatment of LbL-NPs significantly downregulated the BCL-2 level in tumor tissues. BCL-2 expression levels in tumor tissues of CD44-siBCL2-LbL-NP and CD20/44-siBCL2-LbL-NP groups were 3- and 3.7-fold lower than those of the control group, respectively, verifying effective transfection of siBCL2 and successful BCL-2 downregulation in the blood cancer cells by treatment of LbL-NPs (Figure 7a,b and Supporting Figure S5).
Figure 7. Tissue analyses.
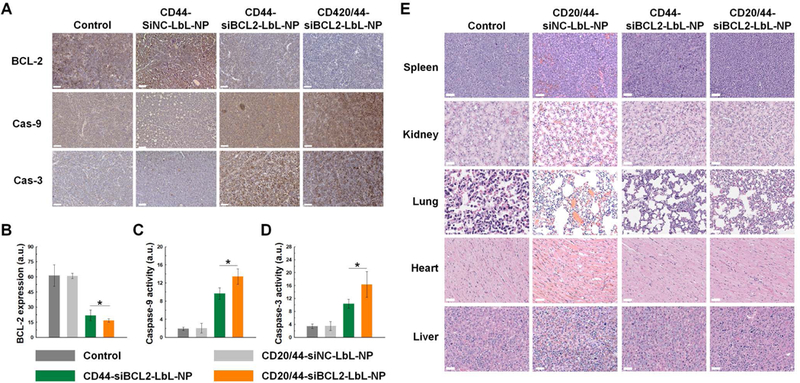
(A) Immunohistochemistry images of tumor sections collected from SCID beige mice intravenously injected with Toledo cells and LbL-NPs (scale bar = 50 μm). (B–D) Quantitative analyses of tissue images. (E) H&E staining images of tissue sections from major organs (scale bar = 50 μm). Error represents SD of 3 technical replicates. P-values were determined by student’s t-test: ***, p ≤ 0.05; **, p ≤ 0.1; *, p ≤ 0.5.
Furthermore, to monitor apoptosis progression in tumor tissues, we stained caspase-9 and -3, which are representative proteases closely related to apoptosis, on the tumor sections by the IHC staining method. The IHC results demonstrated that caspase-9 and -3 were highly upregulated in tumors of CD44-siBCL2-LbL-NP and CD20/44-siBCL2-LbL-NP groups in comparison with those in tumors of control group. The IHC results were quantified and found to indicate that the cleaved caspase-9 levels in tumor tissues of CD44-siBCL2-LbL-NP and CD20/44-siBCL2-LbL-NP groups were 6.1-fold and 7.4-fold higher than in tumors of the control group (Figure 7a,c and Supporting Figure S5). Likewise, caspase-3 levels in tumors of CD44-siBCL2-LbL-NP and CD20/44-siBCL2-LbL-NP groups were 3-fold and 4.4-fold greater than in control groups (Figure 7a,d and Supporting Figure S5). Expression of caspase-9 and -3 were inversely proportional to BCL-2 expression levels, which implies that downregulation of BCL-2 in blood cancer cells can lead to the activation of caspase-9 and -3, thereby inducing apoptosis in the cells.
To assess off-target toxicity, gross histopathology was carried out on the spleen, kidney, heart, lungs and liver. Noticeable histological changes were not found in the major organs, implying biocompatibility of the siRNA formulations (Figure 7e). Besides the tissue analyses, we also evaluated toxicity of LbL-NPs by blood chemistry tests. The test results were in normal ranges, providing no evidence of significant toxicity (Supporting Table S7).
3. Conclusion
LbL nanoparticles serve as a simple precision tool for the delivery of an siRNA therapeutic that acts to downregulate BCL-2 in hematologic malignancies. The LbL approach utilizes an outer layer modified to enable CD20/CD44-dual-targeting with an antibody conjugated hyaluronic acid adsorbed layer. The dual-targeting strategy demonstrates the successful use of antibodies in conjunction with a receptor-binding polysaccharide to target different receptor types and successfully improve siRNA delivery to typically difficult to transfect blood cancer cells. The resulting nanoparticle led to considerable downregulation of BCL-2 expression, and induced apoptosis of blood cancer cells in culture and in animal models. The LbL-NP did not exhibit toxicity in animals at the levels tested, and possessed a significant therapeutic window. While the CD44-targeted version of this nanoparticle improved survival in orthotopic lymphoma mouse models, the dual CD20/CD44 formulation provided a more extensive survival benefit. This targeted RNA delivery technology based on LbL-NPs can provide a versatile means to downregulate any target gene or combination of target genes for treatment of hematologic cancer. This approach opens the possibility to treat hematologic malignancies by silencing genes of interest and downregulating proteins that are otherwise difficult to target in clinic.
