Abstract
Intracerebral hemorrhage (ICH) is a severe neurological disorder with no proven treatment. Our prior research identified a significant association with monocyte level and ICH mortality. To advance our understanding, we sought to identify gene expression after ICH using a swine model to test the hypothesis that ICH would induce peripheral blood mononuclear cell (PBMC) gene expression. In 10 pigs with ICH, two PBMC samples were drawn from each with the first immediately prior to ICH induction and the second six hours later. RNA-seq was performed with subsequent bioinformatics analysis using Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and Ingenuity® Pathway Analysis (IPA). There were 182 significantly upregulated and 153 significantly down-regulated differentially expressed genes (DEGs) after ICH. Consistent with findings in humans, significant GO and KEGG pathways were primarily related to inflammation and the immune response. Five genes, all upregulated post-ICH and known to be associated with monocyte activation, were repeatedly DEGs in the significant KEGG pathways: CD14, TLR4, CXCL8, IL-18, and CXCL2. In IPA, the majority of upregulated disease/function categories were related to inflammation and immune cell activation. TNF and LPS were the most significantly activated upstream regulators, and ERK was the most highly connected node in the top network. ICH induced changes in PBMC gene expression within 6 h of onset related to inflammation, the immune response, and, more specifically, monocyte activation. Further research is needed to determine if these changes affect outcomes and may represent new therapeutic targets.
Keywords: Stroke, Intracerebral hemorrhage, Gene expression, RNA sequencing, Neuroinflammation, Monocyte
Introduction
Intracerebral hemorrhage (ICH) is a severe neurological disorder accounting for 10% of strokes, 50% of stroke mortality, (Mozaffarian et al. 2016) and it has no proven treatment. Inflammatory processes, including white blood cells (WBCs), contribute to brain injury after ICH. (Chen et al. 2015) Increasing evidence suggests that monocytes, a type of WBC, are particularly important for post-ICH inflammatory damage. In a murine study, circulating inflammatory monocytes outnumbered other leukocytes in brain tissue after ICH, monocytes secreted tumor necrosis factor, and mice with fewer inflammatory monocytes had better motor function than controls. (Hammond et al. 2014b) In another report, in which an antibody blocked a cellular adhesion molecule most highly expressed on inflammatory monocytes, there was less recruitment of monocytes into brain tissue and reduced neurobehavioral disability. (Hammond et al. 2014a).
Our group previously found in two independent cohorts of ICH patients that higher monocyte count was associated with more 30-day mortality. Greater WBC count, but not monocyte count, was associated with larger ICH volume. However, after adjustment for covariates the monocyte count, but not total white blood cell count, was independently associated with 30-day ICH mortality. (Adeoye et al. 2014; Walsh et al. 2015) We also reported that M2 monocyte microparticles were significantly increased in the plasma of ICH patients compared to matched controls. (Walsh et al. 2017) Higher levels of chemokine C-C motif ligand 2 (CCL2), the dominant chemokine for monocyte recruitment, were independently associated with poor functional outcome in ICH patients. (Hammond et al. 2014b) In 1302 patients with ICH, higher admission monocyte count, but not neutrophil or lymphocyte count, was an independent predictor of ICH expansion. (Morotti et al. 2016) Altogether, these results suggest that monocytes and inflammatory responses play a pivotal role in ICH injury and consequent patient morbidity and mortality.
To study the molecular mechanisms of ICH-induced inflammatory injury, we analyzed global gene expression profiles in a swine ICH model utilizing RNA-seq of PBMCs. To the best of our knowledge, there have been no prior reports of RNA-seq in swine ICH. Next generation RNA sequencing (RNA-seq) is a powerful method for global, unbiased transcriptomic analysis with advantages compared with other transcriptomics methodology. Microarray technology is limited in measuring the range of gene expression due to background signal for genes with low expression and signal saturation for genes with high expression. However, RNA-seq provides digital sequencing reads that quantify expression accurately over a much larger dynamic range. (Zhao et al. 2014) While techniques like microarrays and polymerase chain reaction utilize nucleic acid probes that are defined in advance, RNA-seq can identify previously unknown transcriptional variations. (Zhang et al. 2015) There are advantages to using the domestic pig to study inflammation, the immune system, and ICH compared with other preclinical models. (Fairbairn et al. 2011) Pigs shared a larger proportion of inducible genes with humans, (Kapetanovic et al. 2012) while the macrophage inducible genes in rodents had more substantial differences. (Schroder et al. 2012) RNA that was extracted from mouse and human macrophages had significant differences despite being cultured under very similar conditions, (Fairbairn et al. 2011) likely due in part to variation in promoters of transcription factors and target genes. (Heinz et al. 2003) When transcriptional regulation was compared in mouse and human macrophages in response to lipopolysaccharide, 10% of genes had absolutely divergent regulation, while about 25% were quantitatively divergent. Those genes showing greatest divergence in mice versus humans were analyzed in pigs, and the transcriptional regulation in pigs vs. humans had substantial resemblance. (Fairbairn et al. 2011) When a number of immune system parameters were compared, pigs were more similar to humans than mice to humans for more than 80% of examined parameters, while mice were more similar for less than 10%.(Schook et al. 2005) The swine ICH model has been reported in many non-RNAseq investigations. (Aviv et al. 2014; Brunberg et al. 2013; Gu et al. 2011; Loftspring et al. 2007; Orakcioglu et al. 2015; Orakcioglu et al. 2012; Wagner et al. 2006; Xi et al. 1998; Zhou et al. 2014) and has advantages versus rodents such as the larger size of the pig brain, approximately 40 times larger than a rat, its gyrencephalic nature, larger white matter composition with better evaluation of damage and edema, (Adeoye et al. 2011; Wagner 2007; Wagner et al. 1996) and ease of experimental reproducibility. (Hua and Xi 2009).
Although there is substantial interest in neuroinflammation as a therapeutic target following ICH, these processes remain poorly understood. Evidence exists for rapid local and systemic ICH inflammatory effects, with changes in human mononuclear gene expression within 3 h, (Sang et al. 2017) and in the perihematomal region of swine ICH, cerebral edema, serum protein accumulation, and metabolic changes within one hour. (Wagner et al. 1998) Peripheral blood mononuclear cell (PBMC) gene expression studies have shown promise in other neurological disorders such as ischemic stroke and seizures. (Tang et al. 2001) Here we present data showing that ICH in pigs induces broad gene expression changes in PBMCs that include molecular pathways involved in monocyte activation and inflammatory responses. ICH promotes rapid alterations in peripheral immune cell function that are implicated in the pathophysiology of ICH.
Methods
Ten female domestic pigs of the American Yorkshire breed, 80 to 90 days old and 36 to 41 kg, were included in the study. Each pig was sedated by intramuscular injection of tiletamine and zolazepam and then endotracheally intubated and placed under isoflurane general anesthesia. To induce ICH as reported previously, (Wagner et al. 1996) a single burr hole craniotomy was performed to pass an angiocatheter and to inject two to four milliliters of autologous blood under pressure control into the frontal white matter. Six hours later, the pig was sacrificed by lethal injection. Peripheral blood was collected immediately prior to ICH induction but after the pig had been placed under general anesthesia for about 2 h and the angiocatheter was in place. The second sample was collected immediately prior to animal sacrifice. The study protocol was approved by the University of Cincinnati Institutional Animal Care and Use Committee, and all aspects of the study were conducted in accordance with the United States Public Health Service’s Policy on Human Care and Use of Laboratory Animals.
