Significance
The field of neutrophil biology has lagged behind in terms of the understanding of heterogeneity and versatility of cellular functions, limiting the development of therapeutic approaches that target aberrant neutrophil phenotypes. Using bulk and single-cell transcriptomic, epigenetic, and functional analyses, this study highlights aspects of neutrophil heterogeneity and their putative role in the pathogenesis of systemic lupus erythematosus and its associated vascular damage.
Keywords: autoimmunity, neutrophils, systemic lupus erythematosus
Abstract
Neutrophil dysregulation is implicated in the pathogenesis of systemic lupus erythematosus (SLE). SLE is characterized by elevated levels of a pathogenic neutrophil subset known as low-density granulocytes (LDGs). The origin and phenotypic, functional, and pathogenic heterogeneity of LDGs remain to be systematically determined. Transcriptomics and epigenetic assessment of lupus LDGs, autologous normal-density neutrophils, and healthy control neutrophils was performed by bulk and single-cell RNA sequencing and assay for transposase-accessible chromatin sequencing. Functional readouts were compared among neutrophil subsets. SLE LDGs display significant transcriptional and epigenetic heterogeneity and comprise 2 subpopulations of intermediate-mature and immature neutrophils, with different degrees of chromatin accessibility and differences in transcription factor motif analysis. Differences in neutrophil extracellular trap (NET) formation, oxidized mitochondrial DNA release, chemotaxis, phagocytosis, degranulation, ability to harm the endothelium, and responses to type I interferon (IFN) stimulation are evident among LDG subsets. Compared with other immune cell subsets, LDGs display the highest expression of IFN-inducible genes. Distinct LDG subsets correlate with specific clinical features of lupus and with the presence and severity of coronary artery disease. Phenotypic, functional, and pathogenic neutrophil heterogeneity are prevalent in SLE and may promote immune dysregulation and prominent vascular damage characteristic of this disease.
Neutrophil dysregulation may play crucial and distinct pathogenic roles in systemic lupus erythematosus (SLE). We previously identified a proinflammatory neutrophil subset known as low-density granulocytes (LDGs) that differs functionally from autologous lupus normal density neutrophils (NDNs) and from healthy control (HC) neutrophils. LDGs induce increased endothelial damage and vascular dysfunction in vitro, through their enhanced ability to synthesize and extrude neutrophil extracellular traps (NETs). NETs are chromatin fibers decorated with immunostimulatory nuclear and granule proteins and oxidized nucleic acids. In SLE, NETs stimulate the production of proinflammatory cytokines and type I interferons (IFNs), promote immune cell maturation, and contribute to tissue damage (1–8). SLE LDG numbers are associated with in vivo vascular inflammation and coronary atherosclerosis, independent of other cardiovascular (CV) risk factors (1, 5, 9, 10), and also with promotion of T cell activation (11). These observations suggest that specifically targeting LDGs could abrogate certain aspects of immune dysregulation, organ damage, and premature atherosclerosis characteristic of SLE.
The origin of lupus LDGs remains unclear, and whether they represent primarily immature neutrophils prematurely released from the bone marrow or a distinct mature neutrophil subset with enhanced proinflammatory capabilities remains to be systematically characterized (1, 2, 6, 12–14). In a previous study in individuals exposed to granulocyte colony-stimulating factor, 2 groups of LDGs were characterized based on the expression level of CD10. These 2 subsets exerted opposing effects on T cells and were found in a variety of conditions, including SLE (15); however, other phenotypic and functional heterogeneity has not been evaluated.
In this study, we examined in detail the transcriptional, epigenetic, and functional profiles of lupus LDGs. We found evidence of phenotypic and functional heterogeneity within LDG subpopulations in SLE, their association with features of clinical disease and CVD, and their contribution to the type I IFN signature characteristic of this disease.
Methods
More details on the methodology of this study are provided in SI Appendix.
Patient Selection.
Peripheral blood was collected by venipuncture from SLE and HC subjects recruited at the NIH Clinical Center in Bethesda, MD. Subjects signed informed consent; all experiments involving human subjects were approved by the National Institute of Arthritis and Musculoskeletal and Skin Diseases/National Institute of Diabetes and Digestive and Kidney Diseases Institutional Review Board (NIH 94-AR-0066).
Comparisons of neutrophil subsets were performed by flow cytometry, single-cell and bulk RNA sequencing and pathway mapping, transposase-accessible chromatin sequencing (ATAC-seq), motif enrichment analysis, endothelium-dependent vasorelaxation assay, mitochondrial (mit) and genomic DNA quantification, chemotaxis and phagocytosis assays, and correlation with demographic and clinical characteristics.
Data Access.
All sequencing data have been deposited in the in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE139360). All other data are available from the authors on request.
Results
SLE LDGs Have a Distinct Transcriptional Profile.
