Abstract
Carboxyl-containing metabolites, such as bile acids and fatty acids, have many important functions and microbiota is involved in the production of them. In the previous study, we found that the chronic kidney disease (CKD) model mice raised under germ-free conditions provided more severe renal damage than the mice with commensal microbiota. However, the precise influence by the microbiome and carboxyl-containing metabolites to the renal functions is unknown. In this study, we aimed to develop a novel chemical isotope labeling-LC-MS/MS method using the 2-picolylamine and its isotopologue and applied the analysis of effects of microbiome and CKD pathophysiology. The developed semi-quantitative method provided the high accuracy not inferior to the absolute quantification. By comparing of four groups of mice, we found that both microbiota and renal function can alter the composition and level of these metabolites in both plasma and intestine. In particular, the intestinal level of indole-3-acetic acid, short-chain fatty acids and n-3 type of polyunsaturated fatty acid, which play important roles in the endothelial barrier function, were significantly lower in germ-free conditions mice with renal failure. Accordingly, it is suggested these metabolites might have a renoprotective effect on CKD by suppressing epithelial barrier disruption.
Subject terms: Metabolomics, Kidney
Introduction
The mammalian gastrointestinal tract contains trillions of microorganisms, referred to collectively as gut microbiota1. The gut microbiota coevolves with the host for a mutually beneficial coexistence by sharing numerous metabolites, including that cannot produce each other. The gut microbiota not only produces certain bile acids as well as short-chain fatty acids (SCFA) and indole-3-acetic acid (IAA) but also affect a fatty acid metabolism of the host2–5. Bile acids, IAA and fatty acids are groups of carboxylic acids and play important roles in many metabolic pathways6–8. The production of these metabolites by microbiomes contributes to the host metabolic phenotype and hence to disease risk3,9,10. Because the kidneys directly impact circulating metabolite levels, declining renal function leads to the retention of enormous metabolites, including microbiota-derived uremic toxins11. Also, some recent studies have shown that gut microbiota contributes to the gut-kidney axis on the various renal disease by affecting the tryptophan12, melamine13, and volatile organic compound metabolism14. Previously, we have shown that some metabolites having a carboxyl group, such as cholic acid (CA), hippuric acid, and, phenaceturic acid, are mainly produced by the microbiota and these plasma levels were greatly elevated in chronic kidney disease (CKD) model mice raised under specific pathogen-free (SPF) conditions which have normal commensal microbiota15. On the other hand, it was not observed in germ-free (GF) mice with renal failure (RF) induced by adenine-feeding. In addition, the renal dysfunction in GF-RF mice was more severe than that in SPF-RF mice. These findings allow us to speculate that carboxyl-containing metabolites play a pathological role in the progression and/or protection of CKD. However, the precise influence of the microbial metabolites having carboxyl groups to the renal functions is obscure. Thus, comprehensive, simultaneous analyses of carboxyl-containing compounds can provide a greater understanding of the relationship between CKD pathology and gut microbiota.
Liquid chromatography/tandem mass spectrometry (LC-MS/MS) is becoming a standard tool for metabolomics study because it offers high sensitivity and selectivity as well as provides structural information useful for compound identification16,17. A large number of microbiota-related uremic solutes have been identified by targeted or nontargeted metabolomics conducted on both animal models and patients with CKD18–21. However, the development of high-coverage method with accurate quantification is challenging because the concentration and physical properties of metabolites are diverse. In addition, matrix effects caused by coexisting components often loses the accuracy and reliability of analytes and affects the result.
To increase coverage and improve the accuracy and precision, chemical isotope labeling (CIL) approaches has been reported22–25. In addition to the high sensitivity by proton affinity group based on the group, matrix effect could be canceled for the simultaneous ionization processing both of light reagent derivative and heavy one at the same time. Accordingly, the differential semi-quantitative analysis of metabolites for two comparative samples can be performed22. CIL also give the more retention in reversed-phase LC which is useful for chromatographic separation. Previously, there are various CIL reagents have been reported26–31. However, these reagents are relatively expensive and time-consuming for reaction, which makes it difficult to use them in routine analyses. Accordingly, the development of a rapid, inexpensive, and high-throughput method is needed for the biological and physiological experiments need many samples.
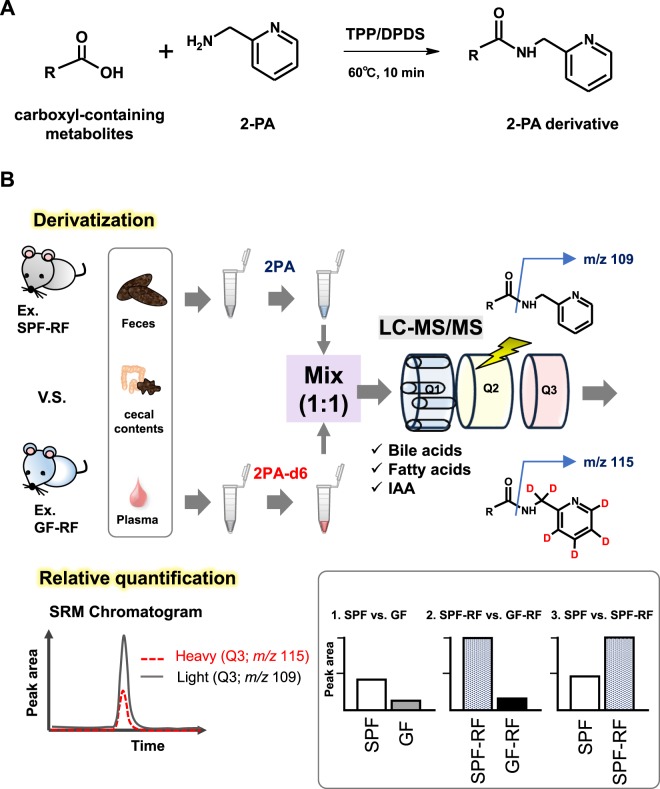
In this study, to perform the comprehensive and accurate analysis of carboxyl-containing metabolites for plasma and intestinal samples in CKD model mice with or without the conditions of germ-free, we developed a CIL-LC-MS/MS method for carboxylic acid tagging with 2-picolylamine (2PA) and 2-picolylamine-d6 (2PA-d6). One of the significances of this approach is 2PA-d6 can easily synthesize through only two steps by inexpensive reagents (Fig. 1) in first. Additionally, 2PA derivatization reaction proceeds as short as 10 min (Fig. 2A). To confirm the performance and applicability of the developed method for biological samples, we applied the analysis of bile acids, fatty acids, and IAA in plasma samples. In next, the accuracy of this method was compared with the absolute quantitative methods due to analysis of propionic acid (C3:0) and butyric acid (C4:0). Finally, to clarify the impact of microbiota in CKD, we compared carboxyl-containing metabolites levels in CKD model mice raised under GF conditions (GF-RF) or a normal microbiota (SPF-RF) (Fig. 2B). The developed CIL-LC-MS/MS-based approach was successfully used for comprehensive relative quantification of carboxyl-containing metabolites in plasma, feces, and cecal contents from mice and suggested that the novel features related to the microbiome and CKD pathology.
Figure 1.
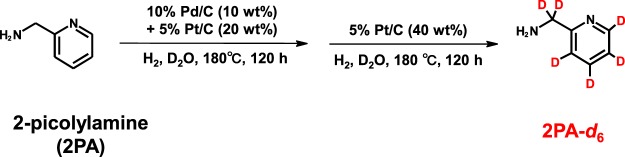
Synthesis of 2-picolylamine-d6 from 2-picolylamine.
Figure 2.
(A) 2PA derivatization reaction of carboxylic acid-containing metabolites. TPP: triphenylphosphine, DPDS: 2,2′-dipyridyl disulfide (B) Workflow of differential analysis of carboxyl-containing metabolites for plasma, feces, and cecal contents from 4 groups of mice.
Results
Selection of derivatization reagents and synthesis of 2PA-d6
A simple, rapid, and inexpensive CIL-LC-MS/MS method for comprehensive profiling of carboxyl-containing metabolites could be useful for the analysis of biological and clinical samples. Because 2PA derivatization has been successfully applied for the analyses of carboxylic acids32–34, 2PA and its labeled compound, 2PA-d6 were chosen as derivatization reagents. Sajiki et al. have reported that post-synthetic H–D exchange methods for various organic compounds in D2O under an atmosphere of H2 in the presence of palladium-on-carbon (Pd/C) and platinum-on-carbon (Pt/C)35,36, and we applied it for the synthesis of 2PA-d6. As a result, we could complete the second step reaction (Fig. 1), the combination of Pd/C and Pt/C and subsequent the reaction with Pt/C.
LC optimization
The chromatographic separation is critical for the analysis of many of the structural isomers of fatty acids and bile acids. Separation efficiency was evaluated under varying ionic strength, pH, and elution gradient conditions using some SCFA as representative isomers. As a result, addition of 0.1% acetic acid (v/v) in mobile phase B provided a shaper symmetric peak and enhanced resolution. In other, 0.3% of acetic acid addition or 0.1% of formic acid addition were not effective for the retention and separation for isomers such as isobutyric acid and butyric acid (Supplementary Fig. S1).
