Abstract
Context:
Next generation sequencing (NGS)-based assays are being increasingly used in the clinical setting for the detection of somatic variants in solid tumors, but limited data are available regarding the interlaboratory performance of these assays.
Objective:
We examined proficiency testing data from the initial College of American Pathologists (CAP) Next-Generation Sequencing Solid Tumor survey to report on laboratory performance.
Design:
CAP proficiency testing results from 111 laboratories were analyzed for accuracy and associated assay performance characteristics.
Results:
The overall accuracy observed for all variants was 98.3%. Rare false-negative results could not be attributed to sequencing platform, selection method, or other assay characteristics. The median and average of the variant allele fractions reported by the laboratories were within 10% of those orthogonally determined by digital polymerase chain reaction (PCR) for each variant. The median coverage reported at the variant sites ranged from 1,922 – 3,297.
Conclusions:
Laboratories demonstrated an overall accuracy of >98% with high specificity when examining 10 clinically relevant somatic single-nucleotide variants (SNVs) with a variant allele fraction of 15% or greater. These initial data suggest excellent performance, but further ongoing studies are needed to evaluate the performance of lower variant allele fractions and additional variant types.
Introduction
The past several years have seen the development or expansion of several national and international efforts that aim to accelerate the implementation of precision medicine1–4. One area of focus is oncology, and molecular alterations are currently being used in clinical practice as biomarkers to assist diagnostic categorization, prognostication, as well as the selection and monitoring of therapies5. Increasingly, massively parallel sequencing, or next generation sequencing (NGS)-based methods, are used to identify deoxyribonucleic acid (DNA) and ribonucleic acid (RNA) biomarkers for clinical management. A central issue for test performance is to ensure robust molecular results to guide clinical care. In this manuscript, we describe the analytic accuracy of clinical NGS-based molecular oncology testing for the detection of somatic variants in solid tumors based on a large inter-laboratory comparison using blinded, engineered specimens.
A previous publication from our group surveyed clinical laboratories regarding specimen requirements, assay characteristics, and other trends in NGS-based oncology testing6. Two key findings from that survey guided the development of this proficiency testing program: 1) the majority of laboratories perform targeted sequencing of tumor-only specimens to detect single-nucleotide variants (SNVs) and small insertions or deletions with a reported 5–10% variant allele fraction as the lower limit of detection, and 2) testing is primarily performed using amplicon-based, predesigned commercial kits using benchtop sequencers. With this guidance, we developed a proficiency-testing program for NGS-based oncology tests designed to detect recurring somatic variants in solid tumor specimens. This and other proficiency testing programs are designed to ensure the accuracy and reliability of patient test results provided by clinical laboratories7–9. Here, we report the performance of laboratories for this initial Next-Generation Sequencing Solid Tumor proficiency testing survey.
Methods
Data were derived from the initial College of American Pathologists (CAP) Next-Generation Sequencing Solid Tumor Survey (NGSST-A 2016). The specimens were sent to laboratories on May 9, 2016, and the laboratories returned the survey results by June 18, 2016. Laboratories were concurrently sent three independent specimens containing linearized plasmids with engineered somatic variants mixed with genomic DNA derived from the GM24385 cell line to achieve variant allele fractions (VAFs) ranging from 15 to 50% (Table 1).
Table 1.
Laboratory performance for detection of somatic variants
| Gene | Transcript | Nucleotide change |
Protein change |
Genomic description (hg19) |
# labs that tested for variant |
# labs that detected variant (%) |
# labs that did not detected variant (%) |
# labs that did not test for variant |
Engineered VAF (%) |
Digital PCR VAF (%) |
Average reported VAF (%) |
Standard deviation VAF |
Median VAF (%) |
Range VAF (%) |
Median coverage |
Range coverage |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AKT1 | NM_005163.2 | c.49G>A | p.E17K | chr14:105,246,551C>T | 102 | 101 (99.0) | 1 (0.98) | 9 | 35 | 35.8 | 36.5 | 8.5 | 35 | 3 – 90 | 2,325X | 47 – 75,886 | |||
