Abstract
Purpose
miR-497-5p can inhibit cervical cancer cell proliferation. However, the underlying mechanism remains to be elucidated.
Methods
Bioinformatics was used to analyze the target genes of miR-497-5p. qRT-PCR and Western blot were used to analyze mRNA and protein expression, respectively. Dual-luciferase reporter assay was used to analyze the direct binding between miR-497-5p and 3ʹ-untranslated region of CBX4. Cell viability was measured with MTT assay. Flow cytometry was performed to detect cell cycle distribution.
Results
Here, using bioinformatics methods we firstly found that miR-497-5p regulated cervical carcinoma proliferation by targeting polycomb chromobox4 (CBX4). Expression of miR-497-5p in cervical carcinoma tissues was negatively correlated with CBX4. A binding region of miR-497-5p in 3ʹ-untranslated region of CBX4 was predicted. Further experiments confirmed that miR-497-5p directly targeted CBX4. Besides, RNA interference of CBX4 inhibited cervical cancer cell proliferation, arrested cells at S phase and reduced the expression of CDK2 and Cyclin A2 proteins. The use of miR-497-5p inhibitor compromised CBX4 interference RNAs induced cycle arrest of cervical cancer cells. Cells co-transfected with miR-497-5p inhibitors and CBX4 interference RNAs had a higher proliferation rate than CBX4 inference RNA-transfected cells.
Conclusion
All together, the present study demonstrates that miR-497-5p inhibits cervical cancer cells proliferation by directly targeting CBX4.
Keywords: MicroRNA-497-5p, CBX4, cervical cancer, carcinoma, proliferation
Introduction
MicroRNA (miRNA or miR) is a class of non-coding RNA with 14–24 nucleotides.1 It is reported that abnormal expression of miRNA is associated with tumorigenesis.1–3 Some miRNA molecules can induce tumorigenesis and promote tumor progression as tumor promoters; on the other hand, some miRNA molecules act as tumor suppressors and inhibit the growth of tumor cells or induce their apoptosis. Therefore, miRNA molecules are considered as novel tumor markers and can be used in the diagnosis, treatment and prognosis prediction of tumors.4 However, one miRNA molecule often has multiple target genes with similar or opposite functions, and the regulatory effects of a miRNA molecule on cell growth are related to many factors, such as cell type.5 Therefore, in order to promote the application of miRNA in cancer diagnosis, treatment and prognosis prediction, it is of great importance to fully understand the targets and regulatory mechanisms of miRNA.
miR-497-5p belongs to miR-15 family and is coded by the first intron of MIR497HG gene (ID: 100506755) on human chromosome 17p13.1. The expression of miR-497-5p is extensive in vivo, and its expression level is closely related to disease development. It is shown that miR-497-5p can promote the differentiation of lung-resident mesenchymal stem cells into myofibroblasts and the formation of pulmonary fibrosis by inhibiting the expression of Reck gene.6 Increased expression of miR-497-5p results in decreased insulin receptor content and induces insulin resistance in HFD-MES rats.7 In addition, changes in the expression of miR-497-5p are related to the occurrence and development of nasopharyngeal carcinoma, non-small cell lung cancer and hepatocellular carcinoma.8–10 The regulation of miR-497-5p on glioma cell growth may be different and may be acted through different mechanisms. For example, Xi et al reported that miR-497-5p targeted and bound with Nrdp1 to promote the proliferation of human glioma U87 cells.11 Low expression of miR-497-5p enhanced growth inhibition and apoptosis of glioma cells induced by TMZ,12 however, Zhao et al demonstrated that miR-497-5p expression decreased glioma cell growth and invasion both in vitro and in vivo through Wnt3a/c-Jun feedback regulation.13 Different from glioma, the function of miR-497-5p in other tumors is growth inhibition. For example, miR-497-5p can target YAP1 to inhibit the proliferation, invasion and migration of thyroid papillary carcinoma and non-small cell lung cancer,14,15 and induce cycle arrest and apoptosis of breast cancer, gastric cancer, lung cancer and cervical cancer by regulating the expression of Bcl-w, Bcl-2, CDK6 or Cyclin E1.16–19 These effects of miR-497-5p make it a potential marker for tumor diagnosis, treatment and prognosis.20–23
Cervical cancer is the fourth most common malignant tumor in the world in terms of morbidity and mortality.24 It has been reported that miR-497-5p is differently expressed in cervical cancer, cervical intraepithelial neoplasia and normal tissues, and it can be used in the clinical diagnosis of cervical cancer.21 By targeting insulin-like growth factor 1 receptor (IGF-1R) and transketolase (TKT), miR-497-5p inhibits cell invasion, migration, and drug resistance of the cervical cancer cells.25,26 Our previous study has also shown that miR-497-5p can target Cyclin E1 to arrest cell cycle of cervical cancer cells.19 However, it still remains unclear whether miR-497-5p can regulate the growth of cervical cancer cells through other mechanisms. In the present study, we investigated the mechanism by which miR-497-5p regulates the growth of cervical cancer cells, in order to promote the application of miR-497-5p in the diagnosis, treatment and prognosis of cervical cancer.
Materials And Methods
Bioinformatics
Target genes of miR-497-5p were firstly predicted using microRNA (http://www.microrna.org/microrna/home.do), TargetScan (http://www.targetscan.org), and miRDB (http://mirdb.org/), and then, intersection analyzed by Venny (version 2.1.0; http://bioinfogp.cnb.csic.es/tools/venny/index.html). After that, PPI Network (https://www.theppinetwork.com/) was used to analyze interactions among different proteins, MCODE APP (http://apps.cytoscape.org/apps/mcode) was used for functional clustering of the proteins. Gene expression data of miR-497-5p and polycomb repressive complex1-associated protein (CBX4) in cancer tissues and tumor-adjacent tissues of cervical cancer patients were downloaded from TCGA Research Network (https://www.cancer.gov/tcga) using the UCSC Xena platform (https://xenabrowser.net/hub/), and used to analyze the genes’ expression relationship on Online software Xena.
