Abstract
Temperature serves as a fundamental signal in biological systems. In some microbial pathogens of humans, mammalian body temperature triggers establishment and maintenance of a developmental program that allows the microbe to survive and thrive in the host. Histoplasma capsulatum is one of a group of fungal pathogens called thermally dimorphic fungi, all of which respond to mammalian body temperature by converting from an environmental mold form that inhabits the soil into a parasitic form that causes disease in the host. It has been known for decades that temperature is a key signal that is sufficient to trigger the switch from the soil to host form (and vice versa) in the laboratory. Recent molecular studies have identified a number of key regulators that are required to specify each of the developmental forms in response to temperature. Here we review the regulatory circuits that govern temperature-dependent dimorphism in Histoplasma.
Introduction
Histoplasma capsulatum is a thermally dimorphic fungal pathogen that is the most common cause of fungal respiratory infections in immunocompetent hosts [1,2]. Histoplasma species are found worldwide [3–6]. Although Histoplasma causes disease even in the setting of a functional immune system, the incidence of Histoplasma in HIV-infected patients has revealed the extent of this global distribution, including North America, Central and South America, Africa, and Asia [1]. Recent analysis suggests that in people with AIDS in Latin America, the incidence of Histoplasma infection is equal to the incidence of tuberculosis [7].
The term “thermally dimorphic fungal pathogen” has been used to describe a group of evolutionarily related fungi, including Histoplasma, that grow in a mold form in the environment but alter their growth program and adopt a parasitic form when exposed to mammalian body temperature [8–10]. Like the majority of these organisms, Histoplasma colonizes the soil as a sporulating mold form, and infection occurs when fungal particles are inhaled. Healthy humans who inhale a large dose of infectious particles and individuals who have defects in cell-mediated immunity are more likely to develop life-threatening respiratory and/or systemic disease [1,11,12]. Even in asymptomatic individuals, the fungus usually disseminates through the bloodstream from the lungs, and can reactivate years later from a latent state if the immune status of the host declines [12].
The environmental form of Histoplasma transitions to the yeast form in response to temperature
Histoplasma grows in the soil as a sporulating mold. The mold form is a multicellular mycelium comprised of an interconnecting network of vegetative hyphae. The hyphae produce asexual spores named micro- or macroconidia based on size [13]. Microconidia range in size from 2–6 μm, whereas macroconidia have been reported to range in size from 8 to 14 μm or 10 to 25 μm, depending on the strain and growth conditions. A number of early studies published several decades ago discuss conidiation conditions, the morphology of the resultant spores, and germination of micro- and macroconidia [13–20]. However, very little is known about the molecular basis of conidial development or germination. There has been a single gene expression profiling study that compared the transcriptome of conidia to that of yeast and hyphae [21], but an analysis of gene expression changes that occur as conidia germinate has not been performed. Additionally, the molecular and developmental relationship between micro- and macroconidia is unclear.
Although little is known about their biology, conidia are thought to play a role in both environmental fungal dispersion and infection of mammals via inhalation. Interestingly, conidial-enriched transcripts include those involved in stress responses [21], perhaps reflecting the challenging conditions encountered by these infectious propagules in either the environment or in the host. Upon disruption of the soil, conidia and/or hyphal fragments are inhaled by the host and then taken up by macrophages and other phagocytic cells [22–24]. Once inside the host, both spores and filaments give rise to yeast cells, which evade killing by phagocytic cells and instead replicate intracellularly within macrophages and monocytes (reviewed in [25]).
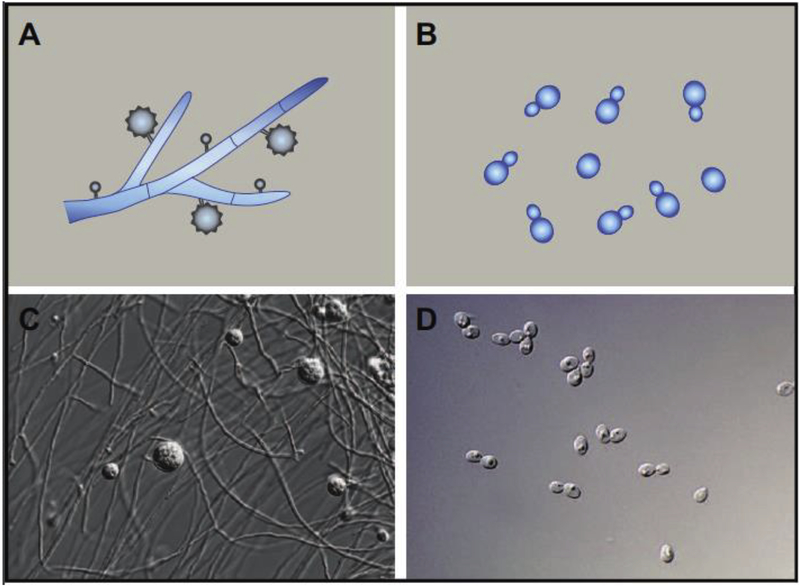
A key aspect of Histoplasma biology is that exposure to mammalian body temperature is sufficient to trigger the developmental transition from hyphal growth to yeast-phase growth [26–28]. This transition can be recapitulated in the laboratory, making trigger Histoplasma a compelling system in which to explore temperature-dependent signaling. During laboratory culture, Histoplasma cells grow in the hyphal form at room temperature and in the yeast form at 37°C (Figure 1), and the transition to the yeast form is accompanied by the expression of virulence factors [23]. Additionally, in vitro studies of conidia show that the majority of spores germinate to give rise to hyphae at room temperature and yeast cells at 37°C [21], again delineating temperature as a critical determinant of developmental fate.