| ALK | NM_004304.4 | c.3824G>A | p.R1275Q | chr2:29,432,664C>T | 90 | 87 (96.7) | 3 (3.3) | 21 | 50 | 48.4 | 49.6 | 5.3 | 50 | 15 – 63 | 2,000X | 102 – 48,763 | |||
| BRAF | NM_004333.4 | c.1799T>A | p.V600E | chr7:140,453,136A>T | 110 | 110 (100) | 0 (0) | 1 | 15 | 14.8 | 14.4 | 2.9 | 15 | 5 – 23 | 1,922X | 60 – 99,999 | |||
| EGFR | NM_005228.3 | c.2155G>A | p.G719S | chr7:55,241,707G>A | 109 | 106 (97.2) | 3 (2.8)* | 1 | 20 | 18.6 | 18.7 | 4.0 | 20 | 8 – 30 | 2064X | 100 – 99,999 | |||
| FBXW7 | NM_033632.3 | c.1394G>A | p.R465H | chr4:153,249,384C>T | 85 | 83 (97.6) | 2 (2.4) | 26 | 50 | 51.3 | 48.6 | 4.6 | 49 | 31 – 64 | 3,297X | 32 – 99,999 | |||
| IDH1 | NM_005896.2 | c.395G>A | p.R132H | chr2:209,113,112C>T | 86 | 84 (97.7) | 2 (2.3)** | 25 | 40 | 41.5 | 40.4 | 5.8 | 41 | 2 – 54 | 2,444X | 106 – 64,237 | |||
| KIT | NM_000222.2 | c.1961T>C | p.V654A | chr4:55,594,258T>C | 102 | 99 (97.1) | 3 (2.9) | 9 | 30 | 28.7 | 25.9 | 4.0 | 26 | 14 – 38 | 2,027X | 272 – 65,134 | |||
| KRAS | NM_004985.3 | c.38G>A | p.G13D | chr12:25,398,281C>T | 111 | 111 (100) | 0 (0) | 0 | 25 | 27.4 | 25.5 | 4.6 | 26 | 10 – 40 | 2,222X | 283 – 48,507 | |||
| NRAS | NM_002524.4 | c.182A>G | p.Q61R | chr1:115,256,529T>C | 110 | 108 (98.2) | 2 (1.8) | 0 | 30 | 31.7 | 30.6 | 4.0 | 30 | 15 – 46 | 2,911X | 68 – 99,999 | |||
| PIK3CA | NM_006218.2 | c.3140A>G | p.H1047R | chr3:178,952,085A>G | 105 | 104 (99.0) | 1 (0.95) | 6 | 20 | 20.4 | 20.2 | 3.7 | 21 | 11 – 31 | 2,000X | 159 – 99,999 | |||
| Total variants | Total variants detected (%) | Total variants not detected (%) | |||||||||||||||||
| 1010 | 993 (98.3) | 17 (1.7) | |||||||||||||||||
Two laboratories failed to detect the EGFR p.G719S variant, but reported p.G719C
One laboratory failed to detect the IDH1 p.R132H variant, but reported IDH1 p.R132S
Definition: VAF – variant allele fraction, PCR - polymerase chain reaction
Specimens were generated by a commercial reference material vendor under Good Laboratory Practice. Specimen 1 contained variants in AKT1, BRAF, FBXW7, IDH1, and KRAS; specimen 2 contained variants in EGFR and NRAS; specimen 3 contained variants in ALK, KIT, and PIK3CA. The synthetic DNA inserts contain a somatic variant with approximately 500 bp of flanking genomic sequence on each side of the variant. The flanking genomic DNA sequence was matched to the diluent genomic DNA (i.e., GM24385). VAFs were orthogonally confirmed by digital PCR by measuring absolute mutant copies and wild type copies. The VAFs were calculated by putting the absolute copies in the following formula for each variant in specimen 1, 2 and 3:
Laboratories were instructed to perform NGS using the methodology routinely performed on clinical samples in their laboratory for the detection of somatic SNVs, insertions, and deletions in solid tumors. This could include targeted gene panels, whole exome, or whole genome sequencing. As is standard for CAP surveys, laboratories were instructed that variant confirmation by a secondary orthogonal methodology must follow the laboratory’s standard procedure for testing clinical specimens, but cannot be referred to another laboratory. Following testing, laboratories reported the variants detected in each specimen by selecting from a master variant list containing 90 variants in 15 genes (AKT1, ALK, BRAF, EGFR, ERBB2, FBXW7, FGFR2, GNAS, IDH1, KIT, KRAS, MET, NRAS, PIK3CA, STK11). In addition, laboratories provided read depth and variant allele fraction for each reported variant as well as the laboratory’s assay performance characteristics.
All figures were generated using the ggplot package (Version 2.2.1. Boston, MA: RStudio; 2016. URL http://github.com/tidyverse/ggplot2/) loaded on R (Version 3.4.1. Vienna, Austria: R Core Team; 2017. URL http://www.R-project.org/). Violin plots were trimmed to the range of each distribution.
Results
Of the 111 laboratories that reported results, 108 performed targeted sequencing of mutation hotspots or cancer genes, two performed whole exome sequencing, and one performed a combination of whole genome, whole exome and targeted sequencing. The number of laboratories whose assay enabled the detection of a particular variant ranged from 85 (77%) for the FBXW7 p.R465H variant to 111 (100%) for the KRAS p.G13D variant (Table 1). The percentage of laboratories that correctly identified each variant ranged from 96.7% (87/90) for the ALK p.R1275Q variant to 100% for the BRAF p.V600E (110/110) and KRAS p.G13D (111/111) variants. The overall accuracy observed for all variants was 98.3% (993/1010).