Cells
Cell lines of human cervical cancer Siha and HeLa cells were gifts from Department of Cell Biology and Genetics, Medical Department, Xi’an Jiaotong University, China). They were cultured in DMEM medium (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (Biological Industries, Kibbutz Beth Haemek, Israel) at 37°C in a humidified atmosphere of 5% CO2. The use of the gifted cell lines was approved by the ethics committee of Medical College of Yan’an University.
Inhibitor, Interference RNA, And Cell Transfection
Inhibitor of hsa-miR-497-5p (miR-497-in; 5ʹ-ACAAACCACAGTGTGCTGCTG-3ʹ) and its negative control (miR-497-inNC; 5ʹ-CAGTACTTTTGTGTAGTACAA-3ʹ) were synthesized by Sangon Biotech, Shanghai, China. In addition, the mimics of hsa-miR-497-5p (miR-497-over), RNA interference of CBX4 (SiCBX4-1and SiCBX4-2) and NC were designed and synthesized by Gene Pharma Biotech, Shanghai, China. Cell transfection was performed with the jetPRIME transfection reagent (Polyplus Co., Illkirch, France). Four hours after transfection, fresh medium was replenished.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from cells using TRIzol (Thermo Fisher Scientific, Waltham, MA, USA). Using the extracted RNA as template, cDNA fragments were synthesized from Oligo dT according to the manual of PrimeScript™ 1st Strand cDNA Synthesis Kit (TaKaRa, Dalian, China). The sequence of reverse transcription primer of U6 was CGCTTCACGAATTTGCGTGTCAT, and that of miR-497-5p was GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACGGTTTGT. The RealStar SYBR Green qPCR Power Mixture (GenStar, Beijing, China) was used to detect the miR-497-5p and CBX4 expression. The primers used in qPCR reactions are listed in Table 1. U6 or GAPDH was used as an internal reference gene. The relative expression levels of miR-497-5p and CBX4 in different samples were calculated by 2−ΔΔCt. Each experiment was performed in triplicate.
Table 1.
Primer Sequences Used In qPCR
| Gene Name | Primer Name | Primer Sequence (from 5ʹ to 3ʹ) |
|---|---|---|
| miR-497-5p | Forward | ATCCAGTGCGTGTCGTG |
| Reverse | TGCTCAGCAGCACACTGT | |
| U6 | Forward | GCTTCGGCAGCACATATACTAAAAT |
| Reverse | CGCTTCACGAATTTGCGTGTCAT | |
| CBX4 | Forward | CTGGTGAAATGGAGAGGC |
| Reverse | GAACGACGGGCAAAGGTAGG | |
| GAPDH | Forward | TGAAGGTCGGAGTCAACGGATT |
| Reverse | CCTGGAAGATGGTGATGGGATT |
Dual-Luciferase Reporter Assay
According to bioinformatics results, wild-type and mutant seed regions of miR-497-5p in the 3ʹ-UTR of CBX4 gene were chemically synthesized in vitro. Then, their two ends were attached with Spe-1 and HindIII restriction sites, and cloned into pMIR-GLO luciferase reporter plasmids. Plasmids (0.8 μg) with wild-type or mutant 3ʹ-UTR sequences were co-transfected with miR-497-5p mimics (30nmol/L; Sangon Biotech, Shanghai, China) into Siha and HeLa cells using jetPRIME. For the control group, Siha and HeLa cells were transfected with miR-negative control (NC). After culturing for 24 hrs, the cells were lysed and analyzed using dual-luciferase reporter assay kit (Promega, Fitchburg, WI, USA) according to the manufacturer’s manual, and luminescence intensity was measured using GloMax 20/20 luminometer (Promega, Fitchburg, WI, USA). The luminescence values of each group of cells were measured. Renilla luminescence activity was used as an internal reference. Each experiment was performed in triplicate.
Western Blotting
Cells were lysed with precooled Radio-Immunoprecipitation Assaylysis buffer supplemented with protease inhibitor (Beyotime Institute of Biotechnology, Shanghai, China) for 30 mins on ice. The supernatant was collected after centrifugation at 14,000 rpm, 4°C for 20 mins. Protein concentration was determined by bicinchoninic acid protein concentration determination kit (RTP7102, Real-Times Biotechnology Co., Ltd., Beijing, China). The samples (20 µg) were subjected to 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred to polyvinylidene difluoride membranes. After blocking with 5% skimmed milk at room temperature for 2 hrs, the membranes were incubated with rabbit anti-human CBX4 (1:1000; Abcam, Cambridge, UK), Cyclin A2 (1:1000; Abcam, Cambridge, UK), CDK2 (1:1000; Abcam, Cambridge, UK) or mouse anti-human β-actin (1:5000; Abcam, Cambridge, UK) monoclonal primary antibodies at 4°C overnight. After extensive washing with phosphate-buffered saline with Tween-20 for 3 times of 15 mins, the membranes were incubated with goat anti-rabbit or goat anti-mouse horseradish peroxidase-conjugated secondary antibody (1:5000; Santa Cruz, Dallas, TX, USA) for 1 hr at room temperature. Then, the membrane was developed with an enhanced chemiluminescence detection kit (Sigma-Aldrich, St. Louis, MO, USA). Image lab v3.0 software (Bio-Rad, Hercules, CA, USA) was used to acquire and analyze imaging signals. The relative contents of target proteins were expressed against β-actin.
MTT Assay
After transfection, cells were seeded into 96-well plates at a density of 2x103 cells per well. Triplicate wells were set up. At 24, 48 and 72 hrs after transfection, 20 µL MTT (5 g/L; Sigma-Aldrich, St. Louis, MO, USA) was added to each well, followed by incubation for 4 hrs at 37°C. DMSO (150 µL per well) was added to dissolve purple crystals. Then, the absorbance of each well was measured at 492 nm with a microplate reader (FLUOstar OPTIMA, BMG, Germany) and cell proliferation curves were plotted.