Figure 1. Histoplasma morphology is regulated by temperature.
Schematic (A) and microscopy image (C) of conidiating hyphae growing at room temperature. Schematic (B) and microscopy image (D) of yeast cells growing at 37°C.
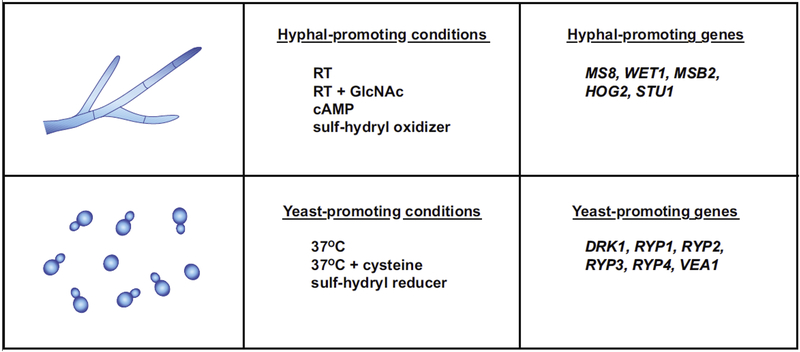
A number of factors in addition to temperature have been noted to promote either hyphal or yeast-phase growth (Figure 2), although the majority of these observations were made in the pre-molecular era and the genetic basis for their effects is unknown. For example, exogenous addition of cystine and cysteine in the culture medium is required to establish yeast-phase growth during the hyphal-to-yeast transition, and further increasing the cysteine levels (or other sulf-hydryl reducing agents such as dithiothreitol (DTT)) can accelerate the transition [29]. Furthermore, addition of DTT has also been shown to trap Histoplasma in the yeast form even when cells are shifted to room temperature [26]. In contrast, addition of the sulf-hydryl oxidizing agent pchloromercuriphenylsulfonic acid (PCMS) was shown to irreversibly trap Histoplasma cells in the hyphal form independent of temperature [26,30,31]. Addition of exogenous cyclic AMP is also thought to promote hyphal growth even at 37°C [28]. Most recently, exposure to the ubiquitous sugar N-Acetylglucosamine (GlcNAc) was shown to robustly accelerate the transition from yeast to hyphae at room temperature in both Histoplasma and the related thermally dimorphic fungus Blastomyces [32]. Interestingly, GlcNAc transporters are required for the yeast-to-hyphae transition even without the addition of exogenous GlcNAc, suggesting that endogenous GlcNAc, which is the building block of the polysaccharide chitin in the fungal cell wall, might be taken up by cells and sensed as a facilitator of hyphal growth.
Figure 2. Conditions and genes that drive phase-specific morphology.
Top panels identify conditions and genes that promote hyphal growth. Bottom panels identify conditions and genes that promote yeast-phase growth.