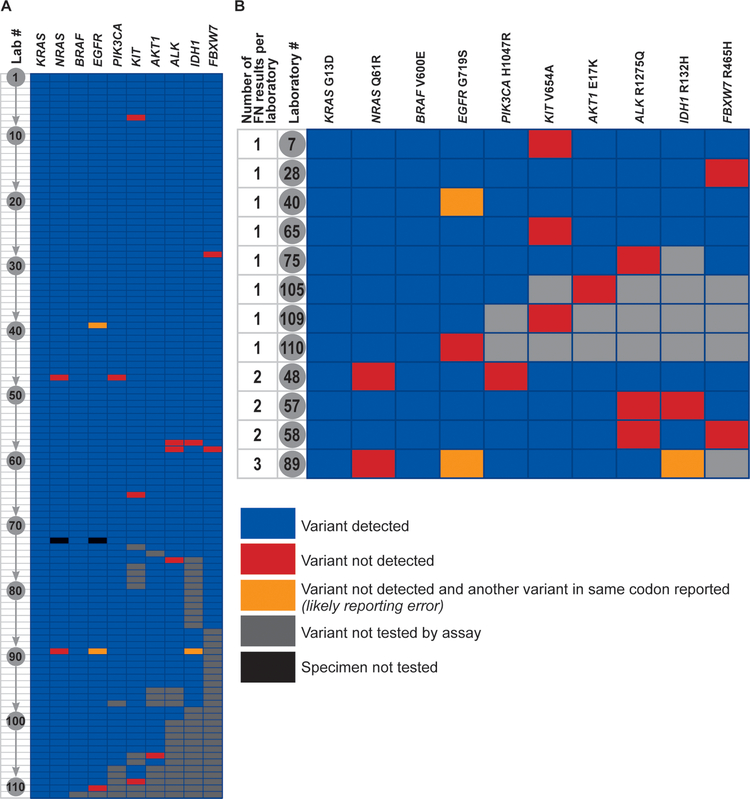
There were 17 false negative results reported by 12 laboratories (Figure 1A, 1B) – 8 laboratories reported a single false negative result, 3 laboratories reported 2 false negative results, and 1 laboratory reported 3 false negative results. There were 3 false positive results in which the laboratory correctly detected the presence of a variant in a gene, but reported it as a different variant from expected (e.g., the laboratory did not report detection of the EGFR p.G719S variant, but instead reported detection of the EGFR p.G719C variant).
Figure 1 –
Performance of individual laboratories. A – Each laboratory is represented as a row numbered from 1 through 111, and each of the 10 variants is represented by a column. Individual cell colors represent the following: blue – variant was detected by the laboratory, red – variant was not detected by the laboratory, yellow – variant was not detected by the laboratory, and the laboratory reported another variant in the same codon (this is likely a post-analytical or reporting error), grey – this particular variant is not tested for by the laboratory’s assay, black – specimen not tested. B – Performance data from the 12 laboratories with false-negative results. FN – False-negative.
Among the 12 laboratories with false negative results, there was a diversity of sequencing platforms and selection methods (Table 2). Likewise, there was no significant association between laboratories with false negative results and the reported lower limit of detection based on allele fraction for SNVs, average coverage, or minimum coverage (Table 3).
Table 2.
Instrument and selection method used by reporting laboratories
| Sequencing Platform | # of labs (%) | # of labs (%)* that did not detect expected mutation | Selection Method for Targeted Sequencing | # of labs (%) | # of labs (%)* that did not detect expected mutation | |
|---|---|---|---|---|---|---|
| Ion Torrent PGM | 49 (44.1) | 4 (33.3) | Ion AmpliSeq Cancer Hotspot Panel v2 | 40 (36.0) | 2 (16.7) | |
| Illumina MiSeq | 34 (30.6) | 4** (33.3) | Custom Enrichment Approach | 34 (30.6) | 5 (41.7) | |
| Illumina HiSeq 2500 | 11 (9.9) | 2 (16.7) | Illumina TruSight Tumor Sequencing Panel | 12 (10.8) | 2 (16.7) | |
| Ion Torrent Proton | 5 (4.5) | Illumina TruSeq Amplicon Cancer Panel | 9 (8.1) | 2** (16.7) | ||
| Illumina NextSeq 500 | 4 (3.6) | 1 (8.3) | Ion AmpliSeq Comprehensive Cancer Panel | 3 (2.7) | ||
| Illumina MiSeqDx | 4 (3.6) | 1 (8.3) | Ion AmpliSeq Colon and Lung Cancer Research Panel | 3 (2.7) | ||
| Illumina HiSeq 3000/4000 | 3 (2.7) | Ion AmpliSeq Oncomine Comprehensive Assay | 2 (1.8) | |||
| Ion Torrent S5/S5 XL | 1 (0.9) | Ion AmpliSeq Oncomine Focus Assay | 2 (1.8) | |||
| RainDance ThunderBolts Cancer Panel | 2 (1.8) | |||||
| Total | 111 | 12 | Multiplicom Tumor Hotspot MASTR Plus Assay | 1 (0.9) | ||
| QIAGEN Human Tumor Actionable Mutations Panel | 1 (0.9) | |||||
| Not Applicable – exome sequencing | 2 (1.8) | 1 (8.3) | ||||
| Total | 111 | 12 |
Percentage is based on the 12 laboratories that did not detect the expected mutation.
Includes results from the laboratory that correctly detected the presence of a variant in a gene, but reported it as a different variant from expected (i.e., the laboratory did not report detection of the EGFR p.G719S variant, but instead reported detection of the EGFR p.G719C variant). This laboratory did not have any other false-positive or false-negative results, and this error likely represents a post-analytical (reporting) error.
