Abstract
Developing alternative MS/MS strategies to distinguish isomeric lipids has become a high impact goal in shotgun lipidomics. Novel approaches have been developed to resolve structural features that are not discernible by traditional shotgun methods and have consequently promoted the discovery of new disease biomarkers. However, these methods have largely been limited to characterizing lipids with low structural complexity. Here, ultraviolet photodissociation (UVPD) strategies for phospholipid characterization are expanded for analysis of cardiolipins (CL), a class of phospholipids that exhibits a higher degree of structural complexity. A hybrid collision induced dissociation/193 nm UVPD (CID/UVPD) approach was implemented to pinpoint the location of both double bond and cyclopropyl unsaturations on the four acyl chains of CLs. This strategy was complemented with CID for the de novo elucidation of unknown CLs in biological extracts.
Introduction
Glycerophospholipids are a structurally complex lipid subclass with an array of cellular roles. Besides comprising the main component of cell membranes, these molecules participate in cellular signaling and regulate membrane protein structure.1–4 Cardiolipins (CL) are a unique phospholipid subset found in bacterial and mitochondrial membranes. In addition to modulating the structure of these membranes with negative curvature, cardiolipins participate in crucial mitochondrial pathways such as electron transport, ATP synthesis, and apoptosis.5–11 Dysregulation of cardiolipin expression and structure has been observed in various diseases including cancer.12–14 The specialized functions of CLs arise from their unusual structures consisting of two phosphatidic acid (PA) moieties crosslinked at the phosphate head by a central glycerol, resulting in distinctive tetraacylated phospholipids with two acidic phosphate groups. These complicated structures present a yet unsolved challenge for structural lipidomics.
Partial structural characterization of CLs has been previously achieved by several tandem mass spectrometry (MS/MS) strategies.12,15–25 For example, collision induced dissociation (CID) of deprotonated CLs results in production of the constituent PA moieties and acyl chains.15–18 Relative abundances of fragment ions corresponding to ketene or fatty acid losses have been used to assign acyl chain stereospecific numbering (sn) positions on the glycerol backbones, while abundances of fragment ions containing the intact PA moieties have been speculated to localize each moiety to either the 1’ or 3’ position on the central glycerol.18–20 Those fragments that correspond to intact PA moieties can be further subjected to another stage of CID (MS3) to identify their acyl chain compositions and regiochemistry.18–20 MS/MS analyses of sodiated cardiolipins in the positive and negative mode have identified the same structural features as those obtained from deprotonated CLs.19–21 In the shotgun analysis of an Escherichia coli (E. coli) CL extract, CID of [M – 2H + Na]− anions produced abundant diacyl glycerol neutral losses that consequently revealed PA composition, and the specific fragment abundances were used to assign each PA to either the 1’ or 3’ position.20 Upon a second stage of CID, these PA product ions revealed the acyl chain composition and sn-positions based on relative fragment ion abundances.20 This MSn strategy has provided the most in-depth structural characterization of CLs to date. Other shotgun MS/MS approaches have expanded CL lipidomics by incorporating quantitation strategies or by utilizing liquid chromatography (LC) to track molecular diversity between samples.12,22–25
Although CID has proven useful in identifying CL acyl chain and PA moiety compositions, other more subtle structural features, such as double bond location and geometry, have not been discerned. Moreover, although relative abundances of acyl chain losses upon CID can be indicative of major sn-isomers present, it does not exclude the presence of minor sn-positional isomers. Taken together, a significant array of structural features cannot be resolved by CID alone. These limitations are not only present in CL analysis but throughout structural lipidomics and has thus promoted interest in the development of alternative methods to determine these important features.26 Techniques that have been used to identify double bond locations of lipids include charge switching derivatization and ion/ion reactions followed by collisional activation,27,28 as well as the use of alternate dissociation methods such as electron induced dissociation (EID) of metal adducted lipids,29 radical directed dissociation (RDD) of derivatized lipids,30,31 and metastable atom-activated dissociation (MAD).32 Xia and coworkers have resolved double bond locations in unsaturated lipids by using CID to rupture oxetane rings formed by the photochemical derivatization of carbon-carbon double bonds (Paternò‐Büchi reaction).33,34 Electron impact excitation of ions from organics (EIEIO)35 and ozone-induced dissociation (OzID)36–38 are two other activation methods that, in addition to elucidating double-bond isomers, are also capable of identifying sn-positional isomers. Sequential CID and OzID (CID/OzID) has been utilized to identify sn-positional isomers, with multiple stages of CID and OzID (CID/OzID2 or (CID/OzID)2) used to unambiguously identify double-bond-positional isomers.37,38 To the best of our knowledge, these techniques have not been applied towards characterization of cardiolipins.
Another activation method that has been developed for the enhanced structural characterization of lipids and other biomolecules is ultraviolet photodissociation (UVPD).39–41 We have previously implemented the use of 193 nm UVPD to identify phosphatidylcholine (PC) double-bond positional isomers as well as demonstrated a targeted hybrid MS3 approach combining collisional activation with UVPD to identify both double bond and sn-positional isomers.42,43 In both approaches, UVPD induced cleavages of carbon-carbon bonds adjacent to double bonds, resulting in the production of diagnostic fragment pairs with a mass difference of 24 Da.42,43 The hybrid approach differentiated sn-positional isomers through the presence of diagnostic fragment ions that localized acyl chains to the sn-1 or sn-2 position on the glycerol backbone of various phospholipid classes.43 Relative quantitation of sn-positional isomers has also been achieved via UVPD by generating and dissociating doubly charged lipid-metal complexes.44 We most recently reported the use of 213 nm UVPD to localize cyclopropyl moieties on acyl chains through the identification of fragment pairs with a diagnostic mass difference of 14 Da.45 In the present study we have focused on the significant challenge of localization of cardiolipin unsaturations through the implementation of a hybrid collisional activation and 193 nm UVPD method.
Experimental Section
Materials.
All CL standards, as well as the E. coli CL extract, were purchased from Avanti Polar Lipids (Alabaster, Alabama), and used without further purification. The general composition of CL molecules is illustrated in Scheme S1, with the structures and molecular weights of all CL standards highlighted in Table S1. The numbering of carbons in fatty acids is displayed in Scheme S2. A detailed description of the nomenclature and shorthand notation for lipids is also provided in Supporting Information. HPLC-grade methanol and acetonitrile were purchased from EMD Millipore (Billerica, MA). HPLC-grade isopropanol and LC/MS-grade formic acid were purchased from Thermo Fisher Scientific (Waltham, MA). Chloroform was purchased from Sigma-Aldrich (St. Louis, MO). A total lipid extract was prepared from a human papillary thyroid carcinoma, procured through the Cooperative Human Tissue Network, utilizing the Bligh and Dyer total lipid extraction method.46 Samples were diluted in 50:50 chloroform-methanol to 10 μM for CL standards and 10 ng/μL for the biological extracts.
Mass Spectrometry.