Demographic and clinical characteristics of subjects studied are presented in SI Appendix, Table S1. Gene expression analysis from RNA-seq was compared in SLE LDGs from 11 patients with active disease and on minimal immunosuppressive medications, autologous SLE NDNs, and matched HC NDNs. Principal component analysis (PCA) indicated that LDGs have a transcriptional profile distinct from autologous SLE and HC NDNs (SI Appendix, Fig. S1A). Indeed, 946 genes were up-regulated and 3,635 genes were down-regulated at the transcriptional level in lupus LDGs compared with HC NDNs, while 991 genes were up-regulated and 2,893 genes were down-regulated in LDGs compared with autologous NDNs (SI Appendix, Figs. S1B and S2). In contrast, lupus and HC NDNs displayed a very similar transcriptional profile, with only 212 genes up-regulated and 168 genes down-regulated in SLE NDNs compared with HC NDNs (SI Appendix, Fig. S1 A and B). Pathways up-regulated in SLE LDGs included those related to neutrophil activation (neutrophil degranulation and antimicrobial peptides) (SI Appendix, Fig. S1C).
SLE patients display elevated levels of type I IFN-stimulated genes (ISGs) in peripheral blood mononuclear cells (PBMCs) (16) and various organs, in association with disease severity (17). To determine whether purified SLE neutrophil subsets also display increased ISG expression, an IFN score based on a panel of 21 ISGs was determined in lupus NDNs and LDGs, as described previously (18, 19). SLE NDNs and LDGs had higher IFN scores compared with HC NDNs (SI Appendix, Fig. S1 D and E). Collectively, SLE LDGs had distinct transcriptional profiles and evidence of enhanced exposure and/or response to type I IFNs.
SLE LDGs Represent a Heterogeneous Neutrophil Population.
Hierarchical clustering of RNA-seq samples (n = 33) revealed that SLE LDGs represent a transcriptionally heterogeneous neutrophil subset (SI Appendix, Fig. S3). Within the 11 SLE subjects studied by RNA-seq, a gene signature displaying a subset of highly up-regulated genes compared with the other samples was observed in 3 of the SLE LDG preparations, while a gene signature composed of predominantly down-regulated genes was observed in the remaining 8 LDG samples (SI Appendix, Fig. S3). One of the mRNA clusters that was increased in LDGs encoded for neutrophil granule proteins, consistent with an immature neutrophil phenotype (6, 13, 14) (SI Appendix, Fig. S4A). An immature neutrophil z-score using 8 of the highest expressing neutrophil granule genes (9) revealed a higher immature neutrophil score in SLE LDGs compared with autologous and HC NDNs (SI Appendix, Fig. S4B). The 3 lupus LDG samples with the most transcriptionally active gene signature showed the highest levels of mRNAs encoding for neutrophil granule proteins compared with the other LDG and NDN samples; therefore, they may represent SLE subjects with higher levels of immature LDGs (LDGimm) (SI Appendix, Figs. S3 and S4 A and B). In contrast, at the transcriptional level, the remaining 8 SLE LDG samples were more consistent with a mature neutrophil phenotype (LDGmat) (SI Appendix, Figs. S3 and S4 A and B). Confirming these findings, LDGimm samples expressed lower levels of the cell surface maturation marker CD10 compared with LDGmat samples on flow cytometry (SI Appendix, Fig. S4C). There were no clinical differences between the patients with the LDGmat and LDGimm signatures (SI Appendix, Table S2). These results suggest that lupus LDGs represent a heterogeneous subset composed of immature and mature neutrophils based on CD10 surface expression and transcriptional analysis.
LDGimm and LDGmat Have Distinct Transcriptional Profiles and Epigenetic Landscapes.
A closer examination of the PCA plot in SI Appendix, Fig. S4A reveals that samples characterized as lupus LDGimm and LDGmat separated into transcriptionally distinct groups (SI Appendix, Fig. S5). LDGimm represent a more transcriptionally active subset compared with LDGmat and HC NDNs, with 4,833 and 2,312 genes up-regulated, respectively (SI Appendix, Fig. S4 D and E). LDGmat represent the bulk of the LDGs and overall are more transcriptionally repressed compared with HC NDNs, with 4,572 genes down-regulated (SI Appendix, Fig. S4F). Pathway analysis of genes up-regulated in LDGimm compared with LDGmat revealed that genes related to chromatin modification, histone acetylation, transcription initiation, and cell cycle progression were increased in LDGimm (SI Appendix, Fig. S4G). In contrast, LDGmat had transcriptional up-regulation of pathways associated with immune responses, including type I IFN signaling and neutrophil activation (SI Appendix, Fig. S4G).
Given that LDGimm had a more transcriptionally active profile and up-regulated expression of genes involved in histone acetylation compared with LDGmat, we carried out ATAC-seq to characterize their chromatin accessibility. Lupus LDGmat, autologous NDNs, and HC NDNs displayed similar numbers of peaks in the promoter regions (SI Appendix, Fig. S4H). In contrast, LDGimm had more peaks in promoter regions than LDGmat (SI Appendix, Fig. S4 H and I). Overall, LDGimm had “open” peaks in 784 unique genes, compared with “open” peaks in 43 genes for LDGmat. Collectively, these results indicate that LDGimm represent a transcriptionally active subset of neutrophils with enhanced chromatin accessibility.