Regulatory circuits that drive yeast-phase growth
Although the dimorphic nature of Histoplasma biology has been known for decades, the molecular regulators and effectors of dimorphism have only started to be elucidated. Both genomic and genetic approaches have successfully identified components of the regulatory circuits that drive morphologic programs in response to temperature. A number of early studies identified phase-specific genes (i.e. those genes whose transcripts show enriched expression in one phase over the other), and made an initial, appealing connection between yeast morphology and the expression of virulence genes [33–36]. More recently, a number of whole-genome expression studies have been performed and up to 20% of the genome exhibits phase-specific gene expression [21,32,37–41]. Additionally, transcriptomics studies have enhanced annotation of the Histoplasma genome by providing experimental evidence that can be used to refine gene predictions and by identifying new genes [40,41]. Core phase-specific genes (those genes exhibiting phase-specific expression in multiple Histoplasma strains) are providing molecular insight on fundamental attributes of hyphae and yeast. Notably, transcripts encoding proteins with predicted signal sequences are more likely to show phase-specific expression. One family of putative secreted factors with homology to cystine-knot proteins is expanded in Histoplasma and shows yeast-specific expression [41]. It will be of interest to determine whether these phase-specific transcripts play a role in the biology of yeast cells during infection. Additionally, this study uncovered phase-dependent variation in transcript architecture, specifically 5’UTR length, implying that regulatory strategies other than simple transcript abundance might influence phase-specific biology. Finally, ribosomal profiling experiments coupled with RNAseq analysis in yeast and hyphae uncovered evidence for transcripts that exhibit phasespecific translation, suggesting that these factors might effect phase-specific biology [41]. Similarly, elucidation of the extracellular proteome of Histoplama yeast cells has uncovered secreted proteins that could be well positioned to influence host biology during infection [42].
Genetics [43] has been a powerful approach to identify regulators of both hyphal and yeast-phase growth in Histoplasma, and Figure 2 summarizes genes that are known to influence Histoplasma morphology. The first regulator of Histoplasma yeast-phase growth was identified by performing a genetic screen in the related fungus Blastomyces. A mutant hunt to identify genes required for expression of a Blastomyces yeast-specific reporter yielded a disruption in a histidine kinase gene that was subsequently named Drk1 (dimorphism-regulating kinase). Drk1 is required for yeast-phase morphology, yeast-phase gene expression, sporulation, and virulence in both Blastomyces and Histoplasma [44]. The precise signal transduced by Drk1 to stimulate yeast-phase growth is unknown, and how Drk1 integrates with other drivers of the yeast phase remains to be seen.
Subsequent to Drk1, two forward genetic screens identified three transcription factors, Ryp1, Ryp2, and Ryp3, that are required for yeast-phase growth in Histoplasma [38,45]. Mutants that lack Ryp1, 2, or 3 are trapped in the hyphal form independent of temperature, and Ryp2 and Ryp3 are also required for spore development and viability [38,39,45]. These key developmental regulators have orthologs in other fungi: Ryp1 orthologs are found throughout the fungal kingdom and include the Candida albicans Wor1, which regulates cell-type specification [46–48]. Ryp2 and Ryp3 are both Velvet family proteins, which regulate developmental transitions in filamentous fungi [49]. A fourth transcriptional regulator of yeast-phase growth, Ryp4 (a Zn(II)2Cys6 transcription factor), was identified because it shows yeast-specific expression and is a direct target of Ryp1, Ryp2, and Ryp3. Transcriptional profiling experiments revealed that Ryp1, Ryp2, Ryp3, and Ryp4 are each required for the vast majority of the normal transcription program at 37°C: in mutants lacking any of the Ryps, the cells fail to induce transcripts that are normally differentially expressed at 37°C and instead inappropriately express hyphal-specific genes [38,39]. Chromatin immunoprecipitation experiments were performed to identify direct targets of the Ryp transcription factors, which are enriched for yeast-specific genes but also include a few hyphal-specific genes, and genes that do not show differential expression between yeast and hyphae. Thus, Ryp factors associate upstream of some yeast-specific genes to enhance their expression at 37°C and also associate upstream of some hyphal-specific genes to repress their expression at 37°C. Notably, the majority of previously identified virulence factors were direct targets of the four Ryp transcription factors, indicating that these transcription factors link morphology and virulence traits in response to temperature.
Biochemical studies revealed that Ryp1, Ryp2, and Ryp3 form a complex [39]. Additionally, Ryp1, Ryp2, Ryp3, and Ryp4 associate with the upstream regulatory regions of Ryp1, Ryp2, and Ryp4, and each of the ryp mutants shows decreased expression of the other Ryp transcripts at 37°C [38,39,45]. A DNA binding motif was defined for Ryp1 and for the Ryp2-Ryp3 heterodimer [39], but the motif recognized by Ryp4 remains unknown. Nonetheless, the Ryp proteins form an interlocking network of transcription factors that regulate each other and common target genes important for yeast-phase growth and virulence. It is likely that the Ryp factors act in a positive-feedback loop at 37°C such that they accumulate and enable yeast-phase growth [39].
Work in Histoplasma and Aspergillus showed that the Velvet domain contained in Ryp2 and Ryp3 is a DNA-binding domain, and that Velvet family proteins can form homo- and heterodimers. [39,50]. Interestingly, there are two other Velvet proteins in Histoplasma, but whether they interact with Ryp2 and/or Ryp3 is unknown. Knockdown of the Velvet protein Vea1 results in the inability to make mating structures (cleistothecia) in Histoplasma. Additionally, silencing of Vea1 resulted in an accelerated switch from yeast to hyphae at room temperature, as well as failure to switch back to the yeast form when these mutant hyphae were transitioned back to 37°C growth. These data suggest that Vea1 may reinforce yeast-phase growth, although unlike ryp2 and ryp3 mutants, the Vea1-silenced strains were not locked in the hyphal form [51].