Spectra were collected on a Thermo Scientific Instruments Orbitrap Fusion Lumos Tribrid mass spectrometer (San Jose, CA) modified to perform 193 nm UVPD with a Coherent Excistar excimer laser (Santa Cruz, CA) as previously described.47 Approximately 10 μL of each sample were loaded into a silver-coated pulled tip glass capillary and directly infused using a Thermo Scientific Nanospray Flex ion source (San Jose, CA) at a spray voltage of 1.1 kV and heated transfer capillary temperature of 275°C. Selected precursor ions were isolated in the negative mode with an isolation width of 0.5 m/z units for doubly charged ions and 1.0 m/z units for singly charged ions. Collision induced dissociation (CID) mass spectra were collected using a normalized collision energy (NCE) between 22–32 to optimize the ion current of the deacylated CL fragments. 193 nm UVPD was performed in the high-pressure trap using 10 laser pulses at 1 mJ per pulse. All spectra were acquired with an AGC target between 1 × 105 and 5 × 105 at a resolution of 60K. 100 transient averages were acquired for each spectrum.
Results/Discussion
Owing to the acidic nature of CLs, they typically do not ionize efficiently in the positive mode, so the negative mode was used for this study. Electrospray ionization (ESI) of CLs yields predominant doubly deprotonated [M-2H]2– and minor [M-H]− anions, as exemplified by the MS1 spectrum for CL (18:1(9Z)/18:1(9Z))/(18:1(9Z)/18:1(9Z)) in Figure S1. The deprotonated species in the 1- and 2- charge states were subjected to CID and UVPD to evaluate the ability to characterize the key features of the structures (acyl chain and PA composition) and localize acyl chain double bond positions. CID of the doubly deprotonated CL promoted loss of one labile fatty acid, as shown in Figure 1A for which prominent complementary fragment ions of m/z 281.25 (18:1 fatty acid anion) and m/z 1173.77 (tri-acylated CL) are observed. Acyl chain composition was also informed through the loss of a neutral ketene that resulted in a doubly charged product at m/z 595.39. Losses of the PA moieties, originating from cleavage between the phosphatidic moiety and either C1’ or C3’ positions on the central glycerol backbone, as originally described by Hsu et al.,1 were also observed, as evidenced by the product ion of m/z 755.52 in Figure 1A. A couple of other ions were observed in the CID mass spectrum; these fragments were consistent with combined losses of one or more acyl chains from the CL or the PA portion (m/z 417.24 and m/z 909.53).
Figure 1:
(A) CID (25 NCE) and (B) UVPD (10 pulses, 1.0 mJ) mass spectra for doubly deprotonated CL (18:1(9Z)/18:1(9Z))/(18:1(9Z)/18:1(9Z)) (m/z 727.51). CID results in an abundant deacylated product of m/z 1173.77, attributed to the loss of an 18:1 fatty acid. (C) CID/UVPD: UVPD of m/z 1173.77 produces diagnostic fragment ions that localize the double bond. The selected precursor ion in each stage is designated with a star. (D) Fragment map for the CL standard.
CID of the 1- charge state of CL (18:1(9Z)/18:1(9Z))/(18:1(9Z)/18:1(9Z)) produced an abundant fragment ion of m/z 699.50 (corresponding to a PA 18:1/18:1 moiety), arising from cleavage of the glycerol backbone that also releases the complementary fragment of m/z 755.52, as shown in Figure S2. Neutral loss of the 18:1 fatty acid also occurred from the deprotonated CL, yielding the ion of m/z 1173.77. Other diagnostic fragments in the CID mass spectrum corresponded to cleavage between the phosphate groups and the C1’ or C3’ glycerol positions along with water loss (m/z 835.49), as well as combinations of fatty acid and phosphatidic acid losses (m/z 417.24).
The CID mass spectra of both singly and doubly deprotonated CL (18:1(9Z)/18:1(9Z))/(18:1(9Z)/18:1(9Z)) were in agreement with previously reported MS/MS spectra of CLs.18 The MS/MS spectra reveal both the acyl chain composition and the sum composition of the constituent PA moieties,1–6 but do not allow differentiation of double bond positions on the acyl chains.
To obtain deeper structural insight, UVPD was used to examine CL (18:1(9Z)/18:1(9Z))/(18:1(9Z)/18:1(9Z)). Several of the fragment ions produced by UVPD are analogous to ones observed upon CID. As shown in Figure S3, UVPD of deprotonated CL yields prominent ions of m/z 699.50 (PA 18:1/18:1) and m/z 755.52, and promotes neutral loss of the fatty acid (m/z 1173.77) and cleavage of the phosphate ester bond to the 3’ glycerol (m/z 835.49 with water loss), all pathways akin to the ones observed upon CID. However, the fragment ion of m/z 741.51 is indicative of cleavage of the C1’- C2’ (or C2’- C3’) bond in the central glycerol, a process not observed upon collisional activation. As also observed upon UVPD of unsaturated phospho- and sphingolipids, UVPD of the deprotonated CL promotes cleavages of C-C bonds vinylic to double bonds, producing a pair of ions of m/z 1317.89 and 1341.88 with a diagnostic mass difference of 24 Da.42,43,48 Although these diagnostic double bond signature ions have low abundances, the Orbitrap mass analyzer allows their detection with exceptional S/N (>50) and with high mass accuracy.
UVPD of doubly deprotonated CL (18:1(9Z)/18:1(9Z))/(18:1(9Z)/18:1(9Z)) resulted in a rich MS/MS spectrum, dominated by an abundant charge reduced precursor, as demonstrated in Figure 1B. Electron photodetachment (EPD) commonly occurs upon UVPD of multicharged anions,49–51 thus accounting for the prevalent charge reduction process. Ions that informed the PA composition (m/z 755.52 and m/z 699.50) and fatty acid composition (m/z 281.25 and m/z 1173.77) in the CID spectrum were also generated via UVPD. Cleavage of other C-C bonds in the central glycerol backbone led to formation of ions of m/z 741.51 and 713.51, as illustrated in the fragmentation map in Figure 1D. Notably, UVPD promoted cleavage of numerous other C-C bonds throughout the acyl chains, as shown in an expanded view of the spectrum from m/z 1170 – 1440 in Figure S4. Fragment ions corresponding to cleavage of the C-C bonds vinylic to the double bonds exhibited a characteristic mass difference of 24 Da, yielding ions of m/z 1316.88 and m/z 1340.87. In summary, 193 nm UVPD of both the singly and doubly deprotonated species yield fragment ions that identify the key structural features of CLs (i.e. PA and fatty acid composition) and at the same time results in additional fragment ions that identify double bond positions.
Direct application of UVPD for the de novo characterization of CLs and differentiation of double bond isomers, however, is a significantly greater challenge. Although UVPD of the deprotonated CLs ions generate fragment ions that readily pinpoint the positions of double bonds, the abundances of singly deprotonated precursors are typically low in complex mixtures, as shown in Figure S5 for a papillary thyroid carcinoma lipid extract. Combined with the low efficiency of producing the diagnostic fragment ions upon UVPD, the mono-deprotonated CLs are poor candidates for double bond characterization. The abundances of the doubly deprotonated CLs are greater and therefore better candidates for MS/MS analysis. Nevertheless, the dense fragmentation patterns, the cleavage of all C-C bonds in the acyl chains complicates de novo interpretation of UVPD mass spectra of CLs.