4. Experimental Section
Cell culture and cell line establishment.
Toledo Non-Hodgkin’s lymphoma and RS4 (11) leukemia cells (ATCC) were cultured in high glucose RPMI-1640 medium supplemented with 10% v/v fetal bovine serum and 1% penicillin/streptomycin. MCF7 breast cancer cells were grown in RPMI-1640 medium supplemented with 10% v/v fetal bovine serum and 1% penicillin/streptomycin, and THP-human monocytic cells were cultured in DMEM medium supplemented with 10% v/v fetal bovine serum and 1% penicillin/streptomycin. Stable GFP- and firefly luciferase-overexpressing Toledo and RS4 (11) were established using lentivirus vectors (Abm) according to the manufacturer’s protocols, and sorted by fluorescence-activated cell sorting in the Flow Cytometry Facility of the Swanson Biotechnology Center (MIT). All cell lines were cultured at 37°C in humidified incubator with 5% CO2. For cell proliferation, cells were seeded at a density of 2.1 × 106 cells in T-75 tissue culture flasks or 2 × 104 cells in 96-well plates. Cells were collected directly from the flask and cell number was determined using Nexcelom Bioscience Cellometer. The dead and viable cells were also determined through Trypan Blue Solution 0.4% (Sigma-Aldrich) in which dead cells take up the blue coloring agent while viable cells remained unstained.
In vivo experiments.
All in vivo experiments were conducted under the supervision of the Division of Comparative Medicine (DCM), Massachusetts Institute of Technology, and in compliance with the Principles of Laboratory Animal Care of the National Institutes of Health. Cell lines were purchased from ATCC and were tested routinely for pathogens before any in vivo experimentation was carried out.
In vitro transfection of blood cancer cells.
We tested transfection of siRNA cocktails into Toledo, RS4 (11) and MCF7 cells. The cells were cultured in 24 well plates and transfected with 1 nM (MCF7) or 100nM (Toledo and RS4(11)) of GAPDH targeting siRNA cocktails formulated with Lipofectamine RNAiMax (Lipo, Thermo Scientific) or Hiperfect (Hiper, Qiagen) according to the manufacturer’s instructions. Transfection of BCL-2 targeting siRNA cocktails into THP cells were also tested using Lipo according to the manufacturer’s instructions. The cells harvested at 24h after transfection, and qPCR analyses were performed. We also investigated on-target and off-target effects of BCL-2 siRNA cocktails compared with single BCL-2 siRNAs. First, we transfected A549 cells with BCL-2 target single siRNAs (siRNA1 and siRNA2), the BCL-2 target siRNA cocktail (cocktail) and the negative control siRNA cocktail (siNC) at 3nM total siRNA concentration using Lipo according to the manufacturer’s instructions. The cells harvested at 24h after transfection, and qPCR analyses were performed to measure BCL2 expression.
For evaluation of off-target effects, we ran an off-target detection software TargetScan (http://www.targetscan.org/vert_72), which is one of the most reliable online tools to predict miRNA-targets and off-targets. Afterwards, we transfected A549 cells with siRNA1, siRNA2, cocktail and siNC at 3nM total siRNA concentration using Lipo, and the expression of multiple off- target candidate genes was measured by RT-PCR. Total RNA were isolated from the cells using a Nucleo Spin 8 RNA (Machery Nagel) and cDNA was synthesized using iScript cDNA Synthesis Kit (BioRad) according to manufacturer’s instructions; qPCR was performed using ssoFast Eva Green Supermix (BioRad) along with selected DNA primer pairs. RNA was isolated 24h after transfection. GAPDH mRNA levels were normalized to the levels of RPL13A.
Synthesis of siRNA-LbL-NP.