Regulatory circuits that drive hyphal growth
To fully understand how temperature regulates dimorphism, it is necessary to investigate both the pathways that drive yeast-phase growth in response to 37°C, as well as those that promote hyphal growth in response to room temperature. Presumably there may also be mechanisms that dampen yeast-phase growth at room temperature and inhibit hyphal growth at 37°C. Furthermore, the identification of regulators whose expression is sufficient to suppress the wild-type morphology program and instead drive the opposing program provides important insight into the gene circuits that control morphology. For example, the putative developmental regulator WET1 is a hyphal-specific gene that shows translational repression in yeast cells. Ectopic expression of WET1 at 37°C is sufficient to promote inappropriate hyphal growth, indicating that restricting the expression of WET1 in wildtype cells is critical for maintenance of the yeast program [41]. How mis-expression of Wet1 disrupts the normal yeast program is unknown.
Given the complexity of hyphal growth, there are likely to be genes that are required for proper hyphal morphology. The hyphal-specific gene MS8 was shown to be required for the normal morphology of hyphal cells: disruption of MS8 has no phenotype at 37°C but, at room temperature, the mutant displays aberrant, “zigzag” hyphae and colonies with altered morphology, size, and pigment [52]. The biochemical function of MS8 is unknown.
Much like the identification of the Ryp genes, genetic approaches are yielding insight into the pathways that drive hyphal growth in response to lower temperature. One pilot screen to identify yeast-locked mutants has been performed, resulting in identification of the signaling mucin Msb2 as required for hyphal growth [53]. Orthologs of the transmembrane protein Msb2 have been studied in Saccharomyces cerevisiae and Candida albicans, in which it stimulates a number of signaling pathways, including the high osmolarity glycerol (HOG) pathway in response to osmotic stress, as well as the filamentous growth pathway in response to nutrient limitation [54–61]. Unlike the Ryp transcription factors, which are required both for yeast-phase morphology and the majority of the gene expression program of cells grown at 37°C, Msb2 is required for hyphal formation in Histoplasma, but dispensable for the vast majority of the room temperature transcriptional program. Specifically, there are ~1870 genes that are differentially expressed by wild-type hyphae at room temperature, and most of these are also expressed by msb2 mutant yeast at room temperature [53]. Notably, approximately 165 genes fail to be induced in the msb2 mutant at room temperature, thereby defining a compact Msb2 regulon of “filament-associated” genes whose expression correlated with the ability to undergo hyphal growth. This gene set includes orthologs of a MAP kinase (Hog2) and an APSES transcription factor (Stu1). Like Msb2, Hog2 and Stu1 are required for efficient hyphal formation at room temperature, and ectopic expression of Stu1 is sufficient to drive hyphal formation even at 37°C in both wild-type cells and the msb2 mutant [53]. These data are consistent with a previous observation that Stu1 is required for the formation of some hyphal cell types and normal mycelia [62]. Since Msb2 is required for transcriptional induction of Hog2 and Stu1 at room temperature, these data suggest that Msb2 may trigger filament formation by signaling through Hog2 and Stu1.
