Abstract
G protein-coupled receptors (GPCRs) represent the largest family of human membrane proteins, as well as drug targets. A recent boom in GPCR structural biology has provided detailed images of receptor ligand binding sites and interactions on the molecular level. An ever-increasing number of ligands is reported that exhibit activity through multiple receptors, binding in allosteric sites, and bias towards different intracellular signalling pathways. Furthermore, a wealth of single point mutants has accumulated in literature and public databases. Integrating these structural and mutagenesis data will help elucidate new GPCR ligand binding sites, and ultimately design drugs with tailored pharmacological activity.
Introduction
The G protein-coupled receptor (GPCR) family comprises about 800 members in human making it the largest membrane protein family [1]. A bit more than half of the GPCRs sense exogenous signals; odours, tastes, light, or pheromones [2]; whereas ~350 receptors are activated by a great variety of endogenous ligands spanning ions, neurotransmitters, lipids, carbohydrates, nucleotides, amino acids, peptides and proteins [3]. GPCRs make up ~19% of targets for marketed drugs and form one of the largest families in clinical trials, however the majority are still unexploited in therapies or trials [4].
GPCRs share a common structural fold of seven transmembrane (7TM) helices that form the machinery for signal transduction across the cell membrane. Crystal structures have revealed common conformational changes during receptor activation, allosteric modulation by ligands, ions and lipids, cholesterol, as well as G protein binding [5–7]. The characterisation of ligand pharmacology has become richer with numerous examples of activity through multiple receptors, binding in allosteric sites, and bias towards different intracellular signalling pathways. This wealth of information has sparked great activity in the GPCR field to understand the underlying structural mechanisms, and to exploit the new templates and principles for drug design.
Here we review the use of structural and mutagenesis data in elucidation of GPCR ligand binding sites at the molecular level. We cover key structural templates obtained during the recent GPCR structural biology boom [8], and the currently available databases containing mutants annotated from literature. To aid future mutagenesis experiments we outline the different strategies and a tool to design new single point mutations. Integration of complementary pharmacological and structural data facilitates new functional and mechanistic insights, and lays a solid foundation for structure-based ligand design.
Receptor-ligand structure complexes
The crystallised GPCRs now cover the classes A, B, C and F. The bound ligands span all activity types (including modulators), although antagonists are most frequent, and include several drugs on the market or in development. The GPCR database, GPCRdb (http://www.gpcrdb.org) [9] features GPCR reference data, analysis tools and visualisation diagrams; including structure statistics showing that in the time of writing, 81 unique ligand-receptor complexes have been crystallised for 37 receptors. The published GPCR-ligand complexes and 6,588 extracted ligand interactions can be browsed and visualised (3D viewer, 2D residue diagrams).