MS/MS optimization
2PA derivatization have a highly ESI-favorable moiety, and reacts easily with carboxylic acids to form the amide derivatives32. The amide group is well-known fragmentable moieties in low-energy collision induced dissociation and therefore, are expected to generate a characteristic product ion suitable for the SRM detection. As shown in Supplementary Table S1, the 2PA-derivatives also provided intense protonated molecules as base peaks in the positive ESI-MS. Using these ions as the precursor ions, the most abundant product ions were detected at m/z 109 for 2PA or m/z 115 for 2PA-d6, except for the propanoic acid-d5 and butyric acid-d7 derivatives, whose product ions were [2PA (C5H4NCH2NH2) + D]+ at m/z 110 similar to the previous report34. Compared to the peak intensities of intact propanoic acid and butyric acid in the negative-ion mode, the 2PA-derivatives yielded the highest ionization efficiency approximately 100-fold times sensitivity (Supplementary Fig. S2).
Evaluation of relative quantification of carboxyl-containing metabolites from standard-mixture and pooled plasma and QC plasma
In processing the method development, we recognized that the much leaching of C3:0 and C4:0 from disposable plastic tubes37,38. As a result of testing many types of tubes (Supplementary Fig. S3), we finally selected the siliconized-plastic tube, that elute less C3:0 and C4:0 than others, as a container for our procedure.
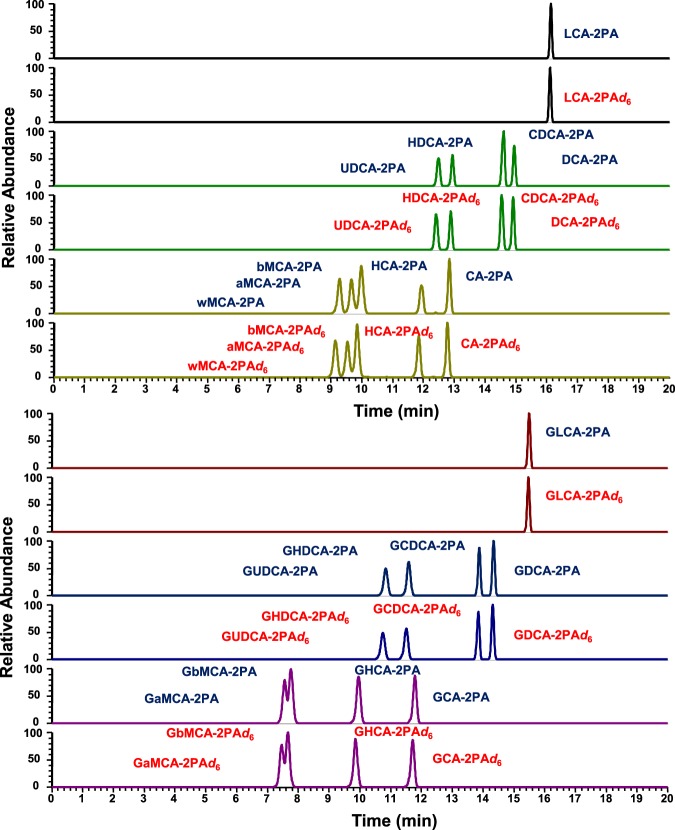
In next, to confirm the quantitative performance of the method, we prepared an equal amount of each samples labeled with light (2PA) or heavy (2PA-d6) labeling reagents and analyzed them as mixture. For testing, five different samples (standard-mixture solutions, human plasma, mouse plasma, human QC plasma, and mouse QC plasma) were used. The detected peak area ratios of each 2PA-d6/2PA-labeled metabolites were investigated, and three independent samples were prepared for each group and analyzed in triplicate. Typical SRM chromatograms were shown in Fig. 3 (of bile acids) and Supplementary Fig. S4 (of fatty acids and IAA). The calculated peak area ratios were summarized in Table 1. However, some bile acids in plasma could not be detected due to low endogenous concentrations, the standard-mixture solutions and both QC plasma made with addition of standards gave peak pairs with almost equally peak area. In addition, most of the fatty acids and IAA also provided the equal ratio. In our approach, because the almost 2PA- and 2PA-d6 derivatives were detected at the same retention times (Figs. 3 and S4), the matrix effect are probably canceled. Accordingly, the errors from theoretical values might be caused by isotopic effect of 6 of deuterium atoms. In summary, this fundamental experiment showed that the quantitative accuracy of the developed method and usefulness as an analytical platform for carboxyl-containing metabolites.
Figure 3.
Selected reaction monitoring (SRM) chromatograms of mixture of 18 bile acid standards. The standard mixture was labeled with either 2PA or 2PA-d6, and 1:1 mixture of the labeled compounds was analyzed by RPLC triple-quadrupole mass spectrometry. LCA: lithocholic acid, UDCA: ursodeoxycholic acid, HDCA: hyodeoxycholic acid, CDCA: chenodeoxycholic acid, DCA: deoxycholic acid, wMCA: ω-muricholic acid, aMCA: α-muricholic acid, bMCA: β-muricholic acid, HCA: hyocholic acid, CA: cholic acid, GLCA: glycolithocholic acid, GUDCA: glycoursodeoxycholic acid, GHDCA: glycohyodeoxycholic acid, GCDCA: glycochenodeoxycholic acid, GDCA: glycodeoxycholic acid, GaMCA: glyco-α-muricholic acid, GbMCA: glyco-β-muricholic acid, GHCA: glycohyocholic acid, GCA: glycocholic acid.
Table 1.
Results of peak area ratio (2PA-d6/ 2PA) (n = 3; mean ± standard deviation, %).
| Analyte |
m/z (2PA) |
m/z (2PA-d6) |
Retention Time (min) |
Std. mixture | Human plasma | Mouse plasma | QC plasma (human) |
QC plasma (mouse) |
|---|---|---|---|---|---|---|---|---|
| Propanoic acid | 165.1 | 171.1 | 2.5 | 0.95 ± 0.11 | 0.92 ± 0.04 | 0.98 ± 0.04 | 0.88 ± 0.04 | 0.87 ± 0.01 |
| Butyric acid | 179.1 | 185.1 | 5.1 | 0.92 ± 0.13 | 0.87 ± 0.04 | 1.04 ± 0.11 | 0.83 ± 0.07 | 0.88 ± 0.02 |
| Indole-3-acetic acid | 266.1 | 272.2 | 11.6 | 0.92 ± 0.16 | 1.33 ± 0.13 | 1.37 ± 0.10 | 1.25 ± 0.17 | 1.16 ± 0.15 |
| Lauric acid | 291.1 | 297.2 | 18.6 | 0.99 ± 0.03 | 0.96 ± 0.03 | 1.29 ± 0.18 | 0.91 ± 0.12 | 1.02 ± 0.09 |
| Stearidonic acid | 367.3 | 373.1 | 19.9 | 1.27 ± 0.07 | 1.18 ± 0.02 | 1.09 ± 0.14 | 1.17 ± 0.13 | 1.22 ± 0.25 |
| Eicosapentaenoic acid | 393.2 | 399.3 | 21.1 | 1.37 ± 0.06 | 1.09 ± 0.03 | 0.99 ± 0.17 | 1.10 ± 0.17 | 1.14 ± 0.30 |
| α-Linolenic acid | 369.3 | 375.2 | 21.2 | 1.37 ± 0.04 | 1.12 ± 0.05 | 0.94 ± 0.15 | 1.12 ± 0.18 | 1.10 ± 0.25 |
| Myristic acid | 319.3 | 325.3 | 21.3 | 0.96 ± 0.05 | 1.03 ± 0.12 | 0.95 ± 0.11 | 0.84 ± 0.19 | 0.80 ± 0.19 |
| γ-Linolenic acid | 369.3 | 375.2 | 21.5 | 1.28 ± 0.05 | 1.08 ± 0.03 | 0.89 ± 0.16 | 1.06 ± 0.18 | 1.06 ± 0.27 |
| Palmitoleic acid | 345.3 | 351.2 | 22.0 | 1.46 ± 0.03 | 1.18 ± 0.05 | 1.04 ± 0.14 | 1.19 ± 0.22 | 1.20 ± 0.26 |
| Docosahexaenoic acid | 419.3 | 425.4 | 22.5 | 1.20 ± 0.06 | 0.84 ± 0.06 | 0.74 ± 0.06 | 0.85 ± 0.17 | 0.86 ± 0.27 |
| Arachidonic acid | 395.3 | 401.3 | 22.8 | 1.21 ± 0.06 | 1.10 ± 0.04 | 0.97 ± 0.06 | 1.11 ± 0.25 | 1.12 ± 0.35 |
| Linoleic acid | 371.3 | 377.3 | 22.9 | 1.05 ± 0.03 | 0.93 ± 0.04 | 0.90 ± 0.11 | 0.93 ± 0.20 | 0.91 ± 0.24 |