Opposing regulatory pathways control dimorphism in response to temperature
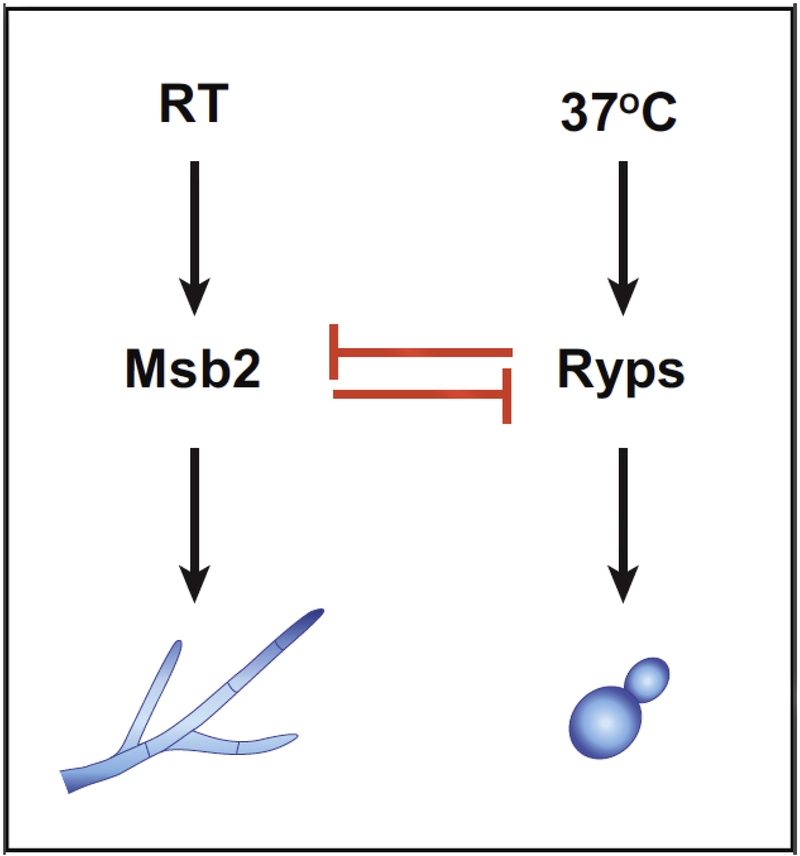
The Msb2 regulon also has implications for the yeast-phase expression program. Interestingly, of the ~1100 yeast-specific genes whose expression is reduced as yeast cells transition from 37°C to room temperature and undergo hyphal morphogenesis, there are 40 genes whose expression is inappropriately maintained in the yeast-locked msb2 mutant at room temperature. These genes include previously identified virulence factors and their expression is particularly interesting since it seems to be uncoupled from temperature but linked to yeast-phase morphology [53]. Notably, this set of 40 genes is enriched for direct transcription targets of the Ryp proteins, suggesting that the Ryp proteins are present in the msb2 mutant even at room temperature. Rodriguez et al. confirmed that whereas the abundance of Ryp proteins at room temperature is markedly decreased in wild-type cells, it is maintained even at room temperature in the msb2 mutant. These data indicate that the Msb2 pathway is required to inhibit Ryp accumulation at room temperature. Conversely, at 37°C, the Ryp3 transcription factor associates with the upstream region of the MSB2 gene and turns down its expression [39,53], indicating that the Ryp circuit antagonizes the Msb2 pathway at high temperature. Thus Histoplasma transitions between two different states in response to temperature by means of opposing regulatory pathways (Figure 3) [53]. Future work will elucidate the mechanisms that allow temperature to toggle the morphology switch in favor of the Ryp pathway at 37°C or the Msb2 pathway at room temperature.
Figure 3. The Msb2 and Ryp pathways oppose each other to regulate morphology in response to temperature.
As described in the text, the Msb2 and Ryp pathways are mutually antagonistic. The Msb2 pathway predominates at room temperature and the Ryp pathway predominates at 37°C.
Conclusions
Genomic and genetic approaches have yielded molecular insight into how temperature regulates morphology and virulence in Histoplasma. Several key molecular regulators that control dimorphism have been identified, and future work is likely to reveal key thermosensors whose activity and/or accumulation are intrinsically responsive to temperature. Ultimately, the decision to switch between yeast and hyphal forms is critical for survival in the environment and in the mammalian host. Thus it is highly likely that multiple, redundant mechanisms integrate a number of key signals to robustly determine the optimal morphology and expression program for a given environment.