Poly-(D,L-lactide-co-glycolide) (PLGA) nanoparticles (NPs) were prepared as we previously described. Briefly, 25 mg of PLGA (acid terminated, lactide:glycolide 50:50, Mw 24,000–38,000, Sigma-Aldrich) in 1.5 mL of sterile acetone. Next, the PLGA solution was added to 1% BSA containing 10mM HEPES buffer (pH 7.4) in a drop-wise fashion during a 2–3 minutes under sonication. After then, the solution covered with a perforated cap was stirred continuously overnight to evaporate the organic solvent. The following day, the solution was diluted in Millipore water to make an 1mg/mL solution and purified using the tangential flow filtration (TFF) to remove excess amount of free polymers with a 115cm2 500kD column.[18] To assemble the siRNA-LNF, equal volume of 0.5mg/mL PLGA nanoparticles and 1mg/mL poly-l-arginine (PLA) solution were mixed through a quick bath sonication process in a glass vial. The mixture was then purified with TFF three to five times and then concentrated. This will result in a solution that has a concentration of 0.4mg/mL of PLGA particles. The PLGA-PLA NP solution was then diluted to a concentration of 0.1mg/mL, in which 0.5mg/mL (73.5μM) of siRNA (siBCL2 or siNC (negative control)) was quickly added on a sonication bath. The sample was then purified and washed three times using TFF and concentrated. At this point, the concentration of PLGA is now 0.05mg/mL. To assemble the next layer, equal volume of 0.5mg/mL of PLA solution and PLGA/PLA/siRNA NPs was mixed on a sonication bath for a brief period. The resulting mixture was purified and concentrated as previously mentioned. Finally, 0.5 mg/mL of hyaluronic acid (HA, Mw 40,000, Lifecore) or HA-methyltetrazine conjugate was mixed with the purified PLGA/PLA/siRNA/PLA-NPs briefly on a sonication bath. This last layer was purified and concentrated using a 115cm2 100kD column. HA-methyltetrazine conjugate was prepared by conjugation of methyltetrazine-amine (TET, Click Chemistry Tools) onto the HA backbone via amide formation in the presence of N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC, Sigma-Aldrich) and N-Hydroxysulfosuccinimide sodium salt (Sulfo-NHS, Sigma-Aldrich). In brief, 60 mg of HA (1.50 μmol) was dissolved in 12 mL of phosphate buffer (PB, 100 mM, pH 6.8). 5.6 mg of TET (23.73 μmol) dissolved in 1 mL of DMSO was added to the HA solution. 9 mg of EDC (47.46 μmol) and 11 mg Sulfo-NHS (47.46 μmol) dissolved in 1 mL of PB (100 mM, pH 6.8) were added to the solution, and stirred at room temperature for 24 h. Unreacted chemical residues were removed by dialysis (MWCO = 12,000–14,000 Da) against distilled water for 3 days, followed by lyophilization to obtain HA-TET.
Synthesis of the CD20/44-siRNA-LbL-NP.
To prepare CD20/CD44 dual-targeted LbL-NPs, anti-CD20 antibody (CD20-Ab, clone 2H7, cat # SAB4700114, Sigma-Aldrich) was conjugated on to the HA-TET layered LbL-NPs via click chemistry. First, CD20-Ab was modified with TCO-NHS ester or ThioLinker-TCO (Click Chemistry Tools) according to the manufacturer’s protocols for modification of lysine of sulfhydryl groups on CD20-Ab, respectively. 30 μg of TCO-conjugated CD20-Ab was added to 5 mL LbL-NP solution (2.5 mg of PLGA-NP), and the solution was stirred for 4–6 h. Unreacted CD20-Ab was removed using the tangential flow filtration (TFF) with a 500kD column.
Characterization and Quantification of the nanoparticles.
To ensure the addition of each polymer layer, the size, zeta potential, and polydispersity index were measured using a Malvern Zetasizer Nano ZS90 particle analyzer (λ = 633 nm, material/dispersant RI 1.590/1.330) after each addition to the nanoparticle. Quantification of the amount of siRNA loaded into the LbL-NP was done using Quant-iT™ RiboGreen® RNA Assay Kit (ThermoFisher Scientific) in which ultrasensitive fluorescent nucleic acid staining is used to quantify the RNA content of a solution. The siRNA-LbL-NPs were treated with heparin in a 1:1 volume ratio for 1 h at 37°C on a sonication bath. After the incubation, serial dilutions of the sample were performed in 1X TE buffer (diluted from the kit’s 20X TE buffer) to a final volume of 100uL on a 96 well plate. Preparation of the dye was followed by kit’s protocol. To each sample, 100uL of the prepared dye was added and incubated at RT for 2–3 minutes protected from light. Finally, the fluorescent was measured using the Tecan Infinite® 200 Pro Plate Reader at an excitation of 480nm and emission of 520nm. To quantify the concentration or RNA in the solution, a standard curve was generated provided by the kit and was used to determine the concentration. To quantify the conjugation efficiency of CD20-Abs onto LbL-NPs, we labeled CD20-Abs (CD20-TCO-NHS or CD20-TCO-ThioLinker) using an Antibody Labeling Kit (ThermoFisher Scientific) according to the manufacturer’s protocols. The conjugation efficiency was calculated based on the fluorescence intensities of CD20-Abs.
