Significance
In this work, we report the use of chloroquine as a treatment against myotonic dystrophy type 1 (DM1), a neuromuscular disease caused by expanded CUG repeats with no available effective treatment. We tested the effect of the drug in 2 different animal models of the disease, Drosophila and mouse, and also in human-derived myoblasts. We demonstrate that chloroquine treatment was able to upregulate Muscleblind levels. These proteins are a key component in the pathogenic mechanism of the disease, as they are sequestered by mutant RNA expressing expanded CUG repeats, and are thus deprived of their normal function. Moreover, we observed that Muscleblind boosting was functional, detecting amelioration of disease-related phenotypes in all 3 models at molecular, cellular, and physiological levels.
Keywords: chloroquine, myotonic dystrophy, muscleblind, therapy
Abstract
Myotonic dystrophy type 1 (DM1) is a life-threatening and chronically debilitating neuromuscular disease caused by the expansion of a CTG trinucleotide repeat in the 3′ UTR of the DMPK gene. The mutant RNA forms insoluble structures capable of sequestering RNA binding proteins of the Muscleblind-like (MBNL) family, which ultimately leads to phenotypes. In this work, we demonstrate that treatment with the antiautophagic drug chloroquine was sufficient to up-regulate MBNL1 and 2 proteins in Drosophila and mouse (HSALR) models and patient-derived myoblasts. Extra Muscleblind was functional at the molecular level and improved splicing events regulated by MBNLs in all disease models. In vivo, chloroquine restored locomotion, rescued average cross-sectional muscle area, and extended median survival in DM1 flies. In HSALR mice, the drug restored muscular strength and histopathology signs and reduced the grade of myotonia. Taken together, these results offer a means to replenish critically low MBNL levels in DM1.
The autosomal dominant disorder myotonic dystrophy type 1 (DM1) is the most prevalent adult-onset muscular dystrophy. DM1 is characterized by severe neuromuscular defects, including myotonia and progressive muscle weakness and wasting (atrophy), leading to disability as the disease progresses, and respiratory distress either from primary muscle failure or from cardiopulmonary complications. DM1 is also characteristically multisystemic and degenerative, affecting body systems such as the heart and the brain (1). Several candidate therapies have been tested in DM1 models but none has reached clinical practice (2) revealing a need to find new drugs against DM1.
The molecular basis of the disease is the pathogenic expansion of an unstable CTG microsatellite in the 3′ UTR of the DM1 protein kinase (DMPK) gene. Transcripts containing expanded repeats form double-stranded structures that accumulate in cell nuclei as ribonuclear foci (3). Mutant DMPK interferes with 2 Muscleblind-like families of proteins that regulate alternative splicing (MBNL1 and MBNL2), with overlapping patterns of expression in skeletal muscle, heart, and central nervous system (CNS), and the CUGBP Elav-like family member 1 (CELF1), a MBNL antagonistic regulator of alternative splicing (AS) that also regulates transcription and translation (4). In affected tissues the MBNL1 and 2 functions are reduced by aberrant binding to CUG expansions, while CELF1 is activated due to increased stabilization because of hyperphosphorylation (5). MBNL proteins are developmental sensors, so MBNL depletion maintains fetal AS patterns in adults that become unfit proteins, giving rise to specific symptoms such as myotonia, muscle weakness, insulin resistance, or cardiac conduction defects (6–8). MBNL proteins control RNA metabolism in additional ways, including fetal-to-adult polyadenylation patterns, stability, differential localization of mRNAs, and miRNA biogenesis (9–12).
Limited availability of MBNL1 and MBNL2 is a primary contributor to DM1 phenotypes. Loss of MBNL1 function accounts for more than 80% of missplicing events and nearly 70% of expression defects in a murine model that expresses 250 CTG repeats in the context of human skeletal actin (HSALR; refs. 13 and 14). Furthermore, overexpression of MBNL1 rescues aberrant AS of muscle transcripts and myotonia in HSALR mice (15). Mbnl1−/− Mbnl2+/− animals consistently reproduce cardinal aspects of DM1, while Mbnl2 knockout (KO) mice or ones that express CUG repeats reproduce CNS phenotypes typical of the disease (16, 17). MBNL1 overexpression is well tolerated in skeletal muscle in mice, and early and long-term overexpression prevents myotonia, myopathy, and AS alterations in DM1 mice (18). Thus, contrary to most genetic diseases in which the phenotypes originate from loss- or gain-of-function mutations in a given gene, most molecular DM1 alterations stem from the depletion of MBNL1 and 2 proteins, which remain encoded in perfectly functional genes. From the therapeutic perspective, this is a particularly favorable situation because protein depletion can be compensated with extra endogenous expression, which will also antagonize excessive CELF1 activity. We have pioneered this approach by identifying miR-23b and miR-218 as translational repressors of all transcript isoforms originated from MBNL1 and 2 genes. miR-23b and miR-218 antagonists raise MBNL1 and 2 protein expression in skeletal muscles and improve DM1-like phenotypes (19). We previously reported abnormal hyperactivation of autophagy in Drosophila and cell models of disease and that inhibiting autophagy restored muscle mass and function in Drosophila (20). This led to the hypothesis that autophagy inhibitory drugs could rescue muscle atrophy in vivo. Chloroquine (CQ) is a well-characterized autophagy blocker so we tested its ability to improve muscle function in Drosophila and mouse models. Subsequent studies led to the discovery that CQ increases Muscleblind levels in a Drosophila model, in patient-derived myoblasts, and in a DM1 murine model, to a sufficient extent to significantly improve various molecular and functional phenotypes associated with the disease.
Results
Autophagy Blocker CQ Rescues CUG RNA Toxicity in Model Fly Muscle.
