Abstract
Purpose
Balanced carriers of structural rearrangements have an increased risk of unbalanced embryos mainly due to the production of unbalanced gametes during meiosis. Aneuploidy for other chromosomes not involved in the rearrangements has also been described. The purpose of this work is to know if the incidence of unbalanced embryos, interchromosomal effect (ICE) and clinical outcomes differ in carriers of different structural rearrangements.
Methods
Cohort retrospective study including 359 preimplantation genetic testing cycles for structural rearrangements from 304 couples was performed. Comparative genomic hybridisation arrays were used for chromosomal analysis. The results were stratified and compared according to female age and carrier sex. The impact of different cytogenetic features of chromosomal rearrangements was evaluated.
Results
In carriers of translocations, we observed a higher percentage of abnormal embryos from day 3 biopsies compared with day 5/6 biopsies and for reciprocal translocations compared with other rearrangements. We observed a high percentage of embryos with aneuploidies for chromosomes not involved in the rearrangement that could be attributed to total ICE (aneuploid balanced and unbalanced embryos). No significant differences were observed in these percentages between types of rearrangements. Pure ICE (aneuploid balanced embyos) was independent of female age only for Robertsonian translocations, and significantly increased in day 3 biopsies for all types of abnormalities. Furthermore, total ICE for carriers of Robertsonian translocations and biopsy on day 3 was independent of female age too. High ongoing pregnancy rates were observed for all studied groups, with higher pregnancy rate for male carriers.
Conclusion
We observed a higher percentage of abnormal embryos for reciprocal translocations. No significant differences for total ICE was found among the different types of rearrangements, with higher pure ICE only for Robertsonian translocations. There was a sex effect for clinical outcome for carriers of translocations, with higher pregnancy rate for male carriers. The higher incidence of unbalanced and aneuploid embryos should be considered for reproductive counselling in carriers of structural rearrangements.
Electronic supplementary material
The online version of this article (10.1007/s10815-019-01593-9) contains supplementary material, which is available to authorized users.
Keywords: Structural rearrangement, PGT-SR, Aneuploid unbalanced embryo, Biopsy, Interchromosomal effect
Introduction
Balanced structural chromosome rearrangements are the most frequent chromosomal abnormalities in the general population, with a prevalence of 0.4% of prenatal samples and 0.2% of newborns [1, 2]. The most common structural chromosome rearrangements are reciprocal translocations, Robertsonian translocations and inversions. Although balanced carriers of these abnormalities are phenotypically normal, they have an increased risk of fertility problems, recurrent miscarriages and producing offspring with congenital abnormalities and cognitive impairment.
The prevalence of such risks among patients with these abnormalities can be up to 25 times higher than in the general population [3–5]. These problems are mainly due to the production of unbalanced gametes during meiosis because of different segregation events in translocation carriers or recombination events in inversion carriers [6, 7]. Unbalanced gametes of these patients will result in embryos with aneuploidy for chromosomes involved in the rearrangements. The type of segregation in reciprocal translocations appears to be determined by the nature of involved chromosomes and the position of breakpoints [8, 9].
Preimplantation genetic testing for structural rearrangements (PGT-SR) can improve reproductive outcomes in these couples, reducing the time to achieve a successful live birth from 4–6 years to < 4 months and decreasing the incidence of miscarriage from > 90 to < 15% [10–12]. Until recently, fluorescence in situ hybridisation (FISH) was the preferred technique for PGT-SR in both polar bodies and interphase nuclei [9, 13–15]. Although FISH diagnosis has improved the reproductive expectations of couples, allowing clinical pregnancy rates to reach ~ 40% [10, 16], the technique has limitations, such as observation of split signals, cross-hybridisation, chromosome polymorphisms, poor fixation quality and loss of micronuclei or chromosomes during fixation [17–19]. Further, FISH identifies only imbalances of translocated chromosomes and does not evaluate all 23 pairs of autosomes for the presence of aneuploidy.
To overcome some of these limitations, several alternative methods have been developed for PGT-SR, such as PCR-based PGT-SR [20]. However, PCR cannot analyse all 24 chromosomes. More recent approaches involve whole genome amplification (WGA), including microarray single-nucleotide polymorphisms (SNPs) [21–25], microarray comparative genome hybridisation (aCGH) [26–29] and next-generation sequencing (NGS) [30, 31].
Chromosomes involved in rearrangements are proposed to interfere with correct segregation of other chromosomes by disrupting chromosome alignment on the spindle during meiosis I. This is known as the interchromosomal effect (ICE) and was first described by Lejeune (1963) [32], who observed an increased rate of carriers of balanced reciprocal translocations among the parents of children with trisomy 21. Several ICE studies have been published, with controversial results for spermatozoa [33–38], oocytes [15, 39–41], cleavage stage embryos and blastocysts [42, 43].
In this study, we retrospectively analysed the results of PGT-SR with aCGH to understand the impact of chromosomes involved in the rearrangement and the position of breakpoints on the incidence of unbalanced embryos and ICE.
Materials and methods
Study population
This multicenter retrospective and observational study compiled 359 PGT-SR cycles performed from June 2011–June 2016 in 304 couples with structural rearrangements. Clinical indications for PGT-SR were the presence of a Robertsonian translocation in 120 couples (n = 143 cycles), reciprocal translocation in 123 couples (n = 150 cycles), pericentric inversion in 23 couples (n = 24 cycles) and polymorphic inversion of chromosome 9 with a previous clinical history of infertility with recurrent miscarriages and/or implantation failures in 38 couples (n = 42 cycles). Couples were included in the study if they carried a structural chromosome rearrangement and the female was ≤ 43 years old. Couples with both partners carrying a balanced structural rearrangement as well as complex translocations involving > 2 chromosomes were excluded.
Ethical approval
The study was approved by the institutional review board at the Instituto Valenciano de Infertilidad (1503-IGX-020-EM).
Oocyte retrieval, culture conditions and embryo biopsy
Patients underwent ovarian stimulation using standardised protocols. When at least two follicles reached 18 mm in diameter, recombinant human chorionic gonadotropin (Ovitrelle, 250 mg; Merck Serono, Geneva, Switzerland) was administered, and oocytes were retrieved 36 h later. Intracytoplasmic sperm injection (ICSI) was performed in all cases [44]. Fertilisation was assessed 17–20 h after microinjection, and embryo cleavage was recorded every 24 h. Embryo culture was performed using IVF/CCM medium (Vitrolife, Göteborg, Sweden) or global sequential culture system (LifeGlobal, Guilford, CT, USA).