To gain further molecular insight into LDG subgrouping, we performed transcription factor (TF) motif enrichment analysis using both ATAC-seq and RNA-seq data. Enriched motifs that are group-specific were identified, with 20 for LDGimm and 7 for LDGmat, that were enriched in neither SLE NDNs nor in Ctrl NDNs (SI Appendix, Fig. S4J). LDGimm also shared fewer enriched motifs with Ctrl NDNs (9 out of 33; 27.3%) compared with LDGmat (10 out of 21; 47.6%). Together with the observation that LDGimm had the highest number of uniquely enriched TF motifs, these results are consistent with the RNA-seq results, in which more up-regulated genes were identified for LDGimm. In addition, of the 20 genes with motifs uniquely enriched in LDGimm, 13 were expressed with reads per kilobase per million mapped reads values > 0.1, and their expressions were different between LDGimm and LDGmat (P = 0.0426, paired t test).
We next performed a bootstrap analysis to assess the nonrandomness and robustness of the number of enriched motifs that are LDG-subgroup specific, against 1,000 sets of bootstrapped motif analysis samples. This analysis, in which the same motif analysis approach was applied to the random datasets, generated a P value of 4.405E-27, indicating a strong statistical significance associated with LDG subgrouping. Altogether, the motif analysis results revealed distinct molecular characteristics of LDGimm and LDGmat, providing additional evidence for LDG subgrouping, with a high level of statistical significance. These results further support LDG heterogeneity, as suggested by the RNA-seq and ATAC-seq analyses.
Sorted CD10− LDGs Have a Similar Transcriptional Profile to LDGimm.
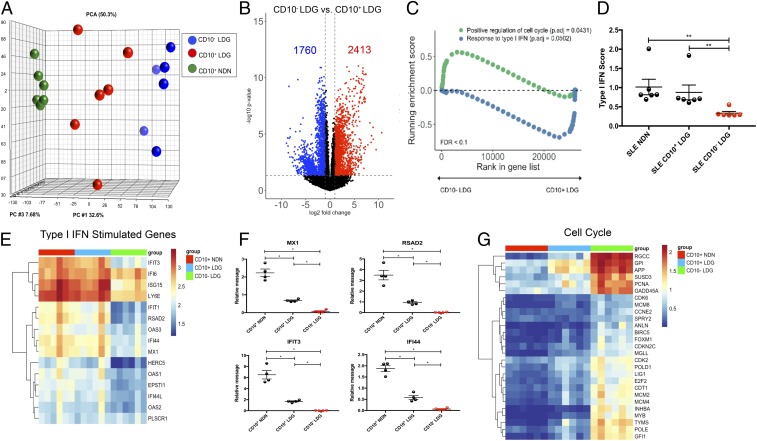
LDGimm and LDGmat were purified by cell sorting into CD10− and CD10+ LDGs and autologous CD10+ NDNs (n = 6/each). RNA-seq analysis was performed, and PCA analysis revealed that CD10− and CD10+ LDGs were transcriptionally distinct compared with autologous CD10+ NDN (Fig. 1A). When this dataset was compared with the previous bulk LDG RNA-seq dataset, purified CD10− LDGs and CD10+ LDGs grouped together with LDGimm and LDGmat, respectively, suggesting that LDGs have transcriptional profiles composed of both CD10− LDGs and CD10+ LDGs (SI Appendix, Fig. S7). Compared with autologous lupus CD10+ NDNs, both CD10− LDGs and CD10+ LDGs displayed profiles of transcriptional activity (SI Appendix, Fig. S7). CD10− LDGs were more transcriptionally active compared with CD10+ LDGs, with 2,413 genes up-regulated and 1,760 genes down-regulated (Fig. 1B). Pathway analysis revealed that CD10− LDGs up-regulated genes involved in cell cycle progression compared with CD10+ LDGs (Fig. 1C). In contrast, CD10+ LDGs displayed elevated transcripts of genes involved in type I IFN signaling compared with CD10− LDGs (Fig. 1C). Compared with CD10− LDGs, autologous lupus NDNs and CD10+ LDGs had enhanced ISG expression (Fig. 1 D–F). CD10− LDGs displayed the highest expression of genes involved in cell cycle progression, followed by CD10+ LDGs, which displayed intermediate expression of cell cycle genes compared with autologous NDNs (Fig. 1G). In summary, the transcriptional profiles of CD10− LDGs and CD10+ LDGs resemble those of LDGimm and LDGmat, respectively, and CD10+ LDGs and autologous NDNs have an enhanced type I IFN gene signature compared with CD10- LDGs.
Fig. 1.
Sorted CD10− LDGs have a similar transcriptional profile to LDGimm. (A) RNA-seq was performed on sorted lupus CD10+ NDNs (green), autologous CD10− LDGs (blue), and CD10+ LDGs (red) (n = 6). (B) Volcano plot of differential gene expression between CD10− and CD10+ LDGs. Up-regulated genes with fold change ≥2 and P < 0.05 are in red, and down-regulated genes with fold change ≤2 and P < 0.05 are in blue. (C) Gene set enrichment analysis of cell cycle genes and ISGs in CD10− and CD10+ LDGs. (D–G) IFN score (D); ISG RNA-seq analysis (E); ISG expression by qRT-PCR, normalized to GAPDH expression and reported as relative message (F); and RNA-seq analysis for cell cycle genes (G) in CD10+ NDN, CD10+ and CD10− LDGs (n = 6/group for D, E, and G and n = 4/group for F). Results represent mean ± SEM from four independent experiments. *P ≤ 0.05; **P < 0.01.
Phenotypic, Functional, and Pathogenic Heterogeneity of Lupus LDGs.