Sample processing and analysis were conducted in the Genomics, Epigenomics, and Sequencing Core at the University of Cincinnati. For each sample, approximately 10 ml of swine peripheral blood was collected and immediately placed into EDTA containing blood collection tubes, inverted multiple times, and processed within one hour. Briefly, Lymphoprep™ (Stemcell Technologies, Cambridge, MA) density gradient medium and SepMate™ (Stemcell Technologies) tubes were used according to the manufacturer’s instructions to isolate PBMCs. The PBMCs were directly lysed with Lysis Buffer from the mirVana miRNA isolation kit (Thermo Fisher, Waltham, MA) to stabilize RNA until the time of RNA extraction as described in the total RNA extraction protocol from the kit. The RNA quality was determined by Bioanalyzer (Agilent, Santa Clara, CA). To isolate the polyA RNA, NEBNext Poly(A) mRNA Magnetic Isolation Module (New England BioLabs, Ipswich, MA) was used with a total of 1 μg of good quality total RNA as input. The NEBNext Ultra Directional RNA Library Prep Kit (New England BioLabs) was used for library preparation. After library quality control and quantification, individually indexed and compatible libraries were proportionally pooled and sequenced using Illumina HiSeq platform. Under the sequencing setting of single read 1 × 51 bp, about 25 million pass filter reads per sample were generated. The RNA-seq data from the study are available through the Gene Expression Omnibus (GEO) of the National Center for Biotechnology Information (NCBI), record number GSE124624.
For bioinformatics analysis of mRNA-seq data, kallisto (Pachter Lab, Pasadena, CA) (Bray et al. 2016) was utilized to quantify the transcript-level abundance. The estimates were reported by transcripts per million reads (TPM) and converted to estimated gene counts. The differentially expressed genes (DEGs) were identified by DESeq2 package (Bioconductor) (Love et al. 2014) in R and annotated with genome assembly Sscrofa11.1 (https://useast.ensembl.org/Sus_scrofa). Pre and post-ICH samples from the same pig were compared as paired samples. The threshold for statistical significance was the absolute value of log 2 fold change (log2fc) > 1 and false discovery rate (FDR) <0.1 (Benjamini-Hochberg adjusted p-values). FDR <0.1 is a conventional threshold for significance when analyzing RNA-seq data as reported in a publication regarding DESeq2 methodology (Love et al. 2014) as well as a number of transcriptional investigations (Bergsveinson et al. 2016; Cabezas-Wallscheid et al. 2014; Nagaraja et al. 2017; Pantazatos et al. 2017). The significant DEGs were applied to Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways by KOBAS 3.0 web server, (Wu et al. 2006) considering significant functional terms and pathways with adjusted P value (Benjamini-Hochberg correction) smaller than 0.1 and including at least 3 genes. To further interpret the gene expression data, Ingenuity® Pathway Analysis (IPA, Qiagen, Redwood City, CA) was used to produce comprehensive analysis of the DEGs in the context of disease functions, predicted upstream regulators, and generated networks of molecular interactions. (Bakshi et al. 2008; Cheong et al. 2016). The log gene expression ratio comparing the second sample to the first sample was used as a value parameter for the analysis.
Results
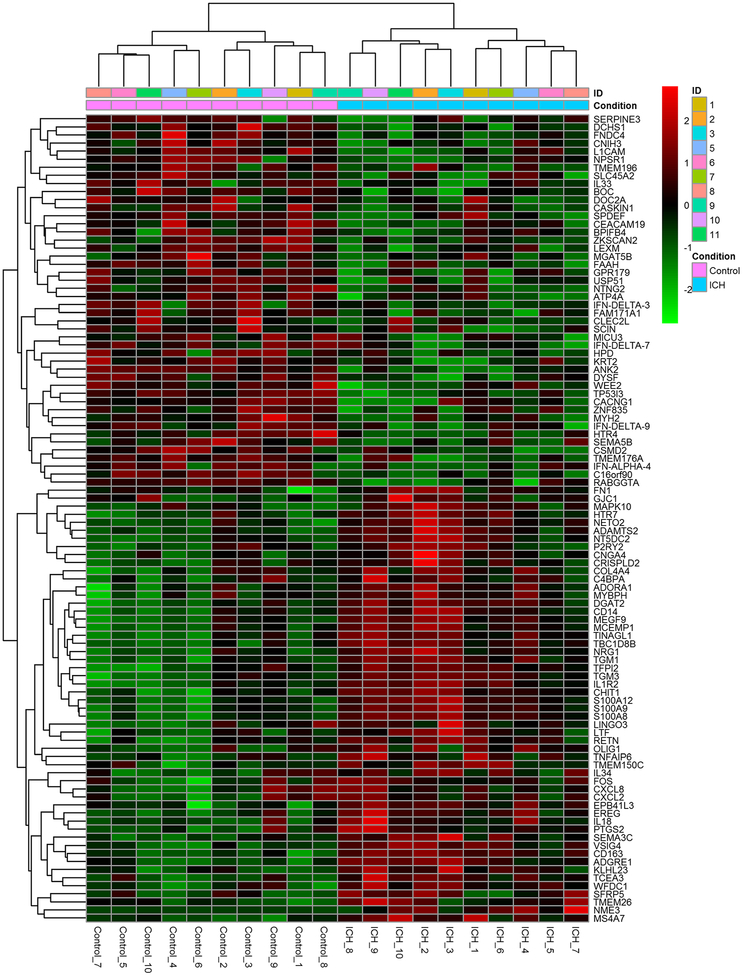
182 genes were identified as significantly upregulated in post-ICH compared to pre-ICH, and 153 genes were significantly down regulated (adjusted P value<0.1). The log normalized gene counts of the 100 most significantly changed DEGs comparing pre and post-ICH samples are shown in Fig. 1. The GO and KEGG enrichment analyses showed that most of the significant pathways were classified into molecular functions associated with inflammation and immune system activation (Table 1). The following four GO terms were statistically significant: inflammatory response, defense response, extracellular space, and immune response. The following seven KEGG pathways were statistically significant: salmonella infection, malaria, pertussis, legionellosis, cytokine-cytokine receptor interaction, amoebiasis, and toll-like receptor signaling pathway.
Fig. 1.
The top 100 annotated differentially expressed genes for which the expression was most changed six hours following versus prior to swine ICH induction (gene counts were log normalized and clustered by correlation)
Table 1.