Flow Cytometry
At 24 hrs after transfection, cells were collected. Cell Cycle Assay Kit (BD Biosciences, Franklin Lakes, NJ, USA) was used to analyze the cell cycle. Briefly, the cells were incubated with 200 μL liquid A for 10 mins, and 150 μL liquid B for another 10 mins. Then, the cells were incubated with 120 μL liquid C in dark for 10 mins before flow cytometry analysis on FACSort (BD Biosciences, Franklin Lakes, NJ, USA). The result was analyzed using ModFit software version 3.2 (Verity Software House, Topsham, ME, USA).
Statistical Analysis
The results were analyzed using SPSS 20.0 statistical software (IBM, Armonk, NY, USA). The data were expressed as means ± standard deviations. Two group comparison was performed by Student’s t-test and multi-group comparison was performed by one-way ANOVA followed by LSD post-hoc test. Correlation between miR-497-5p and CBX4 expression was analyzed using Pearson’s correlation analysis. P < 0.05 indicated statistically significant differences. For all data, * means P<0.05, ** means P<0.01, *** means P<0.001.
Results
Bioinformatics Prediction Shows That miR-497-5p May Regulate The Growth Of Cervical Cancer Cells Through CBX4
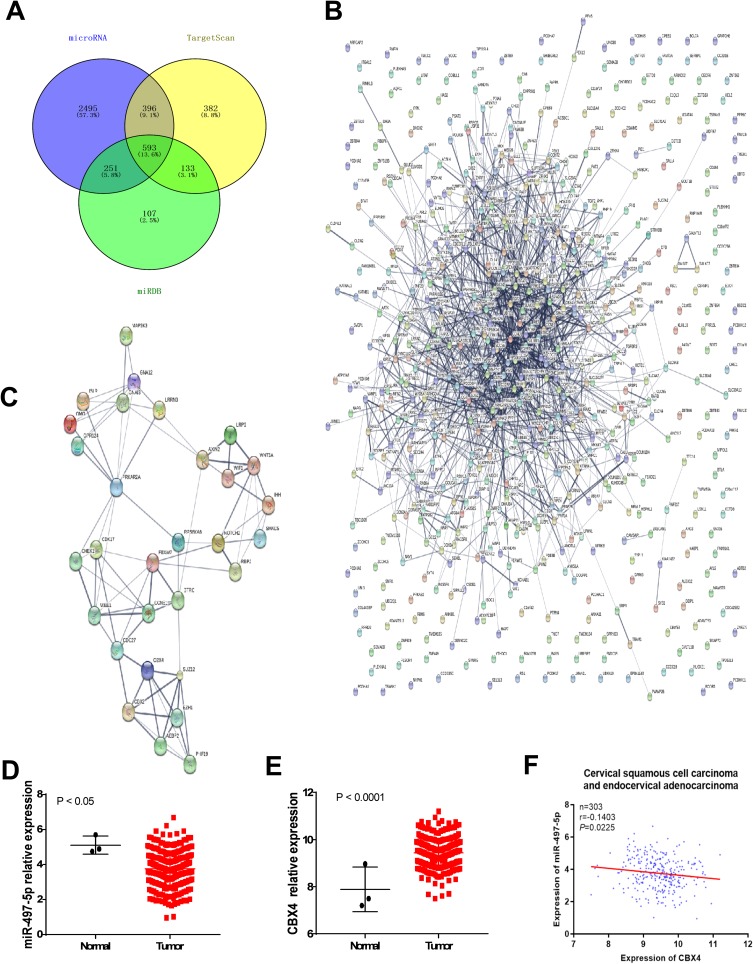
To investigate the mechanism by which miR-497-5p regulates the growth of cervical cancer cells, we used microRNA, TargetScan and miRDB to predict the potential target genes of miR-497-5p. The data showed that proteins coded by 4303 genes could possibly interact with miR-497-5p, among which 593 (13.6%) target genes were found in all three databases (Figure 1A). To further screen the target genes involved in the regulation of cervical cancer cell growth by miR-497-5p, PPI and MCODE APP were used to analyze the interaction network and functional clustering of the 593 target genes. The data showed that potential targets of miR-497-5p could regulate cell activities through multiple PPI networks (Figure 1B and Table 2), and the group with the highest functional clustering score involved 30 genes (Figure 1C). TCGA database was used to evaluate the correlation between the expression of 28 genes (excluding PRKAR2 and AEBP2) and the expression of miR-497-5p. The results shown here are in whole or part based upon data generated by the TCGA Research Network: https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga/using-tcga/citing-tcga. The data showed that four genes (CCNE1, CBX4, GNA12 and CDC27) among the 28 potential target genes were negatively correlated with the expression of miR-497-5p (R<-0.1; Table 3). Further studies showed that miR-497-5p expression in cervical cancer tissues was significantly lower than that in tumor-adjacent tissues (Figure 1D), and CBX4 expression in tumor tissues was significantly higher than that in tumor-adjacent tissues (Figure 1E). Correlation between miR-497-5p and CBX4 expression was analyzed by Pearson’s correlation analysis, which showed that CBX4 expression was negatively correlated with miR-497-5p expression (Figure 1F). These results suggest that miR-497-5p may regulate the growth of cervical cancer cells through CBX4. Finally, CBX4 was selected as the target gene of miR-497-5p for further study.
Figure 1.
Prediction for miR-497-5p regulating genes. (A) an intersection diagram of the number of miR-497-5p interacting genes predicted by microRNA, Target Scan and miRDB software. (B) Protein–protein interaction (PPI) analysis for the 593 miR-497-5p interacting genes commonly predicted by microRNA, Target Scan and miRDB. (C) PPI analysis for the 30 genes that functionally clustered into a group with high cluster score. (D and E) relative expression change analysis of miR-497-5p (D) and CBX4 (E) in cervical squamous cell carcinoma and endocervical adenocarcinoma. Data of TCGA were used. (F) Correlation analysis of miR-497-5p and CBX4 expression in cervical squamous cell carcinoma and endocervical adenocarcinoma. Data of TCGA were used (https://www.cancer.gov/tcga).
Table 2.