Having characterized the transcriptional profiles of CD10− and CD10+ LDGs, we further determined the stage of neutrophil differentiation for the isolated neutrophil subsets. Previous studies reported that LDGs represent a mixed population based on their nuclear morphology (1, 20). We confirmed this in purified subsets, as CD10+ NDNs and CD10+ LDGs displayed multilobulated nuclei characteristic of mature neutrophils (Fig. 2A). In contrast, CD10− LDGs displayed less segmented and more rounded nuclei, consistent with a more immature stage of neutrophil differentiation (Fig. 2A). TF analysis related to myeloid cell development (21) was performed in these cells. CD10− LDGs displayed elevated transcripts for CEBPA and IRF8 compared with CD10+ LDGs and NDNs, suggestive of multipotent granulocyte-macrophage progenitor (GMP) cells (Fig. 2B) (21–23). CD10+ LDGs also displayed enhanced expression of CEBPA and IRF8 compared with CD10+ NDNs but to a lesser degree than CD10− LDGs, supporting their classification as an intermediate-mature subset of neutrophils (Fig. 2B). GFI1 and CEBPE expression in proliferative neutrophil precursors has been reported (21, 24, 25), and CD10− LDGs displayed increased expression of these TFs compared with CD10+ LDGs and CD10+ NDNs (Fig. 2B). CD10+ NDNs and CD10+ LDGs displayed higher expression of the TFs SPI1 and CEBPD, typically associated with mature neutrophils, compared with CD10− LDGs (Fig. 2B). These results suggest that lupus CD10− LDGs express TFs that are representative of GMP and neutrophil precursors, while lupus CD10+ LDGs represent an intermediate stage of neutrophil differentiation that is somewhat more immature than NDNs.
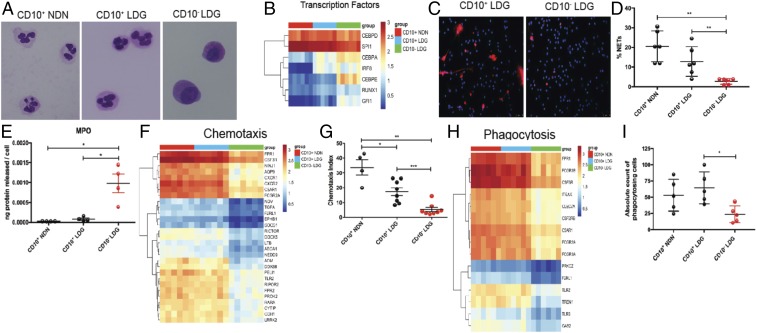
Fig. 2.
Lupus neutrophil subsets differ phenotypically and functionally. (A) Representative images of Giemsa-stained sorted lupus CD10+ NDNs, CD10+ LDGs, and CD10− LDGs (n = 4); original magnification 60×. (B) RNA-seq analysis of CD10+ NDNs, CD10+ LDGs, and CD10− LDGs (n = 6/group) for TFs involved in myeloid development. (C and D) Representative images of cells undergoing NET formation in unstimulated sorted CD10+ LDGs and CD10− LDGs after a 2-h incubation (n = 5/group). MPO is in red and DNA is in blue; original magnification 40×. (E) MPO ELISA of culture supernatants of unstimulated CD10+ NDNs, CD10+ LDGs, and CD10− LDGs (n = 4/group) after a 2-h incubation. (F) RNA-seq analysis for genes involved in chemotaxis in lupus neutrophil subsets. (G) fMLP-induced chemotactic index in lupus neutrophil subsets after a 2-h incubation (n = 4 for CD10+ NDNs; n = 8 for CD10+ and CD10− LDGs). (H) Expression of phagocytosis genes by RNA-seq analysis. (I) Phagocytosis of S. aureus bioparticles for lupus neutrophil subsets (n = 5). Results for all measurements are mean ± SEM. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
We assessed whether the maturation stage of lupus neutrophil subsets associates with their ability to perform canonical neutrophil functions. CD10− SLE LDGs were impaired in their ability to spontaneously form NETs compared with CD10+ SLE LDGs (Fig. 2 C and D), supporting previous observations that immature neutrophils are less effective at netting (26). CD10− LDGs displayed higher spontaneous myeloperoxidase (MPO) release compared with CD10+ NDNs and CD10+ LDGs (Fig. 2E), suggesting an enhanced ability to degranulate. RNA-seq analysis indicated that CD10− LDGs have lower expression of chemotaxis-related genes, including FPR1 (27), NINJ1 (28), CXCR1, and CXC2 (29), and this was recapitulated by observing that their chemotactic activity was decreased compared with that in lupus CD10+ LDGs and NDNs (Fig. 2 F and G). CD10+ LDGs displayed an intermediate capacity to undergo chemotaxis compared with CD10+ NDNs and CD10− LDGs (Fig. 2G), further supporting their intermediate state of maturation. CD10+ LDGs and CD10+ NDNs displayed enhanced expression of genes involved in phagocytosis and an enhanced ability to phagocytose S. aureus particles compared with CD10− LDGs (Fig. 2 H and I). These results suggest that the maturation status of lupus neutrophil subsets affects their ability to perform critical functions including NET formation, chemotaxis, and phagocytosis, while degranulation may be enhanced in the more immature forms.