| Docosapentaenoic acid | 421.3 | 427.4 | 23.3 | 1.06 ± 0.07 | 0.78 ± 0.06 | 0.74 ± 0.13 | 0.78 ± 0.20 | 0.77 ± 0.26 |
| Dihomo-γ-Linolenic acid | 397.3 | 403.4 | 24.0 | 1.46 ± 0.03 | 1.10 ± 0.09 | 0.88 ± 0.08 | 1.13 ± 0.28 | 1.06 ± 0.35 |
| Palmitic acid | 345.3 | 351.2 | 24.4 | 0.96 ± 0.10 | 0.90 ± 0.09 | 0.78 ± 0.07 | 0.92 ± 0.27 | 0.81 ± 0.18 |
| Adrenic acid | 423.4 | 429.4 | 25.0 | 0.92 ± 0.12 | 0.69 ± 0.08 | 0.61 ± 0.10 | 0.70 ± 0.19 | 0.63 ± 0.24 |
| Oleic acid | 373.1 | 379.4 | 25.1 | 1.11 ± 0.10 | 0.79 ± 0.06 | 0.71 ± 0.11 | 0.82 ± 0.22 | 0.74 ± 0.21 |
| Stearic acid | 375.3 | 381.4 | 27.6 | 1.04 ± 0.07 | 1.13 ± 0.29 | 0.83 ± 0.07 | 1.16 ± 0.43 | 0.91 ± 0.24 |
| Arachidic acid | 403.4 | 409.4 | 29.6 | 1.39 ± 0.16 | 1.20 ± 0.27 | 0.83 ± 0.04 | 1.26 ± 0.45 | 0.94 ± 0.23 |
| Nervonic acid | 457.4 | 463.5 | 30.3 | 1.02 ± 0.08 | 0.72 ± 0.09 | 0.60 ± 0.11 | 0.71 ± 0.18 | 0.62 ± 0.15 |
| Lignoceric acid | 459.4 | 465.5 | 31.1 | 1.20 ± 0.28 | 0.75 ± 0.28 | 0.82 ± 0.04 | 0.88 ± 0.29 | 0.79 ± 0.11 |
| Glyco-α/β-muricholic acid | 556.6 | 562.6 | 7.3 | 0.88 ± 0.04 | ND | ND | 0.82 ± 0.06 | 0.88 ± 0.03 |
| ω-muricholic acid | 499.5 | 505.5 | 8.7 | 0.91 ± 0.03 | ND | ND | 0.84 ± 0.06 | 0.85 ± 0.01 |
| α-muricholic acid | 499.5 | 505.5 | 9.1 | 1.00 ± 0.05 | ND | ND | 0.88 ± 0.04 | 0.87 ± 0.04 |
| β-muricholic acid | 499.5 | 505.5 | 9.4 | 0.90 ± 0.03 | ND | 0.82 ± 0.12 | 0.85 ± 0.02 | 0.83 ± 0.04 |
| Glycohyocholic acid | 556.6 | 562.6 | 9.4 | 0.95 ± 0.05 | ND | ND | 0.97 ± 0.06 | 0.89 ± 0.07 |
| Glycoursodeoxycholic acid | 540.6 | 546.6 | 11.1 | 1.04 ± 0.06 | ND | ND | 0.90 ± 0.12 | 0.93 ± 0.02 |
| Glycohyodeoxycholic acid | 540.6 | 546.6 | 11.1 | 1.01 ± 0.04 | ND | ND | 0.91 ± 0.12 | 0.93 ± 0.02 |
| Glycocholic acid | 556.6 | 562.6 | 11.3 | 1.04 ± 0.05 | 1.24 ± 0.31 | ND | 1.05 ± 0.13 | 1.00 ± 0.04 |
| Hyocholic acid | 499.5 | 505.5 | 11.5 | 0.91 ± 0.04 | ND | ND | 0.90 ± 0.10 | 0.88 ± 0.04 |
| Ursodeoxycholic acid | 483.5 | 489.5 | 12.1 | 0.95 ± 0.04 | 0.95 ± 0.11 | ND | 0.86 ± 0.04 | 0.87 ± 0.01 |
| Cholic acid | 499.5 | 505.5 | 12.5 | 0.96 ± 0.04 | 0.87 ± 0.06 | 0.79 ± 0.05 | 0.84 ± 0.03 | 0.89 ± 0.04 |
| Hyodeoxycholic acid | 483.5 | 489.5 | 12.6 | 0.84 ± 0.01 | 0.92 ± 0.16 | ND | 0.82 ± 0.03 | 0.84 ± 0.03 |
| Glycochenodeoxycholic acid | 540.6 | 546.6 | 14.1 | 1.08 ± 0.15 | 0.55 ± 0.20 | ND | 0.87 ± 0.10 | 0.71 ± 0.12 |
| Glycodeoxycholic acid | 540.6 | 546.6 | 14.1 | 0.98 ± 0.04 | 0.93 ± 0.04 | ND | 0.91 ± 0.07 | 0.93 ± 0.03 |
| Chenodeoxycholic acid | 483.5 | 489.5 | 14.1 | 0.97 ± 0.05 | 0.88 ± 0.03 | ND | 0.87 ± 0.03 | 0.91 ± 0.02 |
| Deoxycholic acid | 483.5 | 489.5 | 14.8 | 0.97 ± 0.04 | 0.90 ± 0.06 | 0.99 ± 0.10 | 0.92 ± 0.06 | 0.93 ± 0.04 |
| Glycolithocholic acid | 524.6 | 530.6 | 15.3 | 1.03 ± 0.06 | 0.82 ± 0.10 | ND | 0.86 ± 0.07 | 0.89 ± 0.10 |
| Lithocholic acid | 467.5 | 473.5 | 16.0 | 0.90 ± 0.03 | 0.91 ± 0.04 | ND | 0.89 ± 0.08 | 1.00 ± 0.14 |
ND; Non-detected.
Comparison of absolute quantification method and CIL LC–MS/MS methods
For verification of the developed semi-quantitative method, the comparison with absolute quantitation was performed. In first, to obtain the real concentrations of propanoic acid and butyric acid in pooled plasma samples, we performed absolute quantification based on standard calibration curves. Stable isotopes of propanoic acid-d5 and butyric acid-d7 were used as ISs and calibration curves were plotted using seven calibration concentrations. Correlation coefficients of two analytes were above 0.99 for both analytes, indicating good linearity across the testing range (Supplementary Table S2). A both in human pooled plasma and mouse pooled plasma, the all precision and accuracy values satisfied all criterion for all measured concentrations (Supplementary Table S3). These data suggested that the 2PA-labeling based method could also be applicable for absolute quantification.
Next, to confirm the accuracy of relative quantitative performance of the developed method, we analyzed two SCFAs, propanoic acid and butyric acid in plasma by both absolute quantification and relative quantification, and compared the two results by two GF-to-SPF ratios of two SCFAs. As shown in Fig. 4, the GF-to-SPF ratios of C3:0 and C4:0 by semi-quantitative method were equal to the results by absolute quantitation. This result suggested that the semi-quantitative approach provide the accurate metabolite ratios.
Figure 4.
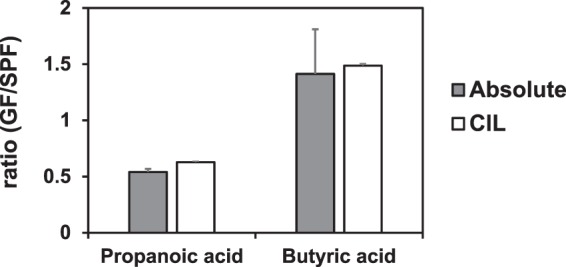
GF-to-SPF ratios of propanoic acid and butyric acid determined by the 2PA derivatization LC-MS/MS method (Absolute) and by the CIL-LC-MS/MS method (CIL). Bars indicate means ± SD. Gray bars indicate the ratio was obtained from absolute quantitation method; Three independent plasma samples were prepared for each group mice and separately labeled by 2PA. GF/SPF ratios of propanoic acid and butyric acid were calculated by mean of the absolute concentrations. White bars indicate the ratio was obtained from CIL-LC-MS/MS method; The plasma samples were obtained by combining equal volume (n = 3) of each group mice. The pooled plasma was labeled with 2PA (for SPF) and 2PA-d6 (for GF), respectively. Then labeled samples were mixed (1:1) and subjected to the differential analysis. GF/SPF ratios of propanoic acid and butyric acid were calculated by the mean of ion peak area in triplicate measurements.