We used a Drosophila model that expresses 480 noncoding CTG repeats throughout the somatic musculature (Mhc-Gal4 UAS-(CTG)480/+) (21). One-day-old adult flies were transferred to tubes containing standard medium supplemented with 10 or 100 μM CQ for 7 d and were compared to DM1 flies taking no drugs and normal controls. Autophagy was detected in somatic muscles of DM model flies using LysoTracker (SI Appendix, Fig. S1 A and B). A strong signal was observed in flies that expressed expanded CTG repeats during myogenesis compared to the controls. These data indicate an abnormal increase of autophagy in muscles expressing toxic RNA, as similarly reported following heat-shock induction of expanded CTG repeats in adult flies (20). We studied caspase-3 and caspase-7 activity and observed no effect on apoptosis levels after CQ treatment. Thus, autophagy block was not compensated by extra activation of apoptosis (SI Appendix, Fig. S1C).
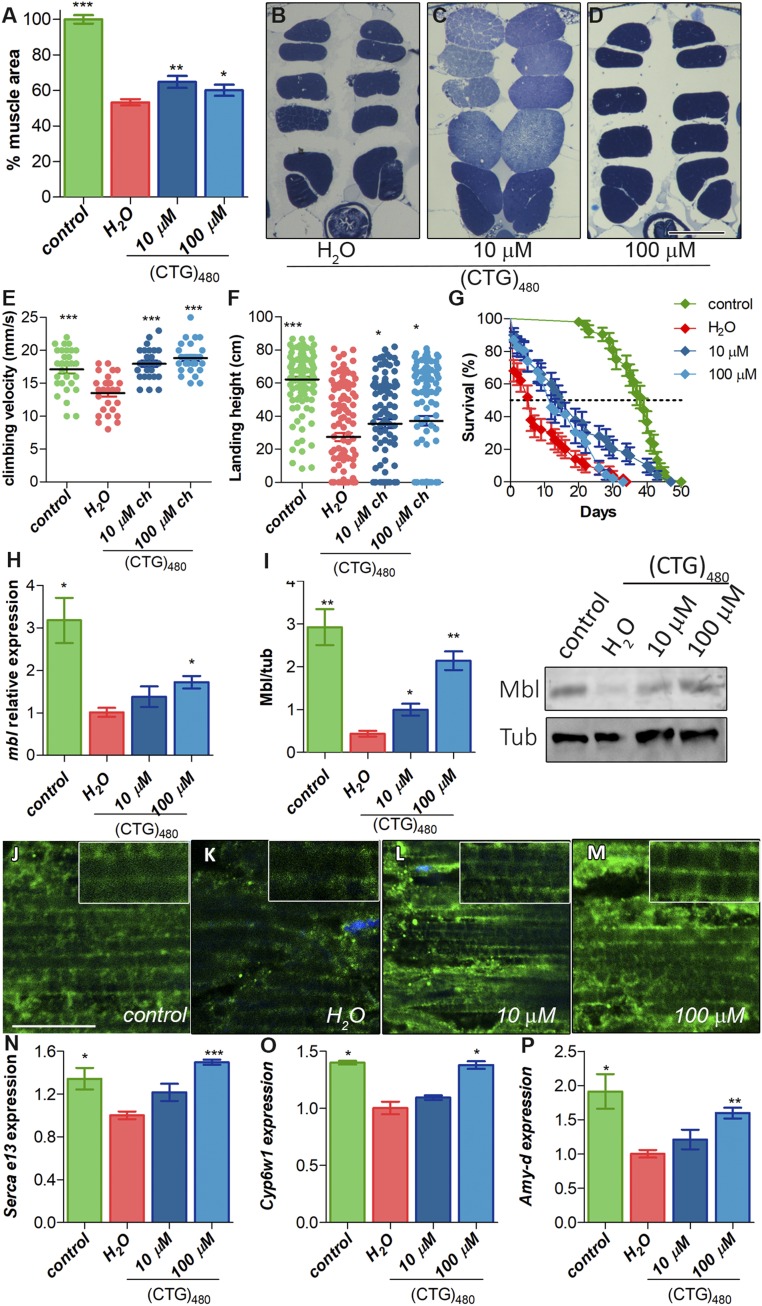
Muscle mass loss is a characteristic phenotype of DM1 patients that model flies clearly reproduce (20). Thus, we studied the effect of CQ treatment on muscle atrophy. We first measured muscle area of indirect flight muscles (IFMs) of untreated DM1 flies and model flies fed with food supplemented with 10 or 100 μM CQ (Fig. 1 A–D). Whereas no effect over the IFM cross-sectional muscle area was detected in control flies at any concentration of the drug (SI Appendix, Fig. S1 D–F), sections of model flies showed a significantly reduced IFM area that reached ∼50% of normal controls. Interestingly, 7-d oral administration of 10 or 100 μM CQ to adult DM1 flies showed a 1.2 and 1.13-fold increase of muscle area, respectively. Therefore, CQ administration to adult flies that already had a 50% reduction in IFM muscles managed to rescue around 25% of the muscle area, clearly indicating a muscle homeostasis contribution to the atrophic phenotype. To test whether increased muscle area correlated to improved functional locomotion, we studied the climbing and flying ability (Fig. 1 E and F). DM1 flies have reductions of 22 and 58% in climbing and flight capacity, respectively. Both phenotypes were significantly rescued upon CQ administration. Furthermore, DM1 patients have reduced life expectancy due to, among other symptoms, muscle wasting and cardiac conduction defects (22). Model flies reproduce this phenotype with a median survival of 6 d compared to normal controls that have a 6.5-fold higher survival (Fig. 1G). Chronic CQ administration was sufficient to significantly increase mean life and also extend maximal lifespan of DM1 flies at the lowest dose of CQ. Taken together these observations confirm that autophagy blocker CQ rescues muscle degeneration and muscle function and manages to extend the lifespan of DM1 model flies.
