Abstract
Functional neuroimaging techniques have transformed our ability to probe the neurobiological basis of behaviour and are increasingly being applied by the wider neuroscience community. However, concerns have recently been raised that the conclusions that are drawn from some human neuroimaging studies are either spurious or not generalizable. Problems such as low statistical power, flexibility in data analysis, software errors and a lack of direct replication apply to many fields, but perhaps particularly to functional MRI. Here, we discuss these problems, outline current and suggested best practices, and describe how we think the field should evolve to produce the most meaningful and reliable answers to neuroscientific questions.
Neuroimaging, particularly using functional MRI (fMRI), has become the primary tool of human neuroscience1, and recent advances in the acquisition and analysis of fMRI data have provided increasingly powerful means to dissect brain function. The most common form of fMRI (known as blood-oxygen-level-dependent (BOLD) fMRI) measures brain activity indirectly through localized changes in blood oxygenation that occur in relation to synaptic signalling2. These changes in signal provide the ability to map activation in relation to specific mental processes, to identify functionally connected networks from resting fMRI3, to characterize neural representational spaces4 and to decode or predict mental function from brain activity5,6. These advances promise to offer important insights into the workings of the human brain but also generate the potential for a ‘perfect storm’ of irreproducible results. In particular, the high dimensionality of fMRI data, the relatively low power of most fMRI studies and the great amount of flexibility in data analysis contribute to a potentially high degree of false-positive findings.
Recent years have seen intense interest in the reproducibility of scientific results and the degree to which some problematic, but common, research practices may be responsible for high rates of false findings in the scientific literature, particularly within psychology but also more generally7–9. There is growing interest in ‘meta-research’ (REF 10) and a corresponding growth in studies investigating factors that contribute to poor reproducibility. These factors include study design characteristics that may introduce bias, low statistical power and flexibility in data collection, analysis and reporting — termed ‘researcher degrees of freedom’ by Simmons et al.8. There is clearly concern that these issues may be undermining the value of science — in the United Kingdom, the Academy of Medical Sciences recently convened a joint meeting with several other funders to explore these issues, and the US National Institutes of Health has an ongoing initiative to improve research reproducibility11.
In this Analysis article, we outline a number of potentially problematic research practices in neuroimaging that can lead to increased risk of false or exaggerated results. For each problematic research practice, we propose a set of solutions. Although most of the proposed solutions are uncontroversial in principle, their implementation is often challenging for the research community, and best practices are not necessarily followed. Many of these solutions arise from the experience of other fields with similar problems (particularly those dealing with similarly large and complex data sets, such as genetics) (BOX 1). We note that, although our discussion here focuses on fMRI, many of the same issues are relevant for other types of neuroimaging, such as structural or diffusion MRI.
Box 1 |. Lessons from genetics.
The study of genetic influences on complex traits has been transformed by the advent of whole-genome methods and by the subsequent use of stringent statistical criteria, independent replication, large collaborative consortia and complete reporting of statistical results. Previously, ‘candidate’ genes would be selected on the basis of known or presumed biology, and a handful of variants genotyped (many of which would go unreported) and tested in small studies. An enormous literature proliferated, but these findings generally failed to replicate74. The transformation brought about by genome-wide association studies (GWAS) applied in very large populations was necessitated by the stringent statistical significance criteria required by simultaneous testing of several hundred thousand genetic loci and an emerging awareness that any effects of common genetic variants are generally very small (<1% phenotypic variance). To realize the very large sample sizes required, large-scale collaboration and data sharing were embraced by the genetics community. The resulting cultural shift has rapidly transformed our understanding of the genetic architecture of complex traits and, in a few years, has produced many hundreds more reproducible findings than in the previous 15 years75. Routine sharing of single-nucleotide polymorphism (SNP)-level statistical results has facilitated routine use of meta-analysis, as well as the development of novel methods of secondary analysis76.
This relatively rosy picture contrasts markedly with the situation in ‘imaging genetics’ — a burgeoning field that has yet to embrace the standards commonly followed in the broader genetics literature and that remains largely focused on individual candidate-gene association studies, which are characterized by numerous researcher degrees of freedom. To illustrate, we examined the first 50 abstracts matching a PubMed search for ‘fMRI’ and ‘genetics’ (excluding reviews, studies of genetic disorders and non-human studies) that included a genetic association analysis (for list of search results, see https://osf.io/spr9a/ ). Of these, the majority (43 out of 50) reported analysis of a single candidate gene or a small number (5 or fewer) of candidate genes; of the remaining 7, only 2 reported a genome-wide analysis, with the rest reporting analyses using biologically inspired gene sets (3) or polygenic risk scores (2). Recent empirical evidence also casts doubt on the validity of candidate-gene associations in imaging genomics. A large GWAS of whole-brain and hippocampal volumes77 identified two genetic associations that were replicated across two large samples that each contained more than 10,000 individuals. Strikingly, the analysis of a set of candidate genes that were previously reported in the literature showed no evidence for any association in this very well-powered study77. The more general lessons for imaging from GWAS seem clear: associations of common genetic variants with complex behavioural phenotypes are generally very small (<1% of phenotypic variance) and thus require large, homogeneous samples to be able to identify them robustly. As the prior odds for an association between any given genetic variant and a novel imaging phenotype are generally low, and given the large number of variants that are simultaneously tested in a GWAS (necessitating a corrected P-value threshold of ~10−8), adequate statistical power can only be achieved by using sample sizes in the many thousands to tens of thousands. Finally, results need to be replicated to ensure robust discoveries.
Low statistical power
The analyses of Button et al.12 provided a wake-up call regarding statistical power in neuroscience, particularly by highlighting the point (that was raised earlier by Ioannidis7) that low power not only reduces the likelihood of finding a true result if it exists but also raises the likelihood that any positive result is false, as well as causing substantial inflation of observed positive effect sizes13. In the context of neuroimaging, Button et al. considered only structural MRI studies. To assess the current state of statistical power in fMRI studies, we performed an analysis of sample sizes and the resulting statistical power of fMRI studies over the past 20 years.
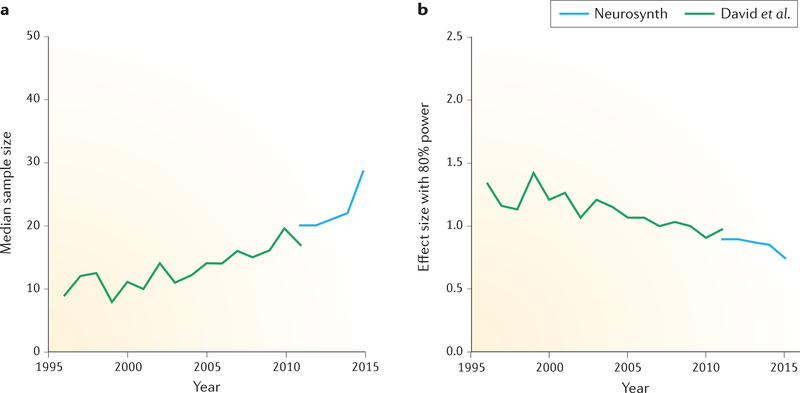
To gain a perspective on how sample sizes have changed during this time period, we obtained sample sizes from fMRI studies using two sources. First, manually annotated sample-size data for 583 studies were obtained from published meta-analyses14. Second, sample sizes were automatically extracted from the Neurosynth database15 for 548 studies that were published between 2011 and 2015 (by searching for regular expressions reflecting sample size, such as ‘13 subjects’ and ‘n = 24’) and then manually annotated to confirm these automatic estimates and to distinguish single-group from multiple-group studies. The data and code that were used to generate all the figures in this paper are available through the Open Science Framework at https://osf.io/spr9a/. FIGURE 1a shows that sample sizes have steadily increased over the past two decades, with the median estimated sample size for a single-group fMRI study in 2015 at 28.5. A particularly encouraging finding from this analysis is that the number of studies with large samples (greater than 100) is rapidly increasing (from 8 in 2012 to 17 in 2015, in the studied sample), suggesting that the field is progressing towards adequately powered research. However, the median group size in 2015 for fMRI studies with multiple groups was 19 subjects, which is below even the absolute minimum sample size of “20 observations per cell” that was proposed by Simonsohn and colleagues8.