Profiling of carboxyl-containing metabolites in plasma, feces, and cecal contents from SPF, SPF-RF, GF, and GF-RF mice
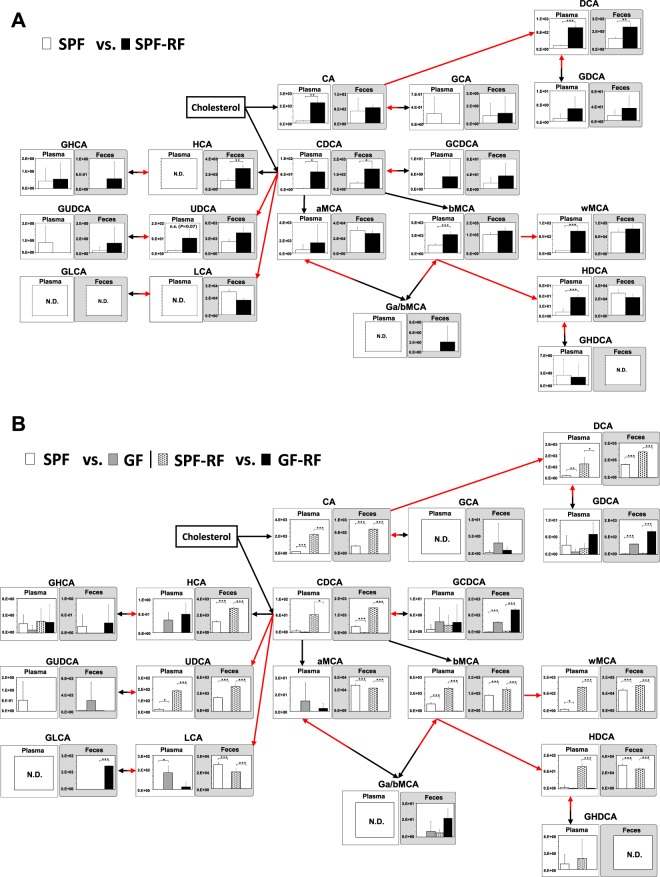
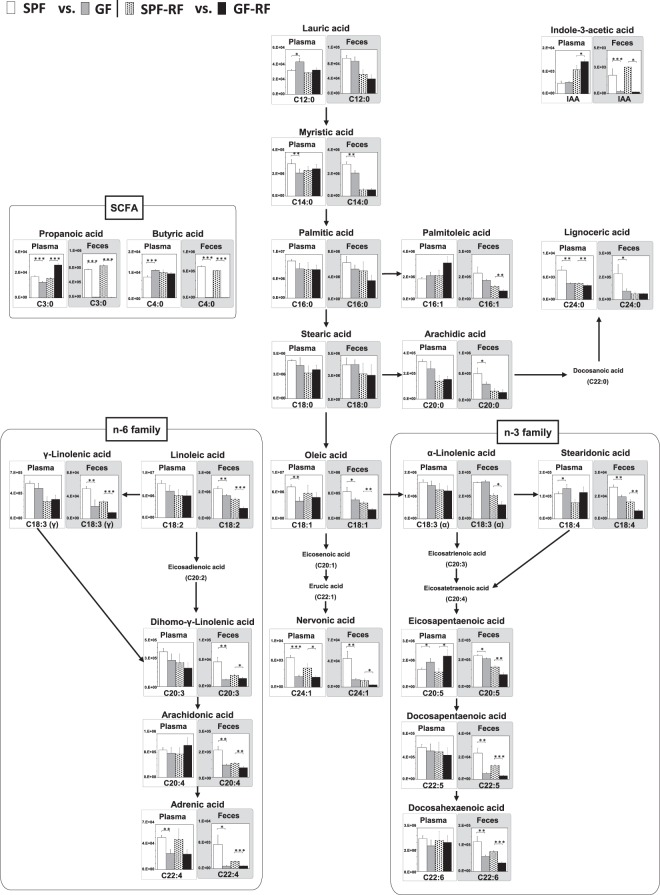
In this study, we focused on microbiota-derived carboxyl-containing metabolites, such as SCFAs, secondary bile acids, and IAA, because they have critical functions for regulating many biological processes9,39–41. Regarding the role of renal disease, SCFAs have been shown to have renoprotective effects on the local and systemic inflammation in CKD and acute kidney injury42,43. Thus, in order to clarify the impact of microbiota in CKD, we prepared the 4 groups of mice (SPF, SPF-RF, GF, and GF-RF) and subsequently, measured 40 carboxyl-containing metabolites in plasma, feces, and cecal contents using a developed CIL-LC-MS/MS method. Hematoxylin and eosin (H&E) and Masson’s trichrome staining indicated that renal inflammation in GF-RF were more severe than that SPF-RF, although the renal fibrosis in GF-RF was less severe than that in SPF-RF (Supplementary Fig. S5). The clinical parameters (e.g. BUN, plasma creatinine) has been described previously15. Reconstruction of bile acid metabolic pathways from 18 bile acids data is shown in Fig. 5 (plasma and feces) and Supplementary Fig. S6 (cecal contents). Although the plasma levels of CA, chenodeoxycholic acid (CDCA), deoxycholic acid, β-muricholic acid, ω-muricholic acid, and hyodeoxycholic acid were greatly elevated in SPF-RF compared with those in SPF (Fig. 5A), they were not or nearly undetectable in GF groups (Fig. 5B). In addition, ursodeoxycholic acid and lithocholic acid were not or nearly undetectable in feces from GF and GF-RF (Fig. 5B). Accordingly, the absence of microbiota resulted in the greatly reduce of these bile acids from GF mice, indicating that these bile acids mostly originated from microbiota metabolism. Although CA and CDCA which are synthesized in the host liver from cholesterol were not detected in GF mouse group, the glycine conjugate (e.g. glycochenodeoxycholic acid) were elevated in the feces (Fig. 5B) and cecal contents (Supplementary Fig. S6). This suggests that glycine-conjugated bile acids were not de-conjugated by intestinal bacteria, hence most of them were expected to be present in the intestine as a conjugate. The alteration of bile acid composition how influence on the renal functions is currently unknown, but it has been reported that secondary bile acid-producing bacteria contribute to the homeostasis of glucose and lipid metabolism44. A further study of the precise effect of bile acids in the blood and intestine on CKD should be conducted.
Figure 5.
Reconstruction of bile acid metabolic pathways from 18 bile acids. Relative quantitative analysis of the bile acids in plasma and feces between (A) SPF and SPF-RF groups. (B) SPF and GF groups, SPF-RF and GF-RF groups. Metabolic pathways are involved in host and microbiota as indicated by black arrows and red arrows, respectively. Bars indicate means ± SD of ion peak area in triplicate measurements. *P < 0.05, **P < 0.01, ***P < 0.001 compared between indicated groups (Student’s t test). The pooled samples were prepared from each groups of mice; N = 4 in SPF control (SPF) and GF control (GF), and N = 3 in SPF renal failure (SPF-RF), and N = 5 GF renal failure (GF-RF) mice.
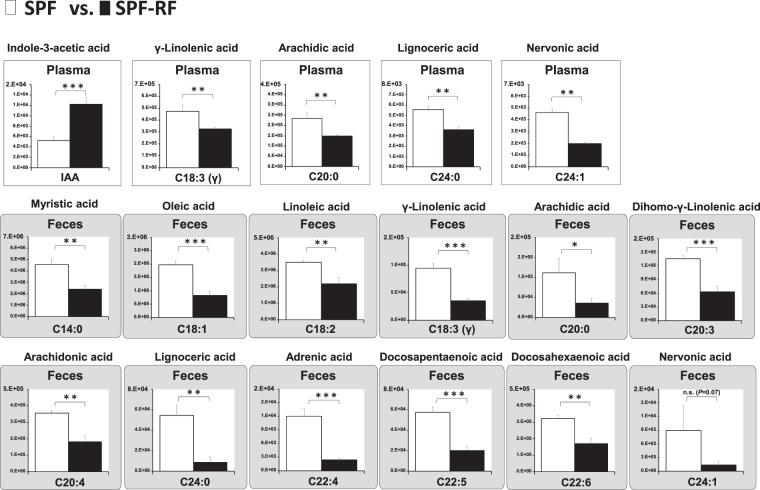
Considering bile acids are secreted into the duodenum to facilitate dietary fat digestion and absorption, it can be presumed that drastic change of bile acids in GF mice affect fatty acid metabolism. Further, Kindt et al. clarify that the gut microbiota promotes hepatic fatty acid desaturation and elongation in mice45. Thus, we hypothesized that GF-RF mice differ from SPF-RF mice in their fatty acid metabolism, leading to the progression of CKD. Also, IAA is uremic solute produced from the dietary tryptophan by gut microbiota, it has been reported to an influence on the host lipid metabolism, via aryl hydrocarbon receptor (AhR) pathway41,46–48.To address this point, we measured IAA and 21 fatty acids, including SCFA, saturated fatty acid, monounsaturated fatty acids, long-chain omega-6 (n-6), and omega-3 (n-3) polyunsaturated fatty acids (PUFA) using a developed method. There was no fatty acid that elevated in SPF-RF plasma compared with those in SPF. On the contrary, a significant decrease of γ-linolenic acid (C18:3 n-6), arachidic acid (C20:0), lignoceric acid (C24:0), and nervonic acid (C24:1) was detected in SPF-RF as compared to controls (Fig. 6). The reason for this decreasing is currently unknown, but the result of γ-linolenic acid is in agreement with the previous study, that was conducted on CKD patients plasma49. Also, the fecal levels of myristic acid (C14:0), oleic acid (C18:1), linoleic acid (C18:2), C18:3 n-6, C20:0, dihomo-γ-linolenic acid (C20:3), arachidonic acid (C20:4), C24:0, adrenic acid (C22:4), docosapentaenoic acid (C22:5), and docosahexaenoic acid (C22:6) were significantly lower in SPF-RF than in SPF (Fig. 6). Notably, these fecal fatty acids and palmitoleic acid (C16:1) were also lower in the GF groups than in the SPF groups, and hence GF-RF mice were the lowest levels among 4 groups (Fig. 7). These results indicating that fatty acid levels in the intestine were affected not only by renal function but also by gut microbiota. This is further supported by the results of cecal contents (Supplementary Fig. S7). The plasma levels of C14:0, C18:1, C24:1, and C22:4 were significantly lower in GF than in SPF. In contrast, lauric acid (C12:0), stearidonic acid (C18:4), and eicosapentaenoic acid (C20:5) were higher in GF than in SPF (Fig. 7). These findings suggest that the absence of microbiota which has an important role in desaturation and elongation of fatty acid caused substantial changes in plasma fatty acid profile. Because a certain gut lactic acid bacterium plays a polyunsaturated fatty acid-saturation metabolism, a part of this result could be due to the absence of these microbes4.