Cytotoxicity Assay.
Cell viability in response to siRNA-LbL-NP treatment was evaluated using PrestoBlue® (Thermofisher Scientific), a cell permeable resazurin-based solution. Upon addition of PrestoBlue®, the solution is modified by the reducing environment of the viable cells in which these cells turn red and become highly fluorescent. The fluorescent values were measured with excitation of 560nm and emission of 590nm using Tecan Infinite® 200 Pro Plate Reader. Cells were seeded in 96-well plates at a density of 2 × 104 cells in 100μL of medium. The cells were then transfected with siRNA –LbL-NP at various concentration. After 3 days of incubation at 37°C, cells were treated with PrestoBlue® and fluorescent was measured at 10min, 20min, 30min, 60min and 90min by a plate reader. Cell viabilities were normalized to the untreated and calculated using a standard curve.
In vitro cellular uptake of LbL-NP.
To evaluate cellular uptake of LbL-NPs, we prepared PLGA-NPs, PLGA/PLA/HA-NPs and PLGA/PLA/HA-CD20-NPs, in which green fluorescent PLGA-NPs (Sigma-Aldrich) were used as a NP core. The NPs were incubated with 5 × 104 cells in 96-well plates for indicated time points (1, 2 or 4 h) at room temperature. The cells were washed three times with 3% FBS containing PBS by repeated centrifugation and resuspension steps, and finally resuspended in PBS for flow cytometry analyses by the Flow Cytometry Facility of the Swanson Biotechnology Center (MIT).
In vitro transfection efficacy siBCL2-LbL-NP.
BCL-2 mRNA expression was quantified using the QuantiGene 2.0 Reagent System (Affymetrix). Initially, Toledo and RS4(11) cells were treated with nanoparticles at various concentrations. After 72 h post-transfection, cells were lysed with 50uL of Lysis Mixture (including Proteinase K) and incubated at 50–55°C for 30 minutes. Ten-fold dilution of the cell lysates were immediately used for QuantiGene 2.0 Assay (Affymetrix) to observe the expression of BCL-2 (Panomics, Inc.) compared to the expression of TBP (Panomics, Inc.) as a control. These particular probes designed by Panomic, Inc. include oligonucleotides with three different synthetic probes- Capture Extenders, Label Extenders, and Blockers- that are conjugated to the wells of the Capture Plate (Affymetrix). Using the QuantiGene assay, a signal amplification was then performed, followed by hybridization to the ends of the oligonucleotides mediated by DNA molecules. This hybridization is then detected by a chemilumigenic 2.0 Substrate that produces a luminescent signal, which quantifies the target of interest. Target RNA was captured during an overnight incubation at 55°C. After an overnight incubation, unbound materials were removed after three washes. The samples were then subsequently treated with RNA amplifier molecules, which includes pre-amplifier hybridization, amplifier hybridization and label probe hybridization. Each hybridization step was followed by three washes to remove unbounded materials. Finally, the samples were exposed to a chemilumigenic 2.0 Substrate and luminescent was measured at an exposure time of 0.2 seconds using a plate reader.
In vivo pharmacokinetics and biodistribution.
To evaluate pharmacokinetics of LbL-NPs, we intravenously injected Alexa fluoro 647-lableled negative control siRNA loaded in CD20/44-LbL-NPs into BALB/c female mice (Taconic) at a siRNA dose of 1 mg/kg. Blood samples were collected at the indicated time points using a retro orbital blood collection method, and fluorescence intensities of Alexa fluoro 647-lableled siRNA were examined to determine serum half-life of siRNA molecules embedded in the LbL-NPs (n = 3 per each time point). Fast and slow half-lives was calculated using a two-compartment model fit.