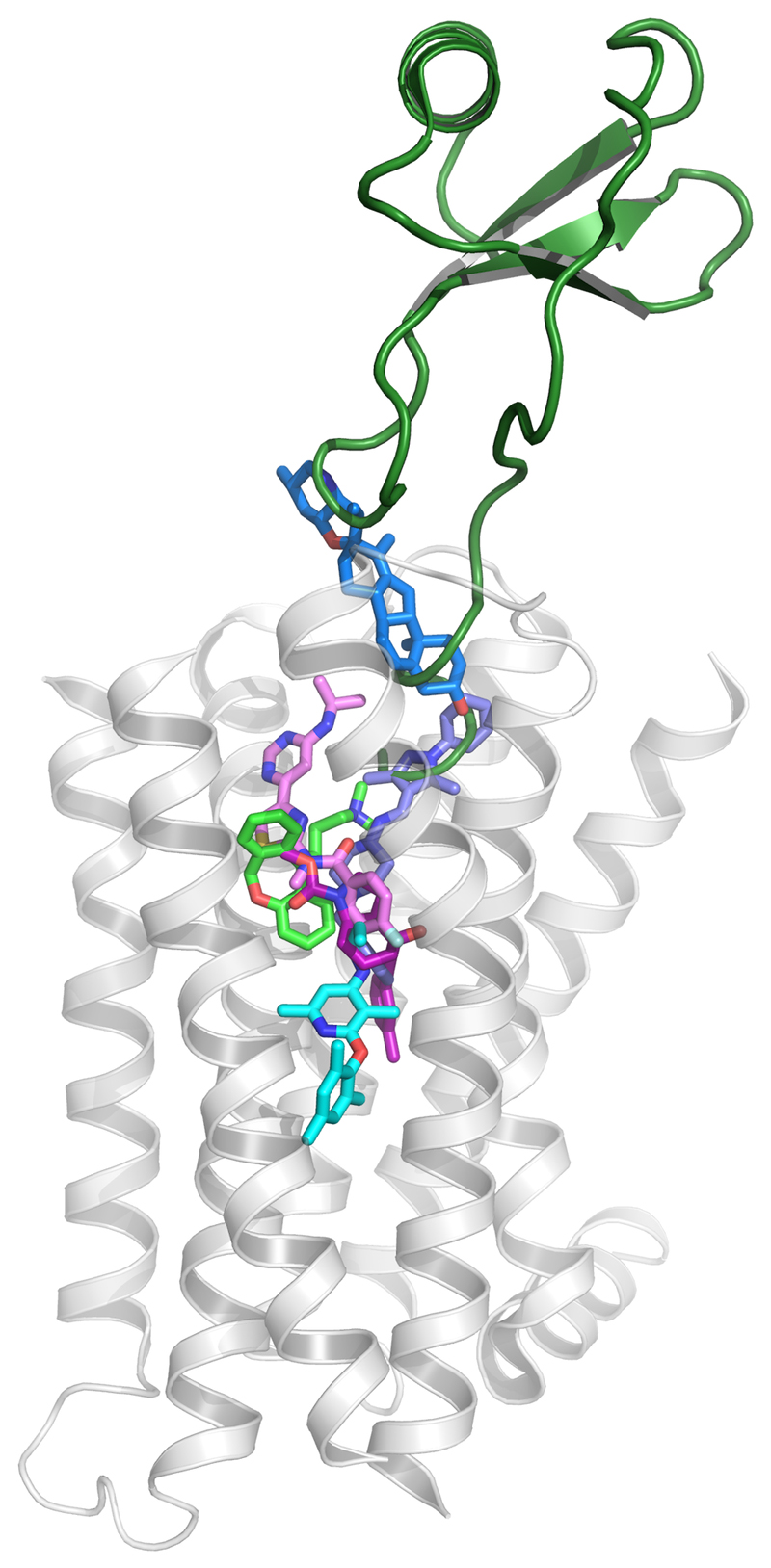
The ligand binding sites largely overlap, but display a variation for the depth of penetration into the transmembrane pocket (Figure 1). Within the largest class, A, the histamine H1 [10] and chemokine CXCR4 [11] receptor ligands display the deepest and most superficial binding, respectively. The class B receptor CRF1 receptor allosteric antagonist CP376395 was found to bind significantly deeper than any previously observed ligand [12]. The class C mGlu1 negative allosteric modulator (NAM) FITM largely overlaps with class A ligands [13], whereas the mGlu5 NAMs, e.g. mavoglurant, utilises what appears to be a subtype-specific pocket [14] that extends deeper to the top of CP376395 in the class B pocket [15]. Finally, the class F receptor, Smoothened (SMO), has been crystallised with multiple ligands – the first [16] binding close to the extracellular surface, but others covering a great vertical span [17].
Figure 1.
Depth of ligand binding in the transmembrane pocket for the GPCR classes A, B, C and F. The deepest and most superficial ligand pair is displayed for each class. The histamine H1 receptor (with light green doxepin, PDB: 3RZE) is displayed as transparent white cartoon, and was used for the superposition of the other structure complexes for; class A: CXCR4-vMIP-II (dark green, PDB: 4RWS), B: CRF1R-CP-376395 (pink, PDB: 4K5Y), C: mGlu1-FITM (pink, PDB: 4OR2) and mGlu5-mavoglurant (magenta, 4OO9), and F: smoothened receptor-SANT-1 (purple, PDB: 4N4W) and smoothened receptor-cyclopamine (blue, PDB: 4O9R).
The vast majority of class A GPCR ligands bind inside the transmembrane bundle, but some ligands have demonstrated atypical sites. Molecular dynamics simulations of ligand entry into the β2-adrenoceptor revealed a transient vestibule on the extracellular surface [18]. This vestibule was later found to constitute an allosteric site for the muscarinic M2 receptor positive allosteric modulator (PAM) LY2119620 [19]. The FFA1 receptor agonist TAK875 extends from the orthosteric site between transmembrane helices 3 and 4 and into the membrane [20]. The P2Y1 antagonist, BPTU [21] and the glucagon receptor antagonist MK-0893 [22] bind on the exterior surface of the helical bundle at the interface with the cell membrane. Finally, several studies including mutagenesis have revealed intracellular ligand binding to several chemokine receptors; including CCR4, CCR5, CXCR1, CXCR2, and CX3CR1 [23].
Ligand interactions have been found to trigger receptor activation micro-switches that shift the equilibrium between receptor functional states [7]. Ligands induce a common activation mechanism, evolving around centrally located residue positions [6]. The active receptor conformation is stabilised by a consensus inter-transmembrane contact network [5], which when mutated perturbs receptor activity [24]. Interestingly, several of the stabilising positions are either the same as or proximal to ligand binding residues.