To alleviate these issues, an alternative MS3 strategy was developed that exploits the loss of labile fatty acid anions upon CID of the [M-2H]2– precursors to produce singly charged, tri-acylated fragment ions that allow the sites of unsaturation to be deciphered by UVPD. This method provides several advantages that allow reliable de novo characterization of CLs at the double bond level. Due to their propensity to ionize in the 2- charge state by ESI, CLs can be readily distinguished in total lipid extracts typically dominated by singly charged phospholipid (phosphatidylethanolamines, phosphatidylglycerols, phosphatidylserines, phosphatidylinositols) anions. Selective dissociation of 2- ions yields singly charged products in the m/z range above the precursor that unambiguously arise from doubly charged precursor ions and thus distinguish informative CL products from those derived from singly charged isobars. In the MS3 approach, CID of the doubly charged CLs in the first stage produces informative fragments that identify the fatty acid and PA composition. UVPD of the resulting deacylated fragment ions yields simple spectra that localize sites of unsaturation through the identification of diagnostic products ions. Moreover, ambiguities that may arise in pinpointing sites of unsaturation in hetero-acylated CLs can be resolved through strategic selection of multiple deacylated fragment ions for UVPD. Furthermore, this MS3 approach provides spectral “clean-up” that simplifies interpretation and increases specificity during shotgun analysis of complex mixtures, as previously demonstrated for lipid A, another structurally complex type of lipid.52
The MS3 (CID/UVPD) method for localizing double bonds was first evaluated with CL (18:1(9Z)/18:1(9Z))/(18:1(9Z)/18:1(9Z)). As described above, CID of the 2- ion generated an abundant fragment of m/z 1173.77, created by the loss of an 18:1 fatty acid chain (Figure 1A). The deacylated product (m/z 1173.77) was isolated and subjected to UVPD, resulting in the cleavage of C-C bonds adjacent to the double bond and yielding the diagnostic pair of ions of m/z 1035.63/1059.63, as shown in Figure 1C. The tandem CID/UVPD method also produced diagnostic ions for a CL possessing distinct acyl chains (16:0/18:1(9Z))/(16:0/18:1(9Z)). In this case, the [M-16:0] or the [M-18:1] products (m/z 1147.75 and m/z 1121.74, respectively) generated upon CID were subjected to UVPD (Figure S6B,C). Each selected deacylation product yielded a unique set of fragment ions corresponding to the double bond position. Distinct acyl chains at the sn-1 and sn-2 position on this CL also provided the opportunity to explore UVPD as a tool to differentiate sn-positional isomers. Inspection of UVPD and CID/UVPD mass spectra, however, did not allow for the confident assignment of this structural feature. Interestingly, sn positional assignment was possible using a two-step (MSn) HCD/UVPD strategy similar to one reported in reference 43. Through HCD of [M + Na]+ ions generated in the positive mode, followed by UVPD of the sodium-adducted headgroup loss products allowed assignment of sn stereochemistry, as shown for (16:0/18:1(9Z))/(16:0/18:1(9Z)) in Figure S7. Exploration of this approach by analysis of charge-inverted cardiolipins merits further study.
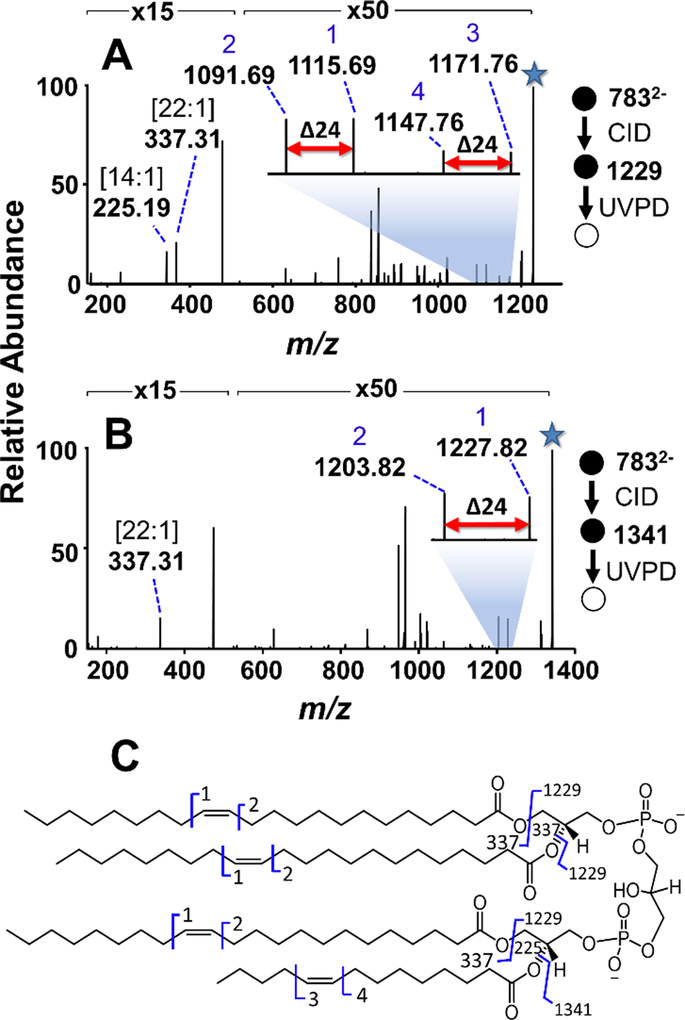
Through the strategic selection and subsequent UVPD of multiple deacylated fragment ions produced by CID in the negative mode, the MS3 strategy is capable of resolving ambiguities that may arise in localization of double bonds in hetero-acylated CLs. To demonstrate this capability, CL (22:1(13Z)/22:1(13Z))/(22:1(13Z)/14:1(9Z)) was subjected to CID/UVPD (Figure 2). CID of the doubly deprotonated CL produces m/z 1341.96 and m/z 1229.83 to identify the acyl chain composition (14:1 and 22:1) and m/z 867.65 and m/z 755.52 that provide the PA sum compositions (PA 44:2 and PA 36:2), as displayed in Figure S8. These four fragment ions complement one another to confirm that this species is CL (22:1/22:1)_(22:1_14:1), but do not elucidate the position of the acyl chain double bonds. UVPD of the more abundant [M-22:1] fragment ion (m/z 1229) results in two diagnostic doublets (m/z 1091.69/1115.69 and m/z 1147.76/1171.76), each characteristic of one double position as shown in Figure 2A. These fragment pairs identify the 22:1 fatty acid as either the 17Δ or 13Δ isomer and the 14:1 fatty acid as either the 5Δ or 9Δ isomer. Accounting for all possible combinations of these four fatty acids, a total of 16 different double bond isomers could be attributed to this spectrum. CID/UVPD of the 14:1 acyl chain loss (m/z 1341) provides one complementary set of diagnostic ions of m/z 1203.82/1227.82 as shown in Figure 2B. The presence of m/z 337.31 (22:1 fatty acid anion) and absence of m/z 225.51 (14:1 fatty acid anion) in the CID/UVPD spectrum confirm that the diagnostic pair m/z 1203.82/1227.82 corresponds to a tri-acylated fragment ion solely composed of 22:1 acyl chains. Thus, m/z 1203.82/1227.82 in Figure 2B and m/z 1091.69/1115.69 in Figure 2A arise from 22:1(13Δ) fatty acids and indicate that the m/z 1147.76/1171.76 diagnostic pair in Figure 2A originated from a 14:1(9Δ) fatty acid. Taken together, UVPD of both [M-22:1] and [M-14:1] reveals that the CL is composed of three 22:1(13Δ) and one 14:1(9Δ) fatty acids.