Acknowledgements
This work was supported by the National Institute of Allergy and Infectious Diseases (R01AI136735, 5R01AI066224, 1R01AI146584). I thank Davina Hocking Murray for assistance with figures and Mark Voorhies and other members of the Sil lab for many helpful discussions.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Bahr NC, Antinori S, Wheat LJ, Sarosi GA: Histoplasmosis infections worldwide: thinking outside of the Ohio River valley. Curr Trop Med Rep 2015, 2:70–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC: Hidden killers: human fungal infections. Sci Transl Med 2012, 4:165rv113. [DOI] [PubMed] [Google Scholar]
- 3.Kasuga T, White TJ, Koenig G, McEwen J, Restrepo A, Castaneda E, Da Silva Lacaz C, Heins-Vaccari EM, De Freitas RS, Zancope-Oliveira RM, et al. : Phylogeography of the fungal pathogen Histoplasma capsulatum. Mol Ecol 2003, 12:3383–3401. [DOI] [PubMed] [Google Scholar]
- 4.Maxwell CS, Sepulveda VE, Turissini DA, Goldman WE, Matute DR: Recent admixture between species of the fungal pathogen Histoplasma. Evol Lett 2018, 2:210–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sepulveda VE, Marquez R, Turissini DA, Goldman WE, Matute DR: Genome Sequences Reveal Cryptic Speciation in the Human Pathogen Histoplasma capsulatum. MBio 2017, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teixeira Mde M, Patane JS, Taylor ML, Gomez BL, Theodoro RC, de Hoog S, Engelthaler DM, Zancope-Oliveira RM, Felipe MS, Barker BM: Worldwide Phylogenetic Distributions and Population Dynamics of the Genus Histoplasma. PLoS Negl Trop Dis 2016, 10:e0004732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adenis AA, Valdes A, Cropet C, McCotter OZ, Derado G, Couppie P, Chiller T, Nacher M: Burden of HIV- associated histoplasmosis compared with tuberculosis in Latin America: a modelling study. Lancet Infect Dis 2018, 18:1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonifaz A, Vazquez-Gonzalez D, Perusquia-Ortiz AM: Endemic systemic mycoses: coccidioidomycosis, histoplasmosis, paracoccidioidomycosis and blastomycosis. J Dtsch Dermatol Ges 2011, 9:705–714; quiz 715. [DOI] [PubMed] [Google Scholar]
- 9.Klein BS, Tebbets B: Dimorphism and virulence in fungi. Curr Opin Microbiol 2007, 10:314–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sil A, Andrianopoulos A: Thermally Dimorphic Human Fungal Pathogens--Polyphyletic Pathogens with a Convergent Pathogenicity Trait. Cold Spring Harb Perspect Med 2014, 5:a019794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Retallack DM, Woods JP: Molecular epidemiology, pathogenesis, and genetics of the dimorphic fungus Histoplasma capsulatum. Microbes Infect 1999, 1:817–825. [DOI] [PubMed] [Google Scholar]
- 12.Kauffman CA: Histoplasmosis: a clinical and laboratory update. Clinical microbiology reviews 2007,20:115–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pine L: Morphological and Physiological Characteristics of Histoplasma capsulatum. Springfield, Ill: Charles C. Thomas; 1960. [Google Scholar]
- 14.Anderson KL, Marcus S: Sporulation characteristics of Histoplasma capsulatum. Mycopathol Mycol Appl 1968, 36:179–187. [DOI] [PubMed] [Google Scholar]
- 15.Artis D, Baum GL: Tuberculate Spore Formation by Thirty-Two Strains of Histoplasma Capsulatum. Mycopathol Mycol Appl 1963, 21:29–35. [DOI] [PubMed] [Google Scholar]
- 16.Garrison RG, Lane JW: Scanning-beam electron microscopy of the conidia of the brown and albino filamentous varieties of Histoplasma capsulatum. Mycopathol Mycol Appl 1973, 49:185–191. [DOI] [PubMed] [Google Scholar]
- 17.Garrison RG, Boyd KS: The fine structure of microconidial germination and vegetative cells of Histoplasma capsulatum. Ann Microbiol (Paris) 1977, 128:135–149. [PubMed] [Google Scholar]
- 18.Garrison RG, Boyd KS: Electron microscopy of yeastlike cell development from the microconidium of Histoplasma capsulatum. J Bacteriol 1978, 133:345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goos RD: Germination of the macroconidia of Histoplasma capsulatum. Mycologia 1964, 56:662–671. [Google Scholar]
- 20.Neilsen GE, Evans RE: A study of the sporulation of Histoplasma capsulatum. Journal of Bacteriology 1964, 68:261–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inglis DO, Voorhies M, Hocking Murray DR, Sil A: Comparative transcriptomics of infectious spores from the fungal pathogen Histoplasma capsulatum reveals a core set of transcripts that specify infectious and pathogenic states. Eukaryot Cell 2013, 12:828–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bullock WE: Interactions between human phagocytic cells and Histoplasma capsulatum. Arch Med Res 1993, 24:219–223. [PubMed] [Google Scholar]
- 23.Eissenberg LG, Goldman WE: Histoplasma variation and adaptive strategies for parasitism: new perspectives on histoplasmosis. Clin Microbiol Rev 1991, 4:411–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woods JP: Knocking on the right door and making a comfortable home: Histoplasma capsulatum intracellular pathogenesis. Curr Opin Microbiol 2003, 6:327–331. [DOI] [PubMed] [Google Scholar]
- 25.Shen Q, Rappleye CA: Differentiation of the fungus Histoplasma capsulatum into a pathogen of phagocytes. Curr Opin Microbiol 2017, 40:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]; **This paper is a comprehensive review of virulence strategies used by Histoplasma to survive and thrive in the host, and is an excellent resource to understand yeast-specific traits.