Table 3.
Reported lower limit of detection (LLOD) based on variant allele fraction (VAF) for SNVs, average coverage, and minimum coverage
| Reported LLOD VAF (%) | # of labs (%) | # of labs (%)* that did not detect expected mutation | Average number of reads covering targeted bases | # of labs (%) | # of labs (%)* that did not detect expected mutation | Minimum number of reads covering targeted bases | # of labs (%) | # of labs (%)* that did not detect expected mutation | ||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 3 (2.7) | 51 – 150 | 2 (1.8) | 1 (8.3) | 0 – 25 | 8 (7.2) | 1 (8.3) | |||
| 2 | 5 (4.5) | 1 (8.3) | 151 – 250 | 3 (2.7) | 26 – 50 | 6 (5.4) | ||||
| 3 | 12 (10.8) | 1 (8.3) | 251 – 350 | 3 (2.7) | 1 (8.3) | 51 – 150 | 23 (20.7) | |||
| 4 | 4 (3.6) | 351 – 500 | 4 (3.6) | 151 – 250 | 13 (11.7) | 2 (16.7) | ||||
| 5 | 70 (63.1) | 8** (66.7) | 501 – 750 | 9 (8.1) | 1 (8.3) | 251 – 350 | 12 (10.8) | 1 (8.3) | ||
| 9 | 1 (0.9) | 751 – 1,000 | 7 (6.3) | 2 (16.7) | 351 – 500 | 17 (15.3) | 4** (33.3) | |||
| 10 | 13 (11.7) | 1 (8.3) | 1,001 – 1,500 | 17 (15.3) | 2** (16.7) | 501 – 750 | 13 (11.7) | |||
| 15 | 2 (1.8) | 1,501 – 2,500 | 18 (16.2) | 751 – 1,000 | 2 (1.8) | |||||
| Not specified | 1 (0.9) | 1 (8.3) | > 2,500 | 39 (35.1) | 3 (25.0) | 1,001 – 1,500 | 4 (3.6) | 2 (16.7) | ||
| Not established | 8 (7.2) | 1 (8.3) | > 2,500 | 3 (2.7) | ||||||
| Total | 111 | 12 | Not specified | 1 (0.9) | 1 (8.3) | No minimum read requirement | 9 (8.1) | 1 (8.3) | ||
| Not specified | 1 (0.9) | 1 (8.3) | ||||||||
| Total | 111 | 12 | ||||||||
| Total | 111 | 12 |
Percentage is based on the 12 laboratories that did not detect the expected mutation.
Includes results from the laboratory that correctly detected the presence of a variant in a gene, but reported it as a different variant from expected (i.e., the laboratory did not report detection of the EGFR p.G719S variant, but instead reported detection of the EGFR p.G719C variant). This laboratory did not have any other false-positive or false-negative results, and this error likely represents a post-analytical (reporting) error.
SNV = single nucleotide variant
Not specified indicates laboratory did not answer the question on the result form.
Not established indicates that the laboratory reported not having established the average number of reads that cover the targeted bases in that laboratory’s assay.
No minimum read requirement indicates that the laboratory reported not having a minimum read requirement for each targeted base in the assay.
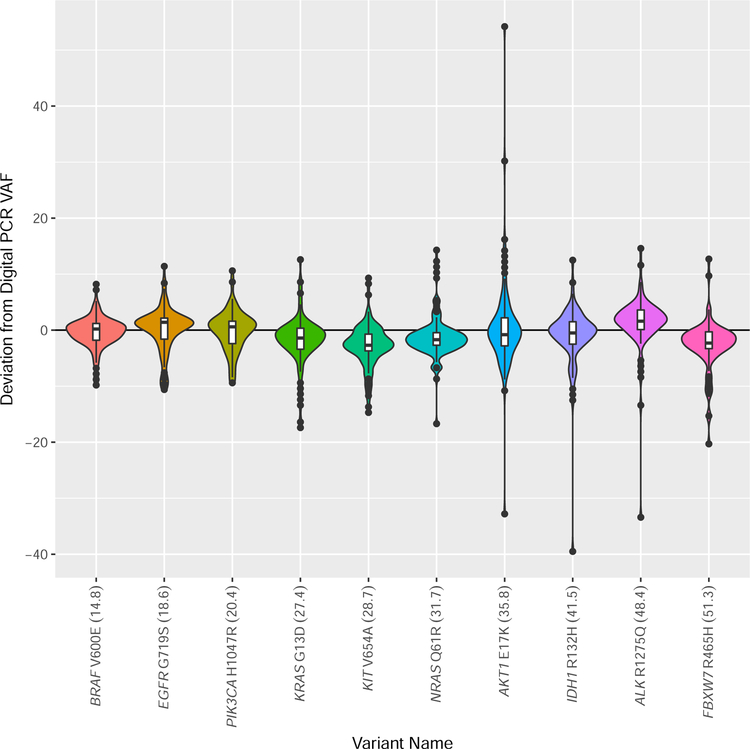
The median and average of the variant allele fractions reported by the laboratories were within 10% of those determined by digital polymerase chain reaction (PCR) for each variant (Table 1, Figure 2). The maximum observed standard deviation was 8.5 (range 2.9 – 8.5) (Table 1), and at least 75% of laboratories reported variant allele fractions within 20% of the engineered value for each variant.