Figure 2:
CID/UVPD (10 pulses, 1.0 mJ) mass spectra of doubly deprotonated CL (22:1(15Z)/22:1(15Z))/(22:1(15Z)/14:1(9Z)) (m/z 783.57). (A) UVPD of m/z 1229.83 resulting from the 14:1(9Z) acyl chain loss from doubly-deprotonated CL. Two pairs of diagnostic ions corresponding to the two different double bond locations for the 22:1 and 14:1 acyl chains are featured. (B) UVPD of m/z 1341.96 resulting from the 22:1(15Z) acyl chain loss from doubly deprotonated CL. One pair of diagnostic ions that localize the double bond of the 22:1 chain to position 15 is highlighted. The selected precursor ion in each stage is designated with a star. (C) Fragment map for the CL standard.
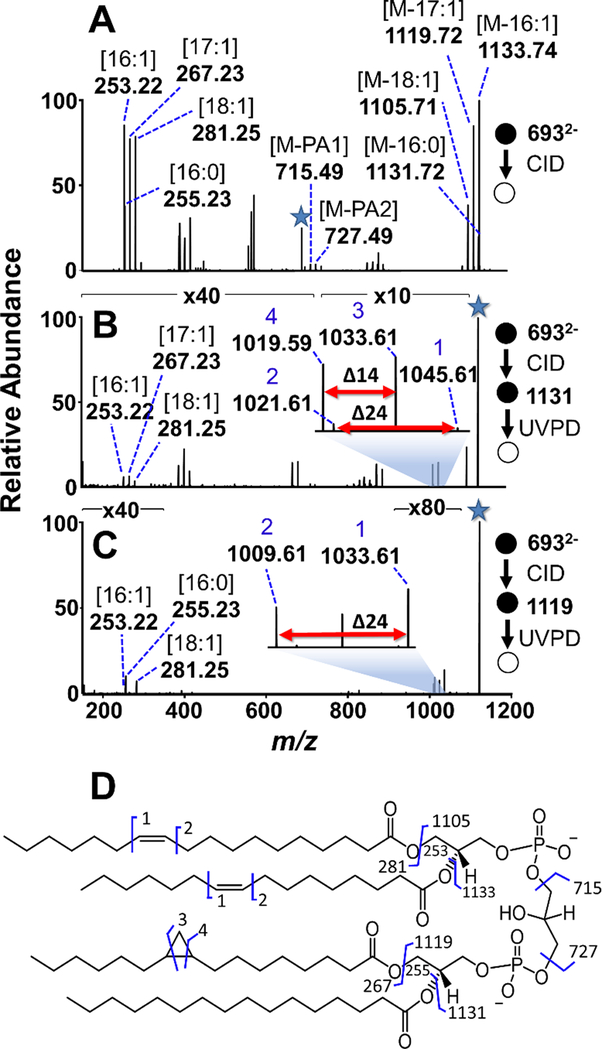
The feasibility of this strategy for the de novo characterization of unknown CLs was evaluated by analyzing a CL extract from E. coli. The fatty acid and PA composition of each CL were determined via CID of the doubly charged CLs with subsequent UVPD of the products generated from fatty acid losses to detail the sites of unsaturation. In addition to double bonds, cyclopropyl unsaturations could be localized as well through the identification of doublets with diagnostic mass differences of 14 Da, as recently reported.45 An example of the results are illustrated in Figure 3 for the characterization of the CL of m/z 693.48 in the E. coli extract. CID of m/z 693.48 (Figure 3A) results in product ions of m/z 715.49 and m/z 727.49, providing the sum composition of the two constituent PA moieties as PA 34:2 and PA 33:1. The CID spectrum also revealed the losses of 16:1, 16:0, 17:1, and 18:1 fatty acids (resulting in product ions of m/z 1131.73, 1133.74, 1119.72, and 1105.71, respectively), indicating that the CL was composed of PA 18:1_16:1 and PA 17:1_16:0. UVPD of the ions of m/z 1131.73 (16:0 loss, Figure 3B) confirmed that the selected tri-acylated CL was composed of 16:1, 17:1, and 18:1 fatty acids, as indicated by the generation of fatty acid ions of m/z 253.22, 267.23, and 281.25, respectively. The spectra presented both a pair of double bond diagnostic ions (m/z 1021.61/1045.61, Δm 24 Da), as well as a pair of cyclopropyl diagnostic ions (m/z 1019.59/1033.61, Δm 14 Da). The cyclopropyl unsaturation is confirmed to be present on the 17:1 fatty acid via UVPD of the deacylated m/z 1133.74 product generated by CID, as shown in Figure 3C. This CID/UVPD spectrum only displays the 16:0, 16:1, and 18:1 fatty acid anions (m/z 255.23, m/z 253.22, and m/z 281.25), as well as one double bond diagnostic pair (m/z 1009.81/1033.61), while the cyclopropyl pair is absent. This outcome provides evidence that the double bond corresponds to 18:1(11Δ) and 16:1(9Δ) and that the cyclopropyl pair in Figure 3B was produced by 17:1(c9Δ). Thus, the ion of m/z 693 in the extract was assigned to CL (16:1(9Δ)_18:1(11Δ))_(16:0_17:1(c9Δ)). All identifications made by this approach for the E. coli CL extract are listed in Table S2.
Figure 3:
(A) CID mass spectrum of the ion of m/z 693.48 from E. coli CL extract. (B) UVPD of the m/z 1131.72 fragment ion resulting from the loss of the 16:0 acyl chain yields pairs of diagnostic ions revealing cyclopropyl and double bond positions. (C) UVPD of the m/z 1119.72 fragment ion, corresponding to the loss of a 17:1 acyl chain, results in one pair of diagnostic ions that localize the double bonds to the 16:1 and 18:1 acyl chains. The selected precursor ion in each stage is designated with a star. (D) Fragmentation map constructed from de novo analysis of the CID and CID/UVPD spectra is consistent with CL (18:1(9Δ)_16:1(9Δ))_(17:1(c9Δ)_(16:0)).
