Significance
Alcohol drinking is a widespread habit in modern society and can cause deleterious metabolic consequences. Recent studies have uncovered the interplay between nutrition, metabolism, and circadian rhythms. Here we explore the effects of alcohol consumption on circadian physiology. We employed unbiased high-throughput proteomics, acetylomics, and circadian transcriptomics to provide a comprehensive analysis of liver metabolism and compared the outcomes of acute and chronic alcohol consumption. We reveal that distinct patterns of alcohol consumption differentially reprogram circadian gene expression in the liver. Particularly, rewiring of circadian SREBP pathways impacts subcellular partitioning of protein acetylation through acetyl-CoA metabolism. These findings contribute to understanding the underlying mechanism of alcoholic liver disease and establishing circadian therapeutic targets and strategies.
Keywords: alcohol, circadian, acetylation, liver, metabolism
Abstract
Binge drinking and chronic exposure to ethanol contribute to alcoholic liver diseases (ALDs). A potential link between ALDs and circadian disruption has been observed, though how different patterns of alcohol consumption differentially impact hepatic circadian metabolism remains virtually unexplored. Using acute versus chronic ethanol feeding, we reveal differential reprogramming of the circadian transcriptome in the liver. Specifically, rewiring of diurnal SREBP transcriptional pathway leads to distinct hepatic signatures in acetyl-CoA metabolism that are translated into the subcellular patterns of protein acetylation. Thus, distinct drinking patterns of alcohol dictate differential adaptation of hepatic circadian metabolism.
Alcohol consumption is a common habit in modern society. Drinking behavior such as habitual drinking, alcohol dependence, and even single-episode binge drinking can cause adverse health consequences and social and economic harm (1). Alcohol-related diseases range from neuropsychiatric disorders to cancers and cardiovascular modalities (1) with significantly different effects depending on the amount and patterns of alcohol consumption (1, 2). Additional factors include age, gender, socioeconomic status, and country (1, 3), underscoring the importance of understanding how acute and chronic patterns of alcohol consumption may induce alcohol-related disease.
Being primarily metabolized in the liver, alcohol is a prominent risk factor for the development of alcoholic liver diseases (ALDs), with degrees of severity ranging from steatosis (fatty liver), alcoholic hepatitis (a combination of steatosis and inflammation), and fibrosis to cirrhosis and hepatocellular carcinoma (2, 4). Multiple underlying molecular mechanisms have been reported in ALD pathogenesis, such as oxidative stress, inflammation, and cellular injury (2, 4, 5). Moreover, additional risk factors are associated with the progression of ALDs, including obesity and smoking (5). Metabolically, ethanol is oxidized to acetaldehyde by NAD+-dependent alcohol dehydrogenase or microsomal CYP2E1 and further oxidized to acetate by NAD+-dependent aldehyde dehydrogenase (2, 6). As a result of a decreased NAD+/NADH ratio, lipid oxidation is attenuated, leading to hepatic steatosis (5, 7). In addition, ethanol-derived acetaldehyde inactivates hepatic Peroxisome Proliferator-Activated Receptor-α (PPAR-α), contributing to the repression of mitochondrial fatty acid oxidation (8).
Emerging evidence suggests a reciprocal link between circadian rhythms and alcohol metabolism (9). Per2-mutant mice display increased alcohol consumption through the glutamatergic system in the brain (10). Moreover, alcohol-induced gut leakiness is aggravated by clock disruption, contributing to progression of ALDs, suggesting that the clock is involved not only in drinking behavior but also in the pathology of alcohol-related diseases (11). Chronic exposure to ethanol disrupts rhythmic expression of Pomc and Per genes in the arcuate nucleus and clock genes in the liver but not in the suprachiasmatic nucleus (12–14). The rhythmicity of NAD+/NADH is also abolished by ethanol consumption, while ethanol induces oscillation of cholesterol and bile acid levels in the liver (14). Interestingly, acetylation of hepatic enzymes involved in ethanol metabolism exhibits circadian rhythmicity, suggesting the importance of acetyl-CoA metabolism in reprogramming of hepatic circadian metabolism (15).
We applied unbiased high-throughput proteomics, acetylomics, and circadian transcriptomics to elucidate the interplay between circadian metabolism and alcohol consumption in the liver. Distinct rhythmic transcriptional pathways are implicated in the acute or chronic consumption of ethanol. Among these pathways, SREBP-driven transcription appears to respond differentially to distinct levels and patterns of alcohol consumption. As a consequence, hepatic rewiring of acetyl-CoA metabolism is converted into specific signatures in the acetylation of cytosolic and mitochondrial proteins. Thus, our study reveals how alcohol-induced differential rewiring of circadian transcription is metabolically translated into distinct patterns of protein acetylation through acetyl-CoA metabolism and thereby provides insights into ALD pathophysiology.
Results
Chronic Alcohol Consumption Disrupts Circadian Metabolism and Behavior.
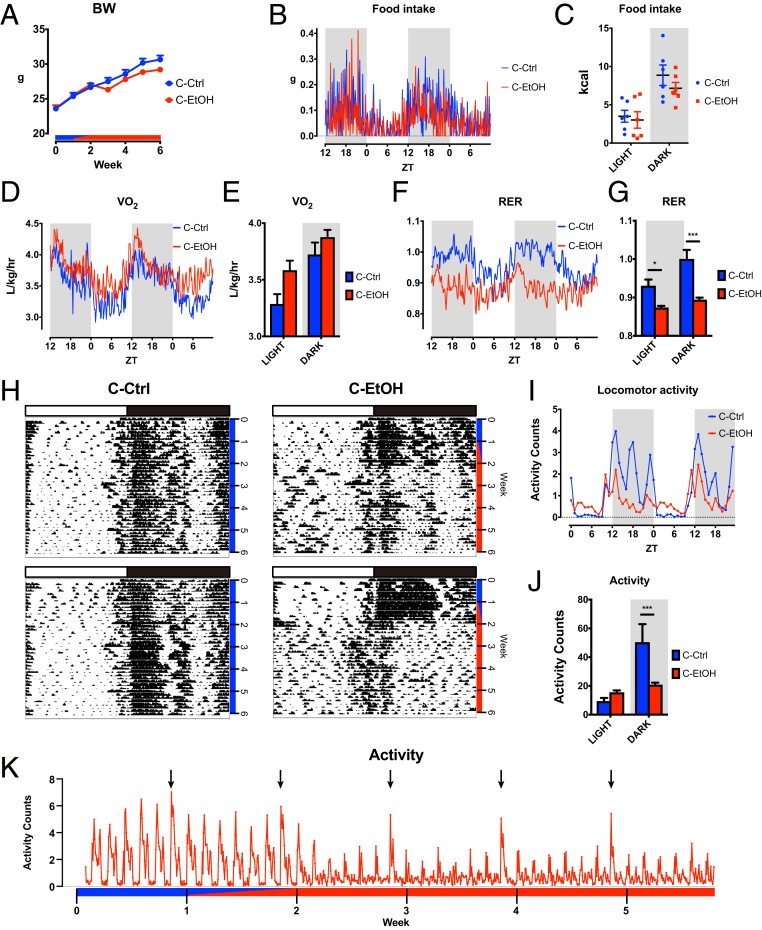
To explore how chronic intake of alcohol alters diurnal metabolism and behavior, 9-wk-old male C57BL/6 mice were randomly assigned to either a control diet group or an alcohol diet group. Mice were acclimatized to tube feeding with control liquid diet for the first week, followed by a transition period of 1 wk, during which the concentration of ethanol was incrementally increased. Subsequently, mice were subjected to ethanol-containing (chronic EtOH) or isocaloric carbohydrate-containing (chronic Ctrl) Lieber-DeCarli liquid diet for 4 wk (16, 17). Body weight of mice subjected to the chronic EtOH diet was slightly, but consistently, lower than that of mice fed a chronic Ctrl diet a week after the start of ethanol feeding (Fig. 1A). Metabolic cage analysis demonstrated that chronic EtOH mice displayed higher oxygen consumption (VO2) and a dampened respiratory exchange ratio (RER) despite comparable food intake and energy expenditure, indicative of a metabolic shift of substrate oxidation from carbohydrate to lipids (Fig. 1 B–G and SI Appendix, Fig. S1 A–D). Intriguingly, the locomotor activity of chronic EtOH mice was disrupted and reduced (Fig. 1 H–K). Specifically, diurnal locomotor rhythmicity became significantly dampened after switching to ethanol feeding, concurrent with a significant decrease (2.5 times) of nighttime activity and a slight increase of daytime activity in chronic EtOH mice (Fig. 1 H–K), in agreement with a previous study (18). Of note, liquid diet was replaced daily at zeitgeber time 10 (ZT10) to ZT12 in the current study, and we observed a small bout of locomotor activity during the food refill, most likely because of experimental intervention (Fig. 1 H and I). These results show that chronic use of alcohol alters circadian metabolism and behavior at the organismal level.