- 26.Maresca B, Medoff G, Schlessinger D, Kobayashi GS: Regulation of dimorphism in the pathogenic fungus Histoplasma capsulatum. Nature 1977, 266:447–448. [DOI] [PubMed] [Google Scholar]
- 27.Pine L, Webster RE: Conversion in strains of Histoplasma capsulatum. J Bacteriol 1962, 83:149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sacco M, Maresca B, Kumar BV, Kobayashi GS, Medoff G: Temperature- and cyclic nucleotide-induced phase transitions of Histoplasma capsulatum. J Bacteriol 1981, 146:117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maresca B, Lambowitz AM, Kumar VB, Grant GA, Kobayashi GS, Medoff G: Role of cysteine in regulating morphogenesis and mitochondrial activity in the dimorphic fungus Histoplasma capsulatum. Proc Natl Acad Sci U S A 1981, 78:4596–4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Medoff G, Kobayashi GS, Painter A, Travis S: Morphogenesis and pathogenicity of Histoplasma capsulatum. Infect Immun 1987, 55:1355–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Medoff G, Sacco M, Maresca B, Schlessinger D, Painter A, Kobayashi GS, Carratu L: Irreversible block of the mycelial-to-yeast phase transition of Histoplasma capsulatum. Science 1986, 231:476–479. [DOI] [PubMed] [Google Scholar]; **This paper represents one of the early seminal studies addressing phase transitions in Histoplasma capsulatum.
- 32.Gilmore SA, Naseem S, Konopka JB, Sil A: N-acetylglucosamine (GlcNAc) triggers a rapid, temperature-responsive morphogenetic program in thermally dimorphic fungi. PLoS Genet 2013, 9:e1003799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abidi FE: Characterization of a yeast phase specific gene (yps 21:E-9) and identification of a DNA binding protein from the dimorphic pathogenic fungus, Histoplasma capsulatum.. Ph.D thesis. University of Minnesota; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keath EJ, Painter AA, Kobayashi GS, Medoff G: Variable expression of a yeast-phase-specific gene in Histoplasma capsulatum strains differing in thermotolerance and virulence. Infect Immun 1989, 57:1384–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Batanghari JW: Calcium Dependence and Binding in Cultures of Histoplasma capsulatum. Infect Immun 1997, 65:5257–5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patel JB, Batanghari JW, Goldman WE: Probing the yeast phase-specific expression of the CBP1 gene in Histoplasma capsulatum. J Bacteriol 1998, 180:1786–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hwang L, Hocking-Murray D, Bahrami AK, Andersson M, Rine J, Sil A: Identifying phase-specific genes in the fungal pathogen Histoplasma capsulatum using a genomic shotgun microarray. Mol Biol Cell 2003, 14:2314–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen VQ, Sil A: Temperature-induced switch to the pathogenic yeast form of Histoplasma capsulatum requires Ryp1, a conserved transcriptional regulator. Proc Natl Acad Sci U S A 2008, 105:4880–4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beyhan S, Gutierrez M, Voorhies M, Sil A: A temperature-responsive network links cell shape and virulence traits in a primary fungal pathogen. PLoS Biol 2013, 11:e1001614. [DOI] [PMC free article] [PubMed] [Google Scholar]; **This study is the most comprehensive analysis of the interlocking network of Ryp factors that drives yeastphase growth in Histoplasma.
- 40.Edwards JA, Chen C, Kemski MM, Hu J, Mitchell TK, Rappleye CA: Histoplasma yeast and mycelial transcriptomes reveal pathogenic-phase and lineage-specific gene expression profiles. BMC Genomics 2013, 14:695. [DOI] [PMC free article] [PubMed] [Google Scholar]; *This study is the first RNAseq transcriptomics analysis of Histoplasma yeast and hyphae.
- 41.Gilmore SA, Voorhies M, Gebhart D, Sil A: Genome-Wide Reprogramming of Transcript Architecture by Temperature Specifies the Developmental States of the Human Pathogen Histoplasma. PLoS Genet 2015, 11:e1005395. [DOI] [PMC free article] [PubMed] [Google Scholar]; *This study is the first ribosomal profiling study of Histoplasma yeast and hyphae as well as extensive RNAseq analysis of phase-specific transcripts.
- 42.Holbrook ED, Edwards JA, Youseff BH, Rappleye CA: Definition of the extracellular proteome of pathogenic-phase Histoplasma capsulatum. J Proteome Res 2011, 10:1929–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sullivan TD, Rooney PJ, Klein BS: Agrobacterium tumefaciens integrates transfer DNA into single chromosomal sites of dimorphic fungi and yields homokaryotic progeny from multinucleate yeast. Eukaryot Cell 2002, 1:895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nemecek JC, Wuthrich M, Klein BS: Global control of dimorphism and virulence in fungi. Science 2006, 312:583–588. [DOI] [PubMed] [Google Scholar]; **This study is a classic example of the fundamental use of genetics to identify the dimorphism-regulating kinase in Blastomyces and Histoplasma.