Figure 1 |. Sample-size estimates and estimated power for functional MRI studies.
a | A summary of 1,131 sample sizes over more than 20 years, obtained from two sources, is shown: 583 sample sizes were obtained by manual extraction from published meta-analyses by David et al.14, and 548 sample sizes were obtained by automated extraction from the Neurosynth database15 with manual verification (for a version with all data points depicted, see Supplementary information S1 (figure)). These data demonstrate that sample sizes have steadily increased over the past two decades, with a median estimated sample size of 28.5 as of 2015. b | Using each of the sample sizes from the left panel, we estimated the standardized effect sizes that would have been required to detect an effect with 80% power for a whole-brain linear mixed-effects analysis using a voxelwise 5% familywise error rate threshold from random field theory16 (for details, see the main text). The median effect size that the studies in 2015 were powered to find was 0.75. Data and code to generate these figures are available at https://osf.io/spr9a/.
To assess the implications of these results for statistical power, for each of the 1,131 sample sizes shown in FIG. 1a, we estimated the standardized effect size that would be required to detect an effect with 80% power (the standard level of power for most fields) for a whole-brain linear mixed-effects analysis using a voxelwise 5% familywise error (FWE) rate threshold from random field theory16 (a standard thresholding level for neuroimaging studies). In other words, we found the minimum effect size that would have been needed in each of these studies for the difference to be considered statistically significant with an 80% probability, given the sample size. We then quantified the standardized effect size using Cohen’s d, which was computed as the mean effect divided by the standard deviation for the data.
To do this, we assumed that each study used a statistical map with t values in an MNI152 template space with a smoothness of three times the voxel size (full width at half maximum), a commonly used value for smoothness in fMRI analysis. The MNI152 template is a freely available template that was obtained from an average T1 scan for 152 subjects with a resolution of 2 mm and a volume within the brain mask of 228,483 voxels, and is used by default in most fMRI analysis software. We assume that, in each case, there would be one active region, with voxelwise standardized effect size d; that is, we assume that, for each subject, all voxels in the active region are, on average, d standardized units higher in their activity than the voxels in the non-active region, and that the active region is 1,600 mm2 (200 voxels). To calculate the voxelwise statistical significance threshold in this model statistical map, we used the function ptoz from the FSL17 (FMRIB Software Library) software package, which computes a FWE threshold for a given volume and smoothness using the Euler characteristic derived from Gaussian random field theory18. This approach ensures that the probability of a voxel in the non-active brain region exceeding this significance threshold is controlled at 5%; the resulting significance threshold, termed zα, is 5.12.
The statistical power is defined as the probability that the local maximum peak of activation in the active region exceeds this significance threshold. This probability was computed using a shifted version of the distribution of local maxima expected under the null hypothesis19, with shift of d·√n to reflect a given effect size d and sample size n. The effect size needed to exceed the significance threshold in each of the studies was found by selecting the effect size d that results in statistical power equal to 0.80, as computed in the previous step.
FIGURE 1b shows the median effect sizes that are needed to establish significance, with 80% power and an α value of 0.05. Despite the decreases in these hypothetical required effect sizes over the past 20 years, in 2015 the median study was only sufficiently powered to detect relatively large effects of greater than ~0.75 (FIG. 1b). Given that many studies will be assessing group differences or brain activity–behaviour correlations (which will inherently have lower power than do average group-activation effects), this represents an optimistic lower bound on the powered effect size.
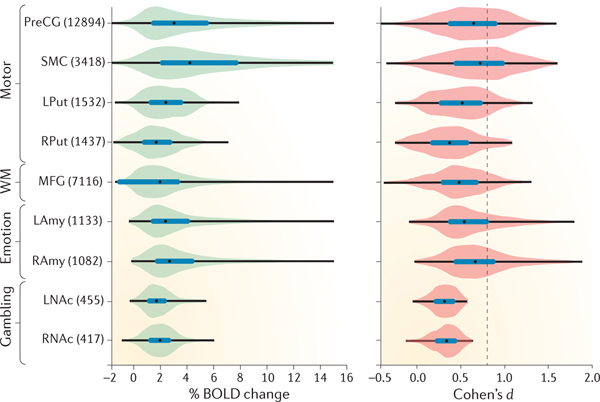
Indeed, the analysis presented in BOX 2 demonstrates that typical effect sizes observed in task-related BOLD imaging studies fall considerably below this level. Briefly, we analysed BOLD data from 186 individuals who were imaged using fMRI while performing motor, emotion, working-memory and gambling tasks as part of the Human Connectome Project (HCP)20. Assessing effect sizes in fMRI requires the definition of an independent region of interest (ROI) that captures the expected activated volume within which the effect size can be measured. Although there are several approaches to defining regions21,22, we created masks defined by the intersection between functional activation (identified through searches on Neurosynth as regions consistently active in studies examining the effects of ‘motor, ‘emotion, ‘gambling’ and ‘working memory’ tasks) and anatomical masks (defined using the Harvard-Oxford probabilistic atlas23, on the basis of the published ROIs from the HCP23). Within these intersection masks, we then determined the average task-related increases in BOLD signal — and the effect size (Cohen’s d) — that were associated with each different task. Additional details are provided in BOX 2. The figure in BOX 2, which lists the resulting BOLD signal changes and inferred effect sizes, demonstrates that realistic effect sizes — that is, BOLD changes that are associated with a range of cognitive tasks — in fMRI are surprisingly small: even for powerful tasks such as the motor task, which evokes median BOLD signal changes greater than 4%, 75% of the voxels in the masks have a standardized effect size d smaller than 1. For tasks evoking weaker activation, such as gambling, only 10% of the voxels in our masks demonstrated standardized effect sizes larger than 0.5. Thus, the average fMRI study remains poorly powered for capturing realistic effects, especially given that data from the HCP are of particularly high quality and thus the present estimates of effect size are probably greater than what would be found with most standard fMRI data sets.
Box 2 |. Effect-size estimates for common neuroimaging experimental paradigms.
The aim of this analysis is to estimate the magnitude of typical effect sizes of blood-oxygen-level-dependent (BOLD) changes in functional MRI (fMRI) signal that are associated with common psychological paradigms. We focus on four experiments conducted by the Human Connectome Project (HCP): an emotion task, a gambling task, a working-memory (WM) task and a motor task (detailed below). We chose data from the HCP for its diverse set of activation tasks and for its large sample size, which enables computation of stable effect-size estimates. The data and code used for this analysis are available at https://osf.io/spr9a/.
Briefly, the processing of data from the HCP was carried out in four main steps:
Step 1: subject selection
The analyses are performed on the 500-subject release of the HCP data, which is freely available at www.humanconnectome.org. We selected 186 independent subjects from the HCP data on the basis that all subjects have results for all four tasks and no subjects are genetically related.
Step 2: group analyses
The first-level analyses, which summarize the relation between the experimental design and the measured time series for each subject, were obtained from the HCP20. The processing and analysis pipelines for these analyses are shared together with the data. Here, we perform second-level analyses — that is, an assessment of the average effect of the task on BOLD signal over subjects — using the FMRIB Software Library (FSL) program flame1 (REF. 17), which performs a linear mixed-effects regression at each voxel, using generalized least squares with a local estimate of random effects variance. This analysis averages over subjects, while separating within-subject and between-subject variability, to ensure control of unobserved heterogeneity.
The following specific contrasts were tested:
Motor: tongue, hand and foot movements versus rest
Emotion: viewing faces with a fearful expression versus viewing neutral faces
Gambling: monetary reward versus punishment
Working memory: ‘2-back’ versus ‘0-back’
Step 3: create masks
The masks used for the analyses are the intersections of anatomical and a priori functional masks for each contrast. The rationale behind this is to find effect sizes in regions that are functionally related to the task but restricted to certain anatomical regions. We created the functional masks using Neurosynth15 by performing forward-inference meta-analysis using the search terms ‘motor’, ‘emotion’, ‘gambling’ and ‘working memory’, with false discovery rate control at 0. 01 (the default threshold on Neurosynth). The resulting functional mask identifies voxels that are consistently found to be activated in studies that mention each of the search terms in their abstract.