Significantly enriched Gene Ontology terms and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways
| Term | Database | ID | Input number | Background number | P Value | Corrected P Value | Input |
|---|---|---|---|---|---|---|---|
| Inflammatory response | Gene Ontology | GO:0006954 | 9 | 138 | 5.77E-05 | 0.073 | ALOX5AP, S100A8, CCR1, CHI3L1 S100A9, CXCL8, S1PR3, IL18, TLR4 |
| Defense response | Gene Ontology | GO:0006952 | 14 | 350 | 9.81E-05 | 0.073 | ALOX5AP, PGLYRP2, S100A8, CCR1 FCN1, CHI3L1, S100A9, LYZ, LTF FCN2, S1PR3, IL18, TLR4, CXCL8 |
| Extracellular space | Gene Ontology | GO:0005615 | 15 | 406 | 1.29E-04 | 0.073 | CXCL8, TINAGL1, S100A8, LOC100153504, CXCL2, CHIT1, CHI3L1 S100A9, LYZ, LTF, MMP8, TIMP1 IL18, A1BG, RETN |
| Immune response | Gene Ontology | GO:0006955 | 13 | 333 | 2.19E-04 | 0.091 | CXCL8, PGLYRP2, TINAGL1, S100A8 CCR1, FCN1, CXCL2, S100A9, LTF FCN, NFIL3, IL18, TLR4 |
| Salmonella infection | KEGG PATHWAY | ssc05132 | 7 | 65 | 2.06E-05 | 0.0023 | CXCL8, CD14, CXCL2, MAPK10 FOS, IL18, TLR4 |
| Malaria | KEGG PATHWAY | ssc05144 | 6 | 46 | 3.06E-05 | 0.0023 | CXCL8, HGF, SDC2 VCAM1, IL18, TLR4 |
| Pertussis | KEGG PATHWAY | ssc05133 | 6 | 62 | 1.41E-04 | 0.0071 | CXCL8, CD14, C4BPA MAPK10, FOS, TLR4 |
| Legionellosis | KEGG PATHWAY | ssc05134 | 5 | 45 | 2.86E-04 | 0.011 | CXCL2, IL18, CXCL8, TLR4, CD14 |
| Cytokine-cytokine receptor interaction | KEGG PATHWAY | ssc04060 | 8 | 186 | 2.03E-03 | 0.061 | CXCL8, CCR1, IL1RAP, ACVR1B CSF3R, IL18, IL1R2, HGF |
| Amoebiasis | KEGG PATHWAY | ssc05146 | 5 | 79 | 3.03E-03 | 0.065 | FN1, CXCL8, IL1R2, TLR4, CD14 |
| Toll-like receptor signaling pathway | KEGG PATHWAY | ssc04620 | 5 | 79 | 0.00303 | 0.065 | MAPK10, CXCL8, FOS, TLR4, CD14 |
(Significance was assessed by the Benjamini-Hochberg corrected P value <0.1)
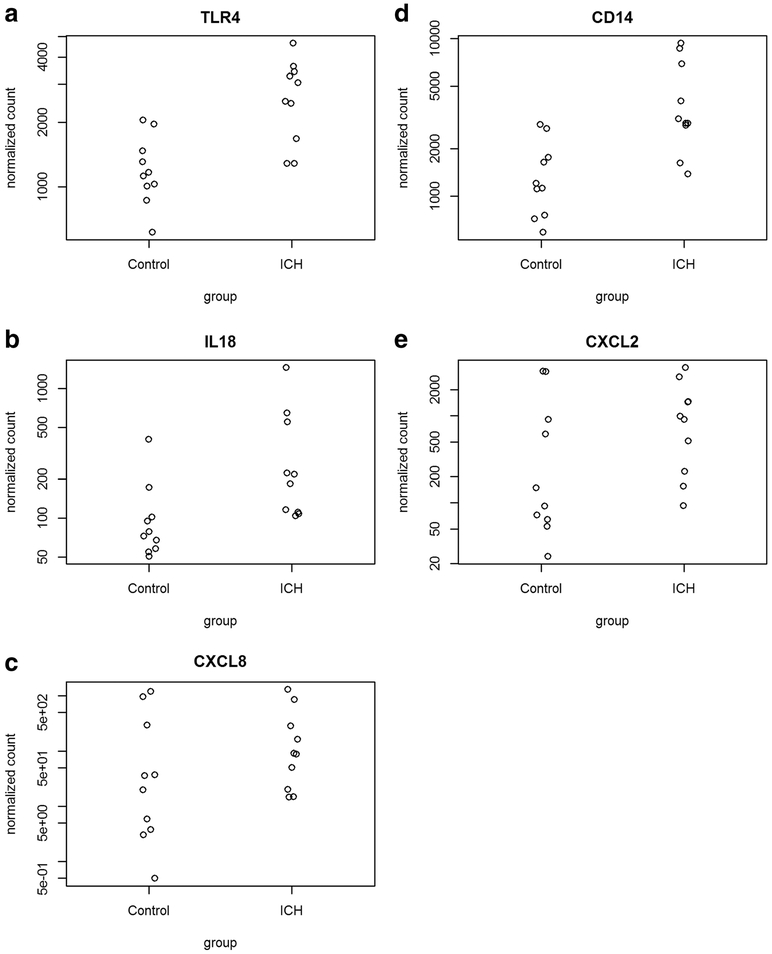
There were five genes, all of which had increased expression post-ICH, that were repeatedly identified as significant DEGs in the statistically significant KEGG pathways (Table 2): CD14 (cluster of differentiation 14), TLR4 (toll-like receptor-4), CXCL8 (CXC motif chemokine-8), IL-18 (inter-leukin 18), and CXCL2 (CXC motif chemokine-2). For these five genes, the gene counts comparing pre and post-ICH samples were determined (Fig. 2). Of the seven significant KEGG pathways, CXCL8 is a component of all. TLR4 is a part of six of the pathways (salmonella infection, malaria, pertussis, legionella, amoebiasis, toll-like receptor signaling). CD14 is present in five of the pathways (salmonella, pertussis, legionella, amoebiasis, and toll-like receptor signaling). IL-18 is a component of four of the pathways (salmonella infection, malaria, legionella, and cytokine-cytokine-receptor signaling). Finally, CXCL2 is found in two of the KEGG pathways (salmonella infection and legionella). Overall, in our study there were 42 statistically significant DEGs in the seven significant KEGG pathways. The five genes discussed above were noted as DEGs in multiple significant KEGG pathways such that these genes (CD14, TLR4, CXCL8, IL-18, and CXCL2) comprised 24 of those 42 DEGs.
Table 2.
Significant differentially expressed genes enriched in multiple functional categories
| Ensembl ID | Base mean | log2 FC | lfcSE | Stat | P value | Adjusted P value | Seq names | Gene name | Description |
|---|---|---|---|---|---|---|---|---|---|
| ENSSSCG00000014369 | 2913.87 | 1.51 | 0.16 | 9.45 | 3.52E-21 | 1.57E-18 | 2 | CD14 | monocyte differentiation antigen CD14 precursor |
| ENSSSCG00000008953 | 273.13 | 1.65 | 0.56 | 2.96 | 3.06E-03 | 1.23E-02 | 8 | CXCL8 | C-X-C motif chemokine ligand 8 |
| ENSSSCG00000008959 | 1034.25 | 1.56 | 0.43 | 3.61 | 3.09E-04 | 1.85E-04 | 8 | CXCL2 | C-X-C motif chemokine 2 precursor |
| ENSSSCG00000005503 | 1994.46 | 1.08 | 0.10 | 10.72 | 7.78E-27 | 8.43E-24 | 1 | TLR4 | toll-like receptor 4 isoform 1 precursor |
| ENSSSCG00000015037 | 243.66 | 1.42 | 0.19 | 7.45 | 9.55E-14 | 1.17E-11 | 9 | IL18 | Interleukin-18 |
FC: fold change, lfcSE: log fold change standard error
Fig. 2.