Tumor Growth and treatment.
To mimic the spread of lymphoma cancer, 5 × 106 firefly luciferase-expressing Toledo Non-Hodgkin’s Lymphoma cells were intravenously injected into the tail veins’ of SCID BEIGE immunocompromised mice (Taconic). Tumors were allowed to form a week after which treatment groups were randomized. siRNA-LbL-NP (2.4mg/kg of siRNA, 5% D-glucose) were intravenously injected twice a week for 5 weeks while control group was left untreated (Supporting Figure S6). Whole-body animal bioluminescent imaging was performed with a CCD camera using a Xenogen IVIS Imagining System (Caliper). Anesthetized mice were placed in the IVIS Imaging System and imaged after a 10–15min intraperitoneal injection of D-luciferin at 150mg/kg body weight. Tumor growth was monitored using bioluminescent imaging twice a week. Imaging and quantification of signals were controlled by the acquisition and analysis software Living Image (Xenogen).
Ex-vivo tumor mRNA and protein quantification of BCL-2.
After euthanasia, inoculated tumors were harvested from SCID BEIGE mice and frozen immediately on dry ice and stored in -80°C until needed. A portion of the tumor was used for western blot while another portion of the tumor was formalin-fixed overnight, paraffin-embedded, and processed immediately by the Histology Facility of the Swanson Biotechnology Center (MIT). Tissue homogenates were prepared according to procedure described by the manufactures (Panomics, Inc.), in which, 5mg of tumor tissues were pulverized under cold condition and combined with 300uL of homogenizing solution that contained 3uL of Proteinase K. Once vortexed to mix, this solution was incubated at 55°C for 30 minutes, given continuous vortex every 10 minutes. The homogenized sample was then separated from debris by centrifugation at 16,000-× g for 15 minutes. The supernatant was collected and transferred to a new tube. The centrifugation was repeated and the homogenate was used immediately in the QuantiGene 2.0 assay, as described above.
Protein expression of BCL-2 was studied using western blot. 5mg of frozen tumor tissue was quickly measured out and homogenized with 300μL of the RIPA buffer using an electric homogenizer. Immediately protease inhibitors were added to the sample and incubated for 2 h at 4°C under constant agitation. After incubation, the samples were centrifuged for 20 minutes at 12,000rpm at 4°C and the supernatant was collected. The sample was then immediately used for western blot as described previously.
To study both the induction of apoptosis through the treatment of the siBCL2-LbL-NP and BCL-2 expression in the tumor model, the paraffin-embedded tumors were sliced at 5μM and stained with specific antibodies for detecting the biomarkers of interest. Anti-Cleaved Caspase 3 (Cell Signaling Technology, 1:200), anti-Caspase 9 p10 (F-7) (Santa Cruz, 1:200) and anti-BCL-2 human specific (Cell Signaling Technology, 1:200) were used. Positive staining was quantified. Vital organs were collected, formalin-fixed overnight, paraffin-embedded and processed for hematoxylin and eosin (H&E) by the Histology Facility of the Swanson Biotechnology Center (MIT).
Statistical Analysis.
The statistical significance of differences among the groups tested was determined with Sigma Plot using a Student’s t-test. A p-value less than 0.05 was considered significant and is specified in the figures with an asterisk. ***, p ≤ 0.05; **, p ≤ 0.1; *, p ≤ 0.5, ns, p > 0.5.