For the anatomical masks, we used the Harvard-Oxford probabilistic atlas23 at P > 0. Regions were chosen for each task, based on the published a priori hypothesized regions from the HCP78. The size of the masks was assessed by the number of voxels in the mask, and the structures contained in the anatomical masks for each of the tasks were as follows:
Motor: precentral gyrus (preCG), supplementary motor cortex (SMC), left putamen (LPut) and right putamen (RPut)
Working memory: middle frontal gyrus (MFG)
Emotion: left amygdala (LAmy) and right amygdala (RAmy)
Gambling: left nucleus accumbens (LNAc) and right nucleus accumbens (RNAc)
Step 4: compute effect size
The intersection masks created above were used to isolate the regions of interest (ROIs) in the second-level-analysed BOLD signal data. From these mask-isolated data sets, the size of the task-related effect (Cohen’s d) was computed for each relevant region (see the figure). The FSL program Featquery computes the percentage BOLD change for each voxel within the masks.
The figure shows the distributions of the observed BOLD signal-change estimates and effect-size estimates for common experimental paradigms, across voxels within each ROI (the numbers in parentheses denote the number of voxels in the ROI). The box plots inside the violins represent the interquartile range (first quartile to third quartile), and the black dots show median values. The results show that, whereas some tasks show very large BOLD signal changes on average, the effect-size estimates computed across subjects are relatively modest, with none reaching the level of d = 0.8, which is customarily taken to define a ‘large’ effect
Solutions.
When possible, all sample sizes should be justified by an a priori power analysis. A number of tools are available to enable power analyses for fMRI; for example, Neuropower24 and Fmripower25 (see Further information). However, one must be cautious in extrapolating from effect sizes that are estimated from small studies, because they are almost certainly inflated. When previous data are not available to support a power analysis, one can instead identify the sample size that would support finding the minimum effect size that would be theoretically informative (for example, on the basis of the results from BOX 2). The use of heuristic sample-size guidelines (for example, those that are based on sample sizes used in previously published studies) is likely to result in a misuse of resources, either by collecting too many or (more likely) too few subjects.
The larger sample sizes that will result from the use of power analysis will have important implications for researchers: given that research funding will probably not increase to accommodate these larger samples, fewer studies may be funded, and researchers with fewer resources may have a more difficult time performing research that meets these standards. This would hit trainees and junior researchers particularly hard, and the community needs to develop ways to address this challenge. We do not believe that the solution is to admit weakly powered studies simply on the basis that the researchers lacked the resources to use a larger sample. This situation is, in many ways, similar to the one that was faced in the field of genetics, which realized more than a decade ago that weakly powered genetic association studies were unreliable; the field moved to the use of much larger samples with high power to detect even very small associations and began to enforce replication. This has been accomplished through the development of large-scale consortia, which have amassed samples in the tens or hundreds of thousands (BOX 1). There are examples of successful consortia in neuroimaging, including the 1000 Functional Connectomes Project and its International Neuroimaging Data-Sharing Initiative (INDI)3,26, and the ENIGMA (Enhancing Neuro Imaging Genetics by Meta-Analysis) consortium27. With such consortia come inevitable challenges of authorship and credit28, but here again we can look to other areas of research that have met these challenges (for an example, see REF. 29).
In some cases, researchers must necessarily use a statistically insufficient sample size in a study, owing to limitations in the specific sample (for example, when studying a rare patient group). In such cases, there are three already commonly used options to improve power. First, researchers can choose to collect a much larger amount of data from each individual and present results at the individual level rather than at the group level30,31 — although the resulting inferences cannot then be generalized to the population as a whole. Second, researchers can use more-liberal statistical thresholding procedures, such as methods controlling the false discovery rate (FDR). However, it should be noted that the resulting higher power comes at the expense of more false-positive results and should therefore be used with caution; any results must be presented with the caveat that they have an inflated false-positive rate. Third, researchers may restrict the search space using a small number of a priori ROIs or an independent ‘functional localizer’ to identify specific ROIs for each individual. It is essential that these ROIs (or a specific functional localizer strategy) be explicitly delineated before any analyses. This is important because it is always possible to develop a post hoc justification for any specific ROI on the basis of previously published papers — a strategy that results in an ROI that seems to be independent but that actually has a circular definition and thus leads to meaningless statistics and inflated type I errors. By analogy to the idea of HARKing (hypothesizing after results are known; in which results of exploratory analyses are presented as having been hypothesized from the beginning)32, we refer to the latter practice as SHARKing (selecting hypothesized areas after results are known). We would only recommend the use of restricted search spaces if the exact ROIs and hypotheses are pre-registered33,34.
Finally, we note the potential for Bayesian methods to make the best use of small, underpowered samples. These approaches stabilize low-information estimates, converging them towards anticipated values that are characterized by prior distributions. Although Bayesian methods have not been widely used in the whole-brain setting, owing to the computational challenge of specifying a joint model over all voxels, newer graphics processing units (GPUs) may provide the acceleration that is needed to make these methods practical35. These methods also require the specification of priors, which often remains a challenge: priors should reflect typical or default knowledge, but if they are poorly set they could overwhelm the data, simply returning the default result.
Flexibility and exploration in data analysis
The typical fMRI analysis workflow contains a large number of pre-processing and analysis operations, each with choices to be made about parameters and/or methods (BOX 3). Carp36 applied 6,912 analysis workflows (using the SPM37 (Statistical Parametric Mapping) and AFNI38 (Analysis of Functional NeuroImages) software packages) to a single data set and quantified the variability in resulting statistical maps. This approach revealed that some brain regions exhibited more substantial variation across the different workflows than did other regions. This issue is not unique to fMRI; for example, similar issues have been raised in genetics39. These ‘researcher degrees of freedom’ can lead to substantial inflation of type I error rates8 — even when there is no intentional ‘P-hacking, and only a single analysis is ever conducted9.
Box 3 |. Flexibility in functional MRI data analysis.
In the early days of functional MRI (fMRI) analysis, it was rare to find two laboratories that used the same software to analyse their data, with most using locally developed custom software. Over time, a small number of open-source analysis packages have gained prominence (Statistical Parametric Mapping (SPM), FMRIB Software Library (FSL), and Analysis of Functional NeuroImages (AFNI) being the most common), and now most laboratories use one of these packages for their primary data processing and analysis. Within each of these packages, there is a great deal of flexibility in how data are analysed; in some cases, there are clear best practices, but in others there is no consensus regarding the optimal approach. This leads to a multiplicity of analysis options. In the table, we outline some of the major choices involved in performing analyses using one of the common software packages (FSL). Even for this non-exhaustive list from a single analysis package, the number of possible analysis workflows — 69,120 — exceeds the number of papers that have been published on fMRI since its inception more than two decades ago.