Figure 6.
A panel of statistically significant fatty acids and IAA in plasma and feces between SPF and SPF-RF groups. Bars indicate means ± SD of ion peak area in triplicate measurements. *P < 0.05, **P < 0.01, ***P < 0.001 compared between indicated groups (Student’s t test).
Figure 7.
Reconstruction of integrate metabolic pathways from 19 fatty acids, SCFA, and IAA. Relative quantitative analysis of the fatty acids and IAA in plasma and feces between SPF and GF groups, SPF-RF and GF-RF groups. Bars indicate means ± SD of ion peak area in triplicate measurements. *P < 0.05, **P < 0.01, ***P < 0.001 compared between indicated groups (Student’s t test).
SCFA and IAA are produced from the dietary fibre and tryptophan by gut microbiota, respectively39,47. As SCFA and IAA are a microbiota-associated metabolite, the plasma C3:0, C4:0, and IAA levels are considered to be governed by microbiota-derived production. However, the present findings revealed that the absence of microbiota did not significant affect plasma these levels in the GF groups, suggesting that host-derived production accounted for a much larger population of the total amounts (Fig. 7). In our previously report that was conducted on CE-TOF MS-based analysis, the plasma C4:0 level was below the limits of detection15. Therefore, the developed CIL-LC-MS based approach provided a more high-sensitive platform for analyses SCFA. In agreement with the previous our studies, C3:0 and C4:0 were nearly undetectable in the feces of GF and GF-RF, suggesting that intestinal SCFA production was ceased in both the GF groups. Likewise, fecal and cecal contents IAA levels were significantly lower in the GF groups than in the SPF groups (Figs. 7 and S7).
Discussion
In this study, to overcome some limitations of current metabolomics methods for carboxyl-containing metabolites, we developed a novel CIL-LC-MS/MS method using the 2PA and its isotopologue as derivatization reagents. This method was a reasonable approach for analyzing the carboxyl submetabolome profiling with many samples, because not only 2PA-d6 could easily be synthesized with inexpensive reagents, but also derivatization reaction was simple and quick. The quantitative performance of it was verified by many of experiments. In all result, this semi-quantitative method provided the high accuracy and high sensitivity as well. In addition, our approach is not affected by matrix effect and favorable due to short sample preparation time and fewer analysis times with mixing different samples.
More recently, several microbiota strains have been shown to be responsible for the production of harmful uremic toxins (e.g. trimethylamine-N-oxide; indoxyl sulfate). These uremic toxins are an independent predictor of mortality in CKD patients50. Further, we have revealed that phenyl sulfate, one of the microbiota-derived uremic toxins, contribute to albuminuria in diabetic kidney disease51. However, in the previous another study, we observed the paradoxical finding that the absence of microbiota exacerbated renal damage in adenine-induced GF-RF mice, despite the attenuating accumulation of such harmful uremic solutes15. Also, it has been reported that GF mice had enhanced renal structural injury and functional decline following ischemia-reperfusion compared with conventional mice52. These findings suggest that some microbiota-derived metabolites have potential protective effects against CKD progression and share a mutually beneficial relationship with their host. There is increasing evidence that microbiota-derived SCFAs have a renoprotective effect39, however, the other carboxyl-containing metabolites how influence on CKD pathology has been little investigated. Using the developed method, we revealed that most bile acid levels in plasma were significantly elevated in SPF-RF mice compared to control mice, as well as the intestinal level of CDCA, HCA, and DCA were increased (Fig. 5A). Although IAA, a circulating uremic toxin, was also elevated in SPF-RF plasma, some fatty acid levels were decreased (Fig. 6). It should be noted that the absence of microbiota caused decreasing bile acids, fatty acids, and IAA levels in the intestine (Fig. 5B, Supplementary Fig. S6, Fig. 7, and Supplementary Fig. S7). In addition, the renal failure enhanced the decrease of the intestinal level of n-3 PUFAs (Figs. 6 and 7). Interestingly, it has also been reported that bile acids, n-3 PUFA, SCFA, and IAA, which are focused in this study, play important roles in the endothelial barrier function, respectively41,53,54. A number of studies have demonstrated the negative effects of CKD on the gut microbiota and intestinal barrier function18,19,55.Recently, it was shown that the metabolome disorders in rat with CKD induced by 5/6 nephrectomy or unilateral ureteral obstruction cause decline in microbial diversity and richness, which were mediated by dysregulations of lipids, amino acids, and bile acids metabolism18,19. Gut dysbiosis, seen commonly in patients with CKD, caused by a decline in microbial diversity and disruption of gut barrier function56. The disrupt of the intestinal epithelial barrier lead to infiltration of various antigen and pro-inflammatory products into the systemic circulation, thus it facilitates the pathogenesis of inflammation in CKD57. Taken together, these observations indicated the decrease of intestinal bile acids, IAA, SCFA, and n-3 PUFAs caused by both renal damage and the absence of microbiota might be also contributed to the exacerbating effects on the intestinal epithelial barrier function that results in CKD progression in GF-RF mice (Supplementary Fig. S8). It was reported that treatment with 5-methoxytryptophan, which plasma levels decrease with progression of CKD, ameliorates renal injury in mouse via the anti-inflammatory and anti-fibrotic effects58. Thus, supplementation with diet or pre-, pro- and symbiotic approach to correct intestinal bile acids, IAA, SCFA, and n-3 PUFAs levels may also lead to potential protective effects against CKD progression by suppressing epithelial barrier disruption.
The primary limitation in this study is that the differences in the plasma metabolites between SPF and SPF-RF (or SPF-RF and GF-RF) may be attributed not only to the decline of renal clearance but also to confounding factors such as body weight, body fluid balance, cage effects59, and dietary intake15. Also, the disruption of the gastrointestinal epithelial barrier function might promote adenine absorption, that results in CKD progression. Thus, to verify our findings in the present study, additional research using other CKD models is required.
In conclusion, we suggest that monitoring a panel of microbiota-derived carboxyl-containing metabolites by the developed CIL-LC-MS/MS method would be useful for obtaining an in-depth appreciation of the compounds involves in the gut–kidney axis and physiological and pathological roles of CKD. Taking the advantages in selectivity and applicability afforded by 2PA into account, the method developed herein is capable of simultaneous analysis of multiple carboxyl-containing compounds. We hope that the outcome of the present study will contribute to a better understanding of the impact of gut microbiota on CKD progression.