Supplementary Material
Acknowledgements
The authors thank the Swanson Biotechnology Center Core Facilities and the David H. Koch Institute, and particularly thank Drs. D.S. Yun and M.R. Griffin for their technical support. This research was supported by the Janssen Incubator Program through Janssen Research and Development, LLC, National Institutes of Health (ECD, NIBIB 1F32EB017614), a Basic Science Research Program (2015R1A6A3A04059033, 2018R1D1A1A02085552) through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Republic of Korea, and a Korea Institute of Science and Technology intramural research grant.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
References
- [1].Siegel RL, Miller KD, Jemal A, CA Cancer J. Clin. 2017, 67, 7. [DOI] [PubMed] [Google Scholar]
- [2].a) Anderson MA, Huang D, Roberts A, Semin. Hematol. 2014, 51, 219; [DOI] [PubMed] [Google Scholar]; b) Shankland KR, Armitage JO, Hancock BW, Lancet 2012, 380, 848; [DOI] [PubMed] [Google Scholar]; c) Davids MS, Letai A, J. Clin. Oncol. 2012, 30, 3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Souers AJ, Leverson JD, Boghaert ER, Ackler SL, Catron ND, Chen J, Dayton BD, Ding H, Enschede SH, Fairbrother WJ, Huang DC, Hymowitz SG, Jin S, Khaw SL, Kovar PJ, Lam LT, Lee J, Maecker HL, Marsh KC, Mason KD, Mitten MJ, Nimmer PM, Oleksijew A, Park CH, Park CM, Phillips DC, Roberts AW, Sampath D, Seymour JF, Smith ML, Sullivan GM, Tahir SK, Tse C, Wendt MD, Xiao Y, Xue JC, Zhang H, Humerickhouse RA, Rosenberg SH, Elmore SW, Nat. Med. 2013, 19, 202. [DOI] [PubMed] [Google Scholar]
- [4].a) Choi KY, Silvestre OF, Huang X, Hida N, Liu G, Ho DN, Lee S, Lee SW, Hong JI, Chen X, Nat. Protoc. 2014, 9, 1900; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Pecot CV, Calin GA, Coleman RL, Lopez-Berestein G, Sood AK, Nat. Rev. Cancer 2011, 11, 59; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Whitehead KA, Langer R, Anderson DG, Nat. Rev. Drug Discov. 2009, 8, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Reed JC, Semin. Hematol. 1997, 34, 9. [PubMed] [Google Scholar]
- [6].a) Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG, Nat. Rev. Genet. 2014, 15, 541; [DOI] [PubMed] [Google Scholar]; b) Kanasty R, Dorkin JR, Vegas A, Anderson D, Nat. Mater. 2013, 12, 967. [DOI] [PubMed] [Google Scholar]
- [7].a) Dreaden EC, Morton SW, Shopsowitz KE, Choi JH, Deng ZJ, Cho NJ, Hammond PT, ACS Nano 2014, 8, 8374; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Deng ZJ, Morton SW, Ben-Akiva E, Dreaden EC, Shopsowitz KE, Hammond PT, ACS Nano 2013, 7, 9571; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Gu L, Deng ZJ, Roy S, Hammond PT, Clin. Cancer Res. 2017, 23, 7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Jackson AL, Linsley PS, Nat. Rev. Drug Discov. 2010, 9, 57. [DOI] [PubMed] [Google Scholar]
- [9].Hannus M, Beitzinger M, Engelmann JC, Weickert MT, Spang R, Hannus S, Meister G, Nucleic Acids Res. 2014, 42, 8049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Choi KY, Saravanakumar G, Park JH, Park K, Colloids Surf. B Biointerfaces 2012, 99, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Smith MR, Oncogene 2003, 22, 7359. [DOI] [PubMed] [Google Scholar]
- [12].Sapra P, Allen TM, Cancer Res. 2002, 62, 7190. [PubMed] [Google Scholar]
- [13].a) Stauder R, Eisterer W, Thaler J, Gunthert U, Blood 1995, 85, 2885; [PubMed] [Google Scholar]; b) Ponta H, Sherman L, Herrlich PA, Nat. Rev. Mol. Cell. Biol. 2003, 4, 33; [DOI] [PubMed] [Google Scholar]; c) Toole BP, Slomiany MG, Semin. Cancer Biol. 2008, 18, 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hashimoto T, Kawazu T, Nagasaki T, Murakami A, Yamaoka T, Sci. Technol. Adv. Mater. 2012, 13, 015009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Morton SW, Poon Z, Hammond PT, Biomaterials 2013, 34, 5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W, Hartsough K, Machemer L, Radka S, Jadhav V, Vaish N, Zinnen S, Vargeese C, Bowman K, Shaffer CS, Jeffs LB, Judge A, MacLachlan I, Polisky B, Nat. Biotechnol. 2005, 23, 1002. [DOI] [PubMed] [Google Scholar]
- [17].Weiner GJ, Semin. Hematol. 2010, 47, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Correa S, Choi KY, Dreaden EC, Renggli K, Shi A, Gu L, Shopsowitz KE, Quadir MA, Ben-Akiva E, Hammond PT, Adv. Funct. Mater. 2016, 26, 991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