It is possible that many of these alternative pipelines could lead to very similar results, although the analyses of Carp36 suggest that many of them may lead to considerable heterogeneity in the results. In addition, there is evidence that choices of pre-processing parameters may interact with the statistical modelling approach (for example, there may be interactions between head motion modelling and physiological noise correction) and that the optimal pre-processing pipeline may differ across subjects (for example, interacting with the amount of head motion)43.
| Processing step | Reason | Options [suboptions] | Number of plausible options |
|---|---|---|---|
| Motion correction | Correct for head motion during scanning | • ‘Interpolation’ [linear or sinc] • ‘Reference volume’ [single or mean] |
4 |
| Slice timing correction | Correct for differences in acquisition timing of different slices | ‘No’, ‘before motion correction’ or ‘after motion correction’ | 3 |
| Field map correction | Correct for distortion owing to magnetic susceptibility | ‘Yes’ or ‘no’ | 2 |
| Spatial smoothing | Increase SNR for larger activations and ensure assumptions of GRF theory | ‘FWHM’ [4 mm, 6 mm or 8 mm] | 3 |
| Spatial normalization | Warps an individual brain to match a group template | ‘Method’ [linear or nonlinear] | 2 |
| High-pass filter | Remove low-frequency nuisance signals from data | ‘Frequency cut-off’ [100 s or 120 s] | 2 |
| Head motion regressors | Remove remaining signals owing to head motion via statistical model | ‘Yes’ or ‘no’ [if yes: 6/12/24 parameters or single time point ‘scrubbing’ regressors] | 5 |
| Haemodynamic response | Account for delayed nature of haemodynamic response to neuronal activity | • ‘Basis function’ [‘single-gamma’ or ‘double-gamma’] • ‘Derivatives’ [‘none’, ‘shift’ or ‘dispersion’] |
6 |
| Temporal autocorrelation model | Model for the temporal autocorrelation inherent in fMRI signals | ‘Yes’ or ‘no’ | 2 |
| Multiple-comparison correction | Correct for large number of comparisons across the brain | ‘Voxel-based GRF’, ‘cluster-based GRF’, ‘FDR’ or ‘non-parametric’ | 4 |
| Total possible workflows | 69,120 |
FDR, false discovery rate; FWHM, full width at half maximum; GRF, Gaussian random field; SNR, signal-to-noise ratio.
Exploration is key to scientific discovery, but rarely does a research paper comprehensively describe the actual process of exploration that led to the ultimate result; to do so would render the resulting narrative far too complex and murky. As a clean and simple narrative has become an essential component of publication, the intellectual journey of the research is often obscured. Instead, reports may engage in HARKing32. Because HARKing hides the number of data-driven choices that are made during analysis, it can strongly overstate the actual evidence for a hypothesis. There is arguably a great need to support the publication of exploratory studies without forcing those studies to masquerade as hypothesis-driven science, while realizing that such exploratory findings (like all scientific results) will ultimately require further validation in independent studies.
Solutions.
We recommend pre-registration of methods and analysis plans. The details to be pre-registered should include planned sample size, specific analysis tools to be used, specification of predicted outcomes, and definition of any specific ROIs or localizer strategies that will be used for analysis. The Open Science Framework and AsPredicted (see Further information) provide established platforms for pre-registration; the former assigns an embargo period during which the registration remains private, obviating some concerns about ideas being disclosed while still under investigation. In addition, some journals now provide the ability to submit a ‘Registered Report’ in which hypotheses and methods are reviewed before data collection, and the study is guaranteed publication regardless of the outcome40 (for examples of such reports, see REFS 41,42; for a list of journals offering the Registered Report format, see https://osf.io/8mpji/wiki/home/ ). Exploratory analyses (including any deviations from planned analyses) should be clearly distinguished from planned analyses in the publication. Ideally, results from exploratory analyses should be confirmed in an independent validation data set.
Although there are concerns regarding the degree to which flexibility in data analysis may result in inflated error rates, we do not believe that the solution is to constrain researchers by specifying a particular set of methods that must be used. Many of the most interesting findings in fMRI have come from the use of novel analysis methods, and we do not believe that there will be a single best workflow for all studies; in fact, there is direct evidence that different studies or individuals will probably benefit from different workflows43. We believe that the best solution is to allow flexibility but require that all exploratory analyses be clearly labelled as such, and strongly encourage validation of exploratory results (for example, through the use of a separate validation data set).
Multiple comparisons
The most common approach to neuroimaging analysis involves mass univariate testing in which a separate hypothesis test is performed for each voxel. In such an approach, the false-positive rate will be inflated if there is no correction for multiple tests. A humorous example of this was seen in the now-infamous ‘dead salmon’ study that was reported by Bennett et at.44 in which ‘activation’ was detected in the brain of a dead salmon but disappeared when the proper corrections for multiple comparisons were performed.
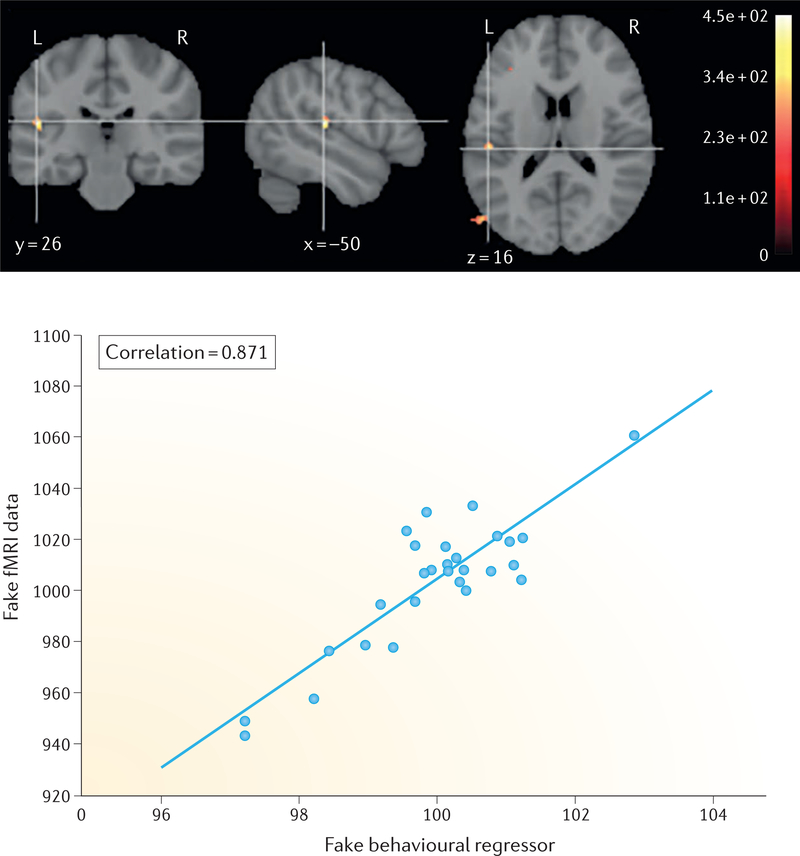
FIGURE 2 presents a similar example in which random data can be analysed (incorrectly) to lead to seemingly impressive results, through a combination of failure to adequately correct for multiple comparisons and circular ROI analysis. We generated random simulated fMRI data for each of 28 simulated participants (based on the median sample size for studies from 2015, as found in the analysis shown in FIG. 1). For each simulated participant, each voxel within an MNI152 mask was assigned a random statistical value from a Gaussian distribution (with a mean ± standard deviation of 1000 ± 100); each value represented a comparison between an ‘activation’ condition and a ‘baseline’ condition. We then spatially smoothed each of the resulting 28 images with a 6 mm Gaussian kernel, based on the common smoothing level of three times the voxel size. A univariate analysis was performed using FSL to assess the correlation between the ‘activation’ in each voxel and the simulated behavioural regressor across subjects, and the resulting statistical map was thresholded at P < 0.001 and with a 10-voxel minimum cluster extent threshold (which is a commonly used heuristic correction that has been shown by Eklund et at.45 to result in highly inflated levels of false positives). This approach revealed a cluster of false-positive activation in the superior temporal cortex in which the simulated fMRI data are highly correlated with the simulated behavioural regressor (FIG. 2).
Figure 2 |. Small samples, uncorrected statistics and circularity can produce misleadingly large effects.
A seemingly impressive brain-behaviour association can arise from completely random data through the use of statistics uncorrected for multiple comparisons and circular region-of-interest analyses that capitalize on the large sampling error that arises from small samples. With the informal P < 0.001 and cluster size k >10 thresholding, the analysis revealed a cluster in the superior temporal gyrus (upper panel); the signal extracted from that cluster (that is, using circular analysis) showed a very strong correlation between the functional MRI (fMRI) data and behavioural data (lower panel). For details of the analysis, see the main text. A computational notebook for this example is available at https://osf.io/spr9a/.