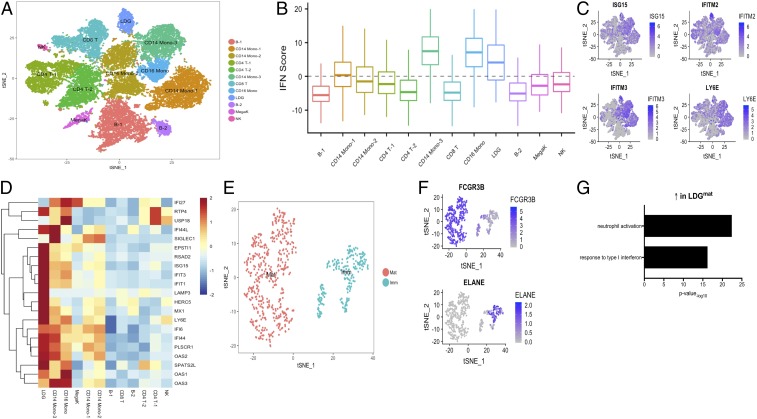
Single-cell (sc)-RNA-seq was performed in SLE PBMCs (n = 3), with a primary focus on uncovering LDG subclusters. We successfully sequenced 26,925 individual cells after combining the samples and found 12 distinct clusters after t-stochastic neighbor embedding (t-SNE) dimension reduction (Fig. 3A): CD4+ and CD8+ T cells, B cells, CD14+ monocytes (3 subclusters), CD16+ monocytes, LDGs, natural killer cells, dendritic cells, and megakaryocytes (Fig. 3A). LDGs were identified by genes highly specific for neutrophils, including FCGR3B (CD16b) (30) and CMTM2 (31) (SI Appendix, Fig. S8). A type I IFN score was calculated for each cluster; LDGs, along with CD14+ and CD16+ monocytes, had the highest IFN score (Fig. 3B). Differential gene analysis revealed multiple up-regulated ISGs in the LDG cluster compared with other cell clusters, including IFITM2, IFITM3, LY6E, and ISG15 (Fig. 3 C and D). These results suggest that LDGs significantly drive the type I IFN signature characteristic of SLE PBMCs.
Fig. 3.
Immature and mature LDGs are identified in lupus PBMCs using single-cell RNA-seq and differ in ISG expression. (A) a t-SNE plot representing gene expression in single cells from SLE PBMCs (n = 3) identifying 12 unique cell clusters. (B) IFN score in each cell cluster. (C) ISGs are highly expressed in LDGs. (D) ISG heatmap showing a high IFN response in LDGs relative to other cell clusters. (E and F) Neutrophils were filtered from PBMCs based on their expression of FCGR3B and ELANE. The t-SNE plot shows 2 transcriptionally distinct clusters based on FCGR3B or ELANE expression, identified as immature (Imm) and mature (Mat), respectively. (G) Pathway analysis indicating that mature neutrophils are activated and respond to type I IFNs.
To further explore neutrophil heterogeneity, we analyzed single cells expressing the neutrophil-specific genes ELANE and FCGR3B. Applying t-SNE to these cells revealed 2 distinct clusters, one with transcriptional characteristics of mature neutrophils (based on CD16b and CD10 gene expression) and the other expressing ELANE, a primary granule gene that identifies immature granulocytes (Fig. 3 E and F). Confirming the bulk RNA-seq data, the mature LDG cluster up-regulated neutrophil activation and type I IFN signaling pathways (Fig. 3G). This analysis confirms and refines the assessment of lupus LDG heterogeneity and identifies mature LDGs as one of the subsets with the highest up-regulation of ISGs compared with other immune cell subsets.
LDG Subsets Associate with Distinct Lupus Clinical Features and CVD.
Confirming that bulk LDGs are increased in SLE, both CD10− (P = 0.003) and CD10+ LDGs (P = 0.04) were higher in circulation in a larger SLE cohort used for data validation (SI Appendix, Table S3) compared with HC (SI Appendix, Fig. S9 A and B). CD10+, but not CD10− or total LDGs, correlated with the Systemic Lupus Collaborating Clinics–American College of Rheumatology damage index, a measure of irreversible organ damage (r = 0.2917, P = 0.019). In the subgroup of Caucasian SLE subjects, percentages of CD10+ LDGs negatively correlated with renal function, as assessed by glomerular filtration rate (r = −0.49, P = 0.015), while CD10− LDG percentage correlated with proteinuria, as assessed by protein:creatinine ratio (r = 0.573, P = 0.01). CD10+ or CD10− LDG percentage did not correlate with disease activity as measured by the SLE Disease Activity Index (SLEDAI; P = 0.45), serum complement C3 or C4 level (P = 0.94 and 0.67, respectively), or anti-dsDNA level (P = 0.56; n = 54). Similarly, percentages of these subsets did not correlate with prednisone dose (P = 0.09). CD10+ LDG percentage negatively correlated and CD10− LDG percentage positively corelated with receipt of prednisone (P = 0.0025).