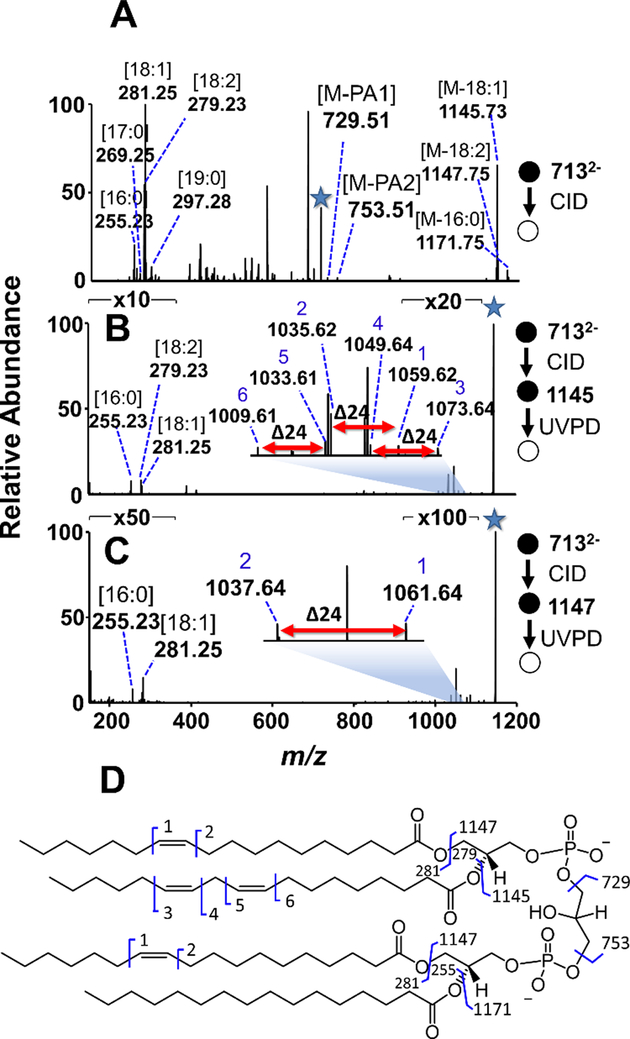
The robustness of the CID/UVPD method for characterization of CLs in more complex mixtures was demonstrated through the analysis of a total lipid extract. The characterization of CLs within biological tissue samples has recently spurred great interest, as CLs have been indicated as possible biomarkers in multiple cancer subtypes. For example, oncocytic thyroid tumors have been shown to present abnormal CL signatures that can serve as biomarkers for this rare subtype of thyroid carcinoma14 and have displayed altered abundances within glioblastoma brain cancer tissues.13 We sought to characterize CLs in a lipid extract of a human papillary thyroid carcinoma tissue via CID/UVPD. Shotgun ESI of a human papillary thyroid carcinoma extract presented several doubly charged ions that distinguished the doubly deprotonated CLs from singly charged lipids in the lipid mixture, as shown in Figure S5. CID/UVPD of the extract revealed a high abundance of CLs incorporating polyunsaturated fatty acids for which the double bond positions could be discerned, as demonstrated in Figure 4 for the ion of m/z 713.49. CID of m/z 713.49 (Figure 4A) revealed the CL to be composed of PA 36:3 and PA 34:1 based on the presence of fragment ions of m/z 753.51 and m/z 729.51, respectively. Although the lower mass region of the CID spectrum showed ions consistent with 16:0, 17:0, 18:1, 18:2, and 19:0 fatty acids, the higher mass region only displayed fatty acid losses corresponding to 16:0, 18:1, and 18:2. This contradiction suggests that the 17:0 and 19:0 fatty acid anions result from co-isolation and activation of isobaric species, a frequent outcome of shotgun methods that leads to chimeric MS/MS spectra. Discounting the 17:0 and 19:0 ions, the other fatty acid losses indicate that the composition is PA 18:1_18:2 and PA 18:1_16:0. In Figure 4B, the fragment ion produced by the loss of an 18:1 fatty acid (m/z 1145.73 in Figure 4A) displayed three sets of double bond diagnostic ions (m/z 1009.61/1033.61, m/z 1035.62/1059.62, and m/z 1049.64/1073.64) upon UVPD. Alternatively, when the ion of m/z 1147.73 in Figure 4A (generated via loss of an 18:2 fatty acid) was subjected to UVPD, two of the diagnostic pairs of ions dropped out of the spectrum (Figure 4C), leaving only one diagnostic pair (m/z 1037.64/1061.64). Because only 16:0 (m/z 255.23) and 18:1 (m/z 281.25) fatty acid anions are observed in the low mass region of this spectrum, the double bond pair in Figure 4C corresponds to 18:1(11Δ). Assuming that the m/z 1035.62/1059.62 diagnostic pair is only produced by the identified 18:1(11Δ) fatty acid, the diagnostic ion pairs of m/z 1091.61/1033.61 and m/z 1049.64/1073.64 in Figure 4B indicate the polyunsaturated fatty acid is 18:2(9Δ,12Δ). When combined, the CID and CID/UVPD mass spectra suggest the ion of m/z 713 is CL (18:1(11Δ)_18:2(9Δ,12Δ))_(18:1(11Δ)_16:0).
Figure 4:
(A) CID mass spectrum of the ion of m/z 713.49 from human papillary thyroid carcinoma lipid extract reveals the fatty acid composition as well as the sum composition of the constituent PA moieties. (B) UVPD of the m/z 1145.73 fragment ion resulting from the loss of a 18:1 acyl chain yields three pairs of diagnostic ions revealing double bond positions and also confirms that the tri-acylated fragment ion contains 16:0, 18:2, and 18:1 acyl chains. (C) UVPD of the m/z 1147.75 ion, corresponding to the loss of a 18:2 acyl chain, results in one pair of diagnostic ions and reveals that the fragment ion contains 16:0 and 18:1 acyl chains. (D) Fragmentation map constructed for the CL. The selected precursor ion in each stage is designated with a star.
The total array of CLs identified in the human papillary thyroid carcinoma extract via the CID/UVPD strategy are listed in Table S3. This approach was not capable of unambiguously assigning double bond positions for the CLs of m/z 711.48 and m/z 725.49 because the hetero-acylated structures determined by CID, shown in Figure S9, would require an MS4 method to assign the 18:1 acyl chain as either 18:1(11Δ) or 18:1(12Δ).