Fig. 1.
CQ treatment rescues DM1-like phenotypes in model flies by Muscleblind up-regulation. (A) Relative quantification of the mean percentage of muscle area per condition (n = 6). (B–D) Representative dorsoventral sections of resin-embedded thoraces of DM1 [Mhc-Gal4 UAS-i(CTG)480] flies treated with the indicated concentrations of CQ or vehicle. (Scale bar, 100 µm.) (E) Quantification of climbing velocity as mean of speed ± SEM (n = 30) comparing control ability (Mhc-Gal4/+; green), DM1 (red), and DM1-treated flies (blue). (F) Graphs of flight assays represent the average landing distance reached by the different experimental groups (n = 100). (G) Survival curves expressed as percentages (50 < n < 80). (H) RT-qPCR amplification of muscleblind from control (green) and DM1 flies (red) treated with CQ (blue bars) after normalization to rp49. (I) Graphs and representative blot image of Muscleblind immunodetection. α-Tubulin protein expression was used as an endogenous control. (J–M) Representative confocal images of longitudinal sections of IFMs showing anti-Mbl staining (green) in control (J), untreated DM1 flies (K), and DM1-treated flies (L and M). Nuclei were counterstained with DAPI (blue). All stainings were performed in parallel and images were acquired using the same settings. (Scale bar, 10 µm.) RT-qPCR analysis to quantify expression of Serca exon 13 (N), Cyp6w1 (O), and Amy-d (P) relative to Rp49 in the indicated conditions (n = 3). All comparisons refer to untreated DM1 flies (H2O in the graphs). *P < 0.05, **P < 0.01, ***P < 0.001 according to Student’s t test.
CQ Enhances Muscleblind Expression in DM1 Fly Muscles.
Considering the critical role of the MBNL proteins in the pathogenesis of DM1, and the possibility that autophagy directly contributes to low MBNL levels by promoting its degradation, we decided to quantify Drosophila Muscleblind (Mbl) in CQ-treated flies. We initially measured mbl transcript levels and normalized expression relative to Rp49 (Fig. 1H). Results revealed a 3-fold decrease of mbl in DM1 flies, and these levels were partially restored, in a dose-dependent manner, upon CQ administration. To confirm that the increase was in somatic muscle, we normalized mbl expression relative to mhc, which encodes a sarcomeric component highly specific of somatic muscle (23) (SI Appendix, Fig. S1G). This normalization further supported the finding that CQ intake enhanced mbl levels in somatic muscle. Mbl protein was around 7 times lower in DM1 flies than in controls, and these levels were partially restored after a 7-d CQ treatment (Fig. 1I). Moreover, we confirmed that Mbl overexpression was CTG repeat dependent. No effect on Mbl levels was observed after CQ treatment of control flies, thus suggesting that up-regulation of Mbl by CQ in DM1 model flies required activation of autophagy by CUG RNA (SI Appendix, Fig. S1H). To investigate whether CQ directly bound CUG repeats, we tested whether the drug had any effect on CUG thermal stability using differential scanning fluorimetry (DSF; ref. 24). The DSF method reports not only RNA structure stability but also its modulation by RNA-binding ligands. We found that in the presence of CQ there was no significant change in fluorescence or the melting temperature of the CUG probe, which indicates the temperature at which 50% of the RNA is unfolded (SI Appendix, Fig. S2). These data indicate that the mechanism by which Mbl accumulates in CQ-treated muscles is not due to the direct binding of CQ to expanded CUG repeats.
We sought independent validation of Mbl expression data in DM1 Drosophila muscles by immunofluorescence of adult thoraces (Fig. 1 J–M). In control flies, we observed a neat signal with a regular transversal banding pattern spanning throughout the IFM fiber width, in agreement with previous reports that localized Mbl in sarcomeric H and Z bands (25). In DM1 fly muscles the signal was greatly reduced, and the banding pattern was obviously disrupted while the protein accumulated in the cell nuclei. In contrast, muscles from treated flies resembled controls in terms of Mbl immunofluorescence intensity and distribution to sarcomeric bands.
It is well known that Mbl regulates AS and transcription of hundreds of mRNAs (12, 14). The boost in Mbl conferred by CQ treatment was enough to reestablish Serca exon 13 inclusion in a dose-dependent manner, which achieved values similar to those in control flies at the highest concentration (Fig. 1N). We also demonstrated that a reduction of around 50% in the expression of Cyp6w1 and Amy-d, both Mbl dependent, was completely rescued upon 100 μM CQ treatment (Fig. 1 O and P).
CQ Improves DM1-Like Phenotypes in Immortalized Patient-Derived Myoblasts.