Mutagenesis literature and databases
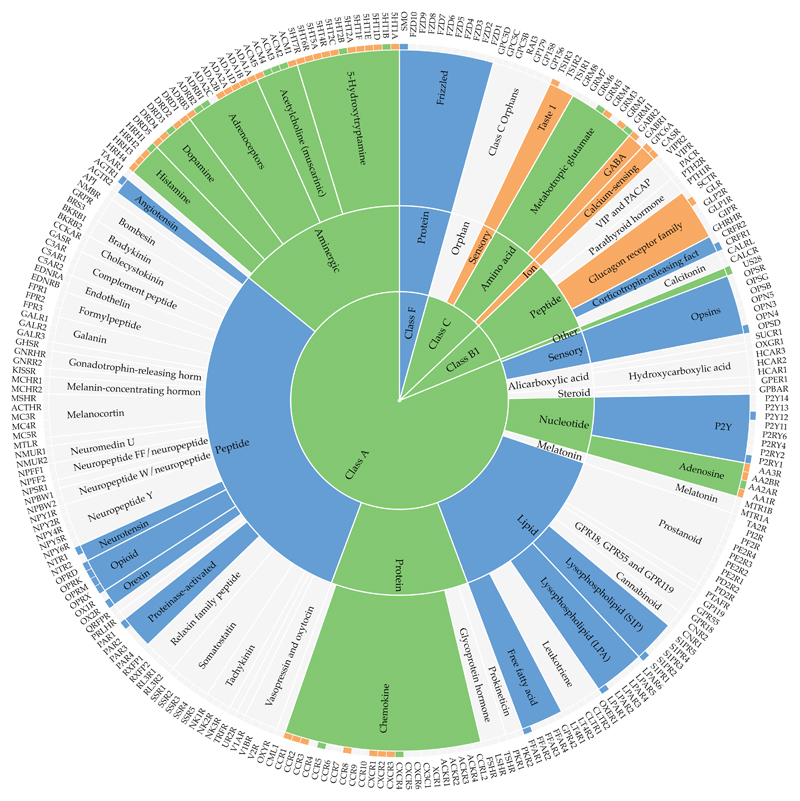
A search in PubMed with the MeSh terms "Mutation" and “Receptors, G-Protein-Coupled” retrieves 10,192 publications, illustrating an ocean of accumulated data. An early study by Rhee et al. provided a web resource containing 390 GPCR manually annotated literature mutations for 38 receptors and comparative sequence alignments [25]. The Protein Mutant Database, not limited to GPCRs, featured 218,873 in vitro and natural protein mutations from 45,239 studies [26], but is no longer accessible. A GPCR-focused annotation was done for the TinyGrap database, which in its latest published release contained 10,500 GPCR mutations from 1400 articles (http://www.cmbi.ru.nl/tinygrap) [27]. Previously, GPCRdb stored data from TinyGrap, own annotation [28] and the software MuteXt [29], but importantly this was limited to only the mutant identity and literature reference. Recently GPCRdb instead shifted to an open community expert-based curation that captures also the effect (qualitative or quantitative) on ligand affinity or potency, as well as influence on receptor surface expression or basal activity [9,30]. Today, GPCRdb contains 5.617 mutations for 24 receptors, as illustrated in Figure 2. The data can be browsed, downloaded, visualised in residue diagrams (snake- and helix box diagrams) or compared in residue tables giving a side-by-side view of receptor subtypes or species (Figure 3).
Figure 2.
Coverage of structural and mutagenesis ligand interaction data in GPCRdb across the GPCR classes, ligand types and receptor families (centre, middle and outer rings, respectively). Colour scheme; blue: structure complex data, orange: mutagenesis data, and grey: both data types.
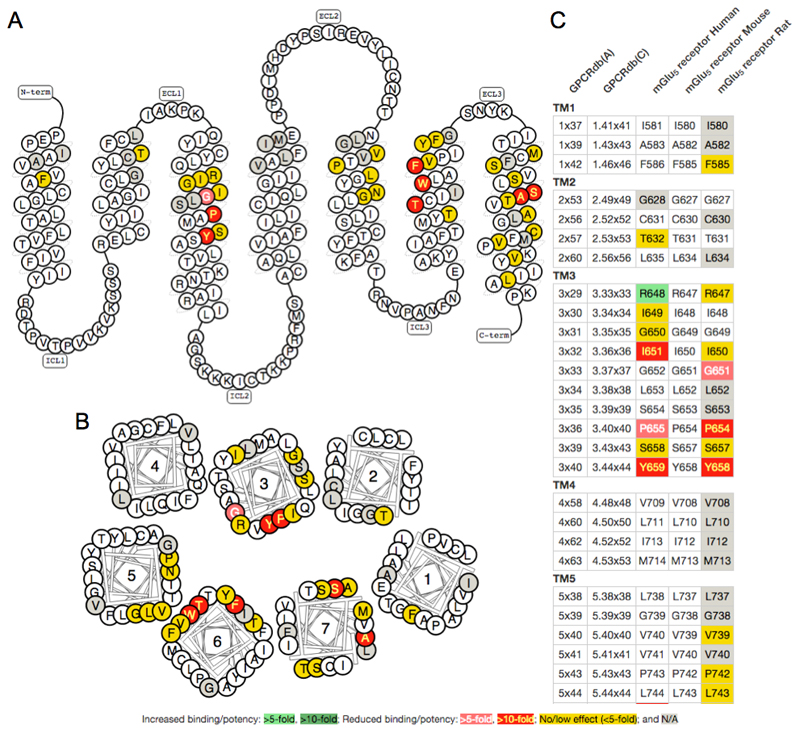
Figure 3.