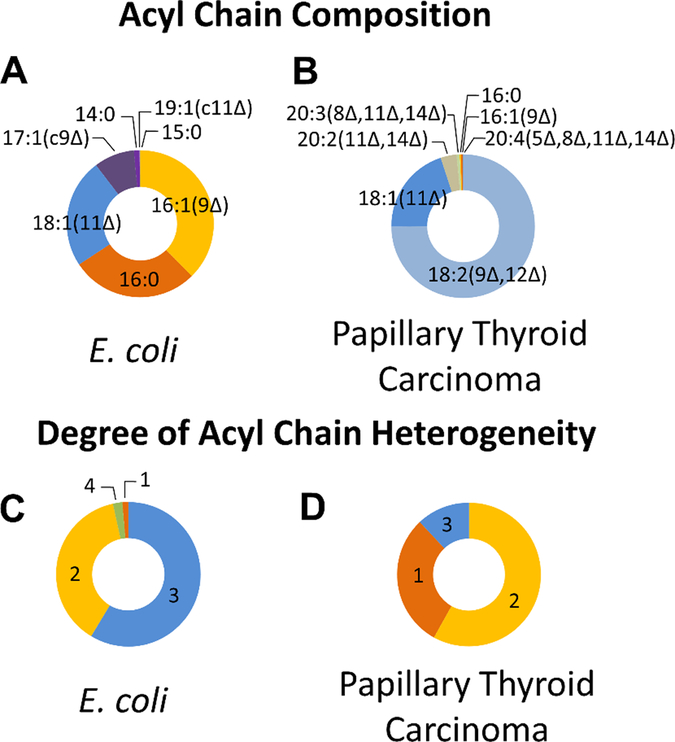
The distributions of acyl chain compositions of the CLs identified in the extracts is summarized in Figure 5, which highlights the differences of prokaryotic and eukaryotic cells. In contrast to prokaryotes, CLs in eukaryotes undergo significant tissue-dependent acyl chain remodeling after initial synthesis.7 This heterogeneity is manifested as variations in the lengths of the acyl chains as well as the diversity of chain lengths within individual CLs. The degree of acyl chain heterogeneity is enumerated based on the number of different chains appended to each CL with values ranging from 1 (all chains of uniform length and double bond position) to 4 (all chains of differing lengths or differing double bond positions). In order to account for this heterogeneity in quantifying fatty acid compositions in Figures 5A and 5B, the abundance of each of the four constituent fatty acid of identified CLs was counted as 25% of the abundance of the molecular ion signal of the CL precursor. For instance, in the case of CL (18:1(11Δ)_18:2(9Δ,12Δ))_(18:1(11Δ)_16:0) in the papillary thyroid carcinoma extract, 50% of the molecular ion signal of m/z 753.51 was attributed to 18:1(11Δ), 25% to 18:2(9Δ,12Δ), and 25% to 16:0. The observed CL composition in the human papillary thyroid carcinoma extract was considerably more homogenous than the distribution of CLs in E.coli. About ~88% of the CLs identified in the papillary thyroid carcinoma cell extract exhibited a degree of heterogeneity of 2 or less (Figure 5D), whereas about ~40% of the E.coli CLs had a heterogeneity of 1 or 2, and about ~60% had a heterogeneity of 3 or 4 (Figure 5C). Furthermore, 18:2(9Δ,12Δ) acyl chains comprised a significant portion (~75%) of the acyl chain composition of papillary thyroid carcinoma extracts (Figure 5B), whereas the acyl chain composition of CLs in E. coli was distributed more evenly among four fatty acids (~38% 16:1(9Δ), ~28% 16:0, ~24% 18:1(11Δ), 9% 17:1(c9Δ)) as shown in Figure 5A. The double bond locations identified through CID/UVPD for the acyl chains in the CLs of E. coli were consistent with fatty acid biosynthesis in which unsaturated fatty acids are synthesized by the elongation of a 10:1(3Δ) acyl chain at the carboxyl end.53,54 This pathway results in all unsaturated fatty acids having the double bond located seven carbons from the methyl end. Monounsaturated 17:c1 and 19:c1 fatty acids (17:1(c9Δ) and 19:1(c11Δ)) were also prominent in the CLs found in E. coli, in agreement with the synthesis of cyclopropane fatty acids through the methylation of unsaturated fatty acids (in this case, 16:1(9Δ) and 18:1(11Δ)). Double bond incorporation in CLs of tumors is less well understood, but it has been shown that the ratios of double bond isomer pairs vary significantly in cancerous tissue and may serve as cancer biomarkers.55 The exclusive identification of 18:1 fatty acids with the double bond at position 11 in the CLs of papillary thyroid carcinoma is a compelling observation that merits further exploration.
Figure 5:
Donut plots depicting the CL compositions for E. coli and human papillary thyroid carcinoma extracts, based on molecular ion abundances in MS1 spectra with features determined through CID and CID/UVPD. Each identified fatty acid was counted as 25% of the abundance of the molecular ion signal of the precursor CL. Acyl chain composition is displayed for CLs in E. coli (A) and PTC (B) extracts. Degree of heterogeneity is displayed for CLs in E. coli (C) and PTC (D) extracts. Homo-acylated CLs are assigned a degree of heterogeneity of 1 and hetero-acylated CLs composed of four distinct acyl chains are assigned a degree of heterogeneity of 4.
Conclusion
Although significant advances have been made in structural lipidomics for the identification of key structural features of lipids, such as double bond positions, these approaches to date have only been applied to characterize sphingo- and phospholipid classes with limited structural complexity. The significant structural complexity exhibited by cardiolipins, exemplified by the heterogeneity of four acyl chains, motivated the development of the new MS3 strategy presented here. We extended UVPD methodologies to a hybrid approach combining collisional activation and 193 nm UVPD to localize unsaturations in CLs presenting a significant degree of acyl chain heterogeneity and a high degree of unsaturation. In the first step of the MS3 method, collisional activation resulted in loss of labile fatty acids, generating de-acylated CL fragment ions with high S/N. Subsequent UVPD resulted in fragment ion pairs displaying a diagnostic mass difference of 24 Da or 14 Da that localized either double bond or cyclopropane unsaturations, respectively. The use of an MS3 workflow offers advantages for shotgun lipidomics in which ionization of isobaric species is common owing to the lack of chromatographic separation. Methodical selection of MS3 precursors for the multi-step characterization of cardiolipins reduced the production of ambiguous chimeric spectra from co-isolated isobaric precursor ions. Moreover, UVPD of multiple de-acylated fragments generated from the same cardiolipin resolved ambiguities that arose from the detection of multiple diagnostic fragment ion pairs that would otherwise confound the localization of unsaturations. This method allowed the de novo characterization of cardiolipins in E. coli and was sufficiently robust to profile the cardiolipins extracted from a papillary thyroid carcinoma tissue.
The development of this hybrid UVPD approach as a quantitative tool is currently stifled, as the abundances of the double bond diagnostic pairs in the MS3 spectra depend on the abundances of CID products in the MS2 spectra which are influenced by the acyl chain identities and sn positions. The possibility of co-isolated isomeric precursor ions, a common occurrence using shotgun methods, is another impediment. The lack of available double bond cardiolipin isomers further undermines the development of relative quantitation strategies. Some of these issues would be bypassed by adaptation of the present method to an LCMS workflow, although not without other significant challenges. Mobile phases (consisting of water, methanol, acetonitrile, isopropanol and ammonium salts) routinely used for separation of CLs result in production of singly charged anions, for which CID produces only low abundance deacylated products. Furthermore, standard C18 chromatography affords unsatisfactory separation of CLs. Given these considerations, integrating the hybrid approach presented here with LC separation of CLs is an ongoing effort with several yet unresolved hurdles.