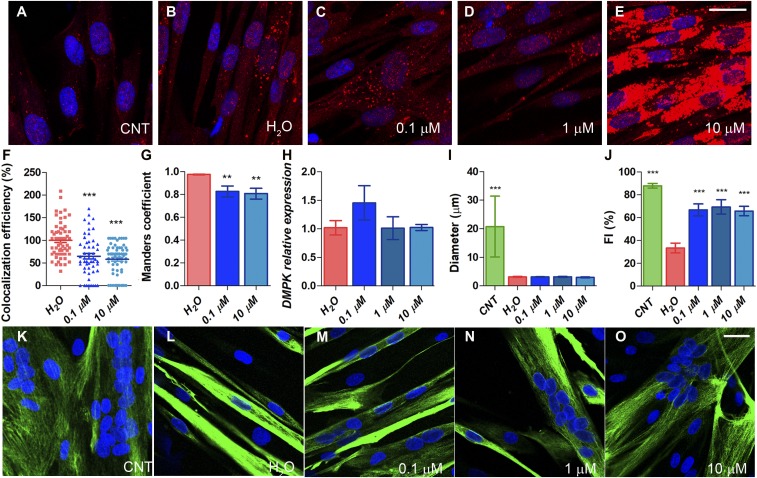
It was previously demonstrated that autophagy was abnormally up-regulated in DM1 myoblasts that express expanded CUG repeats (20, 26). We treated immortalized patient-derived myoblasts (iPDMs) (27) differentiated for 4 d with CQ at concentrations ranging from 0.1 to 10 µM (well below toxic concentration 10, meaning the dose produces a toxic effect in 10% of the population [TC10], to rule out any nonspecific effects related to toxicity; SI Appendix, Fig. S3A). CQ blocks late-stage autophagy by decreasing autophagosome–lysosome fusion, resulting in the accumulation of proteins involved in previous steps in this pathway, such as LC3 (28). We detected LC3 signal and observed a diffuse pattern in immortalized control-derived myoblasts (iCDMs) while strong puncta formation was detected in iPDMs as a consequence of increased autophagic activity (20). A similar signal was observed after 0.1 and 1 µM CQ treatment. However, in cells treated with the highest concentration of CQ, a dramatic increase in the signal was detected, thus confirming an effective blockade of autophagic flux (Fig. 2 A–E). We also analyzed foci number and area in immortalized patient-derived fibroblasts (iPDFs) treated with CQ. (SI Appendix, Fig. S4). In our laboratory conditions, neither of the CQ concentrations (1 and 10 μM) had any effect on ribonuclear foci number or area. MBNL1 sequestration by expanded CUG RNA, however, was significantly rescued according to double immunofluorescence and in situ hybridization data. While we detected dense aggregates of MBNL1 in RNA foci in iPDMs under control conditions, sequestration of MBNL1 was markedly attenuated by CQ as observed in colocalization studies (Fig. 2 F and G and SI Appendix, Fig. S4). Quantitatively, CQ reduced the average percentage of colocalization of MBNL1 and CUG by 35 and 42% at increasing concentrations of CQ. Importantly, we confirmed that DMPK transcript levels remained unchanged after the treatment (Fig. 2H), which was also confirmed by RNA sequencing (RNA-Seq) data (7.35 ± 0.06, 7.36 ± 0.06, 7.39 ± 0.02, counts per million for untreated iPDMs, or treated with 0.1 or 10 µM CQ, respectively).
Fig. 2.
CQ treatment of DM1 iPDMs improves phenotypes. (A–E) Fluorescence images of LC3 immunostaining (red) of iCDMs (control [CNT]) (A) and iPDMs (B) differentiated for 96 h and treated with vehicle or CQ (C–E) for 48 (Scale bar, 20 μm.) (F–G) Analysis of colocalization efficiency (F) and Manders’ coefficient (G) between MBNL1 and (CAG)7 in iPDMs treated with CQ 0.1 and 10 µM. Colocalization efficiencies were normalized to untreated iPDMs (H2O) and data were expressed as percentage (%) in the case of colocalization efficiency. Between 50 and 60 nuclei were analyzed. (H) RT-qPCR analysis to quantify expression of DMPK relative to GAPDH in iPDMs after CQ treatment (n = 3). (I–J) Quantification of myotube diameter and myogenic fusion index of iCDMs (green) and untreated iPDMs (red) and iPDMs treated with CQ (blue) (n = 7 to 10 images per condition). (K–O) Representative confocal images of Desmin-immunostained (green) human myoblasts transdifferentiated for 7 d used for quantifications in I and J. Staining was performed in iCDMs (K), and iPDMs untreated (L) or treated with CQ (M–O) for 48 h. (Scale bar, 1,000 µm.) Nuclei were counterstained with DAPI (blue). **P < 0.01, ***P < 0.001 according to Student’s t test. All comparisons refer to untreated iPDMs (H2O in the graphs).
iPDMs were reported to have impaired fusion capacity in vitro (27). This parameter was specifically quantified in iPDMs differentiated for 7 d and stained for Desmin, which is a marker of terminally differentiated myoblasts (Fig. 2 I–O). iPDM fusion index was about 50%, and myotube diameter about 15%, of normal values (Fig. 2 I and J). Exposure of cells to a range of CQ concentrations did not affect the diameter of the myotubes but strongly rescued fusion index. These data suggest improved differentiation capacity in DM1 cells after CQ treatment.