Cluster Analysis For PPI Network Analyzed Diagram
| Cluster | Score (Density*#Nodes) | Nodes | Edges | Node IDs |
|---|---|---|---|---|
| 1 | 5.379 | 30 | 78 | CHEK1, PHF19, PRKAR2A, CCNE1, FBXW7, OMG, SMAD5, CBX4, IHH, BTRC, GNAI3, CBX2, CDK17, GNA12, WIF1, ISLR, SUZ12, LRRN3, EZH1, GPR124, WEE1, NOTCH2, AEBP2, RBPJ, LRP6, WNT3A, RPS6KA6, CDC27, AXIN2, MAP3K3 |
| 2 | 5 | 5 | 10 | TBPL1, ASH1L, SUPT16H, MED26, TBP |
| 3 | 4 | 4 | 6 | CNTNAP1, SCN2A, NFASC, SCN8A |
| 4 | 3.6 | 6 | 9 | WNT7A, CCND1, FGF2, DLL4, SEMA3A, VEGFA |
| 5 | 3 | 3 | 3 | SMAD7, SMURF1, FKBP1A |
| 6 | 3 | 3 | 3 | RELN, PAFAH1B1, PAFAH1B2 |
| 7 | 3 | 3 | 3 | GALNT7, GALNT1, GALNT13 |
| 8 | 3 | 3 | 3 | SOX6, UBE2J1, SUMO3 |
| 9 | 2.8 | 6 | 7 | SHOC2, RAP2C, PPM1D, PPM1L, CASK, MAP2K1 |
| 10 | 2.364 | 12 | 13 | ATG9A, MYB, VPS4A, PDIK1L, TUBA1A, RPS6KA3, UNC13A, YWHAH, VTI1B, PIM1, PRKG1, WIPI2 |
Notes: Cluster analysis was produced by MCODE of cytoscape software. Parameters are, Scoring (Include Loops: false; Degree Cutoff: 2); Cluster Finding (Node Score Cutoff: 0.2; Haircut: true; Fluff: false; K-Core: 2; Max. Depth from Seed: 100).
Table 3.
Relationship Between Genes’ And miR-497 Expression
| Gene | R Value | Gene | R Value |
|---|---|---|---|
| CHEK1 | −0.01945 | ISLR | 0.324021 |
| PHF19 | 0.050232 | SUZ12 | 0.078904 |
| CCNE1 | −0.1274 | LRRN3 | −0.00977 |
| FBXW7 | −0.00828 | EZH1 | 0.253977 |
| OMG | −0.00805 | GPR124 | 0.221022 |
| SMAD5 | 0.124455 | WEE1 | 0.12194 |
| CBX4 | −0.14025 | NOTCH2 | 0.023932 |
| IHH | 0.203239 | AEBP2 | −0.00545 |
| BTRC | −0.07658 | RBPJ | 0.162236 |
| GNAI3 | −0.02774 | WNT3A | 0.070756 |
| CBX2 | −0.01101 | RPS6KA6 | 0.130981 |
| CDK17 | −0.02892 | CDC27 | −0.12934 |
| GNA12 | −0.17605 | AXIN2 | 0.203712 |
| WIF1 | 0.144997 | MAP3K3 | 0.065202 |
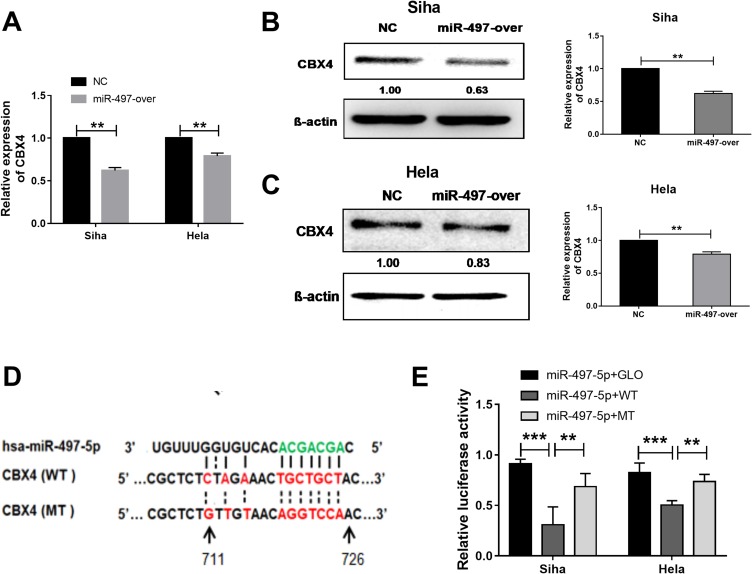
miR-497-5p Inhibits CBX4 Expression By Directly Binding With The 3ʹ-UTR Of CBX4 mRNA
To test whether miR-497-5p regulates CBX4 expression, we transfected Siha and HeLa cells with miR-497-5p mimics and detected CBX4 expression with qPCR and Western blot. The data showed that Siha and HeLa cells transfected with miR-497-5p mimics had reduced CBX4 expression (Figure 2A–C). To examine whether miR-497-5p inhibits CBX4 expression by directly binding with the 3ʹ-UTR of CBX4 mRNA, dual-luciferase reporter assay was performed. The data showed that the luminescence intensity of Siha and HeLa cells in the wild-type group was significantly lower than the negative control group, while that in the mutant group was not significantly different from the negative control group (Figure 2D). The results indicate that miR-497-5p inhibits the expression of CBX4 gene by directly binding with the 3ʹ-UTR of CBX4 mRNA.
Figure 2.
miR-497-5p regulates CBX4 expression by targeting CBX4 mRNA 3ʹUTR region. (A) qRT-PCR examination for CBX4 mRNA expression and (B and C) for CBX protein expression in miR-497-5p mimics (miR-497-over) and negative control miRNAs (NC) transected Siha and HeLa cells. miR-497-5p mimics (miR-497-over) or negative control RNAs (NC) were transfected for 48 hrs. (D) Predicted binding site of miR-497-5p (green) in CBX4 mRNA 3ʹUTR. (E) Dual-Luciferase reporter assay results for wild type and miR-497-5p binding cite mutant CBX4 3ʹUTR. Cells transfected with miR-497-5p mimics and GLO plasmid were used as control treatments. For all data, * means P< 0.05, ** means P<0.01, *** means P<0.001.