Materials and Methods
Chemicals and reagents
CA, CDCA, and palladium-activated carbon (Pd:10%) were purchased from Sigm–Aldrich (St. Louis, MO). LC/MS-grade ammonium acetate (AcONH4) and acetic acid (AcOH), deoxycholic acid, lithocholic acid, IAA, C3:0, C4:0, and platinum-activated carbon (Pt:5%) were purchased from FUJIFILM Wako Pure Chemical Industries (Osaka, Japan). Ursodeoxycholic acid, glycoursodeoxycholic acid were purchased from Nacalai Tesque, Inc. (Kyoto, Japan). Glycocholic acid, glycohyocholic acid, glycochenodeoxycholic acid, glycodeoxycholic acid, glycolithocholic acid, hyocholic acid, hyodeoxycholic acid, α-muricholic acid, β-muricholic acid, ω-muricholic acid, glycohyodeoxycholic acid, glyco-α-muricholic acid, glyco-β-muricholic acid were synthesized in our laboratory using previously reported method60. Propanoic acid-d5 and butyric acid-d7 were obtained from Toronto Research Chemicals (North York, ON, Canada). 2-picolylamine (2PA), 2,2′-dipyridyl disulfide (DPDS) and triphenylphosphine (TPP) were obtained from Tokyo Chemical Co. (Tokyo, Japan). Deuterium oxide (99.9% isotopic purity), LC/MS-grade methanol (MeOH), 2-propanol (IPA), and acetonitrile (MeCN) were obtained from Kanto Chemical (Tokyo, Japan). Human pooled plasma and mouse plasma were obtained from Biopredic International (Rennes, France) and Rockland Immunochemicals Inc. (Limerick, PA, USA), respectively. Saturated/monounsaturated fatty acid mixture and polyunsaturated fatty acid mixture were purchased from Cayman Chemicals (Ann Arbor, MI, USA). Ultra-pure water was purified by a PURELAB ultra water purification system (Organo Co. Ltd, Tokyo). The reagents used for the derivatization were adjusted using MeCN. All stock solutions were stored at −20 °C and avoid light.
Animal treatment and sample collection
The plasma, feces, and cecum from GF and SPF mice were collected at previous work and stored at −80 °C in our laboratory15. Briefly, GF and SPF IQI mice were purchased from Clea Japan (Tokyo, Japan). At 7 weeks of age, each group of mice was randomly divided into control and RF groups. For the GF and SPF control groups, normal sterilized CE-2 diet was continued for 5 weeks (GF, SPF). For the RF groups, sterilized CE-2 diet containing 0.2% adenine (Wako Pure Chemical Industries, Japan) was given for 5 weeks (GF-RF, SPF-RF). In the final 2 weeks of this 5-week period, the RF mice were given a normal diet for 2 days per week not to weaken. At the end of the study, the mice were killed under isoflurane-induced anesthesia, and blood, fresh feces, and cecum contents were collected.
Extraction of metabolites from feces and cecal contents
10 mg of feces or contents in cecum were homogenized in 1.0 mL of MeOH for 2 min and then centrifuged at 15,000 g for 10 min. The obtained supernatants were combined followed by CIL-LC-MS analysis.
Synthesis of 2-picolylamine-d6
2PA-d6 were synthesized from 2PA. Deuterium-labeling (H-D exchange) reaction was examined according to previously reported methods35,61. 2PA (540.7 mg, 5.0 mmol) and 10% Pd/C (54.0 mg, 10 wt % of the substrate) and 5% Pt/C (108.1 mg, 20 wt % of the substrate) in D2O (18 mL) were stirred at 180 °C in a sealed tube under H2 atmosphere for 120 h. After cooling, the reaction mixture was filtered through a filter (Millipore Millex-LG, 0.20 mm) to paper to remove the catalyst. The filtered catalyst was washed with D2O (1 mL). The filtrate and 5% Pt/C (224.9 mg, 40 wt % of the substrate) were stirred at 180 °C in a sealed tube under H2 atmosphere for 120 h. The filtered catalyst was washed with D2O (1 mL) and the filtrate was concentrated in vacuo. Next, the residue was purified by reversed-phase chromatography (H2O/MeOH) to yield 2PA-d6. Chromatographic separation was carried out on DualPore ODS (21 mm i.d. × 75 mm, 12 g, DPS, Tokyo, Japan) at room temperature. UV spectra were recorded on LC-Forte/R system (YMC, Kyoto, Japan). The extracts are evaporated to dryness, redissolved in MeCN. Synthesized structures were confirmed by high-resolution mass spectrometry and 1H NMR spectroscopy. High-resolution mass spectra were recorded on a Q Exactive (Thermo Fisher Scientific, Waltham, MA, USA). 1H NMR spectra were recorded on a JEOL JNM-ECA600 spectrometer. The concentration was determined by LC-UV analysis. UV spectra were recorded on Nexera UHPLC system (Shimadzu, Kyoto, Japan).
Labeling of sample with 2PA and 2PA-d6
2PA or 2PA-d6 was used to label carboxyl-containing metabolites (Fig. 2A). The labeling condition (reaction temperature and time) were according to previous reports32. Briefly, for standard-mixture, 200 µL of MeCN and 10 µmol/L C3:0-d5 and C4:0-d7 mixed solution (30 µL, if absolute quantitation) as internal standards (ISs) added to the samples (20 µL) were placed into a siliconized 1.5-mL sample tube (Watson, Kobe, Japan) and 10 mM TPP in MeCN (10 µL), 10 mM DPDS in MeCN (10 µL) and, 10 mM 2PA in MeCN (10 µL) or 10 mM 2PA-d6 (10 µL) were added for derivatization. The mixture was then kept at 60 °C for ~10 min. The resulting solution was evaporated to dryness by a rotary vacuum evaporator at 40 °C. The dried sample was reconstituted with 50 µL of 50% MeOH in water. Finally, the labeled samples were mixed in equal volume before LC-MS/MS analysis.
For 20 µL aliquots of biological samples, placed into a 1.5-mL sample tube and MeCN (200 μL) was added for deproteinization. The obtained mixture was homogenized for 5 min in an ultrasonic bath. After centrifugation at 15,000 × g for 10 min at 4 °C, 200 µL of supernatant was collected into a new siliconized tube, followed by 2PA or 2PA-d6 derivatization and LC-MS/MS analysis in the same way as described above.
Instrumentation and LC-MS/MS condition
The LC-MS/MS analyses were conducted on a Nexera UHPLC system (Shimadzu, Kyoto, Japan) coupled to a TSQ Quantum Vantage (Thermo Fisher Scientific, Waltham, MA, USA) triple quadrupole mass spectrometer. Chromatographic separation was carried out on Intertsil ODS-3 (2.1 mm i.d. × 100 mm, 2 μm, GL Science, Tokyo, Japan) at 45 °C. The mobile phase was 0.1% AcOH with 10 mM AcONH4 in water (A), and 0.1% AcOH in MeCN/IPA (v/v = 1:1) (B). The flow rate was set at 430 µL/min for analysis of fatty acids or IAA or 450 µL/min for analysis of bile acids. An elution gradient was used as follows: for analysis of fatty acids and IAA, initial elution with 5% B, followed by a linear gradient to 20% B from 1.0–10.0 min, to 60% B from 10.1–15.0 min, to 80% B from 15.1–28.0 min, and, thereafter, to 95% B at 28.1 min, held at 95%B for 32.0 min. Columns were re-equilibrated with 5% B for 3.0 min prior to the next injection. For analysis of bile acids, initial elution with 30% B, followed by a linear gradient to 40% B at 10.0 min, to 70% B from 10.1–15.0 min and, thereafter, to 95% B at 15.1 min, held at 95%B for 17.5 min. Columns were re-equilibrated with 30% B for 2.5 min prior to the next injection.
The mass spectrometer was equipped with a heated electrospray ionization (HESI) source. HESI was performed in positive ion modes. Nitrogen was used for nebulization and desolvation and argon was used as the collision gas. The capillary voltage was set at 3.0 kV. The optimum conditions were: sheath gas pressure 60 psi; auxiliary gas pressure 60 psi; vaporizer temperature 500 °C; capillary temperature 350 °C; declustering voltage −10eV, and collision gas pressure 1.5 mTorr. The selected reaction monitoring (SRM) transitions, S-lens voltage, and collision energy are shown in Supplementary Table 1. The LC-MS/MS system was controlled by Xcalibur software (Thermo Fisher Scientific, Waltham, MA, USA), which was also used to collect data.
Preparation of calibration curve, mixed standard solution and quality control (QC) samples for analysis of C3:0 and C4:0
C3:0, C4:0 and their corresponding ISs (C3:0-d5 and C4:0-d7) were weighed accurately into volumetric flasks and then diluted separately with 50% MeOH in water, to produce stock solutions, which were stored at −20 °C. Before use, stock solutions were serially diluted with MeCN to give a mixed working solution series. Calibration standards were prepared by diluting the ISs with MeCN. QC samples were prepared from working solutions or mixed standard solutions diluted in blank mouse plasma and human plasma (v/v = 1: 9). A 20-µL sample was labeled by 2PA following the same procedure described above for sample labeling. Calibration was performed using nine-point standards across the desired quantification range. Calibration curves were constructed using analyte-to-IS peak area ratios and 1/X weighted linear regressions. All peaks were integrated automatically by the Xcalibur software.
Evaluation of relative quantification of carboxyl-containing metabolites from standard-mixture, pooled plasma, and QC plasma
The mixture solution of 3 carboxyl compounds (bile acid, fatty acid and IAA) with different structures was used to evaluate the accuracy of the relative quantification. The mixed standard solution was combined in a single solution consisting of 19 bile acids mixture (30 µmol/L), 9 saturated-monounsaturated fatty acids mixture (3.75 µg/mL), 10 polyunsaturated fatty acids mixture (3.75 µg/mL), C3:0 (20 µmol/L), C4:0 (20 µmol/L), IAA (50 µmol/L), and stored at −20 °C. Before use, the mixed standard solution was diluted 10-fold with MeCN. QC samples were prepared from working solutions or mixed standard solutions diluted in blank mouse plasma and human plasma (v/v = 1: 9).