Supplementary Material
ACKNOWLEDGMENT
Support from the NIH (R00CA190783 to LSE and R01 GM103655 to JSB) and the Welch Foundation (F-1895 LSE and F-1155 JSB) is gratefully acknowledged. The UT System Proteomics Core Facility Network is gratefully acknowledged.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
References
- (1).Raetz CRH Molecular Genetics of Membrane Phospholipid Synthesis. Annual review of genetics 1986, 20, 253–295. [DOI] [PubMed] [Google Scholar]
- (2).Dowhan W; Bogdanov M Chapter 1 Functional Roles of Lipids in Membranes In New Comprehensive Biochemistry; Elsevier, 2002; Vol. 36, pp 1–35. [Google Scholar]
- (3).Cronan JE Molecular Biology of Bacterial Membrane Lipids. Annual Review of Biochemistry 1978, 47, 163–189. [DOI] [PubMed] [Google Scholar]
- (4).Sandermann H Regulation of Membrane Enzymes by Lipids. Biochimica et Biophysica Acta (BBA) - Reviews on Biomembranes 1978, 515, 209–237. [DOI] [PubMed] [Google Scholar]
- (5).Schlame M Thematic Review Series: Glycerolipids. Cardiolipin Synthesis for the Assembly of Bacterial and Mitochondrial Membranes. J. Lipid Res. 2008, 49, 1607–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Houtkooper RH; Vaz FM Cardiolipin, the Heart of Mitochondrial Metabolism. Cellular and Molecular Life Sciences 2008, 65, 2493–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Maguire JJ; Tyurina YY; Mohammadyani D; Kapralov AA; Anthonymuthu TS; Qu F; Amoscato AA; Sparvero LJ; Tyurin VA; Planas-Iglesias J; He R-R; Klein-Seetharaman J; Bayır H; Kagan VE Known Unknowns of Cardiolipin Signaling: The Best Is yet to Come. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids 2017, 1862, 8–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Klingenberg M Cardiolipin and Mitochondrial Carriers. Biochimica et Biophysica Acta (BBA) - Biomembranes 2009, 1788, 2048–2058. [DOI] [PubMed] [Google Scholar]
- (9).McMillin JB; Dowhan W Cardiolipin and Apoptosis. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids 2002, 1585, 97–107. [DOI] [PubMed] [Google Scholar]
- (10).Claypool SM Cardiolipin, a Critical Determinant of Mitochondrial Carrier Protein Assembly and Function. Biochimica et Biophysica Acta (BBA) - Biomembranes 2009, 1788, 2059–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Claypool SM; Oktay Y; Boontheung P; Loo JA; Koehler CM Cardiolipin Defines the Interactome of the Major ADP/ATP Carrier Protein of the Mitochondrial Inner Membrane. J Cell Biol 2008, 182, 937–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Valianpour F; Wanders RJA; Barth PG; Overmars H; Gennip AH van. Quantitative and Compositional Study of Cardiolipin in Platelets by Electrospray Ionization Mass Spectrometry: Application for the Identification of Barth Syndrome Patients. Clinical Chemistry 2002, 48, 1390–1397. [PubMed] [Google Scholar]
- (13).Kiebish MA; Han X; Cheng H; Chuang JH; Seyfried TN Cardiolipin and Electron Transport Chain Abnormalities in Mouse Brain Tumor Mitochondria: Lipidomic Evidence Supporting the Warburg Theory of Cancer. J Lipid Res 2008, 49, 2545–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Zhang J; Yu W; Ryu S; Lin J; Buentello G; Tibshirani R; Suliburk J; Eberlin LS Cardiolipins Are Biomarkers of Mitochondria-Rich Thyroid Oncocytic Tumors. Cancer Res 2016, 76, 6588–6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Han X; Yang K; Yang J; Cheng H; Gross RW Shotgun Lipidomics of Cardiolipin Molecular Species in Lipid Extracts of Biological Samples. J Lipid Res 2006, 47, 864–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Gao F; McDaniel J; Chen EY; Rockwell HE; Nguyen C; Lynes MD; Tseng Y-H; Sarangarajan R; Narain NR; Kiebish MA Adapted MS/MSALL Shotgun Lipidomics Approach for Analysis of Cardiolipin Molecular Species. Lipids 2018, 53, 133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Han X; Gross RW Structural Determination of Picomole Amounts of Phospholipids via Electrospray Ionization Tandem Mass Spectrometry. J Am Soc Mass Spectrom 1995, 6, 1202–1210. [DOI] [PubMed] [Google Scholar]
- (18).Hsu F-F; Turk J; Rhoades ER; Russell DG; Shi Y; Groisman EA Structural Characterization of Cardiolipin by Tandem Quadrupole and Multiple-Stage Quadrupole Ion-Trap Mass Spectrometry with Electrospray Ionization. Journal of the American Society for Mass Spectrometry 2005, 16, 491–504. [DOI] [PubMed] [Google Scholar]
- (19).Hsu F-F; Turk J Characterization of Cardiolipin as the Sodiated Ions by Positive-Ion Electrospray Ionization with Multiple Stage Quadrupole Ion-Trap Mass Spectrometry. Journal of the American Society for Mass Spectrometry 2006, 17, 1146–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Hsu F-F; Turk J Characterization of Cardiolipin from Escherichia Coli by Electrospray Ionization with Multiple Stage Quadrupole Ion-Trap Mass Spectrometric Analysis of [M − 2H + Na]− Ions. Journal of the American Society for Mass Spectrometry 2006, 17, 420–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Beckedorf AI; Schäffer C; Messner P; Peter‐Katalinić J Mapping and Sequencing of Cardiolipins from Geobacillus Stearothermophilus NRS 2004/3a by Positive and Negative Ion NanoESI-QTOF-MS and MS/MS. Journal of Mass Spectrometry 2002, 37, 1086–1094. [DOI] [PubMed] [Google Scholar]
- (22).Minkler PE; Hoppel CL Separation and Characterization of Cardiolipin Molecular Species by Reverse-Phase Ion Pair High-Performance Liquid Chromatography-Mass Spectrometry. J Lipid Res 2010, 51, 856–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Sparagna GC; Johnson CA; McCune SA; Moore RL; Murphy RC Quantitation of Cardiolipin Molecular Species in Spontaneously Hypertensive Heart Failure Rats Using Electrospray Ionization Mass Spectrometry. J. Lipid Res. 2005, 46, 1196–1204. [DOI] [PubMed] [Google Scholar]
- (24).Oemer G; Lackner K; Muigg K; Krumschnabel G; Watschinger K; Sailer S; Lindner H; Gnaiger E; Wortmann SB; Werner ER; Zschocke J; Keller MA Molecular Structural Diversity of Mitochondrial Cardiolipins. PNAS 2018, 115, 4158–4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Lesnefsky EJ; Stoll MSK; Minkler PE; Hoppel CL Separation and Quantitation of Phospholipids and Lysophospholipids by High-Performance Liquid Chromatography - ScienceDirect. Analytical Biochemistry 2000, 285, 246–254. [DOI] [PubMed] [Google Scholar]