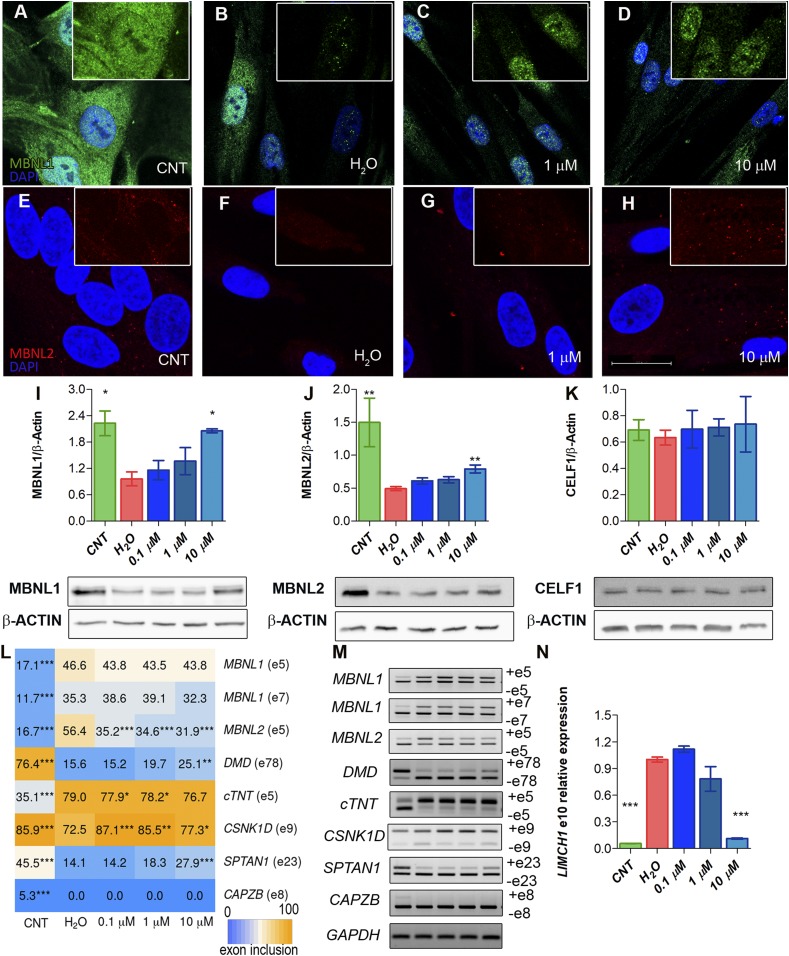
Given our results in the fly model where CQ enhanced Mbl expression, we hypothesized that improvements observed in iPDM could originate from increased MBNL protein levels. MBNL1 and 2 were detected by immunofluorescence in myoblasts differentiated for 4 d (Fig. 3 A–H). In iPDM cells, we found that MBNL1 and 2 expression was substantially lower than in iCDMs, and that the proteins were mainly detected in ribonuclear foci. CQ treatment had a profound and dose-dependent effect on the expression of both proteins and their distribution, which at the highest concentration approached a normal pattern. Importantly, although MBNL proteins were still detected in nuclear foci, they were also broadly dispersed in both cell compartments. Note that MBNL2 increased to similar levels in both the cell nucleus and cytoplasm of treated iPDMs, whereas in iCDMs its expression was mainly cytoplasmic. To confirm these results, we immunodetected MBNL1 and MBNL2 in 2 additional CDM and 2 PDM lines. From these, 1 contained 1,000 and the other 333 CTG repeats (SI Appendix, Fig. S5). Consistently, we observed increased signal in the nucleus for MBNL1 and in the cytoplasm for MBNL2 in a dose-dependent manner after CQ treatment of PDMs.
Fig. 3.
MBNL1 and MBNL2 are overexpressed upon CQ treatment of iPDMs. (A–H) Representative confocal images of MBNL1 (green) and MBNL2 (red) immunostaining in iCDMs (control [CNT]) (A and E), untreated iPDMs (B and F), and iPDMs treated with CQ (C, D, G, and H). Nuclei were counterstained with DAPI. (Scale bar, 20 µm.) (I–K) Western blot quantification and representative blot images of MBNL1 (I), MBNL2 (J), and CELF1 (K) in iCDMs (green bar) and untreated (red bar) or CQ-treated iPDMs (blue bars). β-ACTIN expression was used as an endogenous control. (L) Heatmap representing the quantification of splicing decisions altered in iPDMs. Numbers within the boxes indicate the percentage of inclusion of the indicated exons obtained by semiquantitative RT-PCR. (M) Representative gels used to quantify in L. Percentage of CELF1-regulated CAPZB exon 8 inclusion was determined as a control. (N) RT-qPCR to measure exon 10 inclusion of LIMCH1. GAPDH was used as an internal control. In all analyses, sample size was n = 3, with the exception of MBNL2 Western blot with n = 6. Cells were differentiated for 96 h. All comparisons refer to untreated iPDMs (H2O in the graphs). *P < 0.05, **P < 0.01, ***P < 0.001 according to Student’s t test.
Quantification of MBNL1 and 2 levels demonstrated an increase of up to 100% and 65%, respectively, after the addition of CQ at the indicated concentrations (Fig. 3 I and J). To test whether CQ might have a nonspecific targeted ability to up-regulate protein levels, CELF1 was quantified in parallel experiments and no significant changes were found (Fig. 3K). At the functional level, the CQ-induced MBNL1 increase was sufficient to rescue the inclusion of MBNL2 exon 5, DMD exon 78, and cTNT exon 5, which are altered in DM1 (Fig. 3 L and M and SI Appendix, Fig. S6). However, no effect was detected in the MBNL1 inclusion pattern of exons 5 and 7 that are dependent on MBNL1 levels (29). In the case of the inclusion of SPTAN1 exon 23 and LIMCH1 exon 10, regulated by MBNL2 (16), both were significantly rescued at the highest dose. Exon regulation of CSNK1D, a target of MBNL2, was restored at the 3 tested CQ concentrations (Fig. 3 L–N). The splicing of CAPZB, regulated by CELF1, was also analyzed, and no changes were detected in its exon inclusion (Fig. 3 L and M). CQ therefore not only enhanced MBNL1 and 2 expression levels in iPDMs but rescued its subcellular distribution and molecular function as an AS regulator, thus supporting its potential as a therapy against DM1.