Fig. 1.
Alcohol-induced circadian disruption of metabolism and behavior. (A) Body weight (BW) of chronic ethanol (C-EtOH) fed mice (n = 22) and control (C-Ctrl) mice (n = 14) measured once a week over the time course of the experiment. Blue and red bars denote control and ethanol diet, respectively. (B) Metabolic cage analysis (n = 6 per group) showing the time series of food intake over a 48 h period. (C) The cumulative food intake for individual mice over the light and dark periods. (D) Metabolic cage analysis (n = 6 per group) showing the time series of oxygen consumption (VO2) over a 48-h period. (E) The average of VO2 over the light and dark periods. (F) Metabolic cage analysis (n = 6 per group) showing the time series of RER (= VCO2/VO2) over a 48 h period. (G) The average of RER over the light and dark periods. (H) Representative actograms from chronic Ctrl and chronic EtOH fed mice. White and black bars denote light and dark phases, respectively. Blue and red bars denote administration of chronic Ctrl and chronic EtOH diet, respectively. (I and J) Time series and the average of the quantified locomotor activity from chronic Ctrl (n = 8) and chronic EtOH (n = 16) mice from day 38 to day 40. (K) Time series of activity counts from chronic EtOH mice (n = 16). Arrows represent the time of cage replacement. Blue and red bars denote administration of control and ethanol diets. *P < 0.05 and ***P < 0.001 in ANOVA with the Bonferroni post hoc test.
Specific Hallmarks of Acute and Chronic Ethanol Intake on the Hepatic Circadian Transcriptome.
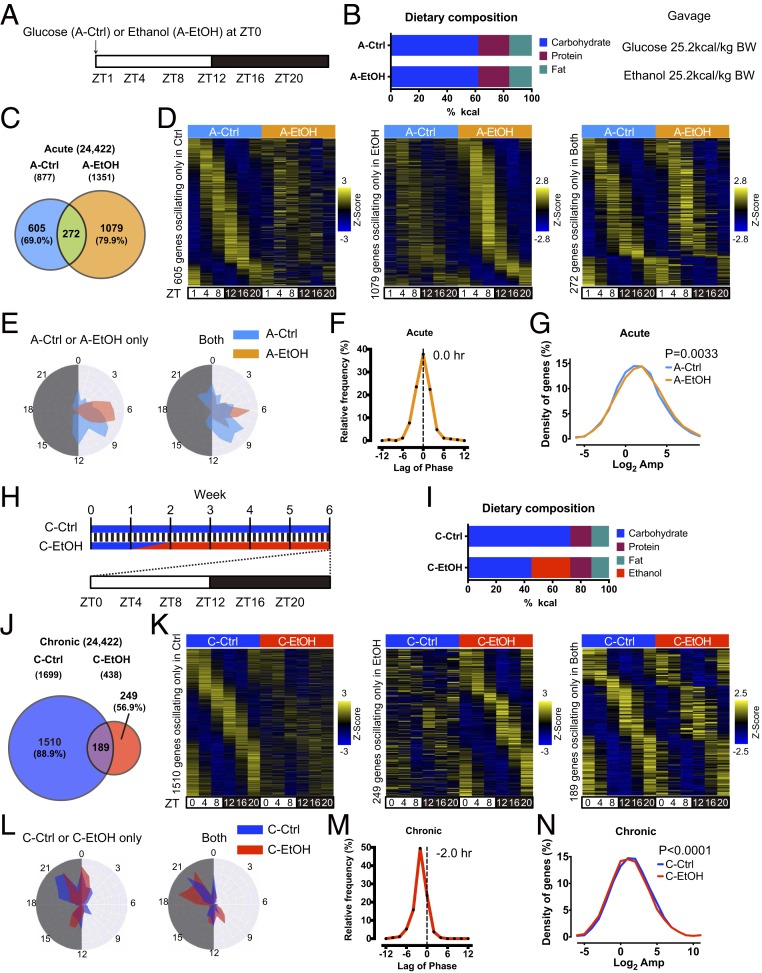
We next investigated whether alcohol alters circadian gene expression profiles in the liver. Recent studies indicated that chronic ethanol feeding may impact circadian rhythms in the liver (13, 14, 19). Yet little is known about how acute versus chronic alcohol intake impacts circadian gene expression on a genome-wide level. To address this question, we conducted a single, acute administration of alcohol (acute EtOH) or isocaloric glucose (acute Ctrl) by oral gavage in ad libitum fed male mice at zeitgeber time 0 (ZT0). Liver samples were subsequently harvested at ZT1, 4, 8, 12, 16, and 20 over the circadian cycle (Fig. 2 A and B). Circadian transcriptome by RNA sequencing (RNA-seq) revealed that ethanol induces a number of de novo oscillatory genes (Fig. 2 C and D and Datasets S1 and S3). The phases of these newly cycling genes clustered at around ZT4 to ZT10 in acute EtOH, while rhythmic expression of genes in acute Ctrl mice was more distributed throughout the light phase, presumably reflecting the robust response to alcohol (Fig. 2E). Although the phase of rhythmic genes oscillating in both groups was redistributed by alcohol, its overall difference was negligible on average (Fig. 2 E and F and SI Appendix, Fig. S2A). Notably, the amplitude of common cyclic transcripts was higher in acute EtOH mice, further supporting the robust effect of alcohol on hepatic response in gene expression (Fig. 2G and SI Appendix, Fig. S2B).
Fig. 2.
Distinct reprogramming of hepatic circadian transcriptome by acute and chronic ethanol feeding. (A) Scheme illustrating the experimental design of the acute ethanol feeding. At ZT0 a single dose of ethanol (A-EtOH, 3.5 g/kg body weight) or isocaloric dextrose (A-Ctrl) was administered. White and black bars represent light and dark phases, respectively. (B) Dietary composition of the acute ethanol feeding. (C) Venn diagram representing the number of hepatic genes rhythmic in A-Ctrl and A-EtOH treated mice. Diurnal genes were selected using BioCycle P value < 0.01. (D) Heat maps showing cyclic genes only in the acute Ctrl group (Left), only in the acute EtOH group (Middle), and in both acute groups (Right). (E) Polar histograms showing the peak phase of rhythmic genes oscillating only in acute Ctrl or acute EtOH (Left) and in both acute conditions (Right). (F) Graph showing the delay in the peak phase of the acute EtOH group compared to that of the acute Ctrl group. The average phase difference is shown in the graph. (G) Graph showing the distribution of amplitude in rhythmic genes oscillating in both acute groups. The P value indicates a significant change between acute Ctrl and acute EtOH. (H) Scheme illustrating the experimental design of the chronic ethanol feeding. Mice were initially fed ad libitum with a control liquid diet for a week to be acclimatized. EtOH was gradually introduced in liquid diet for a week and was maintained with 5% (vol/vol) ethanol for 4 wk (C-EtOH), while C-Ctrl mice were fed the isocaloric control liquid diet for 6 wk. Blue and red bars represent the C-Ctrl and C-EtOH diets, respectively. White and black bars represent light and dark phase, respectively. (I) Dietary composition of the chronic control and ethanol feeding. (J) Venn diagram showing the number of oscillatory genes in chronic Ctrl and chronic EtOH mice. Diurnal genes were selected using BioCycle P value < 0.01. (K) Heat maps showing cyclic genes only in the chronic Ctrl group (Left), only in the chronic EtOH group (Middle), and in both chronic groups (Right). (L) Polar histograms showing the peak phase of rhythmic genes oscillating only in chronic Ctrl or chronic EtOH (Left) and in both chronic groups (Right). (M) Graph showing the delay in the peak phase of the chronic EtOH group compared to that of the chronic Ctrl group. The average phase difference is shown in the graph. (N) Graph showing the distribution of amplitude in the rhythmic genes oscillating in both groups. The P value indicates a significant change between chronic Ctrl and chronic EtOH.