- 45.Webster RH, Sil A: Conserved factors Ryp2 and Ryp3 control cell morphology and infectious spore formation in the fungal pathogen Histoplasma capsulatum. Proc Natl Acad Sci U S A 2008, 105:14573–14578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang G, Wang H, Chou S, Nie X, Chen J, Liu H: Bistable expression of WOR1, a master regulator of white-opaque switching in Candida albicans. Proc Natl Acad Sci U S A 2006, 103:12813–12818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Srikantha T, Borneman AR, Daniels KJ, Pujol C, Wu W, Seringhaus MR, Gerstein M, Yi S, Snyder M, Soll DR: TOS9 regulates white-opaque switching in Candida albicans. Eukaryot Cell 2006, 5:1674–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zordan RE, Miller MG, Galgoczy DJ, Tuch BB, Johnson AD: Interlocking transcriptional feedback loops control white-opaque switching in Candida albicans. PLoS Biol 2007, 5:e256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bayram O, Braus GH: Coordination of secondary metabolism and development in fungi: the velvet family of regulatory proteins. FEMS Microbiol Rev 2012, 36:1–24. [DOI] [PubMed] [Google Scholar]
- 50.Ahmed YL, Gerke J, Park HS, Bayram O, Neumann P, Ni M, Dickmanns A, Kim SC, Yu JH, Braus GH, et al. : The velvet family of fungal regulators contains a DNA-binding domain structurally similar to NF-kappaB. PLoS Biol 2013, 11:e1001750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Laskowski-Peak MC, Calvo AM, Rohrssen J, Smulian AG: VEA1 is required for cleistothecial formation and virulence in Histoplasma capsulatum. Fungal Genet Biol 2012, 49:838–846. [DOI] [PubMed] [Google Scholar]
- 52.Tian X, Shearer G Jr.: The mold-specific MS8 gene is required for normal hypha formation in the dimorphic pathogenic fungus Histoplasma capsulatum. Eukaryot Cell 2002, 1:249–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rodriguez L, Voorhies M, Gilmore SA, Beyhan S, Myint A, Sil A: Opposing signaling pathways regulate morphology in response to temperature in the fungal pathogen Histoplasma capsulatum. PLoS Biol 2019, September 30;17(9):e3000168. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]; **This study defines the role of Msb2 in hyphal growth and shows that the Ryp pathway and the Msb2 pathway antagonize each other to establish temperature regulation of cell shape.
- 54.Bender A, Pringle JR: A Ser/Thr-rich multicopy suppressor of a cdc24 bud emergence defect. Yeast 1992, 8:315–323. [DOI] [PubMed] [Google Scholar]
- 55.Cullen PJ: Post-translational regulation of signaling mucins. Curr Opin Struct Biol 2011, 21:590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cullen PJ, Sabbagh W Jr., Graham E, Irick MM, van Olden EK, Neal C, Delrow J, Bardwell L, Sprague GF Jr.: A signaling mucin at the head of the Cdc42- and MAPK-dependent filamentous growth pathway in yeast. Genes Dev 2004, 18:1695–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.O’Rourke SM, Herskowitz I: A Third Osmosensing Branch in Saccharomyces cerevisiae Requires the Msb2 Protein and Functions in Parallel with the Sho1 Branch. Molecular and Cellular Biology 2002, 22:4739–4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roman E, Cottier F, Ernst JF, Pla J: Msb2 signaling mucin controls activation of Cek1 mitogen- activated protein kinase in Candida albicans. Eukaryot Cell 2009, 8:1235–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pitoniak A, Birkaya B, Dionne HM, Vadaie N, Cullen PJ: The signaling mucins Msb2 and Hkr1 differentially regulate the filamentation mitogen-activated protein kinase pathway and contribute to a multimodal response. Mol Biol Cell 2009, 20:3101–3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tatebayashi K, Tanaka K, Yang HY, Yamamoto K, Matsushita Y, Tomida T, Imai M, Saito H: Transmembrane mucins Hkr1 and Msb2 are putative osmosensors in the SHO1 branch of yeast HOG pathway. EMBO J 2007, 26:3521–3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vadaie N, Dionne H, Akajagbor DS, Nickerson SR, Krysan DJ, Cullen PJ: Cleavage of the signaling mucin Msb2 by the aspartyl protease Yps1 is required for MAPK activation in yeast. J Cell Biol 2008, 181:1073–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Longo LVG, Ray SC, Puccia R, Rappleye CA: Characterization of the APSES-family transcriptional regulators of Histoplasma capsulatum. FEMS Yeast Res 2018, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]