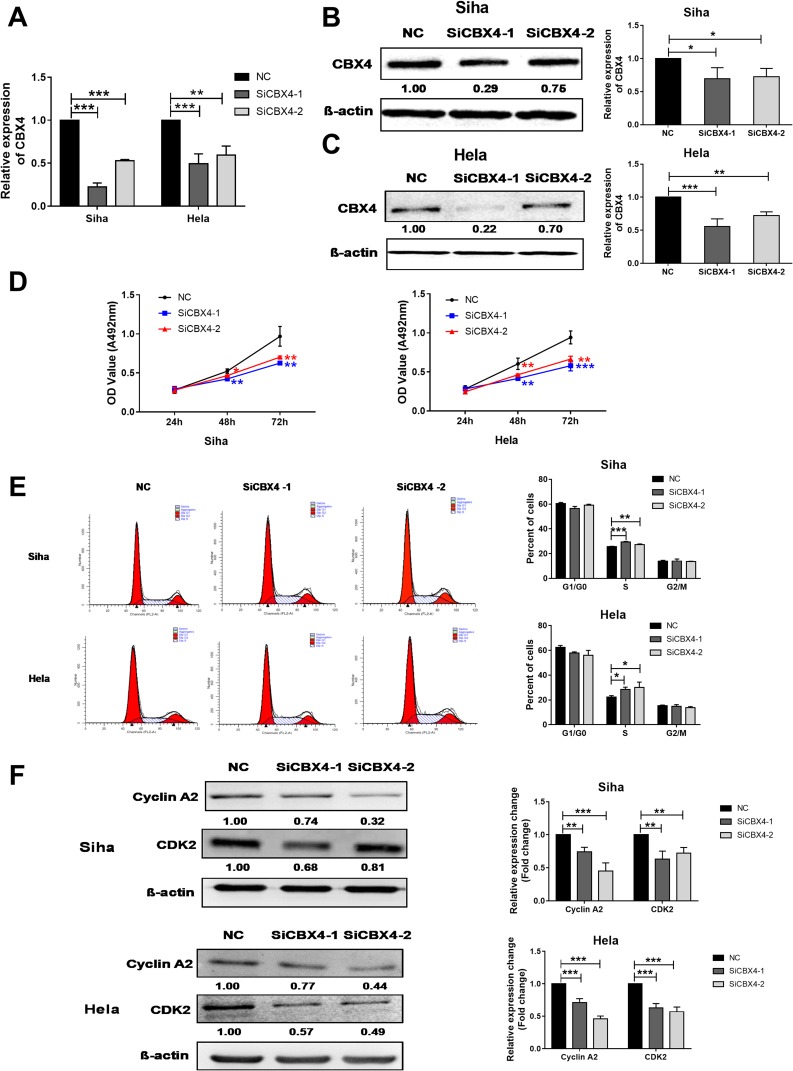
Down-Regulation Of CBX4 Expression Inhibits The Proliferation Of Cervical Cancer Cells By Arresting The Cells In S Phase And Reducing The Expression Of CDK2 And Cyclin A2 Proteins
To inhibit CBX4 expression in cervical cancer cells, two interference RNAs of CBX4, ie SiCBX4-1and SiCBX4-2, were synthesized. The data showed that CBX4 expression in cells transfected with SiCBX4-1 or SiCBX4-2 was significantly lower than that in the negative control group, and the inhibitory rate of SiCBX4-1 was higher than that of SiCBX4-2 (Figure 3A–C). To test the effect of the inhibition of CBX4 expression on cervical cancer cell growth, MTT assay was conducted. The results showed that, at 48 hrs after cell transfection, proliferation of Siha and HeLa cells with silenced expression of CBX4 was significantly lower than that in negative control group (Figure 3D). Flow cytometry showed that the number of S-phase Siha and HeLa cells with silenced expression of CBX4 was significantly higher than that in negative control group (Figure 3E).To determine the expression of cell cycle regulators CDK2 and Cyclin A2, Western blotting was used. The data showed that levels of CDK2 and Cyclin A2 proteins in Siha and HeLa cells with silenced expression of CBX4 were lower than that in negative control group (Figure 3F). These results suggest that down-regulation of CBX4 expression inhibits the proliferation of cervical cancer cells by arresting the cells in S phase and reducing the expression of CDK2and Cyclin A2 proteins.
Figure 3.
CBX4 regulates cervical cancer cells proliferation by regulating of Cyclin A2 and CDK2 expression. (A–C) qRT-PCR (A) and Western blot (B and C) examination for CBX4 RNA interference efficiency. SiCBX4-1 and SiCBX4-2 are interference RNAs used for CBX4 down-regulation. NC is the RNAs used as negative control. (D) MTT examination for the proliferation of Siha (Left) and HeLa (Right) cells pre-transfected with NC, SiCBX4-1 and SiCBX4-2. (E) Cell cycle of Siha and HeLa cells pre-treated with NC or CBX4 siRNA (SiCBX4-1 and SiCBX4-1) were tested using flow cytometry assay (left). The statistic results of three independent experiments is on the right. (F) Influence of CBX4 down-regulation on cell cycle-related protein Cyclin A2 and CDK2 expression. Siha and HeLa cells pre-transfected with NC, SiCBX4-1 or SiCBX4-2 for 48 hrs were used to examine the expression of Cyclin A2 and CDK2 by Western blot (bottom, relative change of gray value). β-Actin was used as the internal control. For all data, * means P< 0.05, ** means P<0.01, *** means P<0.001.