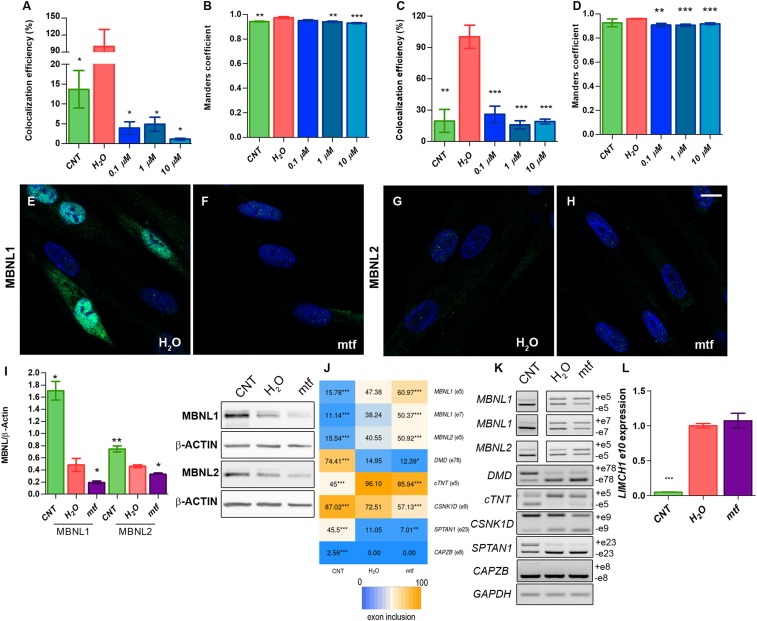
To shed light into the reason why MBNL protein levels went up upon CQ treatment, we performed double staining with LysoTracker and antibodies against MBNL1 or 2 and quantified colocalization of both signals by calculating Manders’ colocalization coefficient and colocalization efficiency, to determine how much and how well the fluorescent signals overlapped (Fig. 4 A–D and SI Appendix, Fig. S7). Importantly, we observed 5 times more colocalization of MBNL1 or 2 with LysoTracker in iPDMs compared to levels detected in iCDMs. These values were dramatically reduced after CQ treatment of iPDMs, which demonstrated that CQ promotes MBNL release from the autophagic pathway preventing degradation and increasing protein levels.
Fig. 4.
Overexpression of MBNLs by CQ is mediated by autophagy blockade in iPDMs. (A–D) Colocalization efficiency and Manders’ coefficient between MBNL1/LysoTracker (A and B) and MBNL2/LysoTracker (C and D). Colocalization efficiencies were normalized to untreated iPDMs (H2O) and data were expressed as percentage (%). No less than 70 nuclei were analyzed. (E–H) Representative confocal images of MBNL1 and MBNL2 (green) immunostaining in untreated iPDMs (E and G) or treated with mtf (F and H). Nuclei were counterstained with DAPI. (Scale bar, 10 µm.) (I) Western blot quantification and representative blot images of MBNL1, MBNL2, and β-ACTIN in iCDMs (control [CNT]; green bar), and untreated (H2O; red bar) or mtf-treated iPDMs (mtf; purple bar). β-ACTIN expression was used as an endogenous control. (J) Heatmap representing the analysis of splicing decisions altered in iPDMs and worsened after mtf treatment. Numbers within boxes indicate the percentage of inclusion of the indicated exons obtained by semiquantitative RT-PCR. (K) Representative gels used to quantify in J. Percentage of CELF1-regulated CAPZB exon 8 inclusion was determined as a control. (L) RT-qPCR to analyze exon 10 inclusion of LIMCH1. GAPDH was used as an internal control. In all analyses sample size was n = 3 and cells were differentiated for 96 h. All comparisons refer to untreated iPDMs (H2O in the graphs). *P < 0.05, **P < 0.01, ***P < 0.001 according to Student’s t test.
Chemical Induction of Autophagy Worsens DM1-Linked Phenotypes in Human Muscle Cells.
To confirm that therapeutic effects observed upon CQ treatment were mediated by MBNL levels through modulation of autophagy, we evaluated the effects of further enhancing autophagy in iPDMs. To this end, we treated cells with 30 mM metformin (mtf), which activates AMPK and consequently leads to autophagy activation via both mTOR inhibition and ULK1 activation (30). After adding mtf, we detected a dramatic decrease in the MBNL1 signal compared to untreated iPDMs. Quantification of the protein by Western blot showed a reduction of 60% after autophagy induction (Fig. 4 E, F, and I). Down-regulation of MBNL2 was not obvious by immunostaining, but Western blot demonstrated a reduction of 35% (Fig. 4 G–I). Analysis of defective-DM1 AS events regulated by MBNL1 and MBNL2 demonstrated that upon mtf treatment missplicing of most alternative exons was higher than in iPDMs treated with water (Fig. 4 J–L and SI Appendix, Fig. S8), except for MBNL2-dependent LIMCH1 exon 10 and cTNT exon 5 inclusion. Overall, we observed a tight correlation between MBNL levels and AS regulation. As a control, we analyzed CELF1-dependent splicing of CAPZB, and we did not observe any effect after mtf treatment.
CQ Treatment Recovers Expression of 59% of Disease-Related Transcripts in iPDMs.