Visualisation of mutations with effect on ligand binding/potency at the mGlu5 receptor. A) Snake and B) helix box diagrams visualise the receptor topology as seen from the side and above, respectively. C) Residue Tables give a side-by-side comparison of receptor species orthologues. The colour scheme indicates the fold effect of mutation on ligand binding, as described in the label. The figures were obtained from the GPCRdb mutation browser [9].
Receptor species orthologues and homologues can be seen as natural multi-point mutations. Evolutionary trace analysis is a technique that identifies residue position pairs that co-evolve and therefore are assumed to be involved in the same biological function. A pioneering such study identified three functional regions for ligand binding, G protein coupling, and signal transduction, respectively, that agreed with over 200 function-altering in vitro mutants [24]. An updated evolutionary tracing could define a minimal common GPCR ligand binding pocket [31]. Several recent analyses paired with mutagenesis experiments have identified ligand binding and efficacy-mediating residues, e.g. for the serotonin 5-HT2A, dopamine D2, and glutamate receptors [32–34]. However, whereas significant for natural ligands and receptor activation, the evolutionary tracing technique cannot generate information about surrogate ligands.
Mutant design strategies
The most common mutagenesis approach is alanine scanning [35]. This reduces the side chain to a methyl moiety, while maintaining the structural integrity of the protein backbone. Glycine has only hydrogen as side chain, but is avoided due to its atypical backbone dihedral angles. Hydrophobic residues, especially leucine, are used for stabilisation of GPCRs [36] by introducing van der Waals contacts between consecutive alpha helix turns [37]. However, alanine or leucine scans do not provide distinct information about ligand interaction types. Furthermore, extensive changes in the nature of the amino acid, especially when mutating large or charged residues, make them more likely to perturb the receptor surface expression or basal activity.
Mutation studies of ligand binding sites are ideally more specific. Alternative interaction types are dissected by several mutations of the same residue, while striving to minimise the indirect effects by choosing the most conservative representative amino acid. For example, a tyrosine residue may be mutated to first phenylalanine and then leucine to differentiate the hydrogen bonding and aromatic interactions, respectively. Prioritisation is typically towards the residues with the strongest ligand interactions, i.e. in the order of charged, polar, aromatic and van der Waals contacts, whereas glycine and proline are avoided as they can perturb the protein backbone structure. A summary of interaction-dissecting mutants for the 20 natural amino acids is provided in Supplementary Table 1.
Mutation design tool for GPCR ligand interactions
New mutagenesis experiments aiming to delineate the location of ligand binding sites or the specific receptor interactions are much more likely to succeed if based on an informed approach. However, many pharmacologists do not have access to the chemical expertise in selection and prioritisation of mutants (both their positions and amino acids) that can provide unambiguous information about the ligand binding mode. Furthermore, manual collation of the wealth of ligand interaction data from structure complexes and mutagenesis literature, even for just one target receptor (family), is a very time-consuming task. To this end, GPCRdb has made available an online tool to design mutations with effect on ligand binding for any receptor of interest. (Figure 4) [9]. It is based on structure-extracted ligand interactions (see Mutagenesis literature and databases) and literature mutants (see Receptor-ligand structure complexes) that had at least a five-fold effect on ligand affinity/potency and are accessible to ligands (update of [38]). This minimises indirect effects caused by interaction networks, e.g. aromatic stacking, or structural perturbation, often from glycine and proline residues.
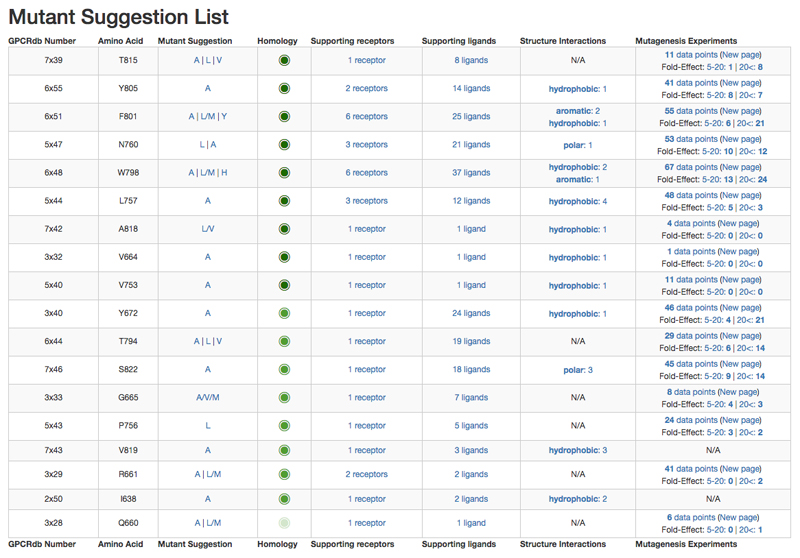
Figure 4.