To explore the long-term effect of alcohol on circadian gene expression, we collected livers at ZT0, 4, 8, 12, 16, and 20 from mice on chronic EtOH diet (Figs. 1 and 2 H and I). In stark contrast to acute EtOH, the majority of rhythmic genes in chronic Ctrl liver ceased oscillation in chronic EtOH liver, while a much smaller number of genes gained oscillation under chronic EtOH, in support of the circadian disruption of metabolism and behavior elicited by chronic alcohol treatment (Figs. 1 and 2 J and K and Datasets S2 and S4). Although the phase distributions of chronic Ctrl– or chronic EtOH–specific rhythmic genes were similar, chronic EtOH appeared to induce phase anticipation within the genes oscillating in both groups (Fig. 2 L and M and SI Appendix, Fig. S2C). Furthermore, the amplitude of the common oscillatory genes was lower under chronic EtOH, emphasizing the observation that chronic intake of ethanol affects diurnal rhythm in the liver. Together, these results show that the diurnal genomic response to ethanol in the liver largely differs depending on the mode of alcohol administration.
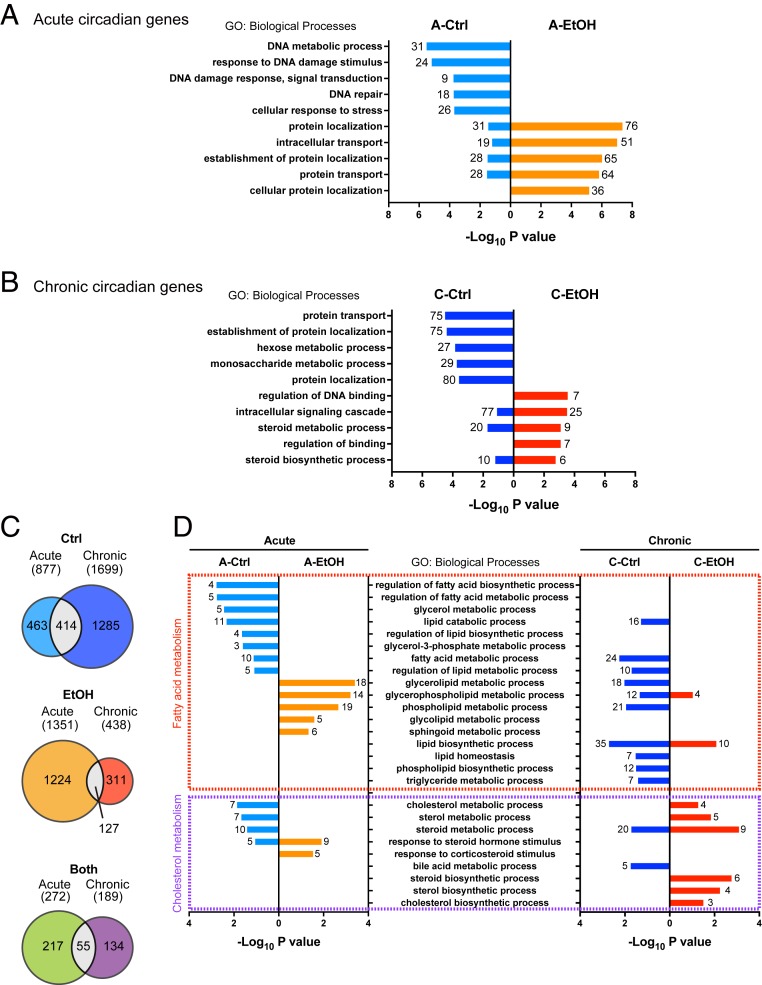
Gene ontology analysis revealed distinct biological processes enriched in rhythmic genes driven by alcohol (Fig. 3 A and B). Specifically, DNA damage response was enriched in acute Ctrl, while protein localization and transport were overrepresented in acute EtOH (Fig. 3A). Interestingly, similar pathways were also identified when comparing chronic Ctrl and EtOH mice, albeit in a reverse manner; protein localization and transport were enriched in chronic Ctrl, whereas DNA damage response was overrepresented in chronic EtOH (Fig. 3B). The distinct pattern of enriched pathways seen even between the 2 Ctrl groups (acute vs. chronic) is likely attributed to the difference in dietary composition as well as the administration of glucose at ZT0 in acute Ctrl. Indeed, a liquid diet alone influences circadian gene expression distinct from a chow diet (18). Supporting this observation, the overlap of diurnal genes rhythmic both in acute and chronic Ctrl is marginal (Fig. 3C). This is also the case with EtOH groups (acute vs. chronic) and rhythmic both in Ctrl and EtOH (acute vs. chronic) (Fig. 3C). On the other hand, genes oscillating in both Ctrl and EtOH were identified with circadian rhythm in both acute and chronic conditions, illustrating the resilient nature of core clock gene expression (SI Appendix, Fig. S2 E and F). Peripheral clocks respond to metabolic perturbations (20–22). Specifically, NAD+ and acetyl-CoA metabolism engages in the regulation of the clock system through acetylation of BMAL1 and histones at promoters of clock-controlled genes (23–26). Since alcohol alters both NAD+ and acetyl-CoA levels in the liver, we explored whether the hepatic clock machinery may be affected by alcohol treatment (14, 27). Acute EtOH appeared to have a minimal effect on the core clock genes and proteins, causing only slight dampening in oscillation (Fig. 2F and SI Appendix, Fig. S3 A–D). On the other hand, chronic EtOH caused phase advance in the expression of clock genes and proteins (Fig. 2M and SI Appendix, Fig. S3 E–H). This phase shift is likely to be independent of feeding activity, as diurnal food intake was virtually equal in both the chronic Ctrl and chronic EtOH mice (Fig. 1 B and C) (28). Of note, chronic ethanol treatment appeared to promote and phase advance acetylation of BMAL1, suggesting attenuation of SIRT1 activity due to the possible consumption of NAD+ by ethanol oxidation, the increased availability of acetyl-CoA, or the combination of both (SI Appendix, Fig. S3 F–H) (23, 24). It is conceivable that acute response of BMAL1 acetylation may have minimal influence on circadian transcription, while repetitious BMAL1 acetylation changes may trigger an adaptive response that could directly impact circadian transcription. Our results support a scenario in which alcohol modulates clock function partly through BMAL1 acetylation.
Fig. 3.
Distinct circadian pathways by acute and chronic alcohol feeding. (A) Gene ontology analysis showing the top 5 biological processes enriched in the rhythmic genes oscillating only in the A-Ctrl and A-EtOH groups, with the number of genes indicated on the graph. (B) Gene ontology analysis showing the top 5 biological processes enriched in the rhythmic genes oscillating only in the C-Ctrl and C-EtOH groups, with the number of genes indicated on the graph. (C) Venn diagram depicting the number of oscillatory genes in the acute and chronic Ctrl groups (Top), acute and chronic EtOH groups (Middle), and both groups in acute and chronic treatments (Bottom). (D) Gene ontology analysis showing the lipid metabolic processes enriched in the rhythmic genes oscillating only in the A-Ctrl, A-EtOH, C-Ctrl, or C-EtOH group.