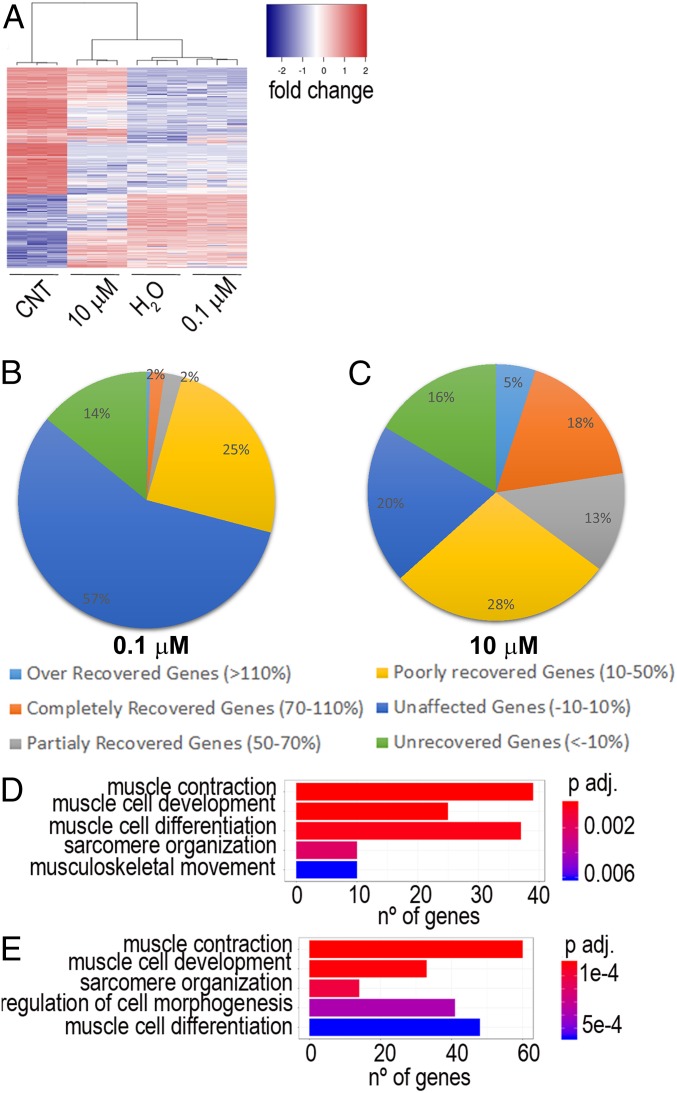
In order to have a comprehensive description of the consequences of CQ treatment on DM1-related gene regulation, we conducted paired-end RNA-Seq from iCDMs and untreated and treated (0.1 and 10 µM CQ) iPDMs differentiated for 96 h (31). Bioinformatics analyses with edgeR software comparing iCDMs and untreated iPDMs identified 3,594 differentially regulated disease-related genes (DRGs). As observed in Fig. 5A there is a strong differential expression in transcripts between iCDMs and iPDMs. Importantly, after 10 µM CQ treatment, the transcript expression profile of the samples is more similar to iCDMs than to iPDMs as indicated by dendrograms. From DRGs, almost 25% were recovered after low-dose treatment and this percentage reaches 59% after exposure to 10 µM CQ. (Fig. 5 B and C). Altogether, we observed a strong correlation between CQ dose and effect on DRG recovery. A gene ontology (GO) enrichment test was also performed, considering all recovered genes in each treatment (% of recovery ranging from 10 to 110%). This test highlighted genes involved in several pathways related to muscle homeostasis and function (Fig. 5 D and E). Considering RNA-Seq data, it is confirmed that the effects of the drug depend on its dose. Globally, our results demonstrate that restoration of muscle functions correlates to MBNL levels and also that CQ can be considered as a potential drug to treat DM1 patients.
Fig. 5.
Global effect of CQ on RNA expression. (A) Heatmap representing transcript expression at the indicated conditions. (B and C) Pie charts showing the percentage of recovery of DRGs after CQ treatment. (D and E) Bar plots based on GO analysis showing the principal biological processes affected by 0.1 µM (D) and 10 µM (E) CQ in DM1. CNT, control.
CQ Raises Mbnl Expression and Ameliorates Molecular Phenotypes in a DM1 Murine Model.
HSALR mice express 250 CTG repeats in the 3′ UTR of the human skeletal actin and reproduce symptoms observed in patients, such as myotonia, splicing defects, and muscle atrophy (13, 32). We treated these mice with 2 different concentrations of i.p. CQ, 25 and 50 mg/kg for 7 d, and compared results with untreated HSALR controls and normal mice with the same genetic background (friend virus B [FVB]). First, we quantified HSALR transgene expression in the gastrocnemius and quadriceps muscles having previously discovered that fluctuations in transgene expression strongly influence phenotypic manifestations in these mice (19). In quadriceps, expression was fairly stable between control and experimental groups, with maximum variations around 2-fold above average. Gastrocnemius, in contrast, revealed bigger changes but toward greater expression of the transgene, which did not impede detection of the typical DM1 phenotypes in the model (SI Appendix, Fig. S9A).
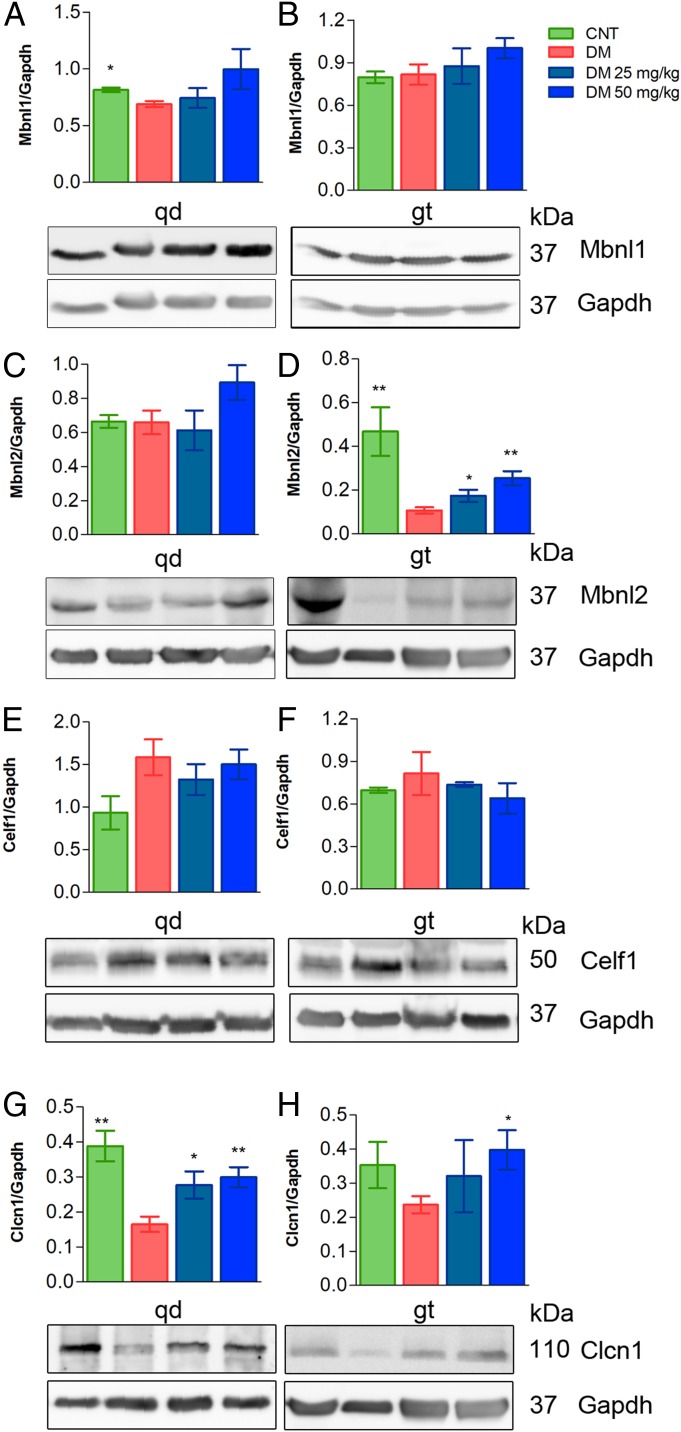