Snapshots from the GPCRdb mutation design tool for the mGluR1 receptor. Suggested mutations are ordered by data inference from same receptor, receptor family, ligand type or GPCR class (displayed as decreasing intensity of green circle); and the sum of supporting structure complex and mutagenesis experiments. Alternative mutant suggestions for the same residue position allow for discrimination of the type of ligand interaction.
The tool accepts either a receptor name or, if available, a structure complex/model in pdb format. The generated mutation suggestions are first ranked by homology of the receptors from which the underlying data was inferred in the order of same: receptor, receptor family, ligand type or GPCR class. Mutation positions are further sorted within each such homology group by decreasing frequency among unique receptor-ligand pairs, i.e. a non-redundant ‘sum of support’ based on all observed interactions at the given position. GPCRdb uses a structure-based residue numbering based on Ballesteros-Weinstein numbers, but adjusting numbers and sequence alignments to account helix bulges and constrictions [39]. Mutant amino acids are suggested based on a molecular interaction-centric substitution table (Supplementary Table 1). In addition, small amino acids (e.g. serine) can be replaced by larger to block the binding site.
Implications to pharmacology and drug design
Pharmacological and structural biology data are complementary and synergistic for functional and mechanistic insight. For example, combined structural and mutagenesis studies have been applied to rationalise subtype-selectivity [14], and constitutive active mutants have been explained in terms of their involvement in interaction networks stabilising an active conformation [6]. The data may also serve to correct or refine the other. The structures provide information about ligand-accessible positions, to filter out mutations that only have an indirect effect on ligand binding. Mutation data can help to refine homology models where the structural information is not sufficient.
The new structures show previously unobtainable details of interactions between GPCRs and ligands, which are fundamental for structure-based drug design. For an increasing number of receptors, there is now sufficient structural information for structure-based ligand optimisation [40], and virtual screening [41,42] of receptor structures or pharmacophores [43]. Furthermore, as demonstrated by three community-wide “GPCR Dock” assessments, the improved structural templates have allowed for binding sites to be closely approximated by docking of ligands into receptor models [44–46].
Conclusions
An unprecedented amount of GPCR ligand binding site data is available from mutagenesis and structures. Our mechanistic understanding of receptor function is increasing rapidly with the characterisation of both single residues and structural sites. The already accumulated data (Figure 2) can be used to direct new mutagenesis experiments to the positions most likely to have an effect on ligand binding. The GPCR transmembrane pocket is generally very well suited for drug design, with a blend of polar (e.g. hydrogen bonding or ionic) and lipophilic areas. Mutants designed to discriminate between these molecular interactions (see Mutant design strategies) can provide more unambiguous elucidation of affinity and selectivity receptor residue hotspots.
We have just started to understand the ligand interactions that trigger the different receptor functional states. With sufficient coverage and resolution this may unlock a rational design of ligands with the exact desired pharmacological activity, i.e. agonism vs. antagonism, as well as potentially biased agonism. Ligand interaction fingerprints offer one such interesting approach, which has been shown to be able to discriminate agonists from antagonists based on their receptor interactions, as well as increase the hit rate from virtual screening [47]. Similar ligands are likely to have similar binding sites. Thus, it would be interesting to implement a mutation design for ligands of interest. Another intriguing outlook is to extend the concept of data-driven mutation design to other functional sites, i.e. the binding sites of G proteins [48,49], β-arrestin [50], and dimerisation interfaces [51].
Supplementary Material
Acknowledgements
The labs of Anke Schiedel, Chris de Graaf and Hugo Gutiérrez-de-Terán are acknowledged for the annotation of mutation data. The authors grateful for the financial support from the European Research Council (DE-ORPHAN 639125), Lundbeck Foundation (R163-2013-16327) and the Danish Council of Independent Research (DFF – 1331-00180), as well as support for meetings and scientific exchange through the COST Action ‘GLISTEN’ (CM1207).