The normalized expression of the genes TLR4, CD14, IL18, CXCL2, and CXCL8 comparing prior to (control) and six hours following ICH
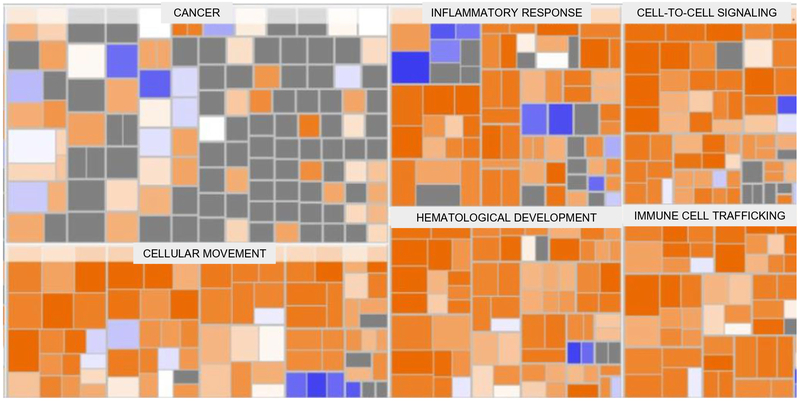
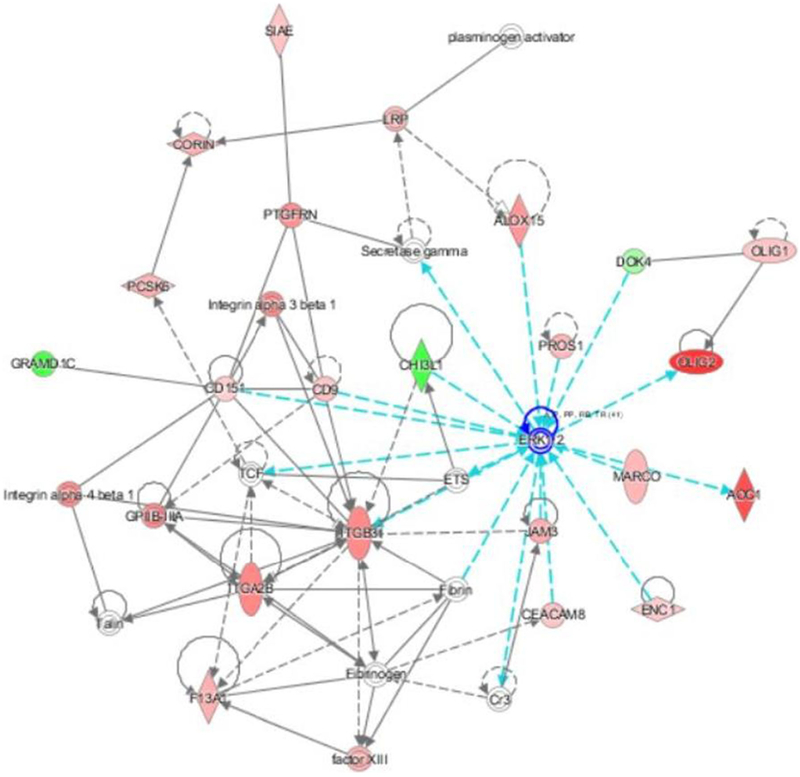
When the RNA-seq findings from the study were analyzed using Ingenuity Pathway Analysis (IPA), the following categories of diseases and functions were the most significantly upregulated: inflammatory response, immune cell trafficking, cellular movement, hematological system development, and cell-to-cell signaling and interaction (Fig. 3). IPA identified two upstream regulators as being activated and with the most significant P values: lipopolysaccharide (P = 1.04E-14) and tumor necrosis factor (P = 1.57E-11) The top network of molecular interactions that was generated by IPA showed extra cellular signal-related kinase (ERK) as the node with the most interactions (Fig. 4).
Fig. 3.
IPA generated heatmap of diseases and functions from the swine ICH RNA-seq data. The majority of the significantly upregulated categories (based on Z-score and with greater upregulation represented by progressively brighter orange to red colors) involve inflammation and activation of the immune system/cellular processes
Fig. 4.
The top IPA generated network based on the swine ICH RNA-seq data. The node with the most interactions with other molecules is extra cellular signal-related kinase (ERK, highlighted in the figure by cyan colored lines)
Discussion
We found in a swine model of ICH that there are significant changes in PBMC gene expression within six hours of ICH induction. Utilizing RNA sequencing, the significant biological pathways primarily identified were those involved in immune system propagation and inflammation, including genes commonly associated with monocyte activation. This is the first study of RNA-seq in swine ICH. Our findings also provide evidence that ICH induces substantial changes in gene expression that are reflected in the peripheral blood, including in the hyperacute period following the hemorrhage. Biomarkers of ICH neuroinflammatory pathophysiology, previously noted in brain tissue from preclinical studies (Wu et al. 2009; Xie et al. 2018) and post-mortem brain tissue from patients (Wu et al. 2010), were found to also exist in the peripheral blood in our investigation, increasing the potential translational relevance of these findings.
CD14 was a statistically significant DEG in our study and one of five DEGs that were repeatedly part of the significant KEGG pathways. CD14 was first identified as a cell surface marker for monocytes. Much of CD14’s known function is related to specifically interacting with TLR4 to facilitate cellular responses to LPS. (Zanoni and Granucci 2013) A CD14 polymorphism had a significantly different genotype distribution between ischemic and hemorrhagic stroke groups. (Das et al. 2017) Much of the other available literature pertaining to CD14 and stroke, and more specifically ICH, has focused on CD14 as a cell surface marker on monocytes. In a recent study of 60 patients with ICH, CD14+ monocytes were the predominant cell to express T cell immunoglobulin and mucin domain 3 (Tim-3), and Tim-3 was more highly expressed in patients with larger ICH volumes. Also, a negative correlation was found between Glasgow Outcome Scale and Tim-3 mRNA (ICH < 30 ml: r = −0.650, P < .001; ICH ≥ 30 ml: r = −0.723, P < .001), suggesting that Tim-3 on CD14+ monocytes contributes to inflammation after ICH and could be a novel treatment target. (Xu et al. 2018).
We also found that the TLR4 gene was significantly overexpressed in swine PBMCs following ICH and had an important role in associated significant KEGG pathways. TLR4 is a cell-surface receptor that activates the immune system during both “sterile” noninfectious inflammation and during bacterial infections. In monocytes from septic patients compared to controls, there was more TLR4 on the cell surface and increased monocyte expression of TLR4 messenger RNA. (Armstrong et al. 2004) TLR4 has been repeatedly identified as a key component of the inflammatory pathway following ICH. Utilizing both murine and human studies, TLR4 inhibition was identified as a promising therapeutic target to promote hematoma absorption and improve neurologic deficits. (Fang et al. 2014) In murine ICH models via TLR4 signaling, extracellular peroxiredoxins increased macrophage inflammation, (Liu et al. 2016) heme induced inflammation in microglia, and autophagy contributed to microglial activation and inflammatory injury. (Yang et al. 2015) TLR4 deficient mice with ICH had decreased perihematomal inflammation, less recruitment of immune cells including monocytes and neutrophils, and improved functional outcome. (Sansing et al. 2011) In 141 patients with ICH, increased expression of TLR4 on the surface of monocytes was associated with poor outcome defined as Modified Rankin Scale >2 at 3 months (5272 arbitrary fluorescence units in poor outcome vs. 2977, P < 0.0001).(Rodriguez-Yanez et al. 2012).
CXCL8 is a gene for which the associated protein is the proinflammatory cytokine IL-8 (Interleukin 8). Stimulation with LPS (Raspe et al. 2013) and with the toxic substance ricin (Gonzalez et al. 2006) resulted in monocyte production of IL-8. Serum concentrations of IL-8 were significantly higher in severe sepsis compared to uncomplicated sepsis both in the emergency department (P = 0.0009) and up to 24 h after presentation (P = 0.011).(Macdonald et al. 2014) In ischemic stroke patients, increased IL-8 serum protein levels and mRNA in blood mononuclear cells has been reported. (Kostulas et al. 1999) In 76 ischemic stroke patients compared with 28 controls, IL-8 was increased in stroke patients’ plasma within 24 h of symptom onset (P < 0.001), and the IL-8 levels were also positively associated with the extent of the ischemic stroke lesion (P < 0.01).(Domac and Misirli 2008) The inhibition of IL-8 as a therapeutic target in stroke has been considered, with inhibitory effects of IL-8 by antiplatelet agents noted in ischemic stroke patients,(Al-Bahrani et al. 2007) reduced inflammation and neurological deficits after treatment with the IL-8 inhibitor reparixin in rats with transient cerebral ischemia, (Villa et al. 2007) and less edema and infarct size in a rabbit model of transient brain ischemia after treatment with an IL-8 neutralizing antibody. (Matsumoto et al. 1997) While data regarding IL-8 and ICH are currently limited, in 94 patients with basal ganglia ICH, a positive correlation was reported between IL-8 and severity of cerebral edema (r = 0.305, P < 0.05).(Wang et al. 2016).