Based on the significant increase of MBNLs in the DM1 fly model and in myoblasts, Mbnl1 and 2 levels were quantified in quadriceps and gastrocnemius. In the case of Mbnl1, despite a trend toward rising levels in drug-treated mice compared to vehicle-only controls (phosphate-buffered saline [PBS]), the differences did not reach statistical significance in either muscle (Fig. 6 A and B). Mbnl2 expression in gastrocnemius, in contrast, did show a significant increase at the 2 tested doses while quadriceps levels remained stable at the low CQ dose and reached borderline significance (P < 0.08) at the highest (Fig. 6 C and D). Additional Mbnl protein levels did not originate from extra transcription or enhanced stability since no changes were found at the mRNA level (SI Appendix, Fig. S9 B and C). Importantly, Celf1 levels remained unchanged after the administration of the drug as similarly observed in the cell model of disease (Fig. 6 E and F).
Fig. 6.
Treatment of HSALR mice increases Mbnl1 and Mbnl2 levels. (A–H) Western blotting analyses of protein extracts from quadriceps (qd) (A, C, E, and G) or gastrocnemius (gt) (B, D, F, and H) of FVB control mice (n = 4, green bars), untreated HSALR model mice (n = 6, red bars), or model mice injected for 7 d with CQ at 25 mg/kg (n = 3, dark blue bars) or 50 mg/kg CQ (n = 5, pale blue bars). Protein levels are relative to Gapdh, which was used as an internal control. Representative blots used for quantification are shown below bar graphs. Because Clcn1 and Mbnl2 proteins were detected in the same blots, the internal control Gapdh was the same (duplicated blot in D and H). All comparisons are relative to HSALR mice treated with the vehicle PBS (DM). *P < 0.05, **P < 0.01 according to Student’s t test. CNT, control.
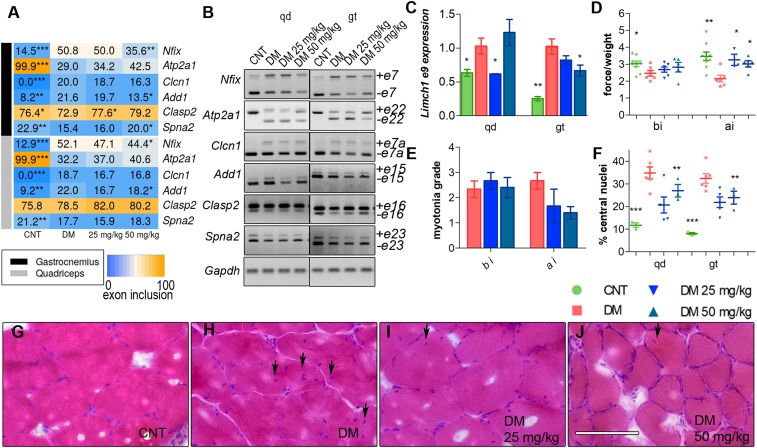
As we detected up-regulation of Mbnl proteins in vivo, we sought to analyze Mbnl-dependent splicing events relevant for HSALR phenotypes. Thus, we studied splicing pattern of Nfix, Atp2a1, and Clcn1, regulated by Mbnl1, and observed that CQ administered at 50 mg/kg significantly ameliorated aberrant exon choices for Nfix (exon 7). Of the splicing events regulated by Mbnl2, we analyzed Add1, Clasp2, Spna2, and Limch1 (Fig. 7 A–C and SI Appendix, Fig. S10). All of the splicing events improved in the 2 muscles studied from treated HSALR mice, except for Clasp2 and Spna2 that did so only in gastrocnemius. To confirm the specificity of CQ toward Mbnl activity, we quantified the inclusion of Capzb exon 8, Mfn2 exon 3, and Ank2 exon 21, which depend on Celf1 (33, 34), and, consistent with previous results, remained unchanged upon CQ treatment (SI Appendix, Fig. S11). Considering the relevance of the myotonic phenotype in the disease and the fact that we could not detect significant changes in exon 7a inclusion in Clcn1 transcripts in our experimental conditions, we quantified Clcn1 protein by Western blot. This confirmed down-regulation of Clcn1 by 60% and 32% in quadriceps and gastrocnemius, respectively, in control HSALR compared to FVB. Importantly, these amounts were reestablished to normal levels upon CQ administration (Fig. 6 G and H). Thus, CQ seems to promote Clcn1 protein levels through processes other than alternative splicing, being transcriptional control of Clcn1 by SP1 (35) one potential mechanism.
Fig. 7.
Injection (i.p.) of CQ improves Mbnl-regulated splicing and muscle function and histopathology of HSALR mice. (A