References
Papers of particular interest, published within the period of review, have been highlighted as:
*Of special interest
**Of outstanding interest
- 1.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 2.Mombaerts P. Genes and ligands for odorant, vomeronasal and taste receptors. Nat Rev Neurosci. 2004;5:263–278. doi: 10.1038/nrn1365. [DOI] [PubMed] [Google Scholar]
- 3.Southan C, Sharman JL, Benson HE, Faccenda E, Pawson AJ, Alexander SP, Buneman OP, Davenport AP, McGrath JC, Peters JA, et al. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. Nucleic Acids Res. 2016;44:D1054–1068. doi: 10.1093/nar/gkv1037. [* New release of the IUPHAR/BPS Guide to Pharmacology database containing more than 1300 protein targets and 6000 ligands.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rask-Andersen M, Masuram S, Schioth HB. The druggable genome: Evaluation of drug targets in clinical trials suggests major shifts in molecular class and indication. Annu Rev Pharmacol Toxicol. 2014;54:9–26. doi: 10.1146/annurev-pharmtox-011613-135943. [DOI] [PubMed] [Google Scholar]
- 5.Venkatakrishnan AJ, Deupi X, Lebon G, Tate CG, Schertler GF, Babu MM. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494:185–194. doi: 10.1038/nature11896. [** A comprehensive GPCR structure comparison revealing ligand and (active) fold-stabilising interactions] [DOI] [PubMed] [Google Scholar]
- 6.Tehan BG, Bortolato A, Blaney FE, Weir MP, Mason JS. Unifying family A GPCR theories of activation. Pharmacol Ther. 2014;143:51–60. doi: 10.1016/j.pharmthera.2014.02.004. [* Structural analysis of active-inactive GPCR pairs unifying class A GPCR activation theories around a conserved central hydrophobic core.] [DOI] [PubMed] [Google Scholar]
- 7.Katritch V, Cherezov V, Stevens RC. Structure-function of the G protein-coupled receptor superfamily. Annu Rev Pharmacol Toxicol. 2013;53:531–556. doi: 10.1146/annurev-pharmtox-032112-135923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooke RM, Brown AJ, Marshall FH, Mason JS. Structures of G protein-coupled receptors reveal new opportunities for drug discovery. Drug Discov Today. 2015;20:1355–1364. doi: 10.1016/j.drudis.2015.08.003. [** Recent review the structural information on GPCR ligand binding sites, and implications to pharmacology and drug discovery.] [DOI] [PubMed] [Google Scholar]
- 9.Isberg V, Mordalski S, Munk C, Rataj K, Harpsoe K, Hauser AS, Vroling B, Bojarski AJ, Vriend G, Gloriam DE. GPCRdb: an information system for G protein-coupled receptors. Nucleic Acids Res. 2016;44:D356–364. doi: 10.1093/nar/gkv1178. [* Introduction, how to and tools for generic residue numbering within and across the GPCR classes, and a solution for transmembrane helix bulges and constrictions.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimamura T, Shiroishi M, Weyand S, Tsujimoto H, Winter G, Katritch V, Abagyan R, Cherezov V, Liu W, Han GW, et al. Structure of the human histamine H1 receptor complex with doxepin. Nature. 2011;475:65–70. doi: 10.1038/nature10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu B, Chien EYT, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, et al. Structures of the CXCR4 Chemokine GPCR with Small-Molecule and Cyclic Peptide Antagonists. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hollenstein K, Kean J, Bortolato A, Cheng RK, Dore AS, Jazayeri A, Cooke RM, Weir M, Marshall FH. Structure of class B GPCR corticotropin-releasing factor receptor 1. Nature. 2013;499:438–443. doi: 10.1038/nature12357. [DOI] [PubMed] [Google Scholar]
- 13.Wu H, Wang C, Gregory KJ, Han GW, Cho HP, Xia Y, Niswender CM, Katritch V, Meiler J, Cherezov V, et al. Structure of a class C GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator. Science. 2014;344:58–64. doi: 10.1126/science.1249489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harpsoe K, Isberg V, Tehan BG, Weiss D, Arsova A, Marshall FH, Brauner-Osborne H, Gloriam DE. Selective Negative Allosteric Modulation Of Metabotropic Glutamate Receptors - A Structural Perspective of Ligands and Mutants. Sci Rep. 2015;5 doi: 10.1038/srep13869. 13869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dore AS, Okrasa K, Patel JC, Serrano-Vega M, Bennett K, Cooke RM, Errey JC, Jazayeri A, Khan S, Tehan B, et al. Structure of class C GPCR metabotropic glutamate receptor 5 transmembrane domain. Nature. 2014;511:557–562. doi: 10.1038/nature13396. [DOI] [PubMed] [Google Scholar]