The problem of multiplicity in neuroimaging analysis was recognized very early, and the past 25 years have seen the development of now well-established and validated methods for correction of FWE and FDR in neuroimaging data46. However, recent work45 has suggested that even some very well-established inferential methods (specifically, certain ones that are based on the spatial extent of activations) can produce inflated type I error rates in certain settings, for instance when the cluster-forming threshold is too low.
There is an ongoing debate between neuroimaging researchers who feel that conventional approaches to multiple-comparison correction are too lax and allow too many false positives47 and those researchers who feel that thresholds are too conservative and risk missing most of the interesting effects48. In our view, the deeper problem is the inconsistent application of principled correction approaches49. Many researchers freely combine different approaches and thresholds in ways that produce a high number of undocumented researcher degrees of freedom8, rendering reported P values uninterpretable.
To assess this more directly, we examined the 100 most recent results for the PubMed query (“fMRI” AND brain AND activation NOT review[PT] AND human[MESH] AND english[la]), performed on 23 May 2016; of these, 66 reported whole-brain task fMRI results and were available in full text (for a full list of these papers and annotations, see mailto:https://osf.io/spr9a/). Only 3 of the 66 analysed papers presented fully uncorrected results, with 4 others presenting a mixture of corrected and uncorrected results; this suggests that corrections for multiple comparisons are now standard. However, there is evidence that researchers may engage in ‘method shopping’ for techniques that provide greater sensitivity, at a potential cost of increased error rates. Notably, 9 of the 66 papers used the FSL or SPM software packages to perform their primary analysis, but then used the AlphaSim or 3dClustSim tools from the AFNI software package (7 papers) or other simulation-based approaches (2 papers) to correct for multiple comparisons. This is concerning, because both FSL and SPM offer well-established methods that use Gaussian random field theory or non-parametric analyses to correct for multiple comparisons. Given the substantial degree of extra work (for example, software installation and file reformatting) that is involved in using multiple software packages, the use of a different tool raises some concern that this might reflect analytic P-hacking. This concern is further amplified by the finding that, until very recently, AlphaSim and its adaptation 3dClustSim had slightly inflated type I error rates45. Sadly, whereas non-parametric methods (such as permutation tests) are known to provide more accurate control over FWE rates than do parametric methods46,50 and are applicable for nearly all models, they were not used in any of these papers.
Solutions.
To balance type I and type II error rates in a principled way, we suggest a dual approach of reporting corrected whole-brain results and (for potential use in later meta-analyses) sharing the unthresholded statistical map (preferably z values) through a repository that allows viewing and downloading (such as NeuroVault51) (for an example of this practice, see REF. 52; for shared data, see http://neurovault.org/collections/122/ ). Any use of non-standard methods for correction of multiple comparisons (for example, using tools from different packages for the main analysis and the multiple-comparison correction) should be justified explicitly (and reviewers should demand such justification). Signals can be detected in the images using either voxelwise or clusterwise inference. With either method, multiple testing can be accounted for with FWE or (typically more sensitive but less specific) FDR error rate measures, although clusterwise and any FDR inferences need to be interpreted with care, as they allow more false-positive voxels than does voxelwise FWE correction.
Alternatively, one can abandon the mass univariate approach altogether. Multivariate methods that treat the entire brain as the measurement (such as the analysis in REF 53) and graph-based approaches that integrate information over all edges (such as the approach in REF 54) avoid the multiple-testing problem. However, these approaches present the challenge of understanding the involvement of individual voxels or edges in an effect55 and raise other interpretation issues.
Software errors
As the complexity of a software program increases, the likelihood of undiscovered bugs quickly reaches certainty56. This implies that the software that is used for fMRI analysis is likely to contain bugs. Most fMRI researchers use one of several open-source analysis packages for pre-processing and statistical analyses; many additional analyses require custom programs. Because most researchers writing custom code are not trained in software engineering, there is insufficient attention to good software-development practices that could help to catch and prevent errors. This issue came to the fore recently, when a 15-year-old bug was discovered in the AFNI program 3dClustSim (and the older AlphaSim), which resulted in slightly inflated type I error rates45,57 (the bug was fixed in May 2015). Although small in this particular case, the impact of such bugs could be widespread; for example, PubMed Central lists 1,362 publications mentioning AlphaSim or 3dClustSim published before 2015 (query [(AlphaSim OR 3DClustSim) AND 1992:2014[DP]] performed on 14 July 2016). Similarly, the analyses presented in a preprint of the present article contained two software errors that led to different results being presented in the final version of the paper. The discovery of these errors led us to perform a code review and to include software tests to reduce the likelihood of remaining errors. Although software errors will happen in commonly used toolboxes as well as in-house code, they are much more likely to be discovered in widely used packages owing to the increased scrutiny of their many more users. It is very likely that consequential bugs exist in custom software that has been built for individual projects but that, owing to the limited user base, those bugs will never be unearthed.
Solutions.
Researchers should avoid the trap of the ‘not invented here’ philosophy: when the problem at hand can be solved using software tools from a well-established project, these should be chosen instead of re-implementing the same method in custom code. Errors are more likely to be discovered when code has a larger user base, and larger projects are more likely to follow better software-development practices. Researchers should learn and implement good programming practices, including the judicious use of software testing and validation. Validation methodologies (such as comparing with another existing implementation or using simulated data) should be clearly defined. Custom analysis code should always be shared on manuscript submission (for an example, see REF. 58). It may be unrealistic to expect reviewers to evaluate code in addition to the manuscript itself, although this is standard in some journals such as the Journal of Statistical Software. However, reviewers should request that the code be made available publicly (so others can evaluate it) and, in the case of methodological papers, that the code is accompanied with a set of automated software tests. Finally, researchers need to acquire sufficient training on the implemented analysis methods, particularly so that they understand the default parameter values of the software (such as cluster-forming thresholds and filtering cut-offs), as well as the assumptions on the data and how to verify those assumptions.
Insufficient study reporting
For the reader of a paper to know whether appropriate analyses have been performed, the methods must be reported in sufficient detail. Some time ago, we published an initial set of guidelines for reporting the methods typically used in an fMRI study59. Unfortunately, reporting standards in the fMRI literature remain poor. Carp60 and Guo et al.61 analysed 241 and 100 fMRI papers, respectively, for the reporting of methodological details, and both found that some important analysis details (such as interpolation methods and smoothness estimates) were rarely described. Consistent with this, in 22 of the 66 papers that we discussed above, it was impossible to identify exactly which multiple-comparison correction technique was used (beyond generic terms such as ‘cluster-based correction’), because no specific method or citation was provided. The Organization for Human Brain Mapping (OHBM) has recently addressed this issue through its 2015–2016 Committee on Best Practices in Data Analysis and Sharing (COBIDAS), which has issued a new, detailed set of reporting guidelines62 (BOX 4).
Box 4 |. Guidelines for transparent methods reporting in neuroimaging.
The Organization for Human Brain Mapping (OHBM) Committee on Best Practices in Data Analysis and Sharing (COBIDAS) report provides a set of best practices for reporting and conducting studies using MRI. It divides practice into seven categories and provides detailed checklists that can be consulted when planning, analysing and writing up a study. The text below lists these categories with summaries of the topics that are covered in the checklists.