IL-18 is a pro-inflammatory cytokine primarily secreted in its active form by macrophages and dendritic cells. A role for IL-18 has been implicated in a number of disease states such as sepsis, inflammatory bowel disease, psoriasis, and emphysema. (Dinarello 2007) In the field of stroke and cardiovascular disease, increased levels of IL-18 mRNA were noted in 70 patients with carotid artery stenosis compared to 75 controls (p = 0.01).(Arapi et al. 2018) In a large case-control study nested within a prospective cohort, with 664 patients with either myocardial infarction (MI) or stroke and 1328 controls, IL-18 level was strongly associated with a number of cardiovascular risk factors. (Jefferis et al. 2013) In 23 patients with ischemic stroke within 24 h of symptom onset, serum IL-18 was higher compared to 15 controls (308.5 ± 138.0 pg/ml vs. 155.7 ± 51.4 pg/ml, P < 0.01), and was also noted to be potentially predictive of stroke outcome. (Zaremba and Losy 2003) Finally, in ICH, a recent murine study found that the anti-inflammatory compound cordycepin alleviated brain edema and neurological deficits while also reducing the release of IL-18.(Cheng et al. 2017) In another rodent study of ICH, those rats treated with P2X7R small interfering RNA had exacerbated brain inflammation and damage potentially secondary to the NLRP3 inflammasome that resulted in cytokine secretion, including IL-18.(Feng et al. 2015).
CXCL2, a gene that encodes the protein MIP-2 (macrophage inflammatory protein 2), is a powerful chemotaxis and cellular activation factor and is secreted in response to infection or injury by a number of cell types. (Qin et al. 2017) It is often increased acutely after injury, resulting in inflammation, and was found to be significantly increased in our study of PBMC gene expression in the first 6 h after swine ICH. MIP-2 was increased in preclinical models of ischemic stroke and had an important role in neuroinflammation and infarct development. (Lee et al. 2015) In the postmortem brains of 36 ICH patients ranging from two to five days after the hemorrhage and compared to six controls, MIP-2 expression was increased in both the area of ICH (P < 0.01) and the contralateral uninjured side (p < 0.05), with greater increase on the side of the brain with ICH. (Wu et al. 2010) In a rat model, MIP-2 levels were higher in the brain within two hours of ICH, peaked at two days, and correlated with degree of brain edema and activation of the inflammatory transcription factor NF-kappaB.(Wu et al. 2009) When ICH induced expression of cytokines in a mouse model of ICH, including MIP-2, the authors concluded that MIP-2 specifically was involved in the degree of hematoma invasion and resultant motor dysfunction, and inhibition of MIP-2 resulted in less motor disability. (Matsushita et al. 2014).
In our data analysis with IPA, lipopolysaccharide (LPS) and tumor necrosis factor (TNF) were the two most significantly activated upstream regulators. IPA considers the expected effects between transcriptional regulators and their target genes, based on a vast database of such cause and effect relationships from the published literature, to identify upstream regulators that would explain the gene expression in a given study. The upstream regulators predicted from our findings, LPS and TNF, supported the concept that ICH induced inflammatory pathways in PBMCs. LPS, also known as endotoxin, is a major component of the outer membrane of gram-negative bacteria and induces a cascade of cellular reactions that potentiates a strong inflammatory response. (Raspe et al. 2013) TNF is a well-known cytokine that induces systemic inflammation and participates in the acute phase response, a complex early defense system that can be activated by a number of events such as infection, inflammation, and trauma. (Bradley 2008). The top network that was generated by IPA based on our findings included extracellular signal-regulated kinase (ERK) as having the most interactions with other molecules (Fig. 4). ERK are protein serine/threonine kinases that mediate a variety of processes such as inflammation, cell differentiation, proliferation, and adhesion. (Roskoski 2012) ERK signaling resulted in the production of inflammatory cytokines like IL-1β and TNF- α in human monocytic cells. (Kurosawa et al. 2000) IL-8 production was dependent on ERK signaling in epithelial cells infected with chlamydia trachomatis. (Buchholz and Stephens 2007) The ERK pathway has been reported as a potential therapeutic target for neurological diseases, including stroke. (Sun and Nan 2017) In rats with ischemic stroke, production of inflammatory cytokines was transcriptionally regulated via ERK, and injection of an ERK inhibitor at 0 and 6 h after stroke reduced infarct volume (11.7% and 15% of total brain volume, respectively, compared with 25% for controls).(Maddahi and Edvinsson 2010) In ICH, nonclinical studies have been reported such as when ICH increased ERK expression in perihematomal tissues in a rat model. (Wen et al. 2017) In astrocytes studied in vitro, hemolysate increased the phosphorylation of ERK 15 fold compared to controls (P < 0.01).(Yang et al. 2016).
Improved understanding of the pathophysiologic mediators of injury and repair post-ICH can lead to identification of novel therapeutic targets. RNA-seq is particularly promising for this type of investigation as it provides a global, unbiased analysis as opposed to only measuring transcriptomic biomarkers that are determined in advance. In addition to the development of novel therapeutic agents, there is also the potential to utilize those treatments that are already in use. For example, it was recently reported from a rodent model of ICH that mannitol and hypertonic saline reduced mortality and hemispheric swelling, reduced markers of inflammatory M1 microglia/macrophage activation (including TNF that was identified as a critical upstream regulator in our study), and increased markers of the anti-inflammatory M2 phenotype. (Schreibman et al. 2018). Our group also recently published a review article regarding the cellular/molecular mechanisms of action of currently approved medications for multiple sclerosis and the potential of these treatments to modulate acute inflammation after ICH. (Napier et al. 2019).
Regarding the validation of the RNA-seq findings by other methods, in previous studies from our core sequencing facility an alternative approach of RT-qPCR was used to validate differentially expressed genes (Sharma et al. 2017), and this showed very high reproducibility between RNA-seq and RT-qPCR results. Since in this current study the same methods and reagents were used under the same experimental conditions, no additional validation of the RNA-seq result was performed.
Strengths of our study include that it is the first report of RNA-seq technology to study gene expression in swine ICH. Further, this preclinical model successfully demonstrated that ICH induces systemic changes in inflammatory gene expression that are potentially novel pathophysiologic biomarkers and treatment targets, and this line of inquiry warrants further investigation. RNA-seq provided a global, unbiased analysis, and advanced bioinformatics techniques benefitted from well-established sources for enrichment analysis. Additional novel methodology includes our focus on the hyperacute period following ICH and the testing of paired samples, i.e. a blood sample from the same animal prior to and then following the induction of ICH, not possible in human subjects, that can allow for identifying RNA-seq differences more specifically from the ICH itself. As discussed previously, there are data to support that the pig is a superior preclinical model to study the immune system than other animals such as rodents.
Limitations of our reported research include that RNA-seq data were not available more than six hours following ICH induction. While the focus of the current work was on the hyperacute period following ICH, analysis of samples collected later could allow for characterization of unique gene expression over a longer period of time. Further, while investigating the gene expression at a number of earlier time points could also strengthen the study, e.g. one, two, and/or three hours from ICH induction, the scope and available resources of the reported research precluded RNA-seq for such a large number of time points and associated blood samples. The pigs were young and lacked comorbidities that might be seen in human ICH patients, and the use of any animal model of ICH, including autologous blood injection into the brain, lacks some of the pathophysiology of naturally occurring ICH. Considering that differences in the inflammatory response have been reported between males and females, possibly related to estrogen level variability, (Casimir and Duchateau 2011; Rathod et al. 2017) a homogenous group of only female pigs were included. Subsequent work could be strengthened by including both male and females.