Figure 2 –
Violin plot demonstrating the deviation of the variant allele fraction (VAF) reported by the participating laboratories from the VAF measured by digital polymerase chain reaction (PCR). The VAF measured by digital PCR for each variant is provided as a percentage in parentheses after each variant name on the x-axis. The width of each violin plot indicates the number of laboratories that reported the different VAFs. The violin plots contain a box that represents the lower and upper quartiles, and the solid line drawn across the box indicates the median value. The interquartile range (IQ) is the difference between the upper (Q3) and lower (Q1) quartiles. The extent of the vertical lines extending from the boxes within each violin plot represent the upper or lower inner fence, where a lower inner fence is defined as Q1–1.5*(IQ) and an upper inner fence as Q3+1.5*(IQ). Outliers, drawn as black dots, are observations that fall outside the upper or lower inner fences.
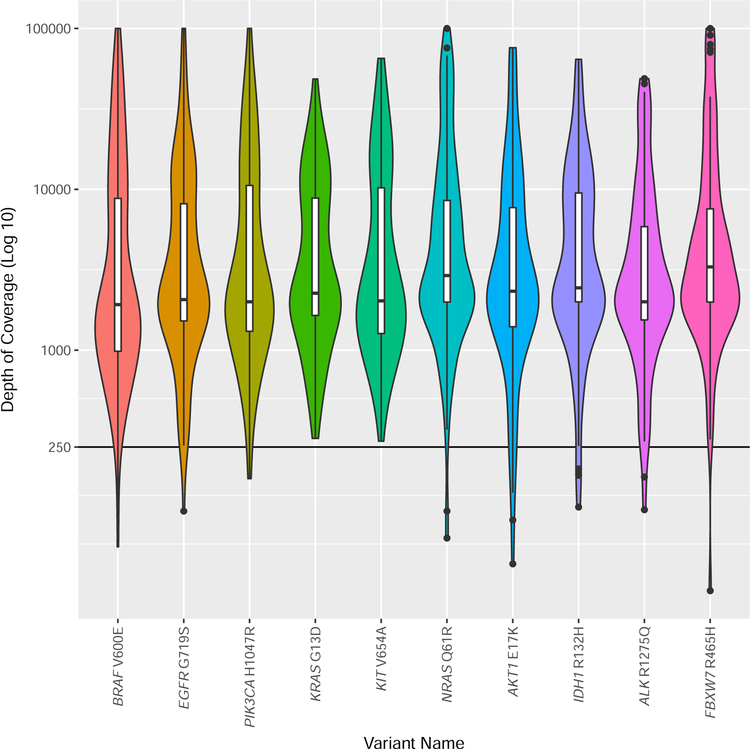
The median coverage reported at the variant sites ranged from 1,922 – 3,297, although a significant range of coverage was reported (Table 1, Figure 3). The coverage data were further evaluated to determine if there were differences between amplicon and hybridization-based capture approaches. Of the 108 laboratories that performed only targeted sequencing of mutation hotspots or cancer genes, 107 laboratories provided sufficient information to determine if they were using amplicon or hybridization-based selection methods. Of these laboratories, 83.2% (89/107) used amplification-based selection methods, and 16.8% (18/107) used hybridization-based selection methods. The median coverage depth reported across all engineered variant positions was 3,445 reads for amplification-based methods (range 100 – 99,999) and 959.5 reads (range 47–4,244) for hybridization-based methods.
Figure 3 –
Violin plot of the depth of coverage reported by the laboratories for each variant. Depth of coverage is plotted as (Log10) of coverage. The width of each violin plot indicates the number of laboratories that reported the different depth of coverage. The violin plots contain a box that represents the lower and upper quartiles, and the solid line drawn across the box indicates the median value. The interquartile range (IQ) is the difference between the upper (Q3) and lower (Q1) quartiles. The extent of the vertical lines extending from the boxes within each violin plot represent the upper or lower inner fence, where a lower inner fence is defined as Q1–1.5*(IQ) and an upper inner fence as Q3+1.5*(IQ). Outliers, drawn as black dots, are observations that fall outside the upper or lower inner fences.
Discussion
Surveys about NGS-based oncology testing practices from our group6 and others10 reveal a couple of observed trends. First, the majority of laboratories are performing targeted sequencing of tumor-only specimens to detect SNVs and small insertions and deletions with a reported lower limited of detection of 5–10% variant allele fraction. Second, the testing is primarily performed using amplicon-based, predesigned commercial kits with benchtop sequencers. These trends were still observed in the current study; however, as was noted previously, there is a significant diversity of practice with respect to both the wet laboratory and bioinformatics processes. These test results from clinical molecular laboratories are used to guide patient care, including assistance with diagnosis, determination of prognosis, as well as the selection and monitoring of therapy. Consequently, evaluation of a set of common, well-characterized specimens is critical to evaluate analytical performance across platforms and approaches.