Embryo biopsy was performed at cleavage or blastocyst stage using laser technology (OCTAX, Herborn, Germany), and one blastomere or a trophectoderm sample was withdrawn from each embryo. At cleavage stage, only embryos with ≥ 5 nucleated blastomeres and < 25% fragmentation were biopsied. Individual blastomeres or trophectoderm samples were placed in 0.2-mL PCR tubes containing 2 μL of PBS. For blastomere and trophectoderm washing and handling, 1% polyvinylpyrrolidone was used. In day 3 biopsies, fresh transfers of normal/balanced embryos were performed. In day 5/6 biopsies, blastocysts were vitrified after biopsy and a deferred transfer was performed. Blastocysts were vitrified using the Cryotop method, as previously described [45].
WGA and aCGH
For WGA amplification, the multiple displacement amplification method for a single cell or a trophectoderm biopsy was performed using a SurePlex WGA Kit (Illumina, Cambridge, UK) according to the manufacturer’s instructions. Amplification quality was ensured by gel electrophoresis (Lonza, Rockland, ME, USA). Sample and control DNA were labelled with Cy3 and Cy5 fluorophores, mixed according to manufacturer’s instructions and hybridised for 6–12 h on 24sure (V2 and V3) for Robertsonian translocations and 24sure+arrays (Illumina, Cambridge, UK) for all other rearrangements. The technique involves competitive hybridisation of differentially labelled test and reference DNA samples. Fluorescence intensity was detected using a two-channel laser scanner (Powerscanner, TECAN, Männedorf, Switzerland), and BlueFuse Multi software (Illumina, Cambridge, UK) was used for data processing.
Statistical analysis
Euploid balanced embyos were defined as embryos without total or partial aneuploidies for any of the chromosomes. Euploid unbalanced embryos were defined as embryos with total or partial aneuploidies only for chromosomes involved in the rearrangement. Aneuploid balanced embryos were defined as embryos with total or partial aneuploidies only for chromosomes not involved in the rearrangement. Aneuploid unbalanced embryos were defined as embryos with total or partial aneuploidies for chromosomes involved and not involved in the rearrangement.
Implantation rate was defined as the percentage of embryos transferred resulting in an implanted gestational sac. Clinical pregnancy rate was calculated as the percentage of clinical pregnancies with a foetal heartbeat. Ongoing clinical pregnancy rate was calculated as the percentage of clinical pregnancies with a foetal heartbeat and evolving to term. Miscarriage rate was defined as the percentage of clinical pregnancies that were spontaneously miscarried before week 12 of pregnancy.
Comparisons of proportions for the incidence of chromosomal abnormalities and clinical outcomes between study groups were conducted using the Fisher or chi-square exact tests. Comparison of means was conducted using the Kruskal-Wallis test (nonparametric ANOVA). Statistical analysis was performed using the R Free software and GraphPad InStat version 3.00 for Windows 95 (GraphPad Software, San Diego California USA). A value of p < 0.05 was considered statistically significant.
Results
Parameters related to PGT-SR cycle
As shown in Table 1, a total of 359 cycles in 304 couples carrying a chromosomal rearrangement were included in the study. Mean female age was similar among groups, although mean age was significantly higher in the group with polymorphic inversions for chromosome 9 compared with the group with Robertsonian translocations (p < 0.05). Mean number of oocytes retrieved per cycle and mean number of previous implantation failures before PGT-SR cycle did not significantly differ among groups. Mean number of embryos biopsied per cycle did not significantly differ in embryos biopsied at a similar stage. As recurrent miscarriage was the main indication in carriers of polymorphic inversions for chromosome 9, this group had a significantly higher mean number of previous miscarriages compared with carriers of Robertsonian translocations (p < 0.05).
Table 1.
Parameters related to PGT-SR cycle
| Day 3 biopsy | Day 5/6 biopsy | |||||
|---|---|---|---|---|---|---|
| Robertsonian translocation | Reciprocal translocation | Pericentric inversion | Polymorphic inversion on chromosome 9 | Robertsonian translocation | Reciprocal translocation | |
| Patients | 112 | 80 | 23 | 38 | 8 | 43 |
| Cycles | 135 | 102 | 24 | 42 | 8 | 48 |
| Mean female age (years, SD) | 34.1 (4.2)a | 35.4 (4.0) | 35.1 (3.8) | 36.1 (4.7)a | 35.9 (5.2) | 35.1 (4.1) |
| Mean MII oocytes (SD) | 11.9 (5.1) | 13.1 (5.9) | 11.5 (4.9) | 11.0 (5.8) | 15.6 (8.2) | 14.0 (8.0) |
| Mean biopsied embryos (SD) | 6.6 (3.7) | 7.1 (4.1) | 5.8 (2.7) | 5.8 (3.2) | 5.5 (5.1) | 3.8 (2.5) |
| Mean previous miscarriages (SD) | 0.6 (0.9)b | 1.0 (1.3) | 0.9 (1.3) | 1.1 (1.0)b | 0.4 (0.7) | 1.0 (1.6) |
| Mean previous implantation failure (SD) | 0.8 (1.6) | 0.8 (1.3) | 0.4 (1.0) | 0.7 (1.3) | 0.6 (1.1) | 0.5 (0.9) |
a,bp < 0.05, for ANOVA comparison between columns in the same row. SD, standard deviation
Incidence of chromosomal abnormalities
Analysis of chromosomal abnormalities is shown in Table 2. No significant differences were observed in the percentage of informative results considering the day of biopsy or type of structural rearrangement (range 97–100%). In carriers of translocations, a higher percentage of abnormal embryos was observed from day 3 biopsies compared with day 5/6 biopsies, with significant differences for reciprocal translocations (p < 0.05). Focusing on day 3 biopsies, the percentage of abnormal embryos for reciprocal translocations was significantly increased compared with the other three groups of rearrangements (p < 0.05). On day 5/6 biopsies, the percentage of abnormal embryos in reciprocal translocations was significantly increased compared with Robertsonian translocations (p < 0.05).
Table 2.