Conclusion
This novel research provides evidence from a swine model that changes in peripheral blood gene expression are induced within 6 h of ICH, and the associated significant biological pathways in PBMCs are primarily those associated with immune system activation, inflammation, and, in particular, activation of monocytes. Our findings contribute to a larger body of literature regarding neuroinflammation following ICH and that monocytes specifically contribute to this process. Further studies in ICH are needed, such as more work in swine ICH in which serial blood samples are collected over a longer range of time following ICH induction, brain tissue is tested in relation to the changes in peripheral blood, and pharmacologic interventions are administered and compared to the gene expression in non-treated experiments. The results reported here would also benefit from comparison to RNA-seq in human ICH. Ultimately, the study of neuroinflammation following ICH, with a particular focus on the effects of monocytes, is deserving of further research in pursuit of a novel therapeutic target for this devastating condition with no proven treatment.
Acknowledgements
The authors would like to acknowledge the financial support for the reported research from the University of Cincinnati Gardner Neuroscience Institute (UCGNI), specifically, the UCGNI Neurobiology Research Center Pilot Research Program.
Footnotes
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s11011-019-00399-z) contains supplementary material, which is available to authorized users.
Disclosure of potential conflicts of interest The authors Walsh, Zhang, Zhu, Wohleb, Woo, and Lu no conflicts of interest to report.
The author Adeoye holds equity in Sense Diagnostics, LLC, Cincinnati, Ohio. For the submitted manuscript, financial support for the swine experiments only was provided by Sense Diagnostics. To further clarify, the financial support for blood sample collection, processing, storage, RNA sequencing, and bioinformatics analysis was not provided by Sense Diagnostics but by the University of Cincinnati Gardner Neuroscience Institute. The research reported in the submitted manuscript was performed as a substudy to the swine intracerebral hemorrhage study performed by Sense Diagnostics for the testing of a non-invasive brain monitor. The reported results in this manuscript, and the RNA-seq data overall, have no bearing on the development of this non-invasive brain monitoring device.
Statement on the welfare of animals All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
All procedures performed in this research involving animals were in accordance with the ethical standards of the University of Cincinnati Institutional Animal Care and Use Committee.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Adeoye O, Clark JF, Khatri P, Wagner KR, Zuccarello M, Pyne-Geithman GJ (2011) Do current animal models of intracerebral hemorrhage mirror the human pathology? Transl Stroke Res 2:17–25. 10.1007/s12975-010-0037-1 [DOI] [PubMed] [Google Scholar]
- Adeoye O et al. (2014) Peripheral monocyte count is associated with case fatality after intracerebral hemorrhage. J Stroke Cerebrovasc Dis 23: e107–e111. 10.1016/j.jstrokecerebrovasdis.2013.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bahrani A, Taha S, Shaath H, Bakhiet M (2007) TNF-alpha and IL-8 in acute stroke and the modulation of these cytokines by antiplatelet agents. Curr Neurovasc Res 4:31–37 [DOI] [PubMed] [Google Scholar]
- Arapi B, Bayoglu B, Cengiz M, Dirican A, Deser SB, Junusbekov Y, Arslan C (2018) Increased expression of interleukin 18 mRNA is associated with carotid artery stenosis. Balkan Med J. 10.4274/balkanmedj.2017.0323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong L, Medford AR, Hunter KJ, Uppington KM, Millar AB (2004) Differential expression of toll-like receptor (TLR)-2 and TLR-4 on monocytes in human sepsis. Clin Exp Immunol 136: 312–319. 10.1111/j.1365-2249.2004.02433.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviv RI et al. (2014) An in vivo, MRI-integrated real-time model of active contrast extravasation in acute intracerebral hemorrhage. AJNR Am J Neuroradiol 35:1693–1699. 10.3174/ajnr.A3939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakshi S, Zhang X, Godoy-Tundidor S, Cheng RY, Sartor MA, Medvedovic M, Ho SM (2008) Transcriptome analyses in normal prostate epithelial cells exposed to low-dose cadmium: oncogenic and immunomodulations involving the action of tumor necrosis factor. Environ Health Perspect 116:769–776. 10.1289/ehp.11215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsveinson J, Friesen V, Ziola B (2016) Transcriptome analysis of beer-spoiling lactobacillus brevis BSO 464 during growth in degassed and gassed beer. Int J Food Microbiol 235:28–35. 10.1016/j.ijfoodmicro.2016.06.041 [DOI] [PubMed] [Google Scholar]
- Bradley JR (2008) TNF-mediated inflammatory disease. J Pathol 214:149–160. 10.1002/path.2287 [DOI] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L (2016) Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34:525–527. 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- Brunberg E, Jensen P, Isaksson A, Keeling LJ (2013) Behavioural and brain gene expression profiling in pigs during tail biting outbreaks -evidence of a tail biting resistant phenotype. PLoS One 8:e66513 10.1371/journal.pone.0066513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz KR, Stephens RS (2007) The extracellular signal-regulated kinase/mitogen-activated protein kinase pathway induces the inflammatory factor interleukin-8 following chlamydia trachomatis infection. Infect Immun 75:5924–5929. 10.1128/iai.01029-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabezas-Wallscheid N et al. (2014) Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell Stem Cell 15:507–522. 10.1016/j.stem.2014.07.005 [DOI] [PubMed] [Google Scholar]
- Casimir GJ, Duchateau J (2011) Gender differences in inflammatory processes could explain poorer prognosis for males. J Clin Microbiol 49:478; author reply 478–479. 10.1128/jcm.02096-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Yang Q, Chen G, Zhang JH (2015) An update on inflammation in the acute phase of intracerebral hemorrhage. Transl Stroke Res 6:4–8. 10.1007/s12975-014-0384-4 [DOI] [PubMed] [Google Scholar]
- Cheng Y et al. (2017) Cordycepin confers neuroprotection in mice models of intracerebral hemorrhage via suppressing NLRP3 inflammasome activation. Metab Brain Dis 32:1133–1145. 10.1007/s11011-017-0003-7 [DOI] [PubMed] [Google Scholar]
- Cheong A et al. (2016) DNA methylome changes by estradiol benzoate and bisphenol a links early-life environmental exposures to prostate cancer risk. Epigenetics 11:674–689. 10.1080/15592294.2016.1208891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Kaul S, Jyothy A, Munshi A (2017) Role of TLR4 (C1196T) and CD14 (C-260T) polymorphisms in development of ischemic stroke, its subtypes and hemorrhagic stroke. J Mol Neurosci: MN 63:300–307. 10.1007/s12031-017-0979-9 [DOI] [PubMed] [Google Scholar]
- Dinarello CA (2007) Interleukin-18 and the pathogenesis of inflammatory diseases. Semin Nephrol 27:98–114. 10.1016/j.semnephrol.2006.09.013 [DOI] [PubMed] [Google Scholar]
- Domac FM, Misirli H (2008) The role of neutrophils and interleukin-8 in acute ischemic stroke. Neurosciences (Riyadh, Saudi Arabia) 13: 136–141 [PubMed] [Google Scholar]