To prepare the 1:1 2PA-d6/2PA-labeled sample for analytical performance evaluation, 40 µL of various samples (the mixed standard solution; blank human plasma; blank mouse plasma, and QC plasma) were divided into two parts: 20 µL was labeled by 2PA-d6 and the other 20 µL was labeled by 2PA following the same procedure described above for sample labeling. After labeling and reconstruction, the labeled samples were mixed in equal volume before LC-MS/MS analysis.
Determination of GF-to-SPF ratios of C3:0 and C4:0 in plasma
The levels of C3:0 and C4:0 in SPF mouse plasma and GF mouse plasma were determined by 2PA derivatized-LC-MS/MS method. The precision and accuracy of the absolute quantification method for C3:0 and C4:0 were confirmed by spiking two or five concentrations of the analyte compounds into mouse pooled plasma and human pooled plasma, respectively. For method validation, six samples with specified concentrations were prepared and analyzed on the same day (intra-day precision). Precision was defined as the relative standard deviation (RSD) of independent measurements, while accuracy was expressed as the relative error (RE), calculated as follows: [(found concentration – endogenous concentration)/spiked concentration – 1] × 100 (%). GF-to-SPF ratios of C3:0 and C4:0 in plasma were calculated by both absolute quantification method and CIL method.
Relative quantitation of carboxyl-containing metabolites in plasma, feces, and cecal contents
The workflow of the experiment for profiling of carboxyl-containing metabolites is shown in Fig. 2B. The plasma, feces, and cecal contents samples were obtained by combining equal volume of each sample from 4 groups of mice, respectively: SPF control (SPF) mice (n = 4), GF control (GF) mice (n = 4), SPF-RF mice (n = 3), and GF-RF mice (n = 5). The protocol was approved by the Institutional Animal Care and Use Committee at Tohoku University. All experiments were performed in accordance with the approved protocol.
Statistical analyses
All statistical analyses were performed using JMP software (ver. 14.1.2; SAS Institute, Cary, NC, USA). Values are presented as mean ± SD. Results were evaluated using Student’s t-test. P values of < 0.05 were statistically significant.
Ethical approval
The protocol was approved by the Institutional Animal Care and Use Committee at Tohoku University.
Supplementary information
Acknowledgements
We would like to thank Saori Shoji (Faculty of Pharmaceutical Sciences, Tohoku University, Sendai, Japan) for the technical establishment of measuring of bile acids.
Author contributions
Y.T., T.A. and N.M. conceived of and supervised the study; Y.K., E.M., M.M. and Y.M. designed and performed the experiments; Y.K. analyzed the data; Y.K. and M.M. wrote the manuscript; and E.M., D.S., H.Y., J.O., H.T. and N.M. reviewed the manuscript. All authors approved the final version of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-55600-1.
References
- 1.Nicholson JK, et al. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–7. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 2.Ridlon JM, Kang D, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 2006;47:241–59. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 3.Koh A, De Vadder F, Kovatcheva-Datchary P, Bäckhed F. From dietary fiber to host physiology: Short-chain fatty acids as key bacterial metabolites. Cell. 2016;165:1332–1345. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 4.Kishino S, et al. Polyunsaturated fatty acid saturation by gut lactic acid bacteria affecting host lipid composition. Proc. Natl. Acad. Sci. 2013;110:17808–17813. doi: 10.1073/pnas.1312937110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spaepen S, Vanderleyden J, Remans R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 2007;31:425–48. doi: 10.1111/j.1574-6976.2007.00072.x. [DOI] [PubMed] [Google Scholar]
- 6.Chiang JYL. Bile acids: regulation of synthesis. J. Lipid Res. 2009;50:1955–1966. doi: 10.1194/jlr.R900010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmitz G, Ecker J. The opposing effects of n-3 and n-6 fatty acids. Prog. Lipid Res. 2008;47:147–155. doi: 10.1016/j.plipres.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 8.Laurans L, et al. Genetic deficiency of indoleamine 2,3-dioxygenase promotes gut microbiota-mediated metabolic health. Nat. Med. 2018;24:1113–1120. doi: 10.1038/s41591-018-0060-4. [DOI] [PubMed] [Google Scholar]
- 9.Just S, et al. The gut microbiota drives the impact of bile acids and fat source in diet on mouse metabolism. Microbiome. 2018;6:1–18. doi: 10.1186/s40168-018-0510-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wahlström A, Sayin SI, Marschall HU, Bäckhed F. Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metab. 2016;24:41–50. doi: 10.1016/j.cmet.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 11.Cosola C, et al. Microbiota metabolites: Pivotal players of cardiovascular damage in chronic kidney disease. Pharmacol. Res. 2018;130:132–142. doi: 10.1016/j.phrs.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 12.Transl J, et al. Aryl hydrocarbon receptor activation mediates kidney disease and renal cell carcinoma. J. Transl. Med. 2019;2:1–14. doi: 10.1186/s12967-019-2054-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jia W, et al. Melamine-induced renal toxicity is mediated by the gut microbiota. Sci. Transl. Med. 2013;5:1–10. doi: 10.1126/scitranslmed.3005114. [DOI] [PubMed] [Google Scholar]
- 14.De Angelis, M. et al. Microbiota and metabolome associated with Immunoglobulin A Nephropathy (IgAN). PLoS One9 (2014). [DOI] [PMC free article] [PubMed]
- 15.Mishima E, et al. Evaluation of the impact of gut microbiota on uremic solute accumulation by a CE-TOFMS-based metabolomics approach. Kidney Int. 2017;92:634–645. doi: 10.1016/j.kint.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 16.Want EJ, et al. Global metabolic profiling procedures for urine using UPLC-MS. Nat. Protoc. 2010;5:1005–1018. doi: 10.1038/nprot.2010.50. [DOI] [PubMed] [Google Scholar]
- 17.Saigusa D, et al. Establishment of protocols for global metabolomics by LC-MS for biomarker discovery. PLoS One. 2016;11:1–18. doi: 10.1371/journal.pone.0160555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen, L. et al. Unilateral ureteral obstruction causes gut microbial dysbiosis and metabolome disorders contributing to tubulointerstitial fibrosis. Exp. Mol. Med. 51 (2019). [DOI] [PMC free article] [PubMed]
- 19.Feng Ya-Long, Cao Gang, Chen Dan-Qian, Vaziri Nosratola D., Chen Lin, Zhang Jun, Wang Ming, Guo Yan, Zhao Ying-Yong. Microbiome–metabolomics reveals gut microbiota associated with glycine-conjugated metabolites and polyamine metabolism in chronic kidney disease. Cellular and Molecular Life Sciences. 2019;76(24):4961–4978. doi: 10.1007/s00018-019-03155-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalim S, Rhee EP. An overview of renal metabolomics. Kidney Int. 2016;91:1–9. doi: 10.1016/j.kint.2016.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shah VO, et al. Plasma Metabolomic Profiles in Different Stages of CKD. Clin. J. Am. Soc. Nephrol. 2013;8:363–370. doi: 10.2215/CJN.05540512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higashi T, Ogawa S. Isotope-coded ESI-enhancing derivatization reagents for differential analysis, quantification and profiling of metabolites in biological samples by LC/MS: A review. J. Pharm. Biomed. Anal. 2016;130:181–193. doi: 10.1016/j.jpba.2016.04.033. [DOI] [PubMed] [Google Scholar]
- 23.Yuan BF, et al. Comprehensive Profiling of Fecal Metabolome of Mice by Integrated Chemical Isotope Labeling-Mass Spectrometry Analysis. Anal. Chem. 2018;90:3512–3520. doi: 10.1021/acs.analchem.7b05355. [DOI] [PubMed] [Google Scholar]
- 24.Guo N, Peng CY, Zhu QF, Yuan BF, Feng YQ. Profiling of carbonyl compounds in serum by stable isotope labeling - Double precursor ion scan - Mass spectrometry analysis. Anal. Chim. Acta. 2017;967:42–51. doi: 10.1016/j.aca.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 25.Lu Y, Yao D, Chen C. 2-Hydrazinoquinoline as a Derivatization Agent for LC-MS-Based Metabolomic Investigation of Diabetic Ketoacidosis. Metabolites. 2013;3:993–1010. doi: 10.3390/metabo3040993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo K, Li L. High-performance isotope labeling for profiling carboxylic acid-containing metabolites in biofluids by mass spectrometry. Anal. Chem. 2010;82:8789–8793. doi: 10.1021/ac102146g. [DOI] [PubMed] [Google Scholar]