Acquisition reporting
Subject preparation: mock scanning; special accommodations; experimenter personnel
MRI system description: scanner; coil; significant hardware modifications; software version
MRI acquisition: pulse sequence type; imaging type; essential sequence and imaging parameters; phase encoding parameters; parallel imaging method and parameters; multiband parameters; readout parameters; fat suppression; shimming; slice order and timing; slice position procedure; brain coverage; scanner-side pre-processing; scan duration; other non-standard procedures; T1 stabilization; diffusion MRI gradient table; perfusion (arterial spin labelling (ASL) MRI or dynamic susceptibility contrast MRI)
Preliminary quality control: motion monitoring; incidental findings
Pre-processing reporting
General: intensity correction; intensity normalization; distortion correction; brain extraction; segmentation; spatial smoothing; artefact and structured noise removal; quality control reports; intersubject registration
Temporal or dynamic: motion correction
Functional MRI: T1 stabilization; slice time correction; function-structure (intra-subject) co-registration; volume censoring; resting-state functional MRI feature
Diffusion: gradient distortion correction; diffusion MRI eddy current correction; diffusion estimation; diffusion processing; diffusion tractography
Perfusion: ASL; dynamic susceptibility contrast MRI
Statistical modelling and inference
Mass univariate analyses: variable submitted to statistical modelling; spatial region modelled; independent variables; model type; model settings; inference (contrast, search region, statistic type, P-value computation, multiple-testing correction)
Functional connectivity: confound adjustment and filtering; multivariate method (for example, independent component analysis); dependent variable definition; functional connectivity measure; effectivity connectivity model; graph analysis algorithm
Multivariate modelling and predictive analysis: independent variables; features extraction and dimension reduction; model; learning method; training procedure; evaluation metrics (discrete response, continuous response, representational similarity analysis, significance); fit interpretation
Results reporting
Mass univariate analysis: effects tested; extracted data; tables of coordinates; thresholded maps; unthresholded maps; extracted data; spatial features
Functional connectivity: independent component analyses; graph analyses (null hypothesis tested)
Multivariate modelling and predictive analysis: optimized evaluation metrics
Data sharing
Define data-sharing plan early: material shared; URL (access information); ethics compliance; documentation; data format
Database for organized data: quality control procedures; ontologies; visualization; de-identification; provenance and history; interoperability; querying; versioning; sustainability plan (funding)
Reproducibility
Documentation: tools used; infrastructure; workflow; provenance trace; literate programming; English language version.
Archiving: tools availability; virtual appliances
Citation: data; workflow
In addition, claims in the neuroimaging literature are often asserted without corresponding statistical support. In particular, failures to observe a statistically significant effect can lead researchers to proclaim the absence of an effect — a dangerous and almost invariably unsupported acceptance of the null hypothesis. ‘Reverse inference’ claims, in which the presence of a given pattern of brain activity is taken to imply a specific cognitive process (for example, “the anterior insula was activated, suggesting that subjects experienced empathy”), are rarely grounded in quantitative evidence63. Furthermore, claims of ‘selective’ activation in one brain region or experimental condition are often made when activation is statistically significant in one region or condition but not in others. This false assertion ignores the fact that “the difference between ‘significant’ and ‘not significant’ is not itself statistically significant” (REF. 64); such claims require appropriate tests for statistical interactions65.
Solutions.
Authors should follow accepted standards for reporting methods (such as the COBIDAS standard for MRI studies), and journals should require adherence to these standards. Every major claim in a paper should be directly supported by appropriate statistical evidence, including specific tests for significance across conditions and relevant tests for interactions. Because the computer code is often necessary to understand exactly how a data set has been analysed, releasing the analysis code is particularly useful and should be standard practice.
Lack of independent replications
There are surprisingly few examples of direct replication in the field of neuroimaging, probably reflecting both the expense of fMRI studies and the emphasis of most top journals on novelty rather than informativeness. Although there are many basic results that are clearly replicable (for example, the presence of activity in the ventral temporal cortex that is selective for faces over scenes, or systematic correlations within functional networks in the resting state), the replicability of weaker and less neurobiologically established effects (for example, group differences and between-subject correlations) is nowhere near as certain. One study66 attempted to replicate 17 studies that had previously found associations between brain structure and behaviour. Only 1 of the 17 attempts showed stronger evidence for an effect as large as the original effect size than for a null effect, and 8 out of 17 showed stronger evidence for a null effect. This suggests that replicability of neuroimaging findings (particularly brain-behaviour correlations) is exceedingly low, as has been demonstrated in other fields, such as cancer biology67 and psychology68.
It is worth noting that, although the cost of conducting a new fMRI experiment is a factor limiting the feasibility of replication studies, there are many findings that can be replicated using publicly available data. Resources such as the FCP-INDI26, the Consortium for Reliability and Reproducibility69, OpenfMRI70 or the HCP20 provide MRI data that are suitable for attempts to replicate many previously reported findings. These resources can also be used to answer questions about sensitivity of a particular finding to the data analysis tools used36. However, even in the cases when replications are possible using publicly available data, they are still few and far between, because the academic community tends to put greater emphasis on novelty of findings rather than on their replicability.
Solutions.
The neuroimaging community should acknowledge replication reports as scientifically important research outcomes that are essential in advancing knowledge. One effort to acknowledge this is the OHBM Replication Award, which is to be awarded for the first time in 2017 for the best neuroimaging replication study in the previous year. In addition, in cases of especially surprising findings, findings that could have influence on public health policy or medical treatment decisions, or findings that could be tested using data from another existing data set, reviewers should consider requesting replication of the finding by the group before accepting the manuscript.
Towards the neuroimaging paper of the future
In this Analysis article, we have outlined a number of problems with current practice and made suggestions for improvements. Here, we outline what we would like to see in the neuroimaging paper of the future, inspired by related work in the geosciences71.
Planning.
The sample size for the study would be determined in advance using formal statistical power analysis. The entire analysis plan, including exclusion and inclusion criteria, software workflows (including contrasts and multiple-comparison methods) and specific definitions for all planned regions of interest, would be formally pre-registered.
Implementation.
All code for data collection and analysis would be stored in a version-control system and would include software tests to detect common problems. The repository would use a continuous integration system to ensure that each revision of the code passes appropriate software tests. The entire analysis workflow (including both successful and failed analyses) would be completely automated in a workflow engine and packaged in a software container or virtual machine to ensure computational reproducibility. All data sets and results would be assigned version numbers to enable explicit tracking of provenance. Automated quality control would assess the analysis at each stage to detect potential errors.
Validation.
For empirical papers, all exploratory results would be validated against an independent validation data set that was not examined before validation. For methodological papers, the approach would follow best practices for reducing overly optimistic results72. Any new method would be validated against benchmark data sets and compared with other state-of-the-art methods.
Dissemination.
All results would be clearly marked as either hypothesis-driven (with a link to the appropriate pre-registration) or exploratory. All analyses performed on the data set (including those analyses that were not deemed useful) would be reported. The paper would be written using a literate programming technique in which the code for figure generation is embedded within the paper and the data depicted in figures are transparently accessible. The paper would be distributed along with the full codebase to perform the analyses and the data necessary to reproduce the analyses, preferably in a container or virtual machine to enable direct reproducibility. Unthresholded statistical maps and the raw data would be shared via appropriate community repositories, and the shared raw data would be formatted according to a community standard, such as the Brain Imaging Data Structure (BIDS)73, and annotated using an appropriate ontology to enable automated meta-analysis.
Conclusion
We have outlined what we see as a set of problems with neuroimaging methodology and reporting, and have suggested approaches to address them. It is likely that the reproducibility of neuroimaging research is no better than that of many other fields in which it has been shown to be surprisingly low. Given the substantial amount of research funds that are currently invested in neuroimaging research, we believe that it is essential that the field address the issues raised here, to ensure that public funds are spent effectively and in ways that advance our understanding of the human brain. We have also laid out what we see as a road map for how neuroimaging researchers can overcome these problems, laying the groundwork for a scientific future that is transparent and reproducible.
Supplementary Material
Acknowledgements
R.A.P., J.D., J.-B.P. and K.J.G. are supported by the Laura and John Arnold Foundation. J.D. has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No 706561. M.R.M. is supported by the Medical Research Council (MRC) (MC UU 12013/6) and is a member of the UK Centre for Tobacco and Alcohol Studies, a UK Clinical Research Council Public Health Research Centre of Excellence. Funding from the British Heart Foundation, Cancer Research UK, the Economic and Social Research Council, the MRC and the National Institute for Health Research, under the auspices of the UK Clinical Research Collaboration, is gratefully acknowledged. C.I.B. is supported by the Intramural Research Program of the US National Institutes of Health (NIH)-National Institute of Mental Health (NIMH) (ZIA-MH002909). T.Y. is supported by the NIMH (R01MH096906). P.M.M. acknowledges personal support from the Edmond J. Safra Foundation and Lily Safra and research support from the MRC, the Imperial College Healthcare Trust Biomedical Research Centre and the Imperial Engineering and Physical Sciences Research Council Mathematics in Healthcare Centre. T.E.N. is supported by the Wellcome Trust (100309/Z/12/Z), NIH-National Institute of Neurological Disorders and Stroke (R01NS075066) and NIH-National Institute of Biomedical Imaging and Bioengineering (NIBIB) (R01EB015611). J.-B.P. is supported by the NIBIB (P41EB019936) and by NIH-National Institute on Drug Abuse (U24DA038653). Data were provided (in part) by the Human Connectome Project, WU-Minn Consortium (principal investigators: D. Van Essen and K. Ugurbil; 1U54MH091657), which is funded by the 16 Institutes and Centers of the NIH that support the NIH Blueprint for Neuroscience Research, and by the McDonnell Center for Systems Neuroscience at Washington University. The authors thank J. Wexler for performing annotation of Neurosynth data, S. David for providing sample-size data, and R. Cox and P. Taylor for helpful comments on a draft of the manuscript.