- 16.Wang C, Wu H, Katritch V, Han GW, Huang XP, Liu W, Siu FY, Roth BL, Cherezov V, Stevens RC. Structure of the human smoothened receptor bound to an antitumour agent. Nature. 2013;497:338–343. doi: 10.1038/nature12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang C, Wu H, Evron T, Vardy E, Han GW, Huang X-P, Hufeisen SJ, Mangano TJ, Urban DJ, Katritch V, et al. Structural basis for Smoothened receptor modulation and chemoresistance to anticancer drugs. Nat Commun. 2014;5 doi: 10.1038/ncomms5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dror RO, Pan AC, Arlow DH, Borhani DW, Maragakis P, Shan Y, Xu H, Shaw DE. Pathway and mechanism of drug binding to G-protein-coupled receptors. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:13118–13123. doi: 10.1073/pnas.1104614108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kruse AC, Ring AM, Manglik A, Hu J, Hu K, Eitel K, Hubner H, Pardon E, Valant C, Sexton PM, et al. Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature. 2013;504:101–106. doi: 10.1038/nature12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.White JF, Noinaj N, Shibata Y, Love J, Kloss B, Xu F, Gvozdenovic-Jeremic J, Shah P, Shiloach J, Tate CG, et al. Structure of the agonist-bound neurotensin receptor. Nature. 2012;490:508–513. doi: 10.1038/nature11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang D, Gao Z-G, Zhang K, Kiselev E, Crane S, Wang J, Paoletta S, Yi C, Ma L, Zhang W, et al. Two disparate ligand-binding sites in the human P2Y1 receptor. Nature. 2015;520:317–321. doi: 10.1038/nature14287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jazayeri A, Doré AS, Lamb D, Krishnamurthy H, Southall SM, Baig AH, Bortolato A, Koglin M, Robertson NJ, Errey JC, et al. Extra-helical binding site of a glucagon receptor antagonist. Nature. 2016;533:274–277. doi: 10.1038/nature17414. [DOI] [PubMed] [Google Scholar]
- 23.Scholten DJ, Canals M, Maussang D, Roumen L, Smit MJ, Wijtmans M, de Graaf C, Vischer HF, Leurs R. Pharmacological modulation of chemokine receptor function. British Journal of Pharmacology. 2012;165:1617–1643. doi: 10.1111/j.1476-5381.2011.01551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madabushi S, Gross AK, Philippi A, Meng EC, Wensel TG, Lichtarge O. Evolutionary trace of G protein-coupled receptors reveals clusters of residues that determine global and class-specific functions. The Journal of biological chemistry. 2004;279:8126–8132. doi: 10.1074/jbc.M312671200. [DOI] [PubMed] [Google Scholar]
- 25.van Rhee AM, Jacobson KA. Molecular Architecture of G Protein-Coupled Receptors. Drug Dev Res. 1996;37:1–38. doi: 10.1002/(SICI)1098-2299(199601)37:1<1::AID-DDR1>3.0.CO;2-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawabata T, Ota M, Nishikawa K. The Protein Mutant Database. Nucleic Acids Research. 1999;27:355–357. doi: 10.1093/nar/27.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edvardsen O, Reiersen AL, Beukers MW, Kristiansen K. tGRAP, the G-protein coupled receptors mutant database. Nucleic Acids Res. 2002;30:361–363. doi: 10.1093/nar/30.1.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vroling B, Sanders M, Baakman C, Borrmann A, Verhoeven S, Klomp J, Oliveira L, de Vlieg J, Vriend G. GPCRDB: information system for G protein-coupled receptors. Nucleic Acids Res. 2011;39:309–319. doi: 10.1093/nar/gkq1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naderi N, Witte R. Automated extraction and semantic analysis of mutation impacts from the biomedical literature. BMC Genomics. 2012;13:S10. doi: 10.1186/1471-2164-13-S4-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Munk C, Isberg V, Mordalski S, Harpsøe K, Rataj K, Hauser A, Kolb P, Bojarski AJ, Vriend G, Gloriam D. GPCRdb: The G protein-coupled receptor database - An introduction. British Journal of Pharmacology. doi: 10.1111/bph.13509. Accepted. [* An introduction to the GPCR database, GPCRdb with best practises guides.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moitra S, Tirupula KC, Klein-Seetharaman J, Langmead CJ. A minimal ligand binding pocket within a network of correlated mutations identified by multiple sequence and structural analysis of G protein coupled receptors. BMC Biophys. 2012;5:13. doi: 10.1186/2046-1682-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sung YM, Wilkins AD, Rodriguez GJ, Wensel TG, Lichtarge O. Intramolecular allosteric communication in dopamine D2 receptor revealed by evolutionary amino acid covariation. Proc Natl Acad Sci U S A. 2016 doi: 10.1073/pnas.1516579113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodriguez GJ, Yao R, Lichtarge O, Wensel TG. Evolution-guided discovery and recoding of allosteric pathway specificity determinants in psychoactive bioamine receptors. Proc Natl Acad Sci U S A. 2010;107:7787–7792. doi: 10.1073/pnas.0914877107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang HJ, Wilkins AD, Lichtarge O, Wensel TG. Determinants of endogenous ligand specificity divergence among metabotropic glutamate receptors. J Biol Chem. 2015;290:2870–2878. doi: 10.1074/jbc.M114.622233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morrison KL, Weiss GA. Combinatorial alanine-scanning. Curr Opin Chem Biol. 2001;5:302–307. doi: 10.1016/s1367-5931(00)00206-4. [DOI] [PubMed] [Google Scholar]