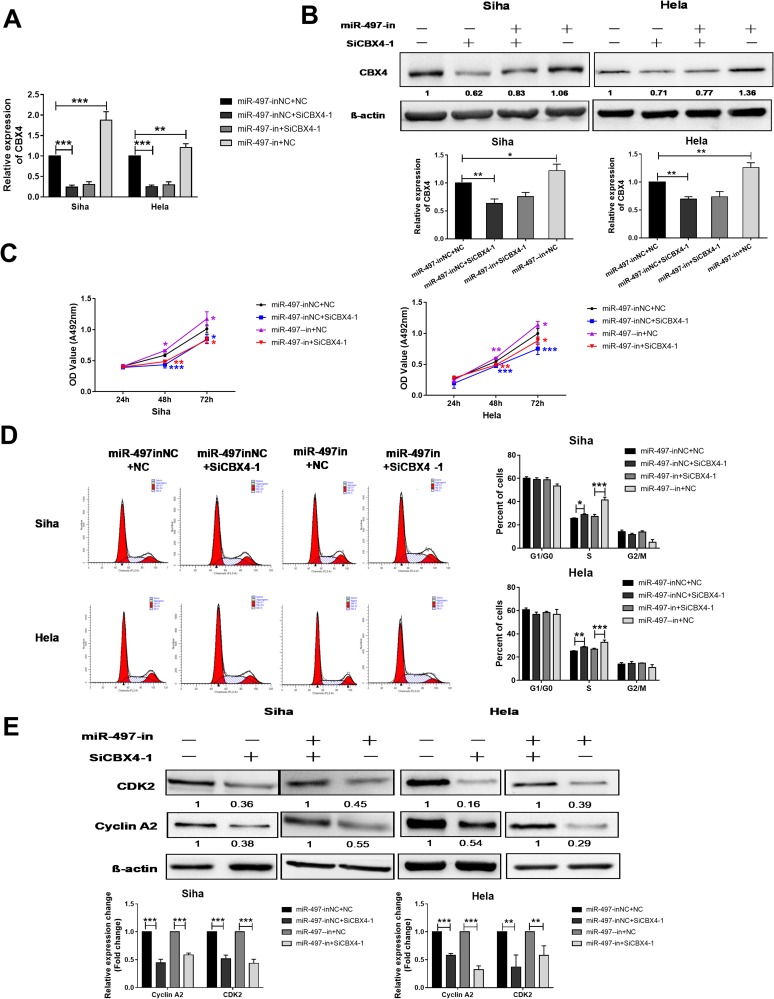
miR-497-5p Is Involved In The Regulation Of Cervical Cancer Cell Proliferation Through Regulating CBX4-CDK2/Cyclin A2 Expression
To test whether miR-497-5p regulates cervical cancer cells growth by regulating CBX4-CDK2/Cyclin A2 expression, we suppressed miR-497-5p expression by miR-497-5p inhibitor (miR-497-in), and silenced CBX4 expression by CBX4 interference RNAs (SiCBX4-1), at the same time. The data showed that suppression of miR-497-5p elevated CBX4 expression when compared with the control group (miR-497-inNC+NC) (Figure 4A). In addition, CBX4 expression in cells with both miR-497-5p inhibitor and CBX4 interference RNAs (miR-497-in+SiCBX4-1 group) was decreased when compared with the control group (miR-497-inNC+NC), suggesting that silenced expression of CBX4 neutralized miR-497-5p inhibitor induced up-regulation of CBX4 (Figure 4B). MTT assay showed that, 48 hrs later after cell transfection, the proliferation of Siha and HeLa cells with the miR-497-5p inhibitor (miR-497-in) was elevated, those with the CBX4 interference RNAs (SiCBX4-1) was reduced, and the proliferation of Siha and HeLa cells with both miR-497-5p inhibitor and CBX4 interference RNAs, miR-497-in+SiCBX4-1 group, was also decreased (Figure 4C). Flow cytometry showed that inhibition of miR-497-5p elevated the number of S-phase Siha and HeLa cells, silencing of CBX4 increased the number of S-phase Siha and HeLa cells, and the increase in the number of S-phase Siha and HeLa cells with inhibition of miR-497-5p was decreased by silencing of CBX4 (Figure 4D). Western blotting showed that the silencing of CBX4 reduced the content of CDK2 and Cyclin A2 proteins, and inhibition of miR-497-5p and silencing of CBX4 at the same time also decreased the expression of CDK2 and Cyclin A2 proteins (Figure 4E). These results indicate that miR-497-5p may regulate cervical cancer cell proliferation through regulating CBX4, CDK2 and Cyclin A2 expression.
Figure 4.
CBX4 is responsible for miR-497-5p regulated cervical cancer proliferation. (A and B) qRT-PCR (A) and Western blot (B) examination for CBX4 expression in Siha and HeLa cells treated with miR-497-5p inhibitors (miR-497-in) and/or CBX4 interference RNAs (SiCBX4-1, SiCBX4-2). (C) MTT examination for Siha and HeLa cells’ proliferation. (D) Flow cytometry examination for Siha and Hela cells, cell cycle changes (left). Cells pre-transfected with different RNAs for 48 hrs were used for above examination. Right is a statistical result of three independent experiments. (E) Western blot examination for CDK2 and Cyclin A2 expression. Siha and HeLa cells treated like A were protein extracted and used for Western blot test. Bottom is the relative blot change of the gray value. For all data, * means P< 0.05, ** means P<0.01, *** means P<0.001.
Discussion
CBX4, also known as hPC2 or NBP16, is a member of the polycomb protein family with SUMO E3 ligase activity. In cell growth, CBX4 can regulate DNA modification, stability and terminal repair through SUMOylation of Dnmt3, BMl1 and Ctlp,27–29 and is possibly related to the maintenance of DNA replication and integrity. It is reported that CBX4 gene expression can promote the growth of breast cancer and hepatocellular carcinoma cells, and it is a potential tumor growth promoter.30–32 Our data showed that reducing the expression of CBX4 gene inhibited the proliferation of Siha and HeLa cells and arrested the cell cycle in S phase. Western blotting confirmed that the expression of CDK2 and Cyclin A2 protein was down-regulated, demonstrating that CBX4 down-regulation inhibits the cell cycle of cervical cancer cells, as well as their proliferation. This result is consistent with the promoting effect of CBX4 on the growth of breast cancer and liver cancer, suggesting that CBX4 may be a therapeutic target for breast cancer, liver cancer and cervical cancer.33 In addition, RNA interference of CBX4 arrested Siha and HeLa cells at S phase, indicating that CBX4 has a positive regulatory effect on S/G2 transition of cervical cancer cells, which is consistent with the regulatory effects of CBX4 on DNA modification, stability and terminal repair.
miR-497-5p can target CBX4 to inhibit the proliferation of cervical cancer cells. When treating cervical cancer cells with miR-497-5p inhibitor, we found that CBX4 gene expression was increased, cell proliferation rate of cervical cancer cells was elevated, and cell cycle was arrested at S-phase, which is consistent with the regulatory effect of miR-497-5p on the cell cycle of ALK-positive cells, melanoma cells and 293T cells.34–36 However, inhibition of miR-497-5p together with down-regulation of CBX4 resulted in decreased proliferation of cervical cancer cells. The number of S-phase cells in this group was lower than that of cells with inhibition of miR-497-5p, and higher than that of control group, suggesting that CBX4 is a key molecule in the regulation of the growth of cervical cancer cells by miR-497-5p, and changes in CBX4 expression directly affect the proliferation of cervical cancer cells.