Based on the possible alteration in lipid metabolic pathways suggested by the metabolic cage analysis, we further focused on the biological processes involved in lipid metabolism (Fig. 3D). Notably, fatty acid metabolism was enriched in acute EtOH and chronic Ctrl liver, whereas cholesterol metabolism was enriched in chronic EtOH liver, suggesting that alcohol regulates lipid metabolism in a distinct manner depending on its dose and treatment period (Fig. 3D). Supporting this notion, a relatively small overlap in cyclic genes was found between acute and chronic EtOH (Fig. 3C). Altogether, these data indicate that distinct ethanol regimens differentially rewire circadian hepatic physiology.
Differential Response of SREBP-Dependent Transcription to Acute and Chronic Ethanol Intake.
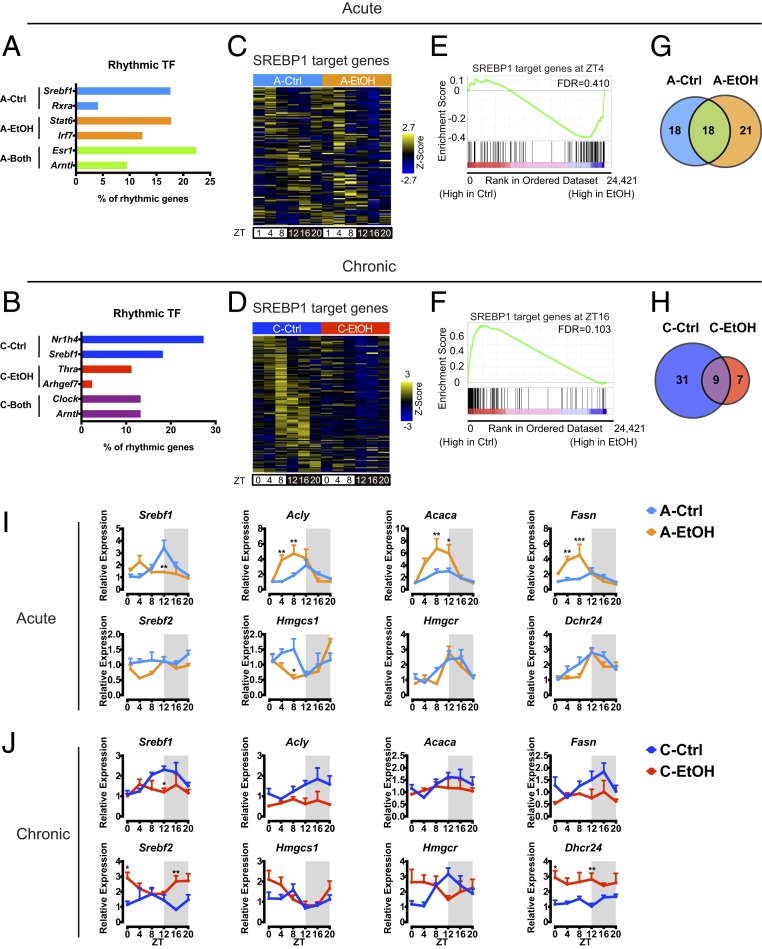
To decipher the transcriptional pathways triggered by ethanol, transcription factor binding site (TFBS) analysis was performed on circadian transcripts using MotifMap, and a further meta-analysis was used to identify associated rhythmic transcription factors (TFs) for each condition (Fig. 4 A and B) (29, 30). Interestingly, SREBP1 and its binding motif were highly enriched in acute and chronic Ctrl conditions (Fig. 4 A and B). SREBPs are basic-helix–loop–helix leucine zipper factors that regulate lipid homeostasis and metabolism (31, 32). While SREBPs have overlapping function, SREBP1 preferentially activates genes involved in fatty acid synthesis, whereas SREBP2 activates genes required for cholesterol biosynthesis.
Fig. 4.
Ethanol differentially alters SREBP-mediated circadian pathway. (A) Analysis of rhythmic TF and corresponding TFBS on hepatic rhythmic genes exclusively in A-Ctrl, exclusively in A-EtOH, and in both acute conditions. Represented as a percentage of TFBS presence among the rhythmic genes for each condition. (B) Analysis of rhythmic TF and corresponding TFBS on hepatic rhythmic genes exclusively in C-Ctrl, exclusively in C-EtOH, and in both chronic conditions. Represented as a percentage of TFBS presence among the rhythmic genes for each condition. (C) Heat map illustrating hepatic SREBP1 target gene expression in acute ethanol feeding. (D) Heat map illustrating hepatic SREBP1 target gene expression in chronic ethanol feeding. (E) Gene set enrichment analysis (GSEA) showing the enrichment of SREBP1 targets in the acute ethanol group at ZT4. (F) GSEA showing the enrichment of SREBP1 targets in the chronic control group at ZT16. (G) Venn diagram depicting the number of oscillatory SREBP1 target genes in acute ethanol feeding. (H) Venn diagram depicting the number of oscillatory SREBP1 target genes in chronic ethanol feeding. (I and J) Representative gene expression profiles of hepatic lipogenic genes in acute and chronic ethanol feeding. Gene expression was normalized to 18S ribosomal RNA and presented as mean + SEM (n = 4 to 6 biological replicates per time point per group). *P < 0.05, **P < 0.01, and ***P < 0.001 in ANOVA with the Bonferroni post hoc test.
Hepatic SREBP1 target genes from published datasets were compared to gene expression profiles in our transcriptomes (Fig. 4 C and D) (32). Interestingly, SREBP1 target genes appeared to be induced by acute EtOH but are repressed by chronic EtOH intake (Fig. 4 C and D). This is strengthened by the gene set enrichment analysis showing that SREBP1 target genes display higher expression under acute EtOH intake in mice at ZT4, while the same set of genes was significantly enriched in chronic Ctrl mice at ZT16 (Fig. 4 E and F). Furthermore, the number of rhythmic SREBP1 target genes was slightly higher in acute EtOH mice but drastically lower in chronic EtOH mice, suggesting that SREBP1 rhythmic activation is severely attenuated by chronic EtOH intake (Fig. 4 G and H). This is exemplified by expression profiles of representative SREBP1 target genes such as Acly, Acaca, and Fasn in acute and chronic EtOH mice (Fig. 4 I and J). Indeed, chronic EtOH significantly mitigated gene expression and nuclear protein levels of SREBP1 in the liver, and similar results were obtained in acute EtOH, suggesting the possible contribution of other transcription factors such as ChREBP in acute EtOH–induced gene induction of fatty acid synthesis (Fig. 4 I and J and SI Appendix, Fig. S4A) (27). SREBP2 target genes involved in cholesterol biosynthesis such as Hmgcr, Hmgcs1, and Dhcr24 were slightly increased in chronic EtOH but displayed comparable expression between acute Ctrl and acute EtOH (Fig. 4 I and J). It is noteworthy that repression of SREBP1 and activation of SREBP2 observed in the chronic EtOH feeding are recapitulated in livers of lung tumor–bearing mice, suggesting common underlying pathophysiology (33). Indeed, as in the case of lung adenocarcinoma (33), plasma cytokine analysis revealed induction of systemic inflammation by chronic alcohol feeding (SI Appendix, Fig. S4 C and D). Additionally, gene expression of liver cytokine receptors was up-regulated by chronic alcohol treatment (SI Appendix, Fig. S4E). These observations suggest that chronic ethanol intake modulates hepatic lipid synthesis partly through proinflammatory pathways. It is of particular interest that bidirectional interactions between lipid metabolism and inflammation have been implicated in the pathogenesis of liver disease (34, 35). Overall, alcohol appears to affect hepatic circadian SREBP1 pathways in opposite manners in acute and chronic treatments.
Ethanol Transcriptional Signatures Linked to Subcellular Proteins Acetylation.