Incidence of chromosomal abnormalities according to type of rearrangement and day of biopsy
| Day 3 biopsy | Day 5/6 biopsy | |||||
|---|---|---|---|---|---|---|
| Robertsonian translocation | Reciprocal translocation | Pericentric inversion | Polymorphic inversion on chromosome 9 | Robertsonian translocation | Reciprocal translocation | |
| Biopsied embryos | 889 | 725 | 140 | 242 | 44 | 180 |
| Informative results (%) | 864 (97.2) | 703 (97.0) | 138 (98.6) | 236 (97.5) | 44 (100.0) | 178 (98.9) |
| Abnormal embryos (%) | 638 (73.8)a | 628 (89.3)a,b,c,d | 94 (68.1)b | 161 (68.2)c | 27 (61.4)e | 143 (80.3)d,e |
| Euploid unbalanced embryos (%) | 114 (13.2)f,g,h | 182 (25.9)f,i,j,k | 9 (6.5)g,i | 7 (3.0)h,j | 7 (15.9)l | 62 (34.8)k,l |
| Aneuploid balanced embryos (%) | 289 (33.4)m,n | 153 (21.8)m,o,p | 50 (36.2)o | 104 (44.1)n,p | 16 (36.4) | 38 (21.3) |
| Aneuploid unbalanced embryos (%) | 118 (13.7)q,r,s | 206 (29.3)q,t,u | 9 (6.5)r,t | 9 (3.8)s,u | 3 (6.8)v | 41 (23.0)v |
| Aneuploid balanced + unbalanced embryos (%) | 407 (47.1) | 359 (51.1) | 59 (42.8) | 113 (47.9) | 19 (43.2) | 79 (44.4) |
| Chaotic embryos | 117 (13.5) | 87 (12.4)w | 26 (18.8) | 41 (17.4) | 1 (2.3) | 2 (1.1)w |
a–wp < 0.05, for Fishers exact test/chi-square test between columns in the same row
In all groups, we observed a high percentage of embryos with aneuploidies for chromosomes not involved in the rearrangement that could be attributed to total ICE (aneuploid balanced and unbalanced embryos). No significant differences were observed in these percentages between types of rearrangements. Pure ICE (aneuploid balanced embyos) was independent of female age only for Robertsonian translocations and significantly increased in day 3 biopsies for all types of abnormalities (Supplementary Table I). Furthermore, total ICE for carriers of Robertsonian translocations and biopsy on day 3 was independent of female age too. The percentage of euploid unbalanced embryos (without ICE) was also significantly higher for reciprocal translocations compared with other groups, both on day 3 and day 5/6 biopsies, followed by Robertsonian translocations, with a low percentage of euploid unbalanced embryos in both types of inversions. However, the pattern for pure ICE observed in aneuploid balanced embryos followed an opposite trend and was significantly higher for Robertsonian translocations and both types of inversions compared with reciprocal translocations for day 3 biopsies, without reaching statistical significance for day 5/6 biopsies. However, no significant differences were found for total ICE. Finally, the percentages of chaotic embryos were significantly higher on day 3 biopsies compared with day 5/6 biopsies (13.7% vs. 1.4%; p < 0.0001), since most chaotic embryos arrest before reaching the blastocyst stage.
Detailed analysis for each group according to maternal age is presented in Supplementary Table I, following a similar pattern as described above. Supplementary Table II shows the analysis according to sex of the carrier, indicating an increase in the percentage of abnormal embryos for female carriers of translocations, but without a sex effect for inversions.
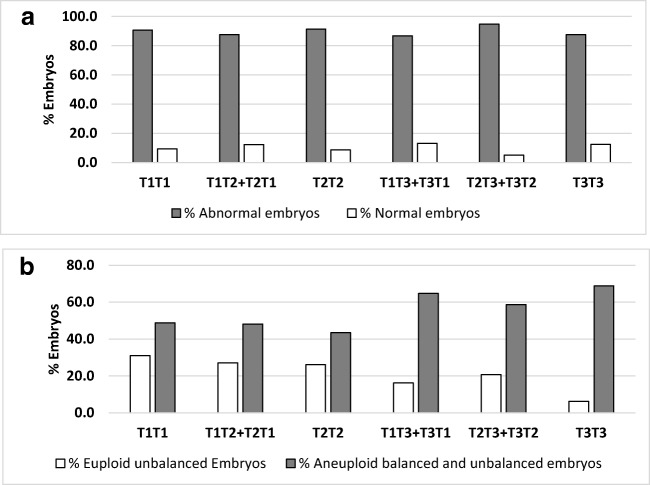
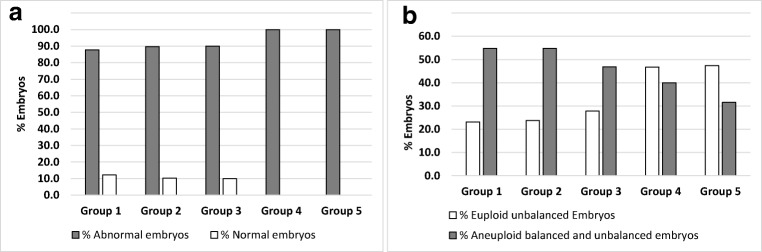
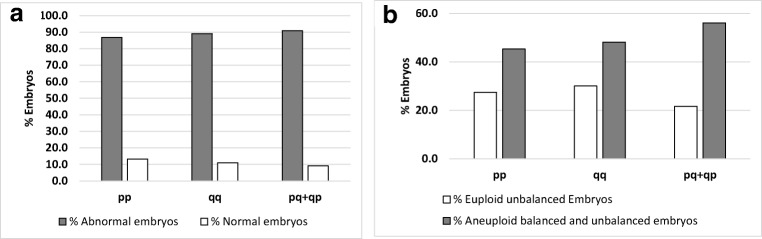
On day 3 biopsies of carriers of reciprocal translocations, we analysed the percentage of normal and abnormal embryos and the percentage of euploid unbalanced (without ICE) and aneuploid balanced/unbalanced (with ICE) embryos according to length of the translocated fragment (Fig. 1), chromosome group involved in the translocation (Fig. 2) and chromosome arms with the breakpoint (Fig. 3). No significant differences were observed for any of these parameters. However, there was a trend towards decreased percentage of euploid unbalanced embryos as the size of one of the chromosomes involved in the rearrangement increases (groups 1, 2 and 3; Fig. 2b).
Fig. 1.
Percentage of normal and abnormal day 3 embryos (a) and percentage of day 3 embryos without ICE (euploid unbalanced) and with ICE (aneuploid balanced and unbalanced) (b) according to translocated fragments length in carriers of reciprocal translocations. Fragment Length: T1 (6–40 Mb), T2 (41–80 Mb), or T3 (81–125 Mb)
Fig. 2.
Percentage of normal and abnormal day 3 embryos (a) and percentage of day 3 embryos without ICE (euploid unbalanced) and with ICE (aneuploid balanced and unbalanced) (b) according to the chromosome group of the human karyotype of translocated chromosomes in carriers of reciprocal translocations. Groups 1, 2 and 3 consists of translocations in which one of the chromosomes involved in the reciprocal translocation belongs to a group of large chromosomes of the karyotype (AA+AB+AC+AD+AE+AF+AG, BC+BD+BE and CC+CD+CE+CF+CG respectively); group 4 consists of translocations in which one of the chromosomes involved in the reciprocal translocation is a medium-size chromosomes (DD+DF) and group 5 consists of translocations in which chromosomes involved in the reciprocal translocation are small chromosomes (EE+EF)
Fig. 3.