- Fairbairn L, Kapetanovic R, Sester DP, Hume DA (2011) The mononuclear phagocyte system of the pig as a model for understanding human innate immunity and disease. J Leukoc Biol 89:855–871. 10.1189/jlb.1110607 [DOI] [PubMed] [Google Scholar]
- Fang H, Chen J, Lin S, Wang P, Wang Y, Xiong X, Yang Q (2014) CD36-mediated hematoma absorption following intracerebral hemorrhage: negative regulation by TLR4 signaling. J Immunol (Baltimore, Md : 1950) 192:5984–5992. 10.4049/jimmunol.1400054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Chen Y, Ding R, Fu Z, Yang S, Deng X, Zeng J (2015) P2X7R blockade prevents NLRP3 inflammasome activation and brain injury in a rat model of intracerebral hemorrhage: involvement of peroxynitrite. J Neuroinflammation 12:190 10.1186/s12974-015-0409-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez TV, Farrant SA, Mantis NJ (2006) Ricin induces IL-8 secretion from human monocyte/macrophages by activating the p38 MAP kinase pathway. Mol Immunol 43:1920–1923. 10.1016/j.molimm.2005.11.002 [DOI] [PubMed] [Google Scholar]
- Gu Y, Hua Y, He Y, Wang L, Hu H, Keep RF, Xi G (2011) Iron accumulation and DNA damage in a pig model of intracerebral hemorrhage. Acta Neurochir Suppl 111:123–128. 10.1007/978-3-7091-0693-8_20 [DOI] [PubMed] [Google Scholar]
- Hammond MD, Ambler WG, Ai Y, Sansing LH (2014a) alpha4 integrin is a regulator of leukocyte recruitment after experimental intracerebral hemorrhage. Stroke 45:2485–2487. 10.1161/strokeaha.114.005551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond MD et al. (2014b) CCR2+ Ly6C(hi) inflammatory monocyte recruitment exacerbates acute disability following intracerebral hemorrhage. J Neurosci 34:3901–3909. 10.1523/jneurosci.4070-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Haehnel V, Karaghiosoff M, Schwarzfischer L, Muller M, Krause SW, Rehli M (2003) Species-specific regulation of Toll-like receptor 3 genes in men and mice. J Biol Chem 278:21502–21509. 10.1074/jbc.M301476200 [DOI] [PubMed] [Google Scholar]
- Hua YGY, Xi G (2009) Blood injection intracerebral hemorrhage pig model Animal models of acute neurological injuries. Humana Press, New York [Google Scholar]
- Jefferis BJ et al. (2013) Prospective study of IL-18 and risk of MI and stroke in men and women aged 60–79 years: a nested case-control study. Cytokine 61:513–520. 10.1016/j.cyto.2012.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapetanovic R, Fairbairn L, Beraldi D, Sester DP, Archibald AL, Tuggle CK, Hume DA (2012) Pig bone marrow-derived macrophages resemble human macrophages in their response to bacterial lipopolysaccharide. J Immunol (Baltimore, Md : 1950) 188:3382–3394. 10.4049/jimmunol.1102649 [DOI] [PubMed] [Google Scholar]
- Kostulas N, Pelidou SH, Kivisakk P, Kostulas V, Link H (1999) Increased IL-1beta, IL-8, and IL-17 mRNA expression in blood mononuclear cells observed in a prospective ischemic stroke study. Stroke 30: 2174–2179 [DOI] [PubMed] [Google Scholar]
- Kurosawa M, Numazawa S, Tani Y, Yoshida T (2000) ERK signaling mediates the induction of inflammatory cytokines by bufalin in human monocytic cells. Am J Physiol Cell Physiol 278:C500–C508. 10.1152/ajpcell.2000.278.3.C500 [DOI] [PubMed] [Google Scholar]
- Lee S et al. (2015) Effect of a broad-specificity chemokine-binding protein on brain leukocyte infiltration and infarct development. Stroke 46: 537–544. 10.1161/strokeaha.114.007298 [DOI] [PubMed] [Google Scholar]
- Liu DL, Zhao LX, Zhang S, Du JR (2016) Peroxiredoxin 1-mediated activation of TLR4/NF-kappaB pathway contributes to neuroinflammatory injury in intracerebral hemorrhage. Int Immunopharmacol 41:82–89. 10.1016/j.intimp.2016.10.025 [DOI] [PubMed] [Google Scholar]
- Loftspring MC, Clark JF, Wagner KR (2007) A novel duplex ELISA method for quantitation of plasma proteins in areas of brain edema. Brain Res 1162:130–132. 10.1016/j.brainres.2007.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald SP, Stone SF, Neil CL, van Eeden PE, Fatovich DM, Arendts G, Brown SG (2014) Sustained elevation of resistin, NGAL and IL-8 are associated with severe sepsis/septic shock in the emergency department. PLoS One 9:e110678 10.1371/journal.pone.0110678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddahi A, Edvinsson L (2010) Cerebral ischemia induces microvascular pro-inflammatory cytokine expression via the MEK/ERK pathway. J Neuroinflammation 7:14 10.1186/1742-2094-7-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T, Ikeda K, Mukaida N, Harada A, Matsumoto Y, Yamashita J, Matsushima K (1997) Prevention of cerebral edema and infarct in cerebral reperfusion injury by an antibody to interleukin-8. Lab Invest 77:119–125 [PubMed] [Google Scholar]
- Matsushita H et al. (2014) Suppression of CXCL2 upregulation underlies the therapeutic effect of the retinoid Am80 on intracerebral hemorrhage in mice. J Neurosci Res 92:1024–1034. 10.1002/jnr.23379 [DOI] [PubMed] [Google Scholar]
- Morotti A et al. (2016) Leukocyte count and intracerebral hemorrhage expansion. Stroke 47:1473–1478. 10.1161/strokeaha.116.013176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, Das S (2016) Heart disease and stroke statistics–2016 update: a report from the American Heart Association. Circulation 133:e38–e60 [DOI] [PubMed] [Google Scholar]
- Nagaraja S et al. (2017) Transcriptional Dependencies in Diffuse Intrinsic Pontine Glioma. Cancer Cell 31:635–652.e636. 10.1016/j.ccell.2017.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napier J, Rose L, Adeoye O, Hooker E, Walsh KB (2019) Modulating acute neuroinflammation in intracerebral hemorrhage: the potential promise of currently approved medications for multiple sclerosis Immunopharmacology and immunotoxicology: 10.1080/08923973.2019.1566361 [DOI] [PubMed] [Google Scholar]
- Orakcioglu B, Uozumi Y, Kentar MM, Santos E, Unterberg A, Sakowitz OW (2012) Evidence of spreading depolarizations in a porcine cortical intracerebral hemorrhage model. Acta Neurochir Suppl 114: 369–372. 10.1007/978-3-7091-0956-4_71 [DOI] [PubMed] [Google Scholar]
- Orakcioglu B, Kentar MM, Schiebel P, Uozumi Y, Unterberg A, Sakowitz OW (2015) Perihemorrhagic ischemia occurs in a volume-dependent manner as assessed by multimodal cerebral monitoring in a porcine model of intracerebral hemorrhage. Neurocrit Care 22:133–139. 10.1007/s12028-014-0027-3 [DOI] [PubMed] [Google Scholar]
- Pantazatos SP, Huang YY, Rosoklija GB, Dwork AJ, Arango V, Mann JJ (2017) Whole-transcriptome brain expression and exon-usage profiling in major depression and suicide: evidence for altered glial, endothelial and ATPase activity. Mol Psychiatry 22:760–773. 10.1038/mp.2016.130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin CC, Liu YN, Hu Y, Yang Y, Chen Z (2017) Macrophage inflammatory protein-2 as mediator of inflammation in acute liver injury. World J Gastroenterol 23:3043–3052. 10.3748/wjg.v23.i17.3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raspe C et al. (2013) Glutamine and alanine-induced differential expression of intracellular IL-6, IL-8, and TNF-alpha in LPS-stimulated monocytes in human whole-blood. Cytokine 62:52–57. 10.1016/j.cyto.2013.02.020 [DOI] [PubMed] [Google Scholar]
- Rathod KS et al. (2017) Accelerated resolution of inflammation underlies sex differences in inflammatory responses in humans. J Clin Invest 127:169–182. 10.1172/jci89429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Yanez M, Brea D, Arias S, Blanco M, Pumar JM, Castillo J, Sobrino T (2012) Increased expression of toll-like receptors 2 and 4 is associated with poor outcome in intracerebral hemorrhage. J Neuroimmunol 247:75–80. 10.1016/j.jneuroim.2012.03.019 [DOI] [PubMed] [Google Scholar]