Glossary
- Linear mixed-effects analysis
An analysis in which some measured independent variables are treated as randomly sampled from the population, in contrast to a traditional fixed-effects analysis, in which all predictors are treated as fixed and known.
- Familywise error (FWE).
The probability of at least one false positive among multiple statistical tests.
- Random field theory
The theory describing the behaviour of geometric points on a random topological space.
- Euler characteristic
A topological measure that is used to describe the set of thresholded voxels in the context of random field theory.
- False discovery rate (FDR).
The expected proportion of false positives among all significant findings when performing multiple statistical tests.
- Functional localizer
An independent scan that is used to identify regions on the basis of their functional response; for example, for the responses of face-responsive regions to faces.
- Bayesian methods
An approach to statistical analysis focusing on updating beliefs via probability distributions and symmetrically comparing candidate models.
- Mass univariate testing
An approach to the analysis of multivariate data in which the same model is fit to each element of the observed data (for example, each voxel).
- Permutation tests
Also known as randomization tests. Approaches for testing statistical significance by comparing to a null distribution that is obtained by rearranging the labels of the observed data.
- ‘Not invented here’ philosophy
The philosophy that any solution to a problem that was developed by someone else is necessarily inferior and must be re-engineered from scratch.
- Interpolation
The operation by which a function is applied to the sampled data to obtain estimates of the data at positions where data have not been sampled.
- Software container
A self-contained software tool that encompasses all of the necessary software and dependencies to run a particular program.
Footnotes
Competing interests statement
The authors declare no competing interests.
SUPPLEMENTARY INFORMATION
See online article: S1 (figure)
FURTHER INFORMATION
AsPredicted: https://aspredicted.org/
Fmripower: http://fmripower.org/
Human Connectome Project: https://www.humanconnectome.org/
NeuroPower: http://neuropowertools.org/
Neurosynth: http://neurosynth.org/
NeuroVault: http://neurovault.org/
Open Science Framework: https://osf.io/
Organization for Human Brain Mapping (OHBM) Committee on Best Practices in Data Analysis and Sharing (COBIDAS): http://www.humanbrainmapping.org/COBIDAS
References
- 1.Poldrack RA & Farah MJ Progress and challenges in probing the human brain. Nature 526, 371–379 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Logothetis NK What we can do and what we cannot do with fMRI. Nature 453, 869–878 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Biswal BB et al. Toward discovery science of human brain function. Proc. Natl Acad. Sci. USA 107, 4734–4739 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kriegeskorte N. et al. Matching categorical object representations in inferior temporal cortex of man and monkey. Neuron 60, 1126–1141 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Norman KA, Polyn SM, Detre GJ & Haxby JV Beyond mind-reading: multi-voxel pattern analysis of fMRI data. Trends Cogn. Sci 10, 424–430 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Poldrack RA Inferring mental states from neuroimaging data: from reverse inference to large-scale decoding. Neuron 72, 692–697 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ioannidis JPA Why most published research findings are false. PLoS Med. 2, e124 (2005). This landmark paper outlines the ways in which common practices can lead to inflated levels of false positives. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simmons JP, Nelson LD & Simonsohn U. False-positive psychology: undisclosed flexibility in data collection and analysis allows presenting anything as significant. Psychol. Sci 22, 1359–1366 (2011). This paper highlights the impact of common ‘questionable research practices’ on study outcomes and proposes a set of guidelines to prevent false-positive findings. [DOI] [PubMed] [Google Scholar]
- 9.Gelman A. & Loken E. The statistical crisis in science. American Scientist 102, 40 (2014). [Google Scholar]
- 10.Ioannidis JPA, Fanelli D, Dunne DD & Goodman SN Meta-research: evaluation and improvement of research methods and practices. PLoS Biol. 13, e1002264 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins FS & Tabak LA Policy: NIH plans to enhance reproducibility. Nature 505, 612–613 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Button KS et al. Power failure: why small sample size undermines the reliability of neuroscience. Nat. Rev. Neurosci 14, 365–376 (2013). This paper sounded the first major alarm regarding low statistical power in neuroscience. [DOI] [PubMed] [Google Scholar]
- 13.Yarkoni T. Big correlations in little studies: inflated fMRI correlations reflect low statistical power — commentary on Vul et al. (2009). Perspect. Psychol. Sci 4, 294–298 (2009). [DOI] [PubMed] [Google Scholar]
- 14.David SP et al. Potential reporting bias in fMRI studies of the brain. PLoS ONE 8, e70104 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yarkoni T, Poldrack RA, Nichols TE, Van Essen DC & Wager TD Large-scale automated synthesis of human functional neuroimaging data. Nat. Methods 8, 665–670 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friston KJ, Frith CD, Liddle PF & Frackowiak RS Comparing functional (PET) images: the assessment of significant change. J. Cereb. Blood Flow Metab. 11, 690–699 (1991). [DOI] [PubMed] [Google Scholar]
- 17.Jenkinson M, Beckmann CF, Behrens TEJ, Woolrich MW & Smith SM FSL. Neuroimage 62, 782–790 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Worsley KJ et al. A unified statistical approach for determining significant signals in images of cerebral activation. Hum. Brain Mapp. 4, 58–73 (1996). [DOI] [PubMed] [Google Scholar]
- 19.Cheng D. & Schwartzman A. Distribution of the height of local maxima of Gaussian random fields. Extremes 18, 213–240 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Essen DC et al. The WU-Minn Human Connectome Project: an overview. Neuroimage 80, 62–79 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tong Y. et al. Seeking optimal region-of-interest (ROI) single-value summary measures for fMRI studies in imaging genetics. PLoS ONE 11, e0151391 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Devlin JT & Poldrack RA In praise of tedious anatomy. Neuroimage 37, 1033–1041 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Desikan RS et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 31, 968–980 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Durnez J. et al. Power and sample size calculations for fMRI studies based on the prevalence of active peaks. Preprint at bioRxiv 10.1101/049429(2016). [DOI] [Google Scholar]
- 25.Mumford JA & Nichols TE Power calculation for group fMRI studies accounting for arbitrary design and temporal autocorrelation. Neuroimage 39, 261–268 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mennes M, Biswal BB, Castellanos FX & Milham MP Making data sharing work: the FCP/ INDI experience. Neuroimage 82, 683–691 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson PM et al. The ENIGMA Consortium: large-scale collaborative analyses of neuroimaging and genetic data. Brain Imaging Behav. 8, 153–182 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rohlfing T. & Poline J-B Why shared data should not be acknowledged on the author byline. Neuroimage 59, 4189–4195 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Austin MA, Hair MS & Fullerton SM Research guidelines in the era of large-scale collaborations: an analysis of Genome-wide Association Study Consortia. Am. J. Epidemiol 175, 962–969 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savoy RL Using small numbers of subjects in fMRI-based research. IEEE Eng. Med. Biol. Mag 25, 52–59 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Poldrack RA et al. Long-term neural and physiological phenotyping of a single human. Nat. Commun 6, 8885 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kerr NL HARKing: hypothesizing after the results are known. Pers. Soc. Psychol. Rev 2, 196–217 (1998). [DOI] [PubMed] [Google Scholar]