Percentage of normal and abnormal day 3 embryos (a) and percentage of day 3 embryos without ICE (euploid unbalanced) and with ICE (aneuploid balanced and unbalanced) (b) according to the chromosome arm containing breakpoints. Translocations with both breakpoints in the short arm of the chromosomes involved in the reciprocal translocation (pp), both breakpoints in the long arm of the chromosomes (qq) or one breakpoint in the short arm and one breakpoint in the long arm (pq+qp) of both translocated chromosomes
Clinical outcome
A summary of clinical outcomes is presented in Table 3. High clinical pregnancy rate per transfer was observed for all groups, independent of the day of biopsy. Miscarriage and ongoing pregnancy rates were also comparable among groups. Clinical outcome according to age group did not show statistical differences for each type of structural rearrangement (Supplementary Table III). Clinical pregnancy rate per transfer was significantly higher for male carriers of translocations, and again, this sex effect was not observed for inversions (Supplementary Table IV).
Table 3.
Clinical outcome according to type of rearrangement and day of biopsy
| Day 3 biopsy | Day 5/6 biopsy | |||||
|---|---|---|---|---|---|---|
| Robertsonian translocation | Reciprocal translocation | Pericentric inversion | Polymorphic inversion on chromosome 9 | Robertsonian translocation | Reciprocal translocation | |
| Cycles with transference (%) | 90 (66.7) | 42 (89.3) | 15 (62.5) | 31 (73.8) | 6 (75.0) | 23 (47.9) |
| Mean transferred embryos (SD) | 1.5 (0.5) | 1.2 (0.4) | 1.4 (0.5) | 1.5 (0.5) | 1.5 (0.5) | 1.1 (0.3) |
| Clinical pregnancy rate/embryo transfer | 48.9 (44/90) | 50.0 (21/42) | 60.0 (9/15) | 71.0 (22/31)a | 83.3 (5/6) | 39.1 (9/23)a |
| Clinical pregnancy rate/patient | 39.3 (44/112)b | 26.3 (21/80)c | 39.1 (9/23) | 57.9 (22/38)c,d | 62.5 (5/8) | 20.9 (9/43)b,d |
| Clinical pregnancy rate/cycle | 32.6 (44/135)e,f | 20.6 (21/102)g | 37.5 (9/24) | 52.4 (22/42)e,g,h | 62.5 (5/8) | 18.8 (9/48)f,h |
| Ongoing clinical pregnancy rate/transfer | 42.2 (38/90) | 50.0 (21/42) | 60.0 (9/15) | 64.5 (20/31) | 50.0 (3/6) | 39.1 (9/23) |
| Implantation rate | 42.4 (59/139) | 45.1 (23/51) | 52.4 (11/21) | 60.0 (27/45)i | 88.9 (8/9) | 34.6 (9/26)i |
| Miscarriage rate | 13.6 (6/44) | 0.0 (0/21)j | 0.0 (0/9) | 9.1 (2/22) | 40.0 (2/5)j | 0.0 (0/9) |
a–jp < 0.05, for Fishers exact test/chi-square test between columns in the same row. SD, standard deviation
Discussion
Our analysis of 359 PGT-SR cycles from 304 couples carrying a chromosomal rearrangement found a higher percentage of abnormal embryos for reciprocal translocations, a higher pure ICE for Robertsonian translocations and a sex effect for clinical outcome for carriers of translocations, with higher pregnancy rate for male carriers. It is important to emphasise the differences in aneuploid balanced embryos because they would be classified as normal/balanced with techniques that only analyse the chromosomes involved in the rearrangement. The size of the study allowed us to perform several sub-analyses considering different types of rearrangements, chromosomes, breakpoints and carrier sex.
Starting with overall incidence of chromosome abnormalities, we observed a higher percentage of abnormal embryos for reciprocal translocations, with more in day 3 biopsies compared with day 5/6 biopsies. This could be explained by natural selection between day 3 and day 5/6 embryos in couples with reciprocal or Robertsonian translocations [46], which could be attributed to delayed development of embryos carrying unbalanced chromosomal translocations as shown by time-lapse imaging [47]. Further, the percentage of chaotic embryos was significantly higher in day 3 biopsies compared with day 5/6 biopsies. We previously reported a similar trend in cases with preimplantation genetic testing for aneuploidies (PGT-A), indicating that chaotic day 3 embryos arrest before reaching the blastocyst stage as a result of the number of aneuploid chromosomes in cell divisions [48]. Previous publications have reported high percentages of abnormal gametes and embryos produced by couples carrying a structural rearrangement. For these couples, PGT-SR is effective to reduce the number of miscarriages and children born with an unbalanced translocation in comparison with spontaneous conception [10, 11, 14, 49], especially with molecular analysis techniques to analyse the whole karyotype [27].
Different frequencies of segregation modes and incidence of unbalanced gametes between male and female carriers of translocations have also been reported recently [50]. In the present study, we found an increase in the percentage of abnormal embryos in female carriers of translocations, but no sex effect for inversions. Regarding the effect of cytogenetic aspects of translocations, many published studies have attempted to determine the relationship between breakpoints, chromosomes involved in the translocation, regions of translocated chromosomes, size of centric and translocated fragments, and symmetry of the quadrivalent during meiosis, with the preferential type of segregation in gametes and embryos of translocation carriers [8, 51]. We found a trend towards higher percentage of abnormal segregation modes with smaller size of translocated fragments and with smaller chromosomes. Quadrivalents with acrocentric chromosomes have been correlated with a higher incidence of unbalanced adjacent-1 products, and asymmetric quadrivalents have been correlated with a higher percentage of unbalanced adjacent-2 products [50].
Focusing on ICE, day 3 biopsies showing aneuploid balanced embryos were significantly higher for Robertsonian translocations and both types of inversions compared with reciprocal translocations, although day 5/6 biopsies showed no significant differences. In sperm, ICE has not been observed for reciprocal translocations [34, 35, 52, 53], but several studies have found ICE in sperm from carriers of Robertsonian translocations. This could be explained by association of asynaptic regions of translocated fragments to other bivalents [54–56].
Most sperm studies using FISH have only analysed a limited number of chromosomes, which could contribute to differences from analysis of 23 chromosome pairs by potentially underestimating ICE in sperm [57]. Godo et al. (2015) [38] found that the frequency of normal/balanced spermatozoa in reciprocal translocations is much lower in aneuploid and diploid sperm than in the population of non-selected spermatozoa, with differences among patients. In fact, our study of reciprocal translocations in day 3 embryos confirmed an increase of ICE only in unbalanced embryos compared with Robertsonian translocations, with increased ICE in balanced/normal embryos. Other studies in oocytes and embryos have found ICE only in Robertsonian translocations [41, 58]. Therefore, ICE may have a mitotic rather than a meiotic origin for embryos [39, 59].
Inter-patient differences in the incidence of ICE could be correlated to size of the rearranged fragment and specific chromosomes involved in the rearrangement [8]. Other studies have associated increased ICE rate to other factors, such as combination with oligoasthenoteratozoospermia or advanced female age [60–64]. In our study, we observed similar percentages of aneuploidy in normal/balanced day 3 embryos for both male and female carriers, and the effect was independent of female age for Robertsonian and reciprocal translocations. However, in inversion carriers, aneuploidy rates were higher in patients 38–42 years of age and thus could be mainly attributed to female age.
Regarding clinical outcome, a recent systematic review showed no benefit in live birth rate in PGT-SR cases with recurrent miscarriages compared with natural conception [65], but only two reviewed studies have compared both natural and PGT-SR cycles [66]. Another limitation of the systematic review is heterogeneity of included studies, some of which collected cumulative live birth rates for different periods of time. Further, PGT-SR technology differed among studies. Ikuma et al. (2015) [66] included FISH on day 3 biopsies, without accounting for other aneuploidies that are detected with aCGH or NGS. Another study also including FISH described successful clinical outcomes compared with previous reproductive history in carriers of translocations with fertility problems [67], emphasising potential population differences in studies of conception in couples that can conceive naturally and PGT-SR studies, in which most couples are infertile due to structural rearrangements. These results suggest cytogenetic differences among translocations and their different impact from gametogenesis to birth.
More recently, a study by Maithripala et al. (2018) [68] including cases with FISH, aCGH and NGS reported live birth rates for PGT-SR and clinical management of 66.6% and 53.3%, respectively. However, this difference was not statistically significant, which could be due to the limited number of cases included in the study and the fact that it was a retrospective study. To our knowledge, there is no prospective study comparing clinical outcomes of natural conceptions vs. PGT-SR with current PGT technology based on trophectoderm biopsies and analysis of 24 chromosomes. Therefore, additional studies are needed to draw conclusions with current PGT-SR protocols.
In addition to considering live birth rates, other critical parameters are relevant, such as decreased miscarriages [66] as well as potential reductions in unbalanced live births despite being neglected in terms of incidence; these are major concerns for most couples. In our study, high ongoing pregnancy rates were achieved for all types of structural rearrangements and biopsy days. Clinical pregnancy rate per transfer was significantly higher for male carriers of translocations, and this sex effect was not observed for inversions. In addition, a sex effect has not been observed previously for reciprocal translocations [50, 69]. Finally, despite no consensus among publications, the length of time to conceive was also dramatically reduced compared with published data for similar populations not undergoing PGT-SR [10]. In summary, these factors, particularly the higher incidence of unbalanced and aneuploid embryos, should be considered for reproductive counselling in carriers of structural rearrangements.
Electronic supplementary materials
Supplementary Table I. Categorization of embryos stratified by maternal age, Supplementary Table II.Categorization of embryos according to sex of the carrier of the rearrangement, Supplementary Table III. Clinical outcome according to maternal age and structural rearrangement, Supplementary Table IV. Clinical outcome according to sex of the carrier of the rearrangement (DOCX 53 kb)
Compliance with ethical standards
The study was approved by the institutional review and ethical board at the Instituto Valenciano de Infertilidad (1503-IGX-020-EM).
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Jacobs PA, Melville M, Ratcliffe S, Keay AJ, Syme J. A cytogenetic survey of 11,680 newborn infants. Ann Hum Genet. 1974;37:359–376. doi: 10.1111/j.1469-1809.1974.tb01843.x. [DOI] [PubMed] [Google Scholar]
- 2.Van Dyke DL, Weiss L, Roberson JR, Babu VR. The frequency and mutation rate of balanced autosomal rearrangements in man estimated from prenatal genetic studies for advanced maternal age. Am J Hum Genet. 1983;35:301–308. [PMC free article] [PubMed] [Google Scholar]
- 3.Campana M, Serra A, Neri G. Role of chromosome aberrations in recurrent abortion: a study of 269 balanced translocations. Am J Med Genet. 1986;24:341–356. doi: 10.1002/ajmg.1320240214. [DOI] [PubMed] [Google Scholar]
- 4.Fryns JP, Van Buggenhout G. Structural chromosome rearrangements in couples with recurrent fetal wastage. Eur J Obstet Gynecol Reprod Biol. 1998;81:171–176. doi: 10.1016/s0301-2115(98)00185-7. [DOI] [PubMed] [Google Scholar]
- 5.Stern C, Pertile M, Norris H, Hale L, Baker HW. Chromosome translocations in couples with in-vitro fertilization implantation failure. Hum Reprod. 1999;14:2097–2101. doi: 10.1093/humrep/14.8.2097. [DOI] [PubMed] [Google Scholar]
- 6.Neri G, Serra A, Campana M, Tedeschi B. Reproductive risks for translocation carriers: cytogenetic study and analysis of pregnancy outcome in 58 families. Am J Med Genet. 1983;16:535–561. doi: 10.1002/ajmg.1320160412. [DOI] [PubMed] [Google Scholar]
- 7.Scriven PN, Handyside AH, Ogilvie CM. Chromosome translocations: segregation modes and strategies for preimplantation genetic diagnosis. Prenat Diagn. 1998;18:1437–1449. [PubMed] [Google Scholar]
- 8.Anton E, Vidal F, Blanco J. Reciprocal translocations: tracing their meiotic behavior. Genet Med. 2008;10:730–738. doi: 10.1097/GIM.0b013e318187760f. [DOI] [PubMed] [Google Scholar]
- 9.Conn CM, Harper JC, Winston RM, Delhanty JD. Infertile couples with Robertsonian translocations: preimplantation genetic analysis of embryos reveals chaotic cleavage divisions. Hum Genet. 1998;102:117–123. doi: 10.1007/s004390050663. [DOI] [PubMed] [Google Scholar]
- 10.Fischer J, Colls P, Escudero T, Munne S. Preimplantation genetic diagnosis (PGD) improves pregnancy outcome for translocation carriers with a history of recurrent losses. Fertil Steril. 2010;94:283–289. doi: 10.1016/j.fertnstert.2009.02.060. [DOI] [PubMed] [Google Scholar]
- 11.Munne S, Sandalinas M, Escudero T, Fung J, Gianaroli L, Cohen J. Outcome of preimplantation genetic diagnosis of translocations. Fertil Steril. 2000;73:1209–1218. doi: 10.1016/s0015-0282(00)00495-7. [DOI] [PubMed] [Google Scholar]
- 12.Verlinsky Y, Tur-Kaspa I, Cieslak J, Bernal A, Morris R, Taranissi M, Kaplan B, Kuliev A. Preimplantation testing for chromosomal disorders improves reproductive outcome of poor-prognosis patients. Reprod BioMed Online. 2005;11:219–225. doi: 10.1016/s1472-6483(10)60961-3. [DOI] [PubMed] [Google Scholar]
- 13.Munne S, Bahce M, Schimmel T, Sadowy S, Cohen J. Case report: chromatid exchange and predivision of chromatids as other sources of abnormal oocytes detected by preimplantation genetic diagnosis of translocations. Prenat Diagn. 1998;18:1450–1458. [PubMed] [Google Scholar]
- 14.Munne S, Morrison L, Fung J, Marquez C, Weier U, Bahce M, Sable D, Grundfeld L, Schoolcraft B, Scott R, Cohen J. Spontaneous abortions are reduced after preconception diagnosis of translocations. J Assist Reprod Genet. 1998;15:290–296. doi: 10.1023/A:1022544511198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verlinsky Y, Evsikov S. Karyotyping of human oocytes by chromosomal analysis of the second polar bodies. Mol Hum Reprod. 1999;5:89–95. doi: 10.1093/molehr/5.2.89. [DOI] [PubMed] [Google Scholar]
- 16.Harper JC, Wilton L, Traeger-Synodinos J, Goossens V, Moutou C, SenGupta SB, Pehlivan Budak T, Renwick P, De Rycke M, Geraedts JP, Harton G. The ESHRE PGD Consortium: 10 years of data collection. Hum Reprod Update. 2012;18:234–247. doi: 10.1093/humupd/dmr052. [DOI] [PubMed] [Google Scholar]
- 17.DeUgarte CM, Li M, Surrey M, Danzer H, Hill D, DeCherney AH. Accuracy of FISH analysis in predicting chromosomal status in patients undergoing preimplantation genetic diagnosis. Fertil Steril. 2008;90:1049–1054. doi: 10.1016/j.fertnstert.2007.07.1337. [DOI] [PubMed] [Google Scholar]
- 18.Munne S. Preimplantation genetic diagnosis of numerical and structural chromosome abnormalities. Reprod BioMed Online. 2002;4:183–196. doi: 10.1016/s1472-6483(10)61938-4. [DOI] [PubMed] [Google Scholar]
- 19.Velilla E, Escudero T, Munne S. Blastomere fixation techniques and risk of misdiagnosis for preimplantation genetic diagnosis of aneuploidy. Reprod BioMed Online. 2002;4:210–217. doi: 10.1016/s1472-6483(10)61808-1. [DOI] [PubMed] [Google Scholar]
- 20.Fiorentino F, Kokkali G, Biricik A, Stavrou D, Ismailoglu B, De Palma R, Arizzi L, Harton G, Sessa M, Pantos K. Polymerase chain reaction-based detection of chromosomal imbalances on embryos: the evolution of preimplantation genetic diagnosis for chromosomal translocations. Fertil Steril. 2010;94:2001-11, 2011.e1-6. doi: 10.1016/j.fertnstert.2009.12.063. [DOI] [PubMed] [Google Scholar]
- 21.Lukaszuk K, Pukszta S, Ochman K, Cybulska C, Liss J, Pastuszek E, Zabielska J, Woclawek-Potocka I. Healthy baby born to a Robertsonian translocation carrier following next-generation sequencing-based preimplantation genetic diagnosis: a case report. AJP Rep. 2015;5:e172–e175. doi: 10.1055/s-0035-1558402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan YQ, Tan K, Zhang SP, Gong F, Cheng DH, Xiong B, Lu CF, Tang XC, Luo KL, Lin G, Lu GX. Single-nucleotide polymorphism microarray-based preimplantation genetic diagnosis is likely to improve the clinical outcome for translocation carriers. Hum Reprod. 2013;28:2581–2592. doi: 10.1093/humrep/det271. [DOI] [PubMed] [Google Scholar]
- 23.Treff NR, Tao X, Schillings WJ, Bergh PA, Scott RT, Jr, Levy B. Use of single nucleotide polymorphism microarrays to distinguish between balanced and normal chromosomes in embryos from a translocation carrier. Fertil Steril. 2011;96:e58–e65. doi: 10.1016/j.fertnstert.2011.04.038. [DOI] [PubMed] [Google Scholar]
- 24.van Uum CM, Stevens SJ, Dreesen JC, Drusedau M, Smeets HJ, Hollanders-Crombach B, Die-Smulders CE, Geraedts JP, Engelen JJ, Coonen E. SNP array-based copy number and genotype analyses for preimplantation genetic diagnosis of human unbalanced translocations. Eur J Hum Genet. 2012;20:938–944. doi: 10.1038/ejhg.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie Yanxin, Xu Yanwen, Wang Jing, Miao Benyu, Zeng Yanhong, Ding Chenhui, Gao Jun, Zhou Canquan. Preliminary analysis of numerical chromosome abnormalities in reciprocal and Robertsonian translocation preimplantation genetic diagnosis cases with 24-chromosomal analysis with an aCGH/SNP microarray. Journal of Assisted Reproduction and Genetics. 2017;35(1):177–186. doi: 10.1007/s10815-017-1045-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alfarawati S, Fragouli E, Colls P, Wells D. First births after preimplantation genetic diagnosis of structural chromosome abnormalities using comparative genomic hybridization and microarray analysis. Hum Reprod. 2011;26:1560–1574. doi: 10.1093/humrep/der068. [DOI] [PubMed] [Google Scholar]
- 27.Fiorentino F, Spizzichino L, Bono S, Biricik A, Kokkali G, Rienzi L, Ubaldi FM, Iammarrone E, Gordon A, Pantos K. PGD for reciprocal and Robertsonian translocations using array comparative genomic hybridization. Hum Reprod. 2011;26:1925–1935. doi: 10.1093/humrep/der082. [DOI] [PubMed] [Google Scholar]
- 28.Li G, Jin H, Xin Z, Su Y, Brezina PR, Benner AT, Kearns WG, Sun Y. Increased IVF pregnancy rates after microarray preimplantation genetic diagnosis due to parental translocations. Syst Biol Reprod Med. 2014;60:119–124. doi: 10.3109/19396368.2013.875241. [DOI] [PubMed] [Google Scholar]
- 29.Tobler KJ, Brezina PR, Benner AT, Du L, Xu X, Kearns WG. Two different microarray technologies for preimplantation genetic diagnosis and screening, due to reciprocal translocation imbalances, demonstrate equivalent euploidy and clinical pregnancy rates. J Assist Reprod Genet. 2014;31:843–850. doi: 10.1007/s10815-014-0230-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bono S, Biricik A, Spizzichino L, Nuccitelli A, Minasi MG, Greco E, Spinella F, Fiorentino F. Validation of a semiconductor next-generation sequencing-based protocol for preimplantation genetic diagnosis of reciprocal translocations. Prenat Diagn. 2015;35:938–944. doi: 10.1002/pd.4665. [DOI] [PubMed] [Google Scholar]
- 31.Wang L, Cram DS, Shen J, Wang X, Zhang J, Song Z, Xu G, Li N, Fan J, Wang S, Luo Y, Wang J, Yu L, Liu J, Yao Y. Validation of copy number variation sequencing for detecting chromosome imbalances in human preimplantation embryos. Biol Reprod. 2014;91:37. doi: 10.1095/biolreprod.114.120576. [DOI] [PubMed] [Google Scholar]
- 32.Lejeune J. Autosomal disorders. Pediatrics. 1963;32:326–337. [PubMed] [Google Scholar]
- 33.Anton E, Vidal F, Blanco J. Interchromosomal effect analyses by sperm FISH: incidence and distribution among reorganization carriers. Syst Biol Reprod Med. 2011;57:268–278. doi: 10.3109/19396368.2011.633682. [DOI] [PubMed] [Google Scholar]
- 34.Anton E, Blanco J, Egozcue J, Vidal F. Sperm FISH studies in seven male carriers of Robertsonian translocation t(13;14)(q10;q10) Hum Reprod. 2004;19:1345–1351. doi: 10.1093/humrep/deh232. [DOI] [PubMed] [Google Scholar]
- 35.Blanco J, Egozcue J, Clusellas N, Vidal F. FISH on sperm heads allows the analysis of chromosome segregation and interchromosomal effects in carriers of structural rearrangements: results in a translocation carrier, t(5;8)(q33;q13) Cytogenet Cell Genet. 1998;83:275–280. doi: 10.1159/000015170. [DOI] [PubMed] [Google Scholar]
- 36.Douet-Guilbert N, Bris MJ, Amice V, Marchetti C, Delobel B, Amice J, Braekeleer MD, Morel F. Interchromosomal effect in sperm of males with translocations: report of 6 cases and review of the literature. Int J Androl. 2005;28:372–379. doi: 10.1111/j.1365-2605.2005.00571.x. [DOI] [PubMed] [Google Scholar]
- 37.Escudero T, Lee M, Carrel D, Blanco J, Munne S. Analysis of chromosome abnormalities in sperm and embryos from two 45,XY,t(13;14)(q10;q10) carriers. Prenat Diagn. 2000;20:599–602. doi: 10.1002/1097-0223(200007)20:7<599::aid-pd883>3.3.co;2-h. [DOI] [PubMed] [Google Scholar]
- 38.Godo A, Blanco J, Vidal F, Sandalinas M, Garcia-Guixe E, Anton E. Altered segregation pattern and numerical chromosome abnormalities interrelate in spermatozoa from Robertsonian translocation carriers. Reprod BioMed Online. 2015;31:79–88. doi: 10.1016/j.rbmo.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 39.Alfarawati S, Fragouli E, Colls P, Wells D. Embryos of robertsonian translocation carriers exhibit a mitotic interchromosomal effect that enhances genetic instability during early development. PLoS Genet. 2012;8:e1003025. doi: 10.1371/journal.pgen.1003025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Durban M, Benet J, Boada M, Fernandez E, Calafell JM, Lailla JM, Sanchez-Garcia JF, Pujol A, Egozcue J, Navarro J. PGD in female carriers of balanced Robertsonian and reciprocal translocations by first polar body analysis. Hum Reprod Update. 2001;7:591–602. doi: 10.1093/humupd/7.6.591. [DOI] [PubMed] [Google Scholar]
- 41.Gutierrez-Mateo C, Gadea L, Benet J, Wells D, Munne S, Navarro J. Aneuploidy 12 in a Robertsonian (13;14) carrier: case report. Hum Reprod. 2005;20:1256–1260. doi: 10.1093/humrep/deh751. [DOI] [PubMed] [Google Scholar]
- 42.Gianaroli L, Magli MC, Ferraretti AP, Munne S, Balicchia B, Escudero T, Crippa A. Possible interchromosomal effect in embryos generated by gametes from translocation carriers. Hum Reprod. 2002;17:3201–3207. doi: 10.1093/humrep/17.12.3201. [DOI] [PubMed] [Google Scholar]
- 43.Tulay P, Gultomruk M, Findikli N, Yagmur E, Bahceci M. Is the interchromosomal effect present in embryos derived from Robertsonian and reciprocal translocation carriers particularly focusing on chromosome 10 rearrangements? Zygote. 2015;23:908–915. doi: 10.1017/S0967199414000628. [DOI] [PubMed] [Google Scholar]
- 44.Rubio C, Mercader A, Alama P, Lizan C, Rodrigo L, Labarta E, Melo M, Pellicer A, Remohi J. Prospective cohort study in high responder oocyte donors using two hormonal stimulation protocols: impact on embryo aneuploidy and development. Hum Reprod. 2010;25:2290–2297. doi: 10.1093/humrep/deq174. [DOI] [PubMed] [Google Scholar]
- 45.Cobo A, Bellver J, Domingo J, Perez S, Crespo J, Pellicer A, Remohi J. New options in assisted reproduction technology: the Cryotop method of oocyte vitrification. Reprod BioMed Online. 2008;17:68–72. doi: 10.1016/s1472-6483(10)60295-7. [DOI] [PubMed] [Google Scholar]
- 46.Beyer Claire E., Willats E. Natural selection between day 3 and day 5/6 PGD embryos in couples with reciprocal or Robertsonian translocations. Journal of Assisted Reproduction and Genetics. 2017;34(11):1483–1492. doi: 10.1007/s10815-017-1009-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amir H, Barbash-Hazan S, Kalma Y, Frumkin T, Malcov M, Samara N, Hasson J, Reches A, Azem F, Ben-Yosef D. Time-lapse imaging reveals delayed development of embryos carrying unbalanced chromosomal translocations. J Assist Reprod Genet. 2019;36:315–324. doi: 10.1007/s10815-018-1361-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodrigo L, Mateu E, Mercader A, Cobo AC, Peinado V, Milan M, Al-Asmar N, Campos-Galindo I, Garcia-Herrero S, Mir P, Simon C, Rubio C. New tools for embryo selection: comprehensive chromosome screening by array comparative genomic hybridization. Biomed Res Int. 2014;2014:517125. doi: 10.1155/2014/517125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Otani T, Roche M, Mizuike M, Colls P, Escudero T, Munne S. Preimplantation genetic diagnosis significantly improves the pregnancy outcome of translocation carriers with a history of recurrent miscarriage and unsuccessful pregnancies. Reprod BioMed Online. 2006;13:869–874. doi: 10.1016/s1472-6483(10)61037-1. [DOI] [PubMed] [Google Scholar]
- 50.Zhang S, Lei C, Wu J, Sun H, Zhou J, Zhu S, Wu J, Fu J, Sun Y, Lu D, Sun X, Zhang Y. Analysis of segregation patterns of quadrivalent structures and the effect on genome stability during meiosis in reciprocal translocation carriers. Hum Reprod. 2018;33:757–767. doi: 10.1093/humrep/dey036. [DOI] [PubMed] [Google Scholar]
- 51.Jalbert P, Sele B, Jalbert H. Reciprocal translocations: a way to predict the mode of imbalanced segregation by pachytene-diagram drawing. Hum Genet. 1980;55:209–222. doi: 10.1007/BF00291769. [DOI] [PubMed] [Google Scholar]