One hundred and eleven laboratories reported results from their NGS-based assay for the identification of somatic variants in solid tumor specimens. The laboratories were provided with three engineered specimens containing a total of 10 recurring somatic variants with a variant allele fraction between 15 – 50%. The overall accuracy observed for all variants was very high at 98.3%, and this high degree of accuracy was observed for each individual variant ranging from 96.7% to 100%. It is difficult to calculate analytic specificity since the exact target region for each assay is not known. However, only three potential false positive results were reported suggesting a high degree of analytic specificity, and as discussed further below, these could be transcription errors.
Twelve of 111 laboratories (10.8%) accounted for the 17-false negative and three concurrent false positive results. All three false positive results were associated with a concurrent false negative result involving the same codon of the same gene, indicating mischaracterization of a single mutation. There is no expected clinical consequence of these errors (e.g., EGFR p.G719C vs p.G719S), and these may be attributed, in part, to the nature of the survey result form, which is not typical of clinical lab reports and is more prone to subtle transcription errors. Consequently, the combined accuracy of the assays for the variants tested may be as high as 98.6% (996/1010).
Prior to this and related studies, discussion about the accuracy and reliability of NGS-based oncology testing was often based on concordance data in which multiple alignment and variant-calling pipelines were applied to exome or genome sequencing data for germline sequencing applications. These studies often demonstrated low to moderate concordance when multiple analysis pipelines were applied to the same exome or genome data11,12. It was unclear at the time, whether these studies could be extrapolated to clinical NGS-based oncology testing due to differences in application (germline vs. somatic), target region (exome or genome vs. targeted panel) and setting (research vs. clinical laboratory). The data presented in this manuscript both directly and rigorously address accuracy and reliability of clinical NGS-based oncology testing, indicating very high inter-laboratory agreement for the detection of somatic SNVs. The robust detection of clinically relevant somatic SNVs in this study is consistent with two prior studies focused on bioinformatics analysis of NGS-based oncology testing within clinical laboratories as well as one analytical validation study performed for the National Cancer Institute (NCI)-MATCH trial. A multi-institutional exchange of FASTQ files between six clinical laboratories showed a high rate of concordance for SNV detection and complete concordance for the detection of clinically significant SNVs13. Likewise, a pilot CAP proficiency testing program in which somatic variants were introduced via an in-silico approach demonstrated 97% accuracy for detection of somatic SNVs and indels with VAFs greater than 15%14. Finally, a validation study of an NGS-based oncology assay performed by four clinical laboratories as part of the NCI-MATCH trial demonstrated a high level of reproducibility in the detection of reportable somatic variants15.
In this study, variants in BRAF, KRAS, and EGFR were detected with an accuracy of 100%, 100%, and 97.2%, respectively. These are consistent with the acceptable proficiency testing results observed for Food and Drug Administration companion diagnostics (FDA-CD) and diverse laboratory-developed tests (LDTs) for BRAF (93.0% FDA-CD, 96.6% LDTs), KRAS (98.8% FDA-CD, 97.4% LDTs), and EGFR (99.1% FDA-CD, 97.6% LDTs)16. Overall, the above studies indicate that clinical laboratories are able to detect clinically significant somatic SNVs from solid tumor specimens with high accuracy and reliability.
The 17 false-negative results were observed for variants with engineered allele fractions ranging from 20–50% (Table 1). Of the 12 laboratories with an error, 11 reported a lower limit of detection for SNVs of ≤15% variant allele fraction, and one laboratory did not specify their lower limit of detection. Since the variants that were not detected had variant allele fractions greater than 18% when measured by digital PCR, at least 11 of the laboratories should have been able to detect these variants based on their reported assay characteristics. Furthermore, there was no clear association between laboratories with an error and sequencing platform, selection method, reported lower limit of detection, or average and minimum number of reads covering the targeted bases. Since laboratories were only asked to report coverage results for positions at which they identified a variant, we are unable to determine if low coverage contributed to these false negative results.
Recent joint consensus recommendations from the Association for Molecular Pathology (AMP), American Society of Clinical Oncology (ASCO), and College of American Pathologists (CAP) indicate that variant allele fraction “should be evaluated and included in the report when appropriate”.10 The average and median variant allele fraction observed for each variant closely approximate the engineered variant allele fraction and that measured by an orthogonal method, digital PCR (Table 1, Figure 2). The observed standard deviation ranged from 2.9 to 8.5 for each variant, and at least 75% of laboratories reported variant allele fractions within 20% of the engineered value for each variant. Collectively, these data indicate that variant allele fraction measured by most laboratories is a reasonable approximation of the actual variant allele fraction. However, a minority of laboratories in this study reported variant allele fractions that deviated significantly from the expected value. This suggests that laboratories that report variant allele fractions from their NGS-based molecular oncology assay should verify the accuracy of their reported variant allele fractions as part of assay validation. For extreme outliers (e.g., reported VAF = 2% for IDH1 p.R132H, when engineered VAF = 40% and observed VAF by digital PCR = 41.5%), it is possible that these represent a transcription error introduced during the reporting process. We also cannot exclude that artificial constructs may have resulted in artifactual low variant allele fractions for some selection methods, although we did not detect a pattern suggesting this interpretation.