We next assessed the stability of the proportion of LDG subsets over time. We obtained longitudinal samples separated by 152 d (5 mo) from 13 SLE patients. By paired analysis, we found no differences in the percentage of CD10+ and CD10− LDGs between day 1 and day 152, suggesting that these populations are stable over time (SI Appendix, Fig. S10). In this additional analysis, we confirmed that there was no correlation between the SLEDAI and the percentage of CD10− LDGs and no difference in the percentage of CD10− LDGs between subjects with active lupus (SLEDAI ≥ 4) and those with inactive lupus (P > 0.05)
We previously reported that SLE LDGs associate with arterial wall inflammation and noncalcified coronary plaque burden (NCB; SI Appendix, Table S3) (9). Reanalysis after separating LDGs into CD10+ and CD10− subsets showed that CD10+ LDG levels positively correlated with NCB severity (r = 0.343, P = 0.04, SI Appendix, Fig. S9C) and negatively correlated with high-density lipoprotein function, as assessed by cholesterol efflux capacity (SI Appendix, Fig. S9D), a variable associated with lower CV risk in the general population. These findings support our previous observations that NETs synthesized by lupus LDGs oxidize high-density lipoprotein and impair its antiatherogenic ability (32).
Supporting that LDGs are vasculopathic, supernatants from lupus CD10+, but not CD10−, LDGs impaired murine aortic endothelium-dependent vasorelaxation (SI Appendix, Fig. S9E). Furthermore, CD10+ LDGs displayed an enhanced ability to externalize oxidized mit-DNA compared with CD10− LDGs (SI Appendix, Fig. S9 F and G), supporting previous observations that lupus LDGs externalize NETs enriched in oxidized mit-DNA with interferogenic properties (8). These results suggest that the intermediate-mature LDG subset promotes vascular damage and is associated with coronary plaque formation.
Discussion
Despite several reports suggesting that LDGs are pathogenic in various inflammatory conditions (2, 9, 33–35), their origin, functionality, and heterogeneity remain to be determined. In the context of SLE, these cells have been linked to enhanced proinflammatory responses and do not appear to have myeloid suppressor cell capabilities (11). Here we report the transcriptional, epigenetic, and functional heterogeneity of SLE LDGs. Hierarchical clustering of bulk mRNA-seq identified 2 subpopulations of lupus LDGs: a small subset that up-regulates genes associated with neutrophil precursors and a more abundant subset characterized by transcriptional repression. The transcriptionally active LDG subset is composed of CD10− immature neutrophils that up-regulate genes related to cell cycle progression and down-regulate immune response genes and is similar to an LDG subset recently described in rheumatoid arthritis (12). SLE CD10− LDGs have nuclear morphology and TF analysis suggestive of a GMP or preNeu development stage (21, 24, 25, 36, 37). Given the immature phenotype, this LDG subtype is impaired in a number of canonical neutrophil functions. In contrast, the majority of lupus LDGs comprises an intermediate-mature, CD10+ subset (38) endowed with several pathogenic features, including NET formation, oxidized nucleic acid release, and promotion of endothelial dysfunction.
The use of sc-RNA-seq allowed us to confirm and refine the analysis of LDG heterogeneity. The immature LDGs were identified based on ELANE expression, while the intermediate-mature LDGs up-regulated FCGR3B. Compared with other myeloid subsets in the lupus mononuclear cell fraction, intermediate-mature LDGs displayed the highest expression of ISGs, suggesting that they contribute to the lupus type I IFN signature. Type I IFNs can activate and prime HC neutrophils to undergo NET formation (3, 39). The enhanced response to type I IFNs by lupus NDNs and intermediate-mature LDGs supports previous findings showing demethylation of ISGs in lupus NDNs and LDGs compared with HC neutrophils (40) and indicating that only more mature forms of neutrophils respond to type I IFNs (26). A limitation of the ATAC-seq analysis was the epigenetic profile diversity in SLE samples, which prevented more in-depth analyses at the individual gene level for each neutrophil subset. Future studies should examine whether ISG promoter regions are more accessible in lupus neutrophils and intermediate-mature LDGs compared with immature LDGs and HC neutrophils.
We previously reported that LDG NETs cause endothelial cell death and dysfunction that may contribute to premature atherosclerosis in SLE (1, 5, 6, 9). Indeed, released products from CD10+ LDGs, but not CD10− LDGs, impair endothelium-dependent vasorelaxation and are enriched for oxidized mit-DNA. It is likely that NETs are the cytotoxic components released by the CD10+ LDGs, because lupus LDG NETs are enriched in oxidized mit-DNA, which induces type I IFN production in a STING-dependent manner (8). This is consistent with a proposed model in which LDGs may further amplify type I IFN responses in the plaque, promoting vascular and myocardial damage (41, 42). Indeed, CD10+ LDGs positively correlate with NCB and negatively correlate with antiatherogenic cholesterol efflux capacity. Given their enhanced ability to degranulate, it is possible that CD10− immature LDGs have other distinct vasculopathic roles through promotion of inflammatory cell recruitment to the arterial wall (43, 44), matrix metalloproteinase release, and plaque instability (43–46). Future studies should investigate how immature LDG subsets contribute to organ damage and immune dysregulation through the mechanisms that we have identified in the intermediate-mature LDG subset.