- Roskoski R Jr (2012) ERK1/2 MAP kinases: structure, function, and regulation. Pharmacol Res 66:105–143. 10.1016/j.phrs.2012.04.005 [DOI] [PubMed] [Google Scholar]
- Sang M et al. (2017) Gene expression profile of peripheral blood mononuclear cells in response to intracerebral hemorrhage. DNA Cell Biol 36:647–654. 10.1089/dna.2017.3650 [DOI] [PubMed] [Google Scholar]
- Sansing LH, Harris TH, Welsh FA, Kasner SE, Hunter CA, Kariko K (2011) Toll-like receptor 4 contributes to poor outcome after intracerebral hemorrhage. Ann Neurol 70:646–656. 10.1002/ana.22528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schook L et al. (2005) Swine in biomedical research: creating the building blocks of animal models. Anim Biotechnol 16:183–190 [DOI] [PubMed] [Google Scholar]
- Schreibman DL, Hong CM, Keledjian K, Ivanova S, Tsymbalyuk S, Gerzanich V, Simard JM (2018) Mannitol and hypertonic saline reduce swelling and modulate inflammatory markers in a rat model of intracerebral hemorrhage. Neurocrit Care 29:253–263. 10.1007/s12028-018-0535-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder K et al. (2012) Conservation and divergence in Toll-like receptor 4-regulated gene expression in primary human versus mouse macrophages. Proc Natl Acad Sci U S A 109:E944–E953. 10.1073/pnas.1110156109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M, Zhang X, Zhang S, Niu L, Ho SM, Chen A, Huang S (2017) Inhibition of endocytic lipid antigen presentation by common lipophilic environmental pollutants. Sci Rep 7:2085 10.1038/s41598-017-02229-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Nan G (2017) The extracellular signal-regulated kinase 1/2 pathway in neurological diseases: a potential therapeutic target (review). Int J Mol Med 39:1338–1346. 10.3892/ijmm.2017.2962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Lu A, Aronow BJ, Sharp FR (2001) Blood genomic responses differ after stroke, seizures, hypoglycemia, and hypoxia: blood genomic fingerprints of disease. Ann Neurol 50:699–707 [DOI] [PubMed] [Google Scholar]
- Villa P et al. (2007) The interleukin-8 (IL-8/CXCL8) receptor inhibitor reparixin improves neurological deficits and reduces long-term inflammation in permanent and transient cerebral ischemia in rats. Mol Med (Cambridge, Mass) 13:125–133. 10.2119/2007-00008.Villa [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner KR (2007) Modeling intracerebral hemorrhage: glutamate, nuclear factor-kappa B signaling and cytokines. Stroke 38:753–758. 10.1161/01.STR.0000255033.02904.db [DOI] [PubMed] [Google Scholar]
- Wagner KR et al. (1996) Lobar intracerebral hemorrhage model in pigs: rapid edema development in perihematomal white matter. Stroke 27: 490–497 [DOI] [PubMed] [Google Scholar]
- Wagner KR, Xi G, Hua Y, Kleinholz M, de Courten-Myers GM, Myers RE (1998) Early metabolic alterations in edematous perihematomal brain regions following experimental intracerebral hemorrhage. J Neurosurg 88:1058–1065. 10.3171/jns.1998.88.6.1058 [DOI] [PubMed] [Google Scholar]
- Wagner KR et al. (2006) Delayed profound local brain hypothermia markedly reduces interleukin-1beta gene expression and vasogenic edema development in a porcine model of intracerebral hemorrhage. Acta Neurochir Suppl 96:177–182 [DOI] [PubMed] [Google Scholar]
- Walsh KB et al. (2015) Monocyte count and 30-day case fatality in intracerebral hemorrhage. Stroke 46:2302–2304. 10.1161/strokeaha.115.009880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh KB, Campos B, Hart K, Thakar C, Adeoye O (2017) M2 monocyte microparticles are increased in intracerebral hemorrhage. Journal of stroke and cerebrovascular diseases. J Stroke Cerebrovasc Dis. 10.1016/j.jstrokecerebrovasdis.2017.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XM et al. (2016) Expressions of serum inflammatory cytokines and their relationship with cerebral edema in patients with acute basal ganglia hemorrhage. Eur Rev Med Pharmacol Sci 20:2868–2871 [PubMed] [Google Scholar]
- Wen Z, Mei B, Li H, Dou Y, Tian X, Shen M, Chen G (2017) P2X7 participates in intracerebral hemorrhage-induced secondary brain injury in rats via MAPKs signaling pathways. Neurochem Res 42: 2372–2383. 10.1007/s11064-017-2257-1 [DOI] [PubMed] [Google Scholar]
- Wu J, Mao X, Cai T, Luo J, Wei L (2006) KOBAS server: a web-based platform for automated annotation and pathway identification. Nucleic Acids Res 34:W720–W724. 10.1093/nar/gkl167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Cong Y, Wang D, Zhao R, Qi J (2009) Correlation of macrophage inflammatory protein-2 expression and brain edema in rats after intracerebral hemorrhage. Int J Clin Exp Pathol 2:83–90 [PMC free article] [PubMed] [Google Scholar]
- Wu H et al. (2010) Dynamic changes of inflammatory markers in brain after hemorrhagic stroke in humans: a postmortem study. Brain Res 1342:111–117. 10.1016/j.brainres.2010.04.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi G et al. (1998) Role of blood clot formation on early edema development after experimental intracerebral hemorrhage. Stroke 29:2580–2586 [DOI] [PubMed] [Google Scholar]
- Xie Y et al. (2018) Sex Differences in Gene and Protein Expression After Intracerebral Hemorrhage in Mice. Transl Stroke Res. 10.1007/s12975-018-0633-z [DOI] [PubMed] [Google Scholar]
- Xu C, Ge H, Wang T, Qin J, Liu, Liu Y (2018) Increased expression of T cell immunoglobulin and mucin domain 3 on CD14(+) monocytes is associated with systemic inflammatory reaction and brain injury in patients with spontaneous intracerebral hemorrhage. J Stroke Cerebrovasc Dis 27:1226–1236. 10.1016/j.jstrokecerebrovasdis.2017.11.041 [DOI] [PubMed] [Google Scholar]
- Yang Z et al. (2015) Toll-like receptor-4-mediated autophagy contributes to microglial activation and inflammatory injury in mouse models of intracerebral haemorrhage. Neuropathol Appl Neurobiol 41:e95–106. 10.1111/nan.12177 [DOI] [PubMed] [Google Scholar]
- Yang XL et al. (2016) Anti-inflammatory effects of fimasartan via Akt, ERK, and NFkappaB pathways on astrocytes stimulated by hemolysate. Inflamm Res 65:115–123. 10.1007/s00011-015-0895-9 [DOI] [PubMed] [Google Scholar]
- Zanoni I, Granucci F (2013) Role of CD14 in host protection against infections and in metabolism regulation. Front Cell Infect Microbiol 3:32 10.3389/fcimb.2013.00032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaremba J, Losy J (2003) Interleukin-18 in acute ischaemic stroke patients. Neurological sciences : official journal of the Italian Neurological Society and of the Italian Society of Clinical Neurophysiology 24:117–124. 10.1007/s10072-003-0096-0 [DOI] [PubMed] [Google Scholar]
- Zhang W et al. (2015) Comparison of RNA-seq and microarray-based models for clinical endpoint prediction. Genome Biol 16:133 10.1186/s13059-015-0694-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Fung-Leung WP, Bittner A, Ngo K, Liu X (2014) Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS One 9:e78644 10.1371/journal.pone.0078644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Xie Q, Xi G, Keep RF, Hua Y (2014) Brain CD47 expression in a swine model of intracerebral hemorrhage. Brain Res 1574:70–76. 10.1016/j.brainres.2014.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]