This study has some limitations. First, the sample size of normal tissue from the TCGA database was small. Second, the correlation between miR-497-5p and CBX4 in cervical squamous cell carcinoma was weak. Further studies are warranted.
In conclusion, the present study demonstrates that miR-497-5p affects the cell cycle of cervical cancer cells by targeting CBX4, and thus inhibits the proliferation of cervical cancer cells. This study provides help in understanding the mechanism by which miR-497-5p regulates the proliferation of cervical cancer cells.
Acknowledgements
The authors would like to thank the National Natural Science Foundation for their funding assistance and support.
Funding Statement
This work was supported by the National Natural Science Foundation of China [Nos. 81660492, 81860444, 81760510, 8176100559], Shaanxi Innovative Talents Promotion Plan-Science and Technology New Star Project [No. 2017KJXX-20], and Yan’an City Science and Technology Research Development Planning Project [No. 2016KS-06] and Scientific Research Plan Projects of Yan‘an university [Nos. YDBK2018-49, YD2015-15, YDT2016-03].
Abbreviations
miRNA, MicroRNA; miR, MicroRNA; CBX4, polycomb chromobox 4.
Data Availability
The analyzed datasets generated during the study are available from the corresponding author on reasonable request.
Author Contributions
ZJ, CY and DJ conceived and designed the present study. ZJ and DJ analyzed the obtained data and drafted the manuscript. CY performed all bioinformatics analysis. WY and JQ, WYF and ZH separately prepared Siha, Hela cells and tested cell proliferation using MTT method. SH performed all Flow cytometry experiments. WYM and XW did western blot experiment. GY and LD constructed plasmids and examined luciferase luciferase activity of the reporter genes. FY and YJ performed real-time quantification PCR. All authors read and approved the manuscript and agree to countable for all aspects of the study in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Feng Y, Bai F, You Y, et al. Dysregulated microRNA expression profiles in gastric cancer cells with high peritoneal metastatic potential. Exp Ther Med. 2018;16(6):4602–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Naorem LD, Muthaiyan M, Venkatesan A. Identification of dysregulated miRNAs in triple negative breast cancer: a meta-analysis approach. J Cell Physiol. 2019;234(7):11768–11779. doi: 10.1002/jcp.v234.7 [DOI] [PubMed] [Google Scholar]
- 3.Hwang J, Min BH, Jang J, et al. MicroRNA expression profiles in gastric carcinogenesis. Sci Rep. 2018;8(1):14393. doi: 10.1038/s41598-018-32782-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petrovic N, Ergun S. miRNAs as potential treatment targets and treatment options in cancer. Mol Diagn Ther. 2018;22(2):157–168. [DOI] [PubMed] [Google Scholar]
- 5.Svoronos AA, Engelman DM, Slack FJ. OncomiR or tumor suppressor? The duplicity of MicroRNAs in cancer. Cancer Res. 2016;76(13):3666–3670. doi: 10.1158/0008-5472.CAN-16-0359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen X, Shi C, Wang C, et al. The role of miR-497-5p in myofibroblast differentiation of LR-MSCs and pulmonary fibrogenesis. Sci Rep. 2017;7:40958. doi: 10.1038/srep40958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, Wang M, Li H, et al. Upregulation of miR-497 induces hepatic insulin resistance in E3 rats with HFD-MetS by targeting insulin receptor. Mol Cell Endocrinol. 2015;416:57–69. doi: 10.1016/j.mce.2015.08.021 [DOI] [PubMed] [Google Scholar]
- 8.Wang S, Mo Y, Midorikawa K, et al. The potent tumor suppressor miR-497 inhibits cancer phenotypes in nasopharyngeal carcinoma by targeting ANLN and HSPA4L. Oncotarget. 2015;6(34):35893–35907. doi: 10.18632/oncotarget.5651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding Q, He K, Luo T, et al. SSRP1 contributes to the malignancy of hepatocellular carcinoma and is negatively regulated by miR-497. Mol Ther. 2016;24(5):903–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Z, Lu Q, Zhu D, Han Y, Zhou X, Ren T. Lnc-SNHG1 may promote the progression of non-small cell lung cancer by acting as a sponge of miR-497. Biochem Biophys Res Commun. 2018;506(3):632–640. doi: 10.1016/j.bbrc.2018.10.086 [DOI] [PubMed] [Google Scholar]
- 11.Gao Y, Sun L, Wu Z, Chen X. [Over-expression of miR-497 promotes the proliferation of U87 glioma cells by targeting neuregulin receptor degradation protein 1]. Xi Bao Yu Fen Zi Mian Yi XueZaZhi. 2017;33(8):1051–1055. [PubMed] [Google Scholar]
- 12.Zhu D, Tu M, Zeng B, et al. Up-regulation of miR-497 confers resistance to temozolomide in human glioma cells by targeting mTOR/Bcl-2. Cancer Med. 2017;6(2):452–462. doi: 10.1002/cam4.2017.6.issue-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao L, Tang X, Luo R, Duan J, Wang Y, Yang B. MicroRNA-490-5P targets CCND1 to suppress cellular proliferation in glioma cells and tissue through cell cycle arrest. Curr Neurovasc Res. 2018;15(3):246–255. [DOI] [PubMed] [Google Scholar]
- 14.Huang C, Ma R, Yue J, Li N, Li Z, Qi D. MiR-497 suppresses YAP1 and inhibits tumor growth in non-small cell lung cancer. Cell Physiol Biochem. 2015;37(1):342–352. [DOI] [PubMed] [Google Scholar]