A recent study using previously acquired gene expression databases suggested that LDGs reflect increased granulopoiesis and not peripheral neutrophil activation and that this is associated with corticosteroid use (47). In the present study and our previous investigations (6, 9), we found no correlation with corticosteroid use and LDG signature, although the percentages of the 2 LDG subsets correlated with current use of corticosteroids, but not with the dose used. It is possible that the release of the subset of the immature LDGs may be exacerbated by steroid use, while the presence of the mature, pathogenic LDG subset was negatively associated with the current use of steroids. Our data assessing purified LDG subsets do not support the hypothesis that the bulk of LDGs are immature neutrophil forms prematurely released from the bone marrow, but rather indicate that most LDGs represent mature proinflammatory neutrophils endowed with pathogenic features. The interpretation of Kegerreis et al. (47) could have relied on the observation that immature LDGs drive granulocyte mRNA expression in the global analysis but do not represent most of the LDGs in SLE. Of note, Kegerreis et al. based their analysis on publicly available databases from different research sources. As such, isolation technique and neutrophil purity may have varied widely among the various samples analyzed and contributed to the differences observed with our analysis, where samples were obtained in a systematic manner and purity and clinical information was obtained following similar parameters.
Overall, our results suggest that a distinct subset of intermediate-mature neutrophils has the highest proinflammatory phenotype within SLE and accounts for the most significant associations with organ damage. This study adds to our understanding of neutrophil heterogeneity in disease states, an area that has lagged behind compared with other immune cell subsets. Future studies should determine whether CD10+ and CD10− LDGs could serve as biomarkers of therapeutic efficacy in type I IFN pathway modulation. Furthermore, this study suggests that specifically targeting intermediate-mature LDG subsets may play important roles in the treatment and/or prevention of lupus vasculopathy and premature CVD.
Supplementary Material
Acknowledgments
This study used the high-performance computational capabilities of the Helix Systems at the NIH (https://hpc.nih.gov). We thank the Office of Science and Technology at the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) for technical support. This study was supported by the Intramural Research program at NIAMS/NIH (ZIA AR041199).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE139360).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1908576116/-/DCSupplemental.
References
- 1.Denny M. F., et al. , A distinct subset of proinflammatory neutrophils isolated from patients with systemic lupus erythematosus induces vascular damage and synthesizes type I IFNs. J. Immunol. 184, 3284–3297 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mistry P., et al. , Dysregulated neutrophil responses and neutrophil extracellular trap formation and degradation in PAPA syndrome. Ann. Rheum. Dis. 77, 1825–1833 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garcia-Romo G. S., et al. , Netting neutrophils are major inducers of type I IFN production in pediatric systemic lupus erythematosus. Sci. Transl. Med. 3, 73ra20 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mistry P., Kaplan M. J., Cell death in the pathogenesis of systemic lupus erythematosus and lupus nephritis. Clin. Immunol. 185, 59–73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carmona-Rivera C., Zhao W., Yalavarthi S., Kaplan M. J., Neutrophil extracellular traps induce endothelial dysfunction in systemic lupus erythematosus through the activation of matrix metalloproteinase-2. Ann. Rheum. Dis. 74, 1417–1424 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Villanueva E., et al. , Netting neutrophils induce endothelial damage, infiltrate tissues, and expose immunostimulatory molecules in systemic lupus erythematosus. J. Immunol. 187, 538–552 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warnatsch A., Ioannou M., Wang Q., Papayannopoulos V., Neutrophil extracellular traps license macrophages for cytokine production in atherosclerosis. Science 349, 316–320 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lood C., et al. , Neutrophil extracellular traps enriched in oxidized mitochondrial DNA are interferogenic and contribute to lupus-like disease. Nat. Med. 22, 146–153 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlucci P. M., et al. , Neutrophil subsets and their gene signature associate with vascular inflammation and coronary atherosclerosis in lupus. JCI Insight 3, 99276 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denny M. F., et al. , Interferon-alpha promotes abnormal vasculogenesis in lupus: A potential pathway for premature atherosclerosis. Blood 110, 2907–2915 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rahman S., et al. , Low-density granulocytes activate T cells and demonstrate a non-suppressive role in systemic lupus erythematosus. Ann. Rheum. Dis. 78, 957–966 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wright H. L., Makki F. A., Moots R. J., Edwards S. W., Low-density granulocytes: Functionally distinct, immature neutrophils in rheumatoid arthritis with altered properties and defective TNF signalling. J. Leukoc. Biol. 101, 599–611 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Carmona-Rivera C., Kaplan M. J., Low-density granulocytes: A distinct class of neutrophils in systemic autoimmunity. Semin. Immunopathol. 35, 455–463 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faurschou M., Borregaard N., Neutrophil granules and secretory vesicles in inflammation. Microbes Infect. 5, 1317–1327 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Marini O., et al. , Mature CD10+ and immature CD10- neutrophils present in G-CSF–treated donors display opposite effects on T cells. Blood 129, 1343–1356 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Baechler E. C., et al. , Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc. Natl. Acad. Sci. U.S.A. 100, 2610–2615 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tucci M., et al. , Glomerular accumulation of plasmacytoid dendritic cells in active lupus nephritis: Role of interleukin-18. Arthritis Rheum. 