- 33.Nosek BA et al. Promoting an open research culture. Science 348, 1422–1425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chambers CD, Dienes Z, McIntosh RD, Rotshtein P. & Willmes K. Registered reports: realigning incentives in scientific publishing. Cortex 66, A1–A2 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Siden P, Eklund A, Bolin D. & Villani M. Fast Bayesian whole-brain fMRI analysis with spatial 3D priors. Neuroimage 146, 211–225 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Carp J. On the plurality of (methodological) worlds: estimating the analytic flexibility of FMRI experiments. Front. Neurosci 6, 149 (2012). This paper reports analyses of a single data set using 6,912 different analysis workflows, highlighting the large degree of variability in results across analyses in some brain regions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Penny WD, Friston KJ, Ashburner JT, Kiebel SJ & Nichols TE Statistical Parametric Mapping: The Analysis of Functional Brain Images (Elsevier Science, 2011). [Google Scholar]
- 38.Cox RW AFNI: what a long strange trip it’s been. Neuroimage 62, 743–747 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heininga VE, Oldehinkel AJ, Veenstra R. & Nederhof E. I just ran a thousand analyses: benefits of multiple testing in understanding equivocal evidence on gene-environment interactions. PLoS ONE 10, e0125383 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chambers CD, Feredoes E, Muthukumaraswamy SD & Etchells JP Instead of ‘playing the game’ it is time to change the rules: Registered Reports at AIMS Neuroscience and beyond. AIMS Neurosci. 1, 4–17 (2014). [Google Scholar]
- 41.Muthukumaraswamy SD, Routley B, Droog W, Singh KD & Hamandi K. The effects of AMPA blockade on the spectral profile of human early visual cortex recordings studied with non-invasive MEG. Cortex 81, 266–275 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Hobson HM & Bishop DVM Mu suppression — a good measure of the human mirror neuron system? Cortex 82, 290–310 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Churchill NW et al. Optimizing preprocessing and analysis pipelines for single-subject fMRI: 2. Interactions with ICA, PCA, task contrast and intersubject heterogeneity. PLoS ONE 7, e31147 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bennett CM, Miller MB & Wolford GL Neural correlates of interspecies perspective taking in the post-mortem Atlantic Salmon: an argument for multiple comparisons correction. Neuroimage 47, S125 (2009). [Google Scholar]
- 45.Eklund A, Nichols TE & Knutsson H. Cluster failure: why fMRI inferences for spatial extent have inflated false-positive rates. Proc. Natl Acad. Sci. USA 113, 7900–7905 (2016). This paper shows that some commonly used methods for cluster-based multiple-comparison correction can exhibit inflated false-positive rates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nichols T. & Hayasaka S. Controlling the familywise error rate in functional neuroimaging: a comparative review. Stat. Methods Med. Res 12, 419–446 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Wager TD, Lindquist M. & Kaplan L. Meta-analysis of functional neuroimaging data: current and future directions. Soc. Cogn. Affect. Neurosci 2, 150–158 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lieberman MD & Cunningham WA Type I and Type II error concerns in fMRI research: re-balancing the scale. Soc. Cogn. Affect. Neurosci 4, 423–428 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bennett CM, Wolford GL & Miller MB The principled control of false positives in neuroimaging. Soc. Cogn. Affect. Neurosci 4, 417–422 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hayasaka S. & Nichols TE Validating cluster size inference: random field and permutation methods. Neuroimage 20, 2343–2356 (2003). [DOI] [PubMed] [Google Scholar]
- 51.Gorgolewski KJ et al. NeuroVault.org: a web-based repository for collecting and sharing unthresholded statistical maps of the human brain. Front. Neuroinform 9, 8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hunt LT, Dolan RJ & Behrens TEJ Hierarchical competitions subserving multi-attribute choice. Nat. Neurosci 17, 1613–1622 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shehzad Z. et al. A multivariate distance-based analytic framework for connectome-wide association studies. Neuroimage 93 (Pt. 1), 74–94 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rubinov M. & Sporns O. Complex network measures of brain connectivity: uses and interpretations. Neuroimage 52, 1059–1069 (2010). [DOI] [PubMed] [Google Scholar]
- 55.Craddock RC, Milham MP & LaConte SM Predicting intrinsic brain activity. Neuroimage 82, 127–136 (2013). [DOI] [PubMed] [Google Scholar]
- 56.Butler RW & Finelli GB The infeasibility of quantifying the reliability of life-critical real-time software. IEEE Trans. Software Eng 19, 3–12 (1993). [Google Scholar]
- 57.Cox RW, Reynolds RC & Taylor PA AFNI and clustering: false positive rates redux. Preprint at bioRxiv 10.1101/065862(2016). [DOI] [Google Scholar]
- 58.Waskom ML, Kumaran D, Gordon AM, Rissman J. & Wagner AD Frontoparietal representations of task context support the flexible control of goal-directed cognition. J. Neurosci 34, 10743–10755 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Poldrack RA et al. Guidelines for reporting an fMRI study. Neuroimage 40, 409–414 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carp J. The secret lives of experiments: methods reporting in the fMRI literature. Neuroimage 63, 289–300 (2012). [DOI] [PubMed] [Google Scholar]
- 61.Guo Q. et al. The reporting of observational clinical functional magnetic resonance imaging studies: a systematic review. PLoS ONE 9, e94412 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nichols TE et al. Best practices in data analysis and sharing in neuroimaging using MRI. Preprint at bioRxiv 10.1101/054262(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Poldrack RA Can cognitive processes be inferred from neuroimaging data? Trends Cogn. Sci 10, 59–63 (2006). [DOI] [PubMed] [Google Scholar]
- 64.Gelman A. & Stern H. The difference between ‘significant’ and ‘not significant’ is not itself statistically significant. Am. Stat 60, 328–331 (2006). [Google Scholar]
- 65.Nieuwenhuis S, Forstmann BU & Wagenmakers E-J Erroneous analyses of interactions in neuroscience: a problem of significance. Nat. Neurosci 14, 1105–1107 (2011). [DOI] [PubMed] [Google Scholar]
- 66.Boekel W. et al. A purely confirmatory replication study of structural brain-behavior correlations. Cortex 66, 115–133 (2015). [DOI] [PubMed] [Google Scholar]
- 67.Begley CG & Ellis LM Drug development: raise standards for preclinical cancer research. Nature 483, 531–533 (2012). [DOI] [PubMed] [Google Scholar]
- 68.Open Science Collaboration. Estimating the reproducibility of psychological science. Science 349, aac4716 (2015). This paper reports a large-scale collaboration that quantified the replicability of research in psychology, showing that less than half of the published findings were replicable. [DOI] [PubMed] [Google Scholar]
- 69.Zuo X-N et al. An open science resource for establishing reliability and reproducibility in functional connectomics. Sci. Data 1, 140049 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Poldrack RA et al. Toward open sharing of task-based fMRI data: the OpenfMRI project. Front. Neuroinform 7, 1–12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gil Y. et al. Toward the geoscience paper of the future: best practices for documenting and sharing research from data to software to provenance. Earth Space Sci. 3, 388–415 (2016). [Google Scholar]
- 72.Boulesteix A-L Ten simple rules for reducing overoptimistic reporting in methodological computational research. PLoS Comput. Biol 11, e1004191 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gorgolewski KJ et al. The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Sci. Data 3, 160044 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Flint J. & Munafò MR Candidate and noncandidate genes in behavior genetics. Curr. Opin. Neurobiol 23, 57–61 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ioannidis JP, Tarone R. & McLaughlin JK The false-positive to false-negative ratio in epidemiologic studies. Epidemiology 22, 450 (2011). [DOI] [PubMed] [Google Scholar]
- 76.Burgess S. et al. Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur. J. Epidemiol 30, 543–552 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stein JL et al. Identification of common variants associated with human hippocampal and intracranial volumes. Nat. Genet 44, 552–561 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Barch DM et al. Function in the human connectome: task-fMRI and individual differences in behavior. Neuroimage 80, 169–189 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.