For each position that a laboratory reported a variant, the total coverage depth at that position was also reported. The median coverage depth for each variant position varied from 1,922 – 3,297, with a range across all variant sites from 32 – 99,999 (with 99,999 being the highest value that could be entered on the result form). Laboratories using hybridization-based selection methods reported several fold lower median coverage depth across all engineered variant positions (3,445 reads for amplification-based methods vs. 959.5 reads for hybridization-based methods). Recent joint consensus recommendations from AMP and CAP suggest a minimum depth of coverage >250 reads.17 Over 98% (974/992) of the total variants reported by all laboratories had coverage exceeding this recommended level.
There are three important caveats regarding coverage depth. First, laboratories can variably define coverage. As an example, coverage depth can refer to the total number of reads covering a site or the total number of unique reads covering a site (if hybrid capture or molecular barcoding is used) with or without the application of other metrics related to base quality, mapping quality, or other quality filters. The proficiency testing survey did not define coverage or inquire as to laboratories definition of coverage, so the provided coverage depths may not be equivalent. A second and related consideration is that the proficiency testing survey did not inquire whether laboratories incorporate library complexity, a measure of the number of unique fragments present in a library, when reporting coverage depth.17,18 Given that the majority of laboratories in this survey used amplification-based methods without molecular barcodes, most laboratories were unable to directly assess library complexity. The minority of laboratories that used hybridization-based methods or molecular barcoding could have incorporated this quality metric, enabling the reporting of independent coverage depth, which is a more robust measure of assay performance. The third consideration is that laboratories were only asked to report coverage depth for sites at which they identified variants. Consequently, we do not know whether low coverage depth could have contributed to the false-negative results.
When considering the implications and generalizability of these data, there are several limitations. First, the laboratories were provided with engineered nucleic acid specimens, i.e., linearized plasmids diluted into genomic DNA. These specimens do not control for many important pre-analytical factors relevant to standard clinical samples such as tissue processing and fixation, selection of a tissue section with optimal neoplastic cellularity, tumor enrichment through microdissection, or nucleic acid extraction methodology.19 Recent work by Sims and colleagues suggests that linearized plasmids diluted in genomic DNA isolated from formalin-fixed, paraffin-embedded cell lines performed similarly to endogenous variants contained within genomic DNA derived from formalin-fixed, paraffin-embedded (FFPE) cell lines.20 One difference is that the diluent genomic DNA used in this study was derived from cells that had not been formalin-fixed and paraffin-embedded. Consequently, these specimens do not exactly recapitulate the tissue specimens routinely processed by clinical laboratories. However, the need to provide homogeneous, well-characterized materials to over a hundred laboratories necessitates some compromise to facilitate these large inter-laboratory comparisons of analytical performance; previous experience with bona fide clinical samples showed variation, due to intratumor heterogeneity, between the sections sent to different participating laboratories. Second, laboratories are asked to return results using a standardized reporting form with structured data that enables high-throughput grading, but is a deviation from their normal reporting procedure. This may introduce transcription and other manual errors that would be less likely with the use of a laboratory information system and other quality control systems during actual clinical testing. Third, the lowest variant allele fraction tested in this survey was approximately 15%. The vast majority of laboratories report lower limits of detection between 5–10% variant allele fraction, and the current study does not evaluate the performance of NGS-based molecular oncology assays <15%. This is important because it becomes more challenging to detect somatic variants as allele fraction decreases.18,21,22 Fourth, the current survey only included somatic SNVs. Insertions, deletions, homopolymer length changes, copy number changes, and structural variants are also commonly detected by NGS-based molecular oncology assays, and these variant types are generally more challenging to detect than SNVs.
In the current manuscript, we present the results of a large, inter-laboratory study about the performance of clinical NGS-based oncology assays used for the detection of somatic variants in solid tumors in 111 participating laboratories. Laboratories demonstrated an overall accuracy of >98% with high specificity when examining 10 clinically relevant somatic SNVs with a variant allele fraction of 15% or greater. These initial data suggest excellent performance, but further work is needed to evaluate the performance of lower variant allele fractions and additional variant types, such as insertions, deletions, homopolymer track length changes, copy number variants, and structural variants. Insertions, deletions, delins, and more complex variants are being incorporated into future CAP proficiency testing surveys and other materials. Likewise, similar efforts using standardized reference samples will be needed to evaluate the performance of other NGS-based oncology assays for new and emerging applications such as cell-free, circulating tumor DNA assays. Recently introduced proficiency testing surveys that began in 2018 will allow the assessment of laboratory performance for detection of cell-free, circulating tumor DNA and for detection for recurring somatic RNA fusions using sequencing-based technologies. These and related efforts should allow for a broader and deeper understanding of the performance of clinical NGS-based oncology assays.