58, 251–262 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Yao Y., et al. , Development of potential pharmacodynamic and diagnostic markers for anti-IFN-α monoclonal antibody trials in systemic lupus erythematosus. Hum. Genomics Proteomics 2009, 374312 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta S., et al. , Distinct functions of autoantibodies against interferon in systemic lupus erythematosus: A comprehensive analysis of anticytokine autoantibodies in common rheumatic diseases. Arthritis Rheumatol. 68, 1677–1687 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bennett L., et al. , Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J. Exp. Med. 197, 711–723 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evrard M., et al. , Developmental analysis of bone marrow neutrophils reveals populations specialized in expansion, trafficking, and effector functions. Immunity 48, 364–379.e8 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Fiedler K., Brunner C., The role of transcription factors in the guidance of granulopoiesis. Am. J. Blood Res. 2, 57–65 (2012). [PMC free article] [PubMed] [Google Scholar]
- 23.Yáñez A., Ng M. Y., Hassanzadeh-Kiabi N., Goodridge H. S., IRF8 acts in lineage-committed rather than oligopotent progenitors to control neutrophil vs monocyte production. Blood 125, 1452–1459 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Hock H., et al. , Intrinsic requirement for zinc finger transcription factor Gfi-1 in neutrophil differentiation. Immunity 18, 109–120 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Yamanaka R., et al. , Impaired granulopoiesis, myelodysplasia, and early lethality in CCAAT/enhancer binding protein epsilon-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 94, 13187–13192 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinelli S., et al. , Induction of genes mediating interferon-dependent extracellular trap formation during neutrophil differentiation. J. Biol. Chem. 279, 44123–44132 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Carp H., Mitochondrial N-formylmethionyl proteins as chemoattractants for neutrophils. J. Exp. Med. 155, 264–275 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahn B. J., et al. , Ninjurin1 is expressed in myeloid cells and mediates endothelium adhesion in the brains of EAE rats. Biochem. Biophys. Res. Commun. 387, 321–325 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Kobayashi Y., The role of chemokines in neutrophil biology. Front. Biosci. 13, 2400–2407 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Unkeless J. C., Shen Z., Lin C. W., DeBeus E., Function of human Fc gamma RIIA and Fc gamma RIIIB. Semin. Immunol. 7, 37–44 (1995). [DOI] [PubMed] [Google Scholar]
- 31.Zhai Y., et al. , Host transcriptional response to influenza and other acute respiratory viral infections—A prospective cohort study. PLoS Pathog. 11, e1004869 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith C. K., et al. , Neutrophil extracellular trap-derived enzymes oxidize high-density lipoprotein: An additional proatherogenic mechanism in systemic lupus erythematosus. Arthritis Rheumatol. 66, 2532–2544 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grayson P. C., et al. ; Rituximab in ANCA-Associated Vasculitis-Immune Tolerance Network Research Group , Neutrophil-related gene expression and low-density granulocytes associated with disease activity and response to treatment in antineutrophil cytoplasmic antibody-associated vasculitis. Arthritis Rheumatol. 67, 1922–1932 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin A. M., et al. , Mast cells and neutrophils release IL-17 through extracellular trap formation in psoriasis. J. Immunol. 187, 490–500 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van der Linden M., et al. , Neutrophil extracellular trap release is associated with antinuclear antibodies in systemic lupus erythematosus and anti-phospholipid syndrome. Rheumatology (Oxford), 10.1093/rheumatology/key067 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Ford A. M., et al. , Regulation of the myeloperoxidase enhancer binding proteins Pu1, C-EBP alpha, -beta, and -delta during granulocyte-lineage specification. Proc. Natl. Acad. Sci. U.S.A. 93, 10838–10843 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lawrence S. M., Corriden R., Nizet V., The ontogeny of a neutrophil: Mechanisms of granulopoiesis and homeostasis. Microbiol. Mol. Biol. Rev. 82, e00057-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fauci A. S. K. D., et al. , Harrison’s Principles of Internal Medicine (McGraw-Hill, New York, NY, ed. 19, 2014). [Google Scholar]
- 39.Gul E., et al. , Type I IFN-related NETosis in ataxia telangiectasia and Artemis deficiency. J. Allergy Clin. Immunol. 142, 246–257 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Coit P., et al. , Epigenome profiling reveals significant DNA demethylation of interferon signature genes in lupus neutrophils. J. Autoimmun. 58, 59–66 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.King K. R., et al. , IRF3 and type I interferons fuel a fatal response to myocardial infarction. Nat. Med. 23, 1481–1487 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knight J. S., et al. , Peptidylarginine deiminase inhibition reduces vascular damage and modulates innate immune responses in murine models of atherosclerosis. Circ. Res. 114, 947–956 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soehnlein O., et al. , Neutrophil secretion products pave the way for inflammatory monocytes. Blood 112, 1461–1471 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gaul D. S., Stein S., Matter C. M., Neutrophils in cardiovascular disease. Eur. Heart J. 38, 1702–1704 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Lee T. D., Gonzalez M. L., Kumar P., Grammas P., Pereira H. A., CAP37, a neutrophil-derived inflammatory mediator, augments leukocyte adhesion to endothelial monolayers. Microvasc. Res. 66, 38–48 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Herman M. P., et al. , Expression of neutrophil collagenase (matrix metalloproteinase-8) in human atheroma: A novel collagenolytic pathway suggested by transcriptional profiling. Circulation 104, 1899–1904 (2001). [DOI] [PubMed] [Google Scholar]
- 47.Kegerreis B. J., et al. , Genomic identification of low-density granulocytes and analysis of their role in the pathogenesis of systemic lupus erythematosus. J. Immunol. 202, 3309–3317 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.