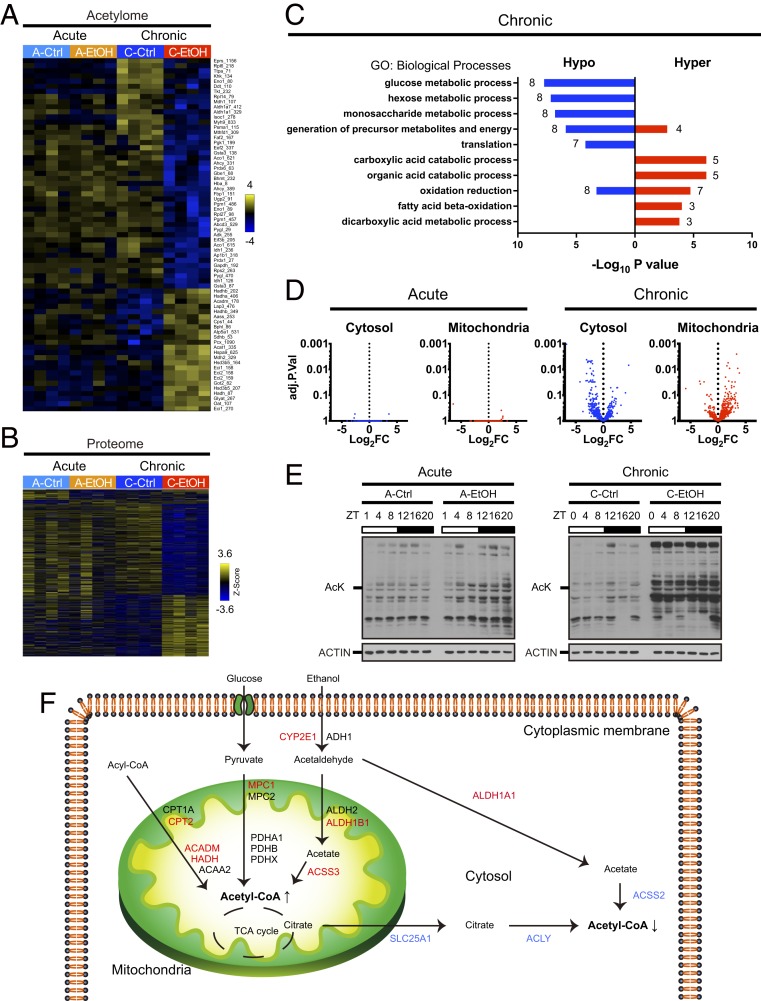
Acetyl-CoA is a major intermediate of metabolism, and its cellular levels reflect the metabolic state of the cell. Importantly, acetyl-CoA serves as a substrate for lysine acetyltransferases (KATs), and fluctuations in its concentration are directly correlated with dynamic protein acetylation (36–39). Our transcriptomics analysis reveals that ethanol reprograms circadian SREBP transcription (Fig. 4 C and D), a pathway responsible for de novo lipid biosynthesis that utilizes acetyl-CoA as a building block. Indeed, ethanol alters transcription of several SREBP targets encoding for rate-limiting enzymes in acetyl-CoA production (such as Acly) and usage (such as Acaca) (Fig. 4 I and J). As acetyl-CoA production may be enhanced by ethanol oxidation through the acetate–acetyl-CoA synthetases (ACSS) pathway, we sought to investigate how acute and chronic ethanol influence global protein acetylation. In order to address this question, we carried out acetylome combined with proteome analyses on liver samples collected at ZT4 after acute and chronic EtOH intake. This is also in light of the differential effect of acute or chronic EtOH intake in modulating BMAL1 acetylation at ZT4 (Fig. 5 A and B, SI Appendix, Fig. S3 F and H, and Datasets S5 and S6) (15). To identify acetylated proteins, we isolated acetylated peptides from the total peptide preparation used for the proteome by immunoprecipitation. Among 2,170 proteins detected in the liver, 11 proteins in acute EtOH and 576 proteins in chronic EtOH were found to be differentially expressed compared to their respective controls (Fig. 5B). Gene ontology (GO) analysis of the differentially expressed proteins revealed that proteins displaying higher expression in chronic EtOH livers (236) belong to processes related to oxidation reduction and cofactor metabolic processes, while proteins with lower expression (340) belong to processes related to oxidation reduction and translation (SI Appendix, Fig. S5A). Specifically, enzymes involved in xenobiotic metabolism appeared to be induced both at the transcript and protein levels in chronic ethanol feeding (SI Appendix, Fig. S5 B and C). Furthermore, 730 proteins were acetylated, with 2,110 unique sites of acetylation (Fig. 5A). Remarkably, while 58 proteins were differentially acetylated, with 69 unique acetylation sites in chronic EtOH liver compared to chronic Ctrl, no proteins with significant changes in acetylation were differentially detected in acute Ctrl and EtOH livers (Fig. 5A). Among the proteins differentially acetylated by chronic ethanol feeding, 38 were hypoacetylated with 45 unique acetylation sites, and 20 were hyperacetylated with 24 unique acetylation sites (Fig. 5A). GO analysis of differentially acetylated proteins revealed that hypoacetylated proteins belong to glucose and glycogen metabolism such as phosphoglycerate kinase 1 (PGK1), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), glycogen phosphorylase (PYGL), and fructose-bisphosphatase 1 (FBP1) (Fig. 5C). Additional hypoacetylated proteins were enzymes involved in methionine and cysteine metabolism such as adenosylhomocysteinase (AHCY), betaine homocysteine S-methyltransferase (BHMT), and methylenetetrahydrofolate dehydrogenase (MTHFD1) (Fig. 5C). Conversely, hyperacetylated proteins were enriched for fatty acid oxidation and included enzymes such as hydroxyacyl-CoA dehydrogenase alpha (HADHA), medium-chain acyl-CoA dehydrogenase (ACADM), and enoyl-CoA delta isomerase 1 (ECI1) (Fig. 5C). These results suggest that chronic exposure to ethanol leads to differential acetylation of enzymes in unique metabolic pathways.
Fig. 5.
Partitioning of metabolic enzymes acetylation and acetyl-CoA metabolism. (A) Heat map illustrating differentially acetylated hepatic proteins at ZT4 (n = 4 per group) by acute or chronic ethanol (EtOH) feeding (P < 0.05). The numbers next to the protein names denote the position of the acetylated amino acid. (B) Heat map illustrating differentially expressed proteins at ZT4 (n = 4 per group) by acute or chronic EtOH feeding (P < 0.05). (C) Gene ontology analysis showing the top 5 biological processes enriched in hypo- and hyperacetylated proteins in chronic EtOH fed mice, with the number of acetylated proteins indicated on the graph. (D) Volcano plots showing the differential acetylation of cytosolic and mitochondrial proteins in acute and chronic EtOH fed mice. (E) Representative immunoblots of hepatic whole-protein lysates with acute (Left) and chronic (Right) EtOH feeding. AcK denotes acetylated lysine. ACTIN was used as a loading control. (F) Scheme depicting the subcellular acetyl-CoA metabolic pathways and corresponding hepatic protein levels. Red and blue denote up-regulated and down-regulated protein expression determined by proteomic analysis, respectively.
Different metabolic pathways take place in distinct subcellular compartments (40). Likewise, rhythmic protein acetylation differs depending on the subcellular compartment and is well correlated with acetyl-CoA synthesis in the respective compartment (15). Since GO analysis demonstrated that hypoacetylated proteins and hyperacetylated proteins were enriched in glucose/glycogen metabolism and fatty acid oxidation, respectively, we hypothesized that protein localization may dictate distinct patterns of acetylation in the context of chronic EtOH diet. To address this question, we annotated cellular localization of acetylated proteins using protein localization databases (41). Intriguingly, cytosolic proteins were prone to be hypoacetylated, while mitochondrial proteins appeared to be hyperacetylated by chronic EtOH; neither cytosolic nor mitochondrial proteins were differentially acetylated by acute EtOH (Fig. 5D). In agreement with this observation, subcellular fractionation of the liver revealed that mitochondrial proteins were indeed hyperacetylated by chronic EtOH, likely reflecting the global increase of protein acetylation observed in whole-liver extracts (Fig. 5E and SI Appendix, Fig. S5D).