- 15.Cheng H, Dong H, Feng J, Tian H, Zhang H, Xu L. miR-497 inhibited proliferation, migration and invasion of thyroid papillary carcinoma cells by negatively regulating YAP1 expression. Onco Targets Ther. 2018;11:4711–4721. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Shen L, Li J, Xu L, et al. miR-497 induces apoptosis of breast cancer cells by targeting Bcl-w. Exp Ther Med. 2012;3(3):475–480. doi: 10.3892/etm.2011.428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu W, Zhu D, Lu S, et al. miR-497 modulates multidrug resistance of human cancer cell lines by targeting BCL2. Med Oncol. 2012;29(1):384–391. doi: 10.1007/s12032-010-9797-4 [DOI] [PubMed] [Google Scholar]
- 18.Furuta M, Kozaki K, Tanimoto K, et al. The tumor-suppressive miR-497-195 cluster targets multiple cell-cycle regulators in hepatocellular carcinoma. PLoS One. 2013;8(3):e60155. doi: 10.1371/journal.pone.0060155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han J, Huo M, Mu M, Liu J, Zhang J. [miR-497 suppresses proliferation of human cervical carcinoma HeLa cells by targeting cyclin E1]. Xi Bao Yu Fen Zi Mian Yi XueZaZhi. 2014;30(6):597–600. [PubMed] [Google Scholar]
- 20.Pang PC, Shi XY, Huang WL, Sun K. miR-497 as a potential serum biomarker for the diagnosis and prognosis of osteosarcoma. Eur Rev Med Pharmacol Sci. 2016;20(18):3765–3769. [PubMed] [Google Scholar]
- 21.Zhang Y, Zhang D, Wang F, Xu D, Guo Y, Cui W. Serum miRNAs panel (miR-16-2*miR-195, miR-2861, miR-497) as novel non-invasive biomarkers for detection of cervical cancer. Sci Rep. 2015;5:17942. doi: 10.1038/srep17942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao X, Zhao Z, Xu W, Hou J, Du X. Down-regulation of miR-497 is associated with poor prognosis in renal cancer. Int J Clin Exp Pathol. 2015;8(1):758–764. [PMC free article] [PubMed] [Google Scholar]
- 23.Xu J, Wang T, Cao Z, et al. MiR-497 downregulation contributes to the malignancy of pancreatic cancer and associates with a poor prognosis. Oncotarget. 2014;5(16):6983–6993. doi: 10.18632/oncotarget.v5i16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 25.Luo M, Shen D, Zhou X, Chen X, Wang W. MicroRNA-497 is a potential prognostic marker in human cervical cancer and functions as a tumor suppressor by targeting the insulin-like growth factor 1 receptor. Surgery. 2013;153(6):836–847. doi: 10.1016/j.surg.2012.12.004 [DOI] [PubMed] [Google Scholar]
- 26.Yang H, Wu XL, Wu KH, et al. MicroRNA-497 regulates cisplatin chemosensitivity of cervical cancer by targeting transketolase. Am J Cancer Res. 2016;6(11):2690–2699. [PMC free article] [PubMed] [Google Scholar]
- 27.Soria-Bretones I, Cepeda-Garcia C, Checa-Rodriguez C, et al. DNA end resection requires constitutive sumoylation of CtIP by CBX4. Nat Commun. 2017;8(1):113. doi: 10.1038/s41467-017-00183-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ismail IH, Gagne JP, Caron MC, et al. CBX4-mediated SUMO modification regulates BMI1 recruitment at sites of DNA damage. Nucleic Acids Res. 2012;40(12):5497–5510. doi: 10.1093/nar/gks222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li B, Zhou J, Liu P, et al. Polycomb protein Cbx4 promotes SUMO modification of de novo DNA methyltransferase Dnmt3a. Biochem J. 2007;405(2):369–378. doi: 10.1042/BJ20061873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meng R, Fang J, Yu Y, et al. miR-129-5p suppresses breast cancer proliferation by targeting CBX4. Neoplasma. 2018;65(4):572–578. doi: 10.4149/neo_2018_170814N530 [DOI] [PubMed] [Google Scholar]
- 31.Zeng JS, Zhang ZD, Pei L, et al. CBX4 exhibits oncogenic activities in breast cancer via Notch1 signaling. Int J Biochem Cell Biol. 2018;95:1–8. doi: 10.1016/j.biocel.2017.12.006 [DOI] [PubMed] [Google Scholar]
- 32.Zheng C, Li J, Wang Q, et al. MicroRNA-195 functions as a tumor suppressor by inhibiting CBX4 in hepatocellular carcinoma. Oncol Rep. 2015;33(3):1115–1122. doi: 10.3892/or.2015.3734 [DOI] [PubMed] [Google Scholar]
- 33.Jiao HK, Xu Y, Li J, et al. Prognostic significance of Cbx4 expression and its beneficial effect for transarterial chemoembolization in hepatocellular carcinoma. Cell Death Dis. 2015;6:e1689. doi: 10.1038/cddis.2015.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoareau-Aveilla C, Quelen C, Congras A, et al. MiR-497 suppresses cycle progression through an axis involving CDK6 in ALK-positive cells. Haematologica. 2019;104(2):347–359. doi: 10.3324/haematol.2018.195131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chai L, Kang XJ, Sun ZZ, et al. MiR-497-5p, miR-195-5p and miR-455-3p function as tumor suppressors by targeting hTERT in melanoma A375 cells. Cancer Manag Res. 2018;3(10):989–1003. doi: 10.2147/CMAR.S163335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jafarzadeh M, Soltani BM, Dokanehiifard S, et al. Experimental evidences for hsa-miR-497-5p as a negative regulator of SMAD3 gene expression. Gene. 2016;586(2):216–221. doi: 10.1016/j.gene.2016.04.003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed datasets generated during the study are available from the corresponding author on reasonable request.