Acknowledgments
Funding:
CS was supported by NIH BD2K Training Grant (Biomedical Data Science Graduate Training at Stanford), PI: Russ Altman, Grant#: 1 T32 LM012409-01
Footnotes
Disclaimers:
The identification of specific products or scientific instrumentation is considered an integral part of the scientific endeavor and does not constitute endorsement or implied endorsement on the part of the author, DoD, or any component agency. The views expressed in this article are those of the author and do not reflect the official policy of the Department of Army/Navy/Air Force, Department of Defense, or U.S. Government.
References
- 1.Allen NE, Sudlow C, Peakman T, Collins R. UK biobank data: come and get it. Sci Transl Med 2014;6(224):224ed224. [DOI] [PubMed] [Google Scholar]
- 2.Laverty H, Goldman M. Editorial: The Innovative Medicines Initiative--collaborations are key to innovating R&D processes. Biotechnol J 2014;9(9):1095–1096. [DOI] [PubMed] [Google Scholar]
- 3.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med 2015;372(9):793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singer DS, Jacks T, Jaffee E. A U.S. “Cancer Moonshot” to accelerate cancer research. Science 2016;353(6304):1105–1106. [DOI] [PubMed] [Google Scholar]
- 5.Bombard Y, Bach PB, Offit K. Translating genomics in cancer care. J Natl Compr Canc Netw 2013;11(11):1343–1353. [DOI] [PubMed] [Google Scholar]
- 6.Nagarajan R, Bartley AN, Bridge JA, et al. A Window into Clinical Next Generation Sequencing-based Oncology Testing Practices. Arch Pathol Lab Med in press. (2017;141(12):1679–1685. [DOI] [PubMed] [Google Scholar]
- 7.Medicare, Medicaid and CLIA programs; revision of the laboratory regulations for the Medicare, Medicaid, and Clinical Laboratories Improvement Act of 1967 programs--HCFA. Final rule with comment period. Fed. Reg 1990;55(50):9538–9610. [PubMed] [Google Scholar]
- 8.Regulations for implementing the Clinical Laboratory Improvement Amendments of 1988: a summary. MMWR Recommendations and reports : Morbidity and mortality weekly report Recommendations and reports / Centers for Disease Control 1992;41(RR-9):1–17. [PubMed] [Google Scholar]
- 9.CLIA program; approval of the College of American Pathologists--HCFA. Notice. Fed. Reg 1995;60(27):7774–7780. [PubMed] [Google Scholar]
- 10.Li MM, Datto M, Duncavage EJ, et al. Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn 2017;19(1):4–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Rawe J, Jiang T, Sun G, et al. Low concordance of multiple variant-calling pipelines: practical implications for exome and genome sequencing. Genome Med 2013;5(3):28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cornish A, Guda C. A Comparison of Variant Calling Pipelines Using Genome in a Bottle as a Reference. BioMed Res Int 2015;2015:456479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davies KD, Farooqi MS, Gruidl M, et al. Multi-Institutional FASTQ File Exchange as a Means of Proficiency Testing for Next-Generation Sequencing Bioinformatics and Variant Interpretation. J Mol Diagn 2016;18(4):572–579. [DOI] [PubMed] [Google Scholar]
- 14.Duncavage EJ, Abel HJ, Merker JD, et al. A Model Study of In Silico Proficiency Testing for Clinical Next-Generation Sequencing. Arch Pathol Lab Med 2016;140(10):1085–1091. [DOI] [PubMed] [Google Scholar]
- 15.Lih CJ, Harrington RD, Sims DJ, et al. Analytical Validation of the Next-Generation Sequencing Assay for a Nationwide Signal-Finding Clinical Trial: Molecular Analysis for Therapy Choice Clinical Trial. J Mol Diagn 2017;19(2):313–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim AS, Bartley AN, Bridge JA, et al. Comparison of Laboratory-Developed Tests and FDA-Approved Assays for BRAF, EGFR, and KRAS Testing. JAMA Oncol. 2018;4(6):838–841.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jennings LJ, Arcila ME, Corless C, et al. Guidelines for Validation of Next-Generation Sequencing-Based Oncology Panels: A Joint Consensus Recommendation of the Association for Molecular Pathology and College of American Pathologists. J Mol Diagn 2017;19(3):341–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cottrell CE, Al-Kateb H, Bredemeyer AJ, et al. Validation of a next-generation sequencing assay for clinical molecular oncology. J Mol Diagn 2014;16(1):89–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.CLSI. Molecular Diagnostic Methods for Solid Tumors (Nonhematological Neoplasms). 1st ed. In. CLSI guideline MM23 Wayne, PA: Clinical and Laboratory Standards Institute; 2015. [Google Scholar]
- 20.Sims DJ, Harrington RD, Polley EC, et al. Plasmid-Based Materials as Multiplex Quality Controls and Calibrators for Clinical Next-Generation Sequencing Assays. J Mol Diagn 2016;18(3):336–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013;31(3):213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu H, DiCarlo J, Satya RV, Peng Q, Wang Y. Comparison of somatic mutation calling methods in amplicon and whole exome sequence data. BMC Genomics 2014;15:244. [DOI] [PMC free article] [PubMed] [Google Scholar]