- 36.Heydenreich F, Vuckovic Z, Matkovic M, Veprintsev D. Stabilization of G protein-coupled receptors by point mutations. Frontiers in Pharmacology. 2015;6 doi: 10.3389/fphar.2015.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo P, Baldwin RL. Origin of the different strengths of the (i,i+4) and (i,i+3) leucine pair interactions in helices. Biophys Chem. 2002;96:103–108. doi: 10.1016/s0301-4622(02)00010-8. [DOI] [PubMed] [Google Scholar]
- 38.Gloriam DE, Foord SM, Blaney FE, Garland SL. Definition of the G protein-coupled receptor transmembrane bundle binding pocket and calculation of receptor similarities for drug design. J Med Chem. 2009;52:4429–4442. doi: 10.1021/jm900319e. [DOI] [PubMed] [Google Scholar]
- 39.Isberg V, de Graaf C, Bortolato A, Cherezov V, Katritch V, Marshall FH, Mordalski S, Pin JP, Stevens RC, Vriend G, et al. Generic GPCR residue numbers - aligning topology maps while minding the gaps. Trends Pharmacol Sci. 2015;36:22–31. doi: 10.1016/j.tips.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Congreve M, Dias JM, Marshall FH. Structure-based drug design for G protein-coupled receptors. Prog Med Chem. 2014;53:1–63. doi: 10.1016/B978-0-444-63380-4.00001-9. [DOI] [PubMed] [Google Scholar]
- 41.Kolb P, Rosenbaum DM, Irwin JJ, Fung JJ, Kobilka BK, Shoichet BK. Structure-based discovery of beta2-adrenergic receptor ligands. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:6843–6848. doi: 10.1073/pnas.0812657106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carlsson J, Coleman RG, Setola V, Irwin JJ, Fan H, Schlessinger A, Sali A, Roth BL, Shoichet BK. Ligand discovery from a dopamine D3 receptor homology model and crystal structure. Nat Chem Biol. 2011;7:769–778. doi: 10.1038/nchembio.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fidom K, Isberg V, Hauser AS, Mordalski S, Lehto T, Bojarski AJ, Gloriam DE. A new crystal structure fragment-based pharmacophore method for G protein-coupled receptors. Methods. 2015;71:104–112. doi: 10.1016/j.ymeth.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 44.Michino M, Abola E, Brooks CL, 3rd, Dixon JS, Moult J, Stevens RC. Community-wide assessment of GPCR structure modelling and ligand docking: GPCR Dock 2008. Nat Rev Drug Discov. 2009;8:455–463. doi: 10.1038/nrd2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kufareva I, Rueda M, Katritch V, Stevens RC, Abagyan R. Status of GPCR modeling and docking as reflected by community-wide GPCR Dock 2010 assessment. Structure. 2011;19:1108–1126. doi: 10.1016/j.str.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kufareva I, Katritch V, Participants of GD. Stevens RC, Abagyan R. Advances in GPCR modeling evaluated by the GPCR Dock 2013 assessment: meeting new challenges. Structure. 2014;22:1120–1139. doi: 10.1016/j.str.2014.06.012. [* GPCR-ligand complex modelling competition yielding high accuracy for close homology targets, and correct fold for distant homology targets.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Graaf C, Kooistra AJ, Vischer HF, Katritch V, Kuijer M, Shiroishi M, Iwata S, Shimamura T, Stevens RC, de Esch IJ, et al. Crystal structure-based virtual screening for fragment-like ligands of the human histamine H(1) receptor. Journal of Medicinal Chemistry. 2011;54:8195–8206. doi: 10.1021/jm2011589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rasmussen SG, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Standfuss J, Edwards PC, D'Antona A, Fransen M, Xie G, Oprian DD, Schertler GF. The structural basis of agonist-induced activation in constitutively active rhodopsin. Nature. 2011;471:656–660. doi: 10.1038/nature09795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kang Y, Zhou XE, Gao X, He Y, Liu W, Ishchenko A, Barty A, White TA, Yefanov O, Han GW, et al. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature. 2015;523:561–567. doi: 10.1038/nature14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang J, Chen S, Zhang JJ, Huang XY. Crystal structure of oligomeric beta1-adrenergic G protein-coupled receptors in ligand-free basal state. Nat Struct Mol Biol. 2013;20:419–425. doi: 10.1038/nsmb.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.