- (26).Siegel TP; Ekroos K; Ellis SR Reshaping Lipid Biochemistry by Pushing Barriers in Structural Lipidomics. Angewandte Chemie International Edition 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Yang K; Dilthey BG; Gross RW Identification and Quantitation of Fatty Acid Double Bond Positional Isomers: A Shotgun Lipidomics Approach Using Charge-Switch Derivatization. Anal Chem 2013, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Randolph CE; Foreman DJ; Betancourt SK; Blanksby SJ; McLuckey SA Gas-Phase Ion/Ion Reactions Involving Tris-Phenanthroline Alkaline Earth Metal Complexes as Charge Inversion Reagents for the Identification of Fatty Acids. Anal. Chem 2018, 90, 12861–12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Yoo HJ; Håkansson K Determination of Double Bond Location in Fatty Acids by Manganese Adduction and Electron Induced Dissociation. Anal. Chem 2010, 82, 6940–6946. [DOI] [PubMed] [Google Scholar]
- (30).Pham HT; Ly T; Trevitt AJ; Mitchell TW; Blanksby SJ Differentiation of Complex Lipid Isomers by Radical-Directed Dissociation Mass Spectrometry. Anal. Chem 2012, 84, 7525–7532. [DOI] [PubMed] [Google Scholar]
- (31).Pham HT; Trevitt AJ; Mitchell TW; Blanksby SJ Rapid Differentiation of Isomeric Lipids by Photodissociation Mass Spectrometry of Fatty Acid Derivatives. Rapid Communications in Mass Spectrometry 2013, 27, 805–815. [DOI] [PubMed] [Google Scholar]
- (32).Deimler RE; Sander M; Jackson GP Radical-Induced Fragmentation of Phospholipid Cations Using Metastable Atom-Activated Dissociation Mass Spectrometry (MAD-MS). Int J Mass Spectrom 2015, 390, 178–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Ma X; Xia Y Pinpointing Double Bonds in Lipids by Paternò-Büchi Reactions and Mass Spectrometry. Angewandte Chemie 2014, 126, 2630–2634. [DOI] [PubMed] [Google Scholar]
- (34).Stinson CA; Xia Y A Method of Coupling the Paternò–Büchi Reaction with Direct Infusion ESI-MS/MS for Locating the CC Bond in Glycerophospholipids. Analyst 2016, 141, 3696–3704. [DOI] [PubMed] [Google Scholar]
- (35).Campbell JL; Baba T Near-Complete Structural Characterization of Phosphatidylcholines Using Electron Impact Excitation of Ions from Organics. Anal. Chem 2015, 87, 5837–5845. [DOI] [PubMed] [Google Scholar]
- (36).Thomas MC; Mitchell TW; Harman DG; Deeley JM; Nealon JR; Blanksby SJ Ozone-Induced Dissociation: Elucidation of Double Bond Position within Mass-Selected Lipid Ions. Anal. Chem 2008, 80, 303–311. [DOI] [PubMed] [Google Scholar]
- (37).Pham HT; Maccarone AT; Thomas MC; Campbell JL; Mitchell TW; Blanksby SJ Structural Characterization of Glycerophospholipids by Combinations of Ozone- and Collision-Induced Dissociation Mass Spectrometry: The next Step towards “Top-down” Lipidomics. Analyst 2013, 139, 204–214. [DOI] [PubMed] [Google Scholar]
- (38).Marshall DL; Pham HT; Bhujel M; Chin JSR; Yew JY; Mori K; Mitchell TW; Blanksby SJ Sequential Collision- and Ozone-Induced Dissociation Enables Assignment of Relative Acyl Chain Position in Triacylglycerols. Anal. Chem 2016, 88, 2685–2692. [DOI] [PubMed] [Google Scholar]
- (39).Brodbelt JS Photodissociation Mass Spectrometry: New Tools for Characterization of Biological Molecules. Chem. Soc. Rev 2014, 43, 2757–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Reilly JP Ultraviolet Photofragmentation of Biomolecular Ions. Mass Spectrom Rev 2009, 28, 425–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Ly T; Julian RR Ultraviolet Photodissociation: Developments towards Applications for Mass-Spectrometry-Based Proteomics. Angewandte Chemie International Edition 2009, 48, 7130–7137. [DOI] [PubMed] [Google Scholar]
- (42).Klein DR; Brodbelt JS Structural Characterization of Phosphatidylcholines Using 193 Nm Ultraviolet Photodissociation Mass Spectrometry. Anal. Chem 2017, 89, 1516–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Williams PE; Klein DR; Greer SM; Brodbelt JS Pinpointing Double Bond and sn-Positions in Glycerophospholipids via Hybrid 193 Nm Ultraviolet Photodissociation (UVPD) Mass Spectrometry. J. Am. Chem. Soc 2017, 139, 15681–15690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Becher S; Esch P; Heiles S Relative Quantification of Phosphatidylcholine Sn-Isomers Using Positive Doubly Charged Lipid–Metal Ion Complexes. Anal. Chem 2018, 90, 11486–11494. [DOI] [PubMed] [Google Scholar]
- (45).Blevins MS; Klein DR; Brodbelt JS Localization of Cyclopropane Modifications in Bacterial Lipids via 213 Nm Ultraviolet Photodissociation Mass Spectrometry. Analytical Chemistry 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Bligh EG; Dyer WJ A Rapid Method of Total Lipid Extraction and Purification. Can. J. Biochem. Physiol 1959, 37, 911–917. [DOI] [PubMed] [Google Scholar]
- (47).Klein DR; Holden DD; Brodbelt JS Shotgun Analysis of Rough-Type Lipopolysaccharides Using Ultraviolet Photodissociation Mass Spectrometry. Anal. Chem 2016, 88, 1044–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Klein DR; Feider CL; Garza KY; Lin JQ; Eberlin LS; Brodbelt JS Desorption Electrospray Ionization Coupled with Ultraviolet Photodissociation for Characterization of Phospholipid Isomers in Tissue Sections. Anal. Chem 2018, 90, 10100–10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Guan Z; Kelleher NL; O’Connor PB; Aaserud DJ; Little DP; McLafferty FW 193 Nm Photodissociation of Larger Multiply-Charged Biomolecules. International Journal of Mass Spectrometry and Ion Processes 1996, 157–158, 357–364. [Google Scholar]
- (50).Antoine R; Joly L; Tabarin T; Broyer M; Dugourd P; Lemoine J Photo-Induced Formation of Radical Anion Peptides. Electron Photodetachment Dissociation Experiments. Rapid Communications in Mass Spectrometry 2007, 21, 265–268. [DOI] [PubMed] [Google Scholar]
- (51).Madsen JA; Cullen TW; Trent MS; Brodbelt JS IR and UV Photodissociation as Analytical Tools for Characterizing Lipid A Structures. Anal. Chem 2011, 83, 5107–5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Crittenden CM; Herrera CM; Williams PE; Ricci DP; Swem LR; Trent MS; Brodbelt JS Mapping Phosphate Modifications of Substituted Lipid A via a Targeted MS3 CID/UVPD Strategy. Analyst 2018, 143, 3091–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Magnuson K; Jackowski S; Rock CO; Cronan JE Regulation of Fatty Acid Biosynthesis in Escherichia Coli. Microbiology and Molecular Biology Reviews 1993, 57, 522–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Feng Y; Cronan JE Escherichia Coli Unsaturated Fatty Acid Synthesis: Complex Transcription of the FabA Gene and in Vivo Idenification of the Essential Reaction Catalyzed by FabB. Journal of Biological Chemistry 2009, 284, 29526–29535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Zhang W; Donghui Z; Chen Q; Wu J; Ouyang Z; Xia Y Online Photochemical Derivatization Enables Comprehensive Mass Spectrometric Analysis of Unsaturated Phospholipid Isomers. Nature Communications 2019, 10, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.