- 27.Han J, Lin K, Sequeira C, Borchers CH. An isotope-labeled chemical derivatization method for the quantitation of short-chain fatty acids in human feces by liquid chromatography-tandem mass spectrometry. Anal. Chim. Acta. 2015;854:86–94. doi: 10.1016/j.aca.2014.11.015. [DOI] [PubMed] [Google Scholar]
- 28.Leng J, et al. A highly sensitive isotope-coded derivatization method and its application for the mass spectrometric analysis of analytes containing the carboxyl group. Anal. Chim. Acta. 2013;758:114–121. doi: 10.1016/j.aca.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 29.Cai T, Ting H, Xin-Xiang Z, Jiang Z, Jin-Lan Z. HPLC-MRM relative quantification analysis of fatty acids based on a novel derivatization strategy. Analyst. 2014;139:6154–6159. doi: 10.1039/C4AN01314J. [DOI] [PubMed] [Google Scholar]
- 30.Zhu QF, et al. Analysis of liposoluble carboxylic acids metabolome in human serum by stable isotope labeling coupled with liquid chromatography–mass spectrometry. J. Chromatogr. A. 2016;1460:100–109. doi: 10.1016/j.chroma.2016.07.017. [DOI] [PubMed] [Google Scholar]
- 31.Zhu Y, Deng P, Zhong D. Derivatization methods for LC-MS analysis of endogenous compounds. Bioanalysis. 2015;7:2557–2581. doi: 10.4155/bio.15.183. [DOI] [PubMed] [Google Scholar]
- 32.Higashi T, et al. Simple and practical derivatization procedure for enhanced detection of carboxylic acids in liquid chromatography–electrospray ionization-tandem mass spectrometry. J. Pharm. Biomed. Anal. 2010;52:809–818. doi: 10.1016/j.jpba.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 33.Joo KM, et al. A rapid and highly sensitive UPLC-MS/MS method using pre-column derivatization with 2-picolylamine for intravenous and percutaneous pharmacokinetics of valproic acid in rats. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2013;938:35–42. doi: 10.1016/j.jchromb.2013.08.022. [DOI] [PubMed] [Google Scholar]
- 34.NAGATOMO R, OKADA Y, Ichimura M, Tsuneyama K, INOUE K. Application of 2-Picolylamine Derivatized Ultra-high Performance Liquid Chromatography Tandem Mass Spectrometry for the Determination of Short-chain Fatty Acids in Feces Samples. Anal. Sci. 2018;34:1031–1036. doi: 10.2116/analsci.18SCP10. [DOI] [PubMed] [Google Scholar]
- 35.Esaki H, et al. General method of obtaining deuterium-labeled heterocyclic compounds using neutral D2O with heterogeneous Pd/C. Tetrahedron. 2006;62:10954–10961. doi: 10.1016/j.tet.2006.08.088. [DOI] [Google Scholar]
- 36.Sajiki H, et al. Aromatic ring favorable and efficient H-D exchange reaction catalyzed by Pt/C. Tetrahedron Lett. 2005;46:6995–6998. doi: 10.1016/j.tetlet.2005.08.067. [DOI] [Google Scholar]
- 37.McDonald GR, et al. Bioactive contaminants leach from disposable laboratory plasticware. Science (80-.). 2008;322:917. doi: 10.1126/science.1162395. [DOI] [PubMed] [Google Scholar]
- 38.Stewart J, Drexler DM, Leet JE, McNaney CA, Herbst JJ. Labware additives identified to be selective monoamine oxidase-b inhibitors. J. Biomol. Screen. 2014;19:1409–1414. doi: 10.1177/1087057114551523. [DOI] [PubMed] [Google Scholar]
- 39.Maslowski KM, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuno T, Hirayama-Kurogi M, Ito S, Ohtsuki S. Reduction in hepatic secondary bile acids caused by short-term antibiotic-induced dysbiosis decreases mouse serum glucose and triglyceride levels. Sci. Rep. 2018;8:1–7. doi: 10.1038/s41598-017-17765-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krishnan S, et al. Gut Microbiota-Derived Tryptophan Metabolites Modulate Inflammatory Response in Hepatocytes and Macrophages. Cell Rep. 2018;23:1099–1111. doi: 10.1016/j.celrep.2018.03.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vaziri ND, et al. High amylose resistant starch diet ameliorates oxidative stress, inflammation, and progression of chronic kidney disease. PLoS One. 2014;9:e114881. doi: 10.1371/journal.pone.0114881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andrade-Oliveira V, et al. Gut Bacteria Products Prevent AKI Induced by Ischemia-Reperfusion. J. Am. Soc. Nephrol. 2015;26:1877–1888. doi: 10.1681/ASN.2014030288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuno T, Hirayama-Kurogi M, Ito S, Ohtsuki S. Reduction in hepatic secondary bile acids caused by short-term antibiotic-induced dysbiosis decreases mouse serum glucose and triglyceride levels. Sci. Rep. 2018;8:1253. doi: 10.1038/s41598-018-19545-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kindt A, et al. The gut microbiota promotes hepatic fatty acid desaturation and elongation in mice. Nat. Commun. 2018;9:3760. doi: 10.1038/s41467-018-05767-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poesen R, et al. The influence of dietary protein intake on mammalian tryptophan and phenolic metabolites. PLoS One. 2015;10:1–12. doi: 10.1371/journal.pone.0140820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Itoh Y, Ezawa A, Kikuchi K, Tsuruta Y, Niwa T. Protein-bound uremic toxins in hemodialysis patients measured by liquid chromatography/tandem mass spectrometry and their effects on endothelial ROS production. Anal. Bioanal. Chem. 2012;403:1841–1850. doi: 10.1007/s00216-012-5929-3. [DOI] [PubMed] [Google Scholar]
- 48.Tanos, R., Murray, I. A., Smith, P. B., Patterson, A. & Perdew, G. H. Role of the Ah Receptor in Homeostatic Control of Fatty Acid Synthesis in the Liver. 129, 372–379 (2012). [DOI] [PMC free article] [PubMed]
- 49.Stenvinkel P, Heimbürger O, Shearer GC, Carrero JJ, Barany P. Plasma Fatty Acids in Chronic Kidney Disease: Nervonic Acid Predicts Mortality. J. Ren. Nutr. 2011;22:277–283. doi: 10.1053/j.jrn.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 50.Ramezani A, et al. Role of the Gut Microbiome in Uremia: A Potential Therapeutic Target. Am. J. Kidney Dis. 2016;67:483–98. doi: 10.1053/j.ajkd.2015.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kikuchi K, et al. Gut microbiome-derived phenyl sulfate contributes to albuminuria in diabetic kidney disease. Nat. Commun. 2019;10:1835. doi: 10.1038/s41467-019-09735-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jang HR, et al. Early exposure to germs modifies kidney damage and inflammation after experimental ischemia-reperfusion injury. Am. J. Physiol. Renal Physiol. 2009;297:F1457–65. doi: 10.1152/ajprenal.90769.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Veldhoen M, Brucklacher-Waldert V. Dietary influences on intestinal immunity. Nat. Rev. Immunol. 2012;12:696–708. doi: 10.1038/nri3299. [DOI] [PubMed] [Google Scholar]
- 54.Agostoni C, et al. Fatty Acids in Nephrotic Syndrome and Chronic Kidney Disease. J. Ren. Nutr. 2017;28:145–155. doi: 10.1053/j.jrn.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 55.Sabatino A, et al. Alterations of intestinal barrier and microbiota in chronic kidney disease. Nephrol. Dial. Transplant. 2015;30:924–933. doi: 10.1093/ndt/gfu287. [DOI] [PubMed] [Google Scholar]
- 56.Nallu A, Sharma S, Ramezani A, Muralidharan J, Raj D. Gut microbiome in chronic kidney disease: challenges and opportunities. Transl. Res. 2017;179:24–37. doi: 10.1016/j.trsl.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vaziri ND, et al. Disintegration of colonic epithelial tight junction in uremia: A likely cause of CKD-associated inflammation. Nephrol. Dial. Transplant. 2012;27:2686–2693. doi: 10.1093/ndt/gfr624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen DQ, et al. Identification of serum metabolites associating with chronic kidney disease progression and anti-fibrotic effect of 5-methoxytryptophan. Nat. Commun. 2019;10:1–15. doi: 10.1038/s41467-018-07882-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Laukens D, et al. Heterogeneity of the gut microbiome in mice: Guidelines for optimizing experimental design. FEMS Microbiol. Rev. 2015;40:117–132. doi: 10.1093/femsre/fuv036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goto J, et al. Studies on steroids: CCXXXVIII. Determination of bile acids in liver tissue by gas chromatography-mass spectrometry with negative ion chemical ionization detection. J. Chromatogr. A. 1988;452:119–129. doi: 10.1016/S0021-9673(01)81441-2. [DOI] [PubMed] [Google Scholar]
- 61.Sajiki H, et al. Complete replacement of H2 by D2 via Pd/C-catalyzed H/D exchange reaction. Org. Lett. 2004;6:3521–3523. doi: 10.1021/ol048591b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.