Next, we focused on the metabolic pathways involved in acetyl-CoA production in cytosol and mitochondria. Thus, we extracted proteins involved in acetyl-CoA metabolism in each subcellular compartment from our proteome dataset. Remarkably, expression of proteins belonging to the mitochondrial acetyl-CoA pathway, especially enzymes such as ACSS3 responsible for acetyl-CoA production from ethanol-derived acetate, was increased after chronic EtOH exposure and paralleled the increase of protein acetylation in this compartment (Fig. 5F). Conversely, enzymes in the cytosolic acetyl-CoA pathway, most of which are SREBP targets, were repressed by chronic EtOH feeding, which correlated with the decrease of protein acetylation levels in the cytosol (Fig. 5F). For instance, ACLY, which converts citrate to acetyl-CoA in the cytosol, was decreased after chronic EtOH feeding (Fig. 5F). Moreover, ACSS2, which is responsible for the generation of cytosolic pools of acetate-derived acetyl-CoA, was also reduced by chronic exposure to EtOH. Of note, changes in protein levels within the acetyl-CoA pathway were not observed in acute EtOH feeding. These results indicated that chronic alcohol treatment alters hepatic acetyl-CoA production in the cytosol and mitochondria through the circadian SREBP1 pathway, which is likely to be translated to a distinct pattern of protein acetylation in each subcellular compartment.
Discussion
Circadian homeostasis governs a wide variety of metabolic processes by integrating multiple tiers of regulations such as the sleep–wake cycle, feeding–fasting rhythm, and tissue-intrinsic clock oscillation (42–44). Dietary challenges have been shown not only to operate tissue-specific metabolic cycles but also to alter behavioral and bioenergetic rhythms, resulting in activation of de novo alternative circadian pathways in a tissue-specific manner (20, 22, 45, 46).
Alcohol is associated with adverse consequences such as cancers and psychiatric disorders (1). While the majority of alcohol is oxidized in the liver, alcohol and its metabolized acetaldehyde influence a wide range of organs, including the brain. Supporting this notion, locomotor activity in chronic EtOH mice was disrupted, altering behavioral rhythms. In the liver, alcohol causes serious health consequences such as alcoholic steatohepatitis, liver cirrhosis, and hepatocellular carcinoma (2). Interestingly, we reveal that the amount and pattern of alcohol intake differentially reprogram hepatic gene expression. Specifically, acute administration of EtOH doses drives the lipogenic program that is classically linked to alcoholic fatty liver. This is in agreement with the induction of fatty acid synthesis by SREBP1 and its implication in ALD (47, 48). In contrast, chronic treatment with EtOH appears to silence SREBP1 and fatty acid synthesis, consistent with previous observations (13, 17). Hence, ALD is likely to comprise heterogeneous pathophysiologies depending on the levels and patterns of alcohol consumption. Although blood alcohol concentration (BAC) was not determined in this study, BACs under analogous animal models have been demonstrated elsewhere (13, 49). Additionally, female mice were not included in the study, as is the case with most of the circadian studies (20, 22, 50). Since female mice were more susceptible to alcoholic liver injury, future studies will address gender-specific effects of ethanol on hepatic circadian homeostasis (51).
Histone and nonhistone proteins undergo various modifications linked to the availability of metabolic substrates, highlighting the pivotal role of metabolism in cellular physiology (52, 53). Posttranslational protein modifications display circadian fluctuation (15, 54–56). Acetyl-CoA is required for protein acetylation, leading to functional changes in target proteins, such as intracellular localization, enzyme activity, stability, and interaction with other proteins (40, 57, 58). N-epsilon-acetylation is a reversible posttranslational modification which is either mediated by lysine N-epsilon-acetyltransferase (KAT) on specific proteins or achieved by a spontaneous nonenzymatic reaction of acetyl-CoA with solvent-accessible lysines of abundant proteins. Regardless of the mechanism, acetyl-CoA levels dictate the loads of protein acetylation. Moreover, acetyl-CoA is not diffusible across the mitochondrial membrane, thereby requiring the localized and compartmentalized synthesis within different subcellular compartments such as the mitochondria and cytosol (40). Previous studies have illustrated partitioning of nuclear and cytosolic pools of acetyl-CoA, even though the nuclear membrane allows acetyl-CoA to freely translocate (59). The subcellular acetyl-CoA concentration may be reflective of the acetylation level of proteins in each organelle. While mitochondrial and cytosolic acetyl-CoA levels were not determined (59), expression profiles of acetyl-CoA–producing enzymes in each compartment indicated repression of cytosolic acetyl-CoA production and induction of the mitochondrial counterpart, possibly leading to the imbalance of protein acetylation between the 2 organelles. Supporting this notion, inhibition of acetyl-CoA–producing enzymes such as ATP-citrate lyase (ACLY) and acetyl-CoA synthetase 1 (ACSS2) leads to histone hypoacetylation (38, 39, 60, 61). Likewise, suppression of acetyl-CoA consumption through the inhibition of acetyl-CoA carboxylase leads to hyperacetylation of histone and nonhistone proteins (37, 62). Intriguingly, cytosolic acetyl-CoA–producing and -consuming enzymes in the liver are controlled by SREBP1, a well-documented circadian transcription factor that regulates lipid metabolism. While feeding induces cytosolic acetyl-CoA levels, fasting increases mitochondrial acetyl-CoA production (15, 50, 63, 64). Thus, a chronic EtOH–induced subcellular pattern of protein acetylation appears to recapitulate the fasted state, implicating a paradoxically nutrient-depleted condition despite comparable calorie intake, likely reflecting a “malnutrition” state that may be involved in the pathophysiology of ALD.
Acetylation of proteins is known to alter enzymatic activities (65). For example, PYGL, the rate-limiting enzyme involved in glycogen catabolism, is activated after chronic alcohol consumption through deacetylation (66) and could contribute to glycogen storage depletion. On the other hand, hyperacetylation at Lys406 of HADHA, a beta-oxidation enzyme, is associated with decreased enzymatic activity, possibly contributing to inhibition of fatty acid oxidation in mitochondria (67). While the precise effect of site-specific acetylation on the activity or stability of metabolic enzymes remains to be elucidated, GO analysis reveals overrepresentation of key pathways involved in energy metabolism and ALD pathogenesis. For example, misregulation of hepatic methionine metabolism and fatty acid oxidation are the result of alcohol abuse and major contributors in ALDs (68, 69).
The design of our study focused on how alcohol may influence circadian function rather than parallel alcohol-induced organ injury. This approach has allowed us to minimize secondary events such as inflammation and fibrosis. Hence, we favored animal models with marginal elevation of ALT and minimal steatosis. The dose of acute alcohol administration used parallels that in previous studies (51), and while it did not reportedly cause ALT elevation, it was sufficient to increase blood alcohol concentration (49, 70). Indeed, we observed a robust induction in the expression of hepatic genes involved in fatty acid synthesis. On the other hand, changes in gene expression by acute EtOH treatment likely manifest response to alcohol, thus presumably correlating with blood alcohol concentration. Yet it is difficult to distinguish the acute response to EtOH intake from modulation of circadian gene expression. Thus, we reasoned that glucose administration is an appropriate control for the binge model in order to take the responsive effect of caloric intake into account, based upon previous literatures (71–73).
In conclusion, various alcohol consumptions differentially reprogram hepatic circadian gene expression and trigger partitioning of subcellular protein acetylation. The opposite effect of acute and chronic ethanol feeding on the diurnal regulation of SREBP pathways suggests a transcriptional adaptation to chronic ethanol ingestion, whereas acute exposure reflects more a stress response. Supporting this notion, no significant changes in protein acetylation have been observed in acute ethanol livers. Altogether, these findings contribute to further understanding the differential underlying mechanism of alcoholic liver injury and could lead to the identification of circadian therapeutic targets and strategies.