- 52.Chelli MH, Ferfouri F, Boitrelle F, Albert M, Molina-Gomes D, Selva J, Vialard F. High-magnification sperm selection does not decrease the aneuploidy rate in patients who are heterozygous for reciprocal translocations. J Assist Reprod Genet. 2013;30:525–530. doi: 10.1007/s10815-013-9959-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Estop AM, Cieply K, Munne S, Surti U, Wakim A, Feingold E. Is there an interchromosomal effect in reciprocal translocation carriers? Sperm FISH studies. Hum Genet. 2000;106:517–524. doi: 10.1007/s004390000275. [DOI] [PubMed] [Google Scholar]
- 54.Kirkpatrick G, Ren H, Liehr T, Chow V, Ma S. Meiotic and sperm aneuploidy studies in three carriers of Robertsonian translocations and small supernumerary marker chromosomes. Fertil Steril. 2015;103:1162-9.e7. doi: 10.1016/j.fertnstert.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 55.Luciani JM, Guichaoua MR, Mattei A, Morazzani MR. Pachytene analysis of a man with a 13q;14q translocation and infertility. Behavior of the trivalent and nonrandom association with the sex vesicle. Cytogenet Cell Genet. 1984;38:14–22. doi: 10.1159/000132023. [DOI] [PubMed] [Google Scholar]
- 56.Navarro J, Vidal F, Benet J, Templado C, Marina S, Egozcue J. XY-trivalent association and synaptic anomalies in a male carrier of a Robertsonian t(13;14) translocation. Hum Reprod. 1991;6:376–381. doi: 10.1093/oxfordjournals.humrep.a137343. [DOI] [PubMed] [Google Scholar]
- 57.Rogenhofer N, Durl S, Ochsenkuhn R, Neusser M, Aichinger E, Thaler CJ, Muller S. Case report: elevated sperm aneuploidy levels in an infertile Robertsonian translocation t(21;21) carrier with possible interchromosomal effect. J Assist Reprod Genet. 2012;29:343–346. doi: 10.1007/s10815-012-9720-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pujol A, Durban M, Benet J, Boiso I, Calafell JM, Egozcue J, Navarro J. Multiple aneuploidies in the oocytes of balanced translocation carriers: a preimplantation genetic diagnosis study using first polar body. Reproduction. 2003;126:701–711. doi: 10.1530/rep.0.1260701. [DOI] [PubMed] [Google Scholar]
- 59.Ghevaria H, SenGupta S, Shmitova N, Serhal P, Delhanty J. The origin and significance of additional aneuploidy events in couples undergoing preimplantation genetic diagnosis for translocations by array comparative genomic hybridization. Reprod BioMed Online. 2016;32:178–189. doi: 10.1016/j.rbmo.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 60.Moosani N, Pattinson HA, Carter MD, Cox DM, Rademaker AW, Martin RH. Chromosomal analysis of sperm from men with idiopathic infertility using sperm karyotyping and fluorescence in situ hybridization. Fertil Steril. 1995;64:811–817. doi: 10.1016/s0015-0282(16)57859-5. [DOI] [PubMed] [Google Scholar]
- 61.Pang MG, Hoegerman SF, Cuticchia AJ, Moon SY, Doncel GF, Acosta AA, Kearns WG. Detection of aneuploidy for chromosomes 4, 6, 7, 8, 9, 10, 11, 12, 13, 17, 18, 21, X and Y by fluorescence in-situ hybridization in spermatozoa from nine patients with oligoasthenoteratozoospermia undergoing intracytoplasmic sperm injection. Hum Reprod. 1999;14:1266–1273. doi: 10.1093/humrep/14.5.1266. [DOI] [PubMed] [Google Scholar]
- 62.Rubio C, Bellver J, Rodrigo L, Castillon G, Guillen A, Vidal C, Giles J, Ferrando M, Cabanillas S, Remohi J, Pellicer A, Simon C. In vitro fertilization with preimplantation genetic diagnosis for aneuploidies in advanced maternal age: a randomized, controlled study. Fertil Steril. 2017;107:1122–1129. doi: 10.1016/j.fertnstert.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 63.Rubio C, Gil-Salom M, Simon C, Vidal F, Rodrigo L, Minguez Y, Remohi J, Pellicer A. Incidence of sperm chromosomal abnormalities in a risk population: relationship with sperm quality and ICSI outcome. Hum Reprod. 2001;16:2084–2092. doi: 10.1093/humrep/16.10.2084. [DOI] [PubMed] [Google Scholar]
- 64.Xie Y, Xu Y, Wang J, Miao B, Zeng Y, Ding C, Gao J, Zhou C. Preliminary analysis of numerical chromosome abnormalities in reciprocal and Robertsonian translocation preimplantation genetic diagnosis cases with 24-chromosomal analysis with an aCGH/SNP microarray. J Assist Reprod Genet. 2018;35:177–186. doi: 10.1007/s10815-017-1045-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Iews M, Tan J, Taskin O, Alfaraj S, AbdelHafez FF, Abdellah AH, Bedaiwy MA. Does preimplantation genetic diagnosis improve reproductive outcome in couples with recurrent pregnancy loss owing to structural chromosomal rearrangement? A systematic review. Reprod BioMed Online. 2018;36:677–685. doi: 10.1016/j.rbmo.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 66.Ikuma S, Sato T, Sugiura-Ogasawara M, Nagayoshi M, Tanaka A, Takeda S. Preimplantation genetic diagnosis and natural conception: a comparison of live birth rates in patients with recurrent pregnancy loss associated with translocation. PLoS One. 2015;10:e0129958. doi: 10.1371/journal.pone.0129958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Keymolen K, Staessen C, Verpoest W, Liebaers I, Bonduelle M. Preimplantation genetic diagnosis in female and male carriers of reciprocal translocations: clinical outcome until delivery of 312 cycles. Eur J Hum Genet. 2012;20:376–380. doi: 10.1038/ejhg.2011.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maithripala S, Durland U, Havelock J, Kashyap S, Hitkari J, Tan J, Iews M, Lisonkova S, Bedaiwy MA. Prevalence and treatment choices for couples with recurrent pregnancy loss due to structural chromosomal anomalies. J Obstet Gynaecol Can. 2018;40:655–662. doi: 10.1016/j.jogc.2017.09.024. [DOI] [PubMed] [Google Scholar]
- 69.Kato K, Aoyama N, Kawasaki N, Hayashi H, Xiaohui T, Abe T, Kuroda T. Reproductive outcomes following preimplantation genetic diagnosis using fluorescence in situ hybridization for 52 translocation carrier couples with a history of recurrent pregnancy loss. J Hum Genet. 2016;61:687–692. doi: 10.1038/jhg.2016.39. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table I. Categorization of embryos stratified by maternal age, Supplementary Table II.Categorization of embryos according to sex of the carrier of the rearrangement, Supplementary Table III. Clinical outcome according to maternal age and structural rearrangement, Supplementary Table IV. Clinical outcome according to sex of the carrier of the rearrangement (DOCX 53 kb)