Methods
Detailed experimental procedures are outlined in SI Appendix, SI Text.
Animals and Diets.
Male wild-type C57BL/6J mice (Jackson Laboratory, 000664) were housed with ad libitum food and water access on a 12 h light/12 h dark cycle. Mice were maintained with regular chow diet (2020X Teklad Global Extruded Rodent Diet) before beginning the ethanol feeding study. All experiments were performed in accordance with the Institutional Animal Care and Use Committee guidelines at the University of California, Irvine.
Acute ethanol feeding.
Male wild-type C57BL/6J mice at 12 to 24 wk of age were randomly divided into an ethanol group and a control group; 3.5 g/kg of ethanol or an isocaloric dose of dextrose (7.2 g/kg) were delivered to mice by oral gavage at ZT0. Tissues were collected on the same day as the gavage at ZT1, 4, 8, 12, 16, and 20 (n = 5 per time point for each group). All mice were individually housed 1 wk prior to harvesting tissue.
Chronic ethanol feeding.
Male wild-type C57BL/6J mice at 9 wk of age were randomly divided into an ethanol group and a control group. The control mice were pair fed with ethanol-treated mice a control liquid diet without ethanol for 6 wk (Bioserv, Low Fat Lieber-DeCarli, F1340SP). The ethanol-treated mice were fed a control liquid diet in the first week, a liquid diet with ethanol incrementally increased (Bioserv, Low Fat Lieber-DeCarli, F1341SP, 1% ethanol for 3 d, 2% for the 2 d, 3.3% for the 2 d) with maltose dextrin (Bioserv, 3653) in the second week, and a liquid diet containing 5% ethanol for the following 4 wk. Tissues were collected at ZT0, ZT4, ZT8, ZT12, ZT16, and ZT20 (n = 4 to 6 per group for each time point).
RNA Extraction and Quantitative Real-Time PCR Analysis.
Frozen liver tissues were homogenized in TRIzol Reagent (Invitrogen). Total RNA was isolated by precipitation with isopropanol and ethanol; 1 µg RNA was reverse transcribed to cDNA using an iScript complementary DNA (cDNA) synthesis kit (Bio-Rad Laboratories, 1708840), according to the manufacturer’s protocol.
cDNA was used for quantitative real-time PCR using SsoAdvanced SYBR Green Supermix (Bio-Rad Laboratories, 1725270). Gene expression was normalized to 18S ribosomal RNA. Primer sequences used for gene expression analysis are listed in Dataset S7.
Data Availability.
The RNA-seq data reported in this paper are available in the Gene Expression Omnibus. The accession number is GSE132103.
Supplementary Material
Acknowledgments
We thank all members of the P.S.-C. laboratory for stimulating discussion and technical assistance. We also thank Dr. Tsukamoto (University of Southern California) for insight and support. We also thank Melanie Oakes, Seung-Ah Chung, and Yuzo Kanomata at the Genomics High-Throughput Facility at the University of California, Irvine. J.G. was supported by a postdoctoral fellowship from the University of California, Irvine, Hitachi-Nomura fund. K.K. was supported by a postdoctoral fellowship from the Japan Society for the Promotion of Science. C.M.G. was supported by the National Cancer Institute of the US NIH (NIH T32 2T32CA009054-36A1) and by European Research Council (ERC MSCA-IF-2016 MetEpiClock 749869). Work in the P.S.-C. laboratory was supported by INSERM, NIH, in part by the Pilot Project Program of the National Institute on Alcohol Abuse and Alcoholism-funded Southern California Research Center for ALPD and Cirrhosis (P50AA011999). Work by N.C. and P.B. was in part supported by Defense Advanced Research Projects Agency grant D17AP00002 and NIH grant GM123558 to P.B. These authors would also like to acknowledge computing support by Yuzo Kanomata.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. B.G. is a guest editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE132103).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1911189116/-/DCSupplemental.
References
- 1.Gilmore W., et al. , Alcohol: Taking a population perspective. Nat. Rev. Gastroenterol. Hepatol. 13, 426–434 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Louvet A., Mathurin P., Alcoholic liver disease: Mechanisms of injury and targeted treatment. Nat. Rev. Gastroenterol. Hepatol. 12, 231–242 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Babor T. F., et al. , Alcohol: No ordinary commodity–A summary of the second edition. Addiction 105, 769–779 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Seitz H. K., et al. , Alcoholic liver disease. Nat. Rev. Dis. Primers 4, 16 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Gao B., Bataller R., Alcoholic liver disease: Pathogenesis and new therapeutic targets. Gastroenterology 141, 1572–1585 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cederbaum A. I., Alcohol metabolism. Clin. Liver Dis. 16, 667–685 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baraona E., Lieber C. S., Effects of ethanol on lipid metabolism. J. Lipid Res. 20, 289–315 (1979). [PubMed] [Google Scholar]
- 8.Galli A., Pinaire J., Fischer M., Dorris R., Crabb D. W., The transcriptional and DNA binding activity of peroxisome proliferator-activated receptor alpha is inhibited by ethanol metabolism. A novel mechanism for the development of ethanol-induced fatty liver. J. Biol. Chem. 276, 68–75 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Udoh U. S., Valcin J. A., Gamble K. L., Bailey S. M., The molecular circadian clock and alcohol-induced liver injury. Biomolecules 5, 2504–2537 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spanagel R., et al. , The clock gene Per2 influences the glutamatergic system and modulates alcohol consumption. Nat. Med. 11, 35–42 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Summa K. C., et al. , Disruption of the circadian clock in mice increases intestinal permeability and promotes alcohol-induced hepatic pathology and inflammation. PLoS One 8, e67102 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen C. P., Kuhn P., Advis J. P., Sarkar D. K., Chronic ethanol consumption impairs the circadian rhythm of pro-opiomelanocortin and period genes mRNA expression in the hypothalamus of the male rat. J. Neurochem. 88, 1547–1554 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Filiano A. N., et al. , Chronic ethanol consumption disrupts the core molecular clock and diurnal rhythms of metabolic genes in the liver without affecting the suprachiasmatic nucleus. PLoS One 8, e71684 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou P., Ross R. A., Pywell C. M., Liangpunsakul S., Duffield G. E., Disturbances in the murine hepatic circadian clock in alcohol-induced hepatic steatosis. Sci. Rep. 4, 3725 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mauvoisin D., et al. , Circadian and feeding rhythms orchestrate the diurnal liver acetylome. Cell Rep. 20, 1729–1743 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu M., et al. , Regulation of hepatic lipin-1 by ethanol: Role of AMP-activated protein kinase/sterol regulatory element-binding protein 1 signaling in mice. Hepatology 55, 437–446 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leung T. M., et al. , Argininosuccinate synthase conditions the response to acute and chronic ethanol-induced liver injury in mice. Hepatology 55, 1596–1609 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou P., et al. , Dissociation between diurnal cycles in locomotor activity, feeding behavior and hepatic PERIOD2 expression in chronic alcohol-fed mice. Alcohol 49, 399–408 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Udoh U. S., et al. , Genetic deletion of the circadian clock transcription factor BMAL1 and chronic alcohol consumption differentially alter hepatic glycogen in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 314, G431–G447 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eckel-Mahan K. L., et al. , Reprogramming of the circadian clock by nutritional challenge. Cell 155, 1464–1478 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kohsaka A., et al. , High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 6, 414–421 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Tognini P., et al. , Distinct circadian signatures in liver and gut clocks revealed by ketogenic diet. Cell Metab. 26, 523–538.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Asher G., et al. , SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134, 317–328 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Nakahata Y., et al. , The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134, 329–340 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakahata Y., Sahar S., Astarita G., Kaluzova M., Sassone-Corsi P., Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 324, 654–657 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramsey K. M., et al. , Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 324, 651–654 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marmier S., et al. , Novel role for carbohydrate responsive element binding protein in the control of ethanol metabolism and susceptibility to binge drinking. Hepatology 62, 1086–1100 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Mukherji A., et al. , Shifting eating to the circadian rest phase misaligns the peripheral clocks with the master SCN clock and leads to a metabolic syndrome. Proc. Natl. Acad. Sci. U.S.A. 112, E6691–E6698 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daily K., Patel V. R., Rigor P., Xie X., Baldi P., MotifMap: Integrative genome-wide maps of regulatory motif sites for model species. BMC Bioinf. 12, 495 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie X., Rigor P., Baldi P., MotifMap: A human genome-wide map of candidate regulatory motif sites. Bioinformatics 25, 167–174 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eberlé D., Hegarty B., Bossard P., Ferré P., Foufelle F., SREBP transcription factors: Master regulators of lipid homeostasis. Biochimie 86, 839–848 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Horton J. D., Goldstein J. L., Brown M. S., SREBPs: Activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109, 1125–1131 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masri S., et al. , Lung adenocarcinoma distally rewires hepatic circadian homeostasis. Cell 165, 896–909 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fon Tacer K., Kuzman D., Seliskar M., Pompon D., Rozman D., TNF-alpha interferes with lipid homeostasis and activates acute and proatherogenic processes. Physiol. Genomics 31, 216–227 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Vida M., et al. , Chronic administration of recombinant IL-6 upregulates lipogenic enzyme expression and aggravates high-fat-diet-induced steatosis in IL-6-deficient mice. Dis. Model. Mech. 8, 721–731 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chow J. D., et al. , Genetic inhibition of hepatic acetyl-CoA carboxylase activity increases liver fat and alters global protein acetylation. Mol. Metab. 3, 419–431 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rios Garcia M., et al. , Acetyl-CoA carboxylase 1-dependent protein acetylation controls breast cancer metastasis and recurrence. Cell Metab. 26, 842–855.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Wellen K. E., et al. , ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076–1080 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao S., et al. , ATP-citrate lyase controls a glucose-to-acetate metabolic switch. Cell Rep. 17, 1037–1052 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pietrocola F., Galluzzi L., Bravo-San Pedro J. M., Madeo F., Kroemer G., Acetyl coenzyme A: A central metabolite and second messenger. Cell Metab. 21, 805–821 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Thul P. J., et al. , A subcellular map of the human proteome. Science 356, eaal3321 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Koronowski K. B., et al. , Defining the independence of the liver circadian clock. Cell 177, 1448–1462.e14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schibler U., Sassone-Corsi P., A web of circadian pacemakers. Cell 111, 919–922 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Welz P. S., et al. , BMAL1-driven tissue clocks respond independently to light to maintain homeostasis. Cell 177, 1436–1447.e12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Asher G., Schibler U., Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 13, 125–137 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Dyar K. A., et al. , Atlas of circadian metabolism reveals system-wide coordination and communication between clocks. Cell 174, 1571–1585.e11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crabb D. W., Liangpunsakul S., Alcohol and lipid metabolism. J. Gastroenterol. Hepatol. 21 (suppl. 3), S56–S60 (2006). [DOI] [PubMed] [Google Scholar]
- 48.You M., Fischer M., Deeg M. A., Crabb D. W., Ethanol induces fatty acid synthesis pathways by activation of sterol regulatory element-binding protein (SREBP). J. Biol. Chem. 277, 29342–29347 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Livy D. J., Parnell S. E., West J. R., Blood ethanol concentration profiles: A comparison between rats and mice. Alcohol 29, 165–171 (2003). [DOI] [PubMed] [Google Scholar]
- 50.Kinouchi K., et al. , Fasting imparts a switch to alternative daily pathways in liver and muscle. Cell Rep. 25, 3299–3314.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ghosh Dastidar S., Warner J. B., Warner D. R., McClain C. J., Kirpich I. A., Rodent models of alcoholic liver disease: Role of binge ethanol administration. Biomolecules 8, E3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Katada S., Imhof A., Sassone-Corsi P., Connecting threads: Epigenetics and metabolism. Cell 148, 24–28 (2012). [DOI] [PubMed] [Google Scholar]
- 53.Sabari B. R., Zhang D., Allis C. D., Zhao Y., Metabolic regulation of gene expression through histone acylations. Nat. Rev. Mol. Cell Biol. 18, 90–101 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Masri S., et al. , Circadian acetylome reveals regulation of mitochondrial metabolic pathways. Proc. Natl. Acad. Sci. U.S.A. 110, 3339–3344 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robles M. S., Humphrey S. J., Mann M., Phosphorylation is a central mechanism for circadian control of metabolism and physiology. Cell Metab. 25, 118–127 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Wang J., et al. , Nuclear proteomics uncovers diurnal regulatory landscapes in mouse liver. Cell Metab. 25, 102–117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choudhary C., Weinert B. T., Nishida Y., Verdin E., Mann M., The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell Biol. 15, 536–550 (2014). [DOI] [PubMed] [Google Scholar]
- 58.Menzies K. J., Zhang H., Katsyuba E., Auwerx J., Protein acetylation in metabolism - metabolites and cofactors. Nat. Rev. Endocrinol. 12, 43–60 (2016). [DOI] [PubMed] [Google Scholar]
- 59.Bulusu V., et al. , Acetate recapturing by nuclear acetyl-CoA synthetase 2 prevents loss of histone acetylation during oxygen and serum limitation. Cell Rep. 18, 647–658 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sahar S., et al. , Circadian control of fatty acid elongation by SIRT1 protein-mediated deacetylation of acetyl-coenzyme A synthetase 1. J. Biol. Chem. 289, 6091–6097 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takahashi H., McCaffery J. M., Irizarry R. A., Boeke J. D., Nucleocytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Mol. Cell 23, 207–217 (2006). [DOI] [PubMed] [Google Scholar]
- 62.Galdieri L., Vancura A., Acetyl-CoA carboxylase regulates global histone acetylation. J. Biol. Chem. 287, 23865–23876 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shi L., Tu B. P., Acetyl-CoA and the regulation of metabolism: Mechanisms and consequences. Curr. Opin. Cell Biol. 33, 125–131 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wellen K. E., Thompson C. B., A two-way street: Reciprocal regulation of metabolism and signalling. Nat. Rev. Mol. Cell Biol. 13, 270–276 (2012). [DOI] [PubMed] [Google Scholar]
- 65.Hirschey M. D., et al. , SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 464, 121–125 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang T., et al. , Acetylation negatively regulates glycogen phosphorylase by recruiting protein phosphatase 1. Cell Metab. 15, 75–87 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thapa D., et al. , The protein acetylase GCN5L1 modulates hepatic fatty acid oxidation activity via acetylation of the mitochondrial β-oxidation enzyme HADHA. J. Biol. Chem. 293, 17676–17684 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lu S. C., Tsukamoto H., Mato J. M., Role of abnormal methionine metabolism in alcoholic liver injury. Alcohol 27, 155–162 (2002). [DOI] [PubMed] [Google Scholar]
- 69.Kharbanda K. K., Alcoholic liver disease and methionine metabolism. Semin. Liver Dis. 29, 155–165 (2009). [DOI] [PubMed] [Google Scholar]
- 70.Carson E. J., Pruett S. B., Development and characterization of a binge drinking model in mice for evaluation of the immunological effects of ethanol. Alcohol Clin. Exp. Res. 20, 132–138 (1996). [DOI] [PubMed] [Google Scholar]
- 71.Banerjee A., Abdelmegeed M. A., Jang S., Song B. J., Increased sensitivity to binge alcohol-induced gut leakiness and inflammatory liver disease in HIV transgenic rats. PLoS One 10, e0140498 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bertola A., Mathews S., Ki S. H., Wang H., Gao B., Mouse model of chronic and binge ethanol feeding (the NIAAA model). Nat. Protoc. 8, 627–637 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen P., et al. , Microbiota protects mice against acute alcohol-induced liver injury. Alcohol Clin. Exp. Res. 39, 2313–2323 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data reported in this paper are available in the Gene Expression Omnibus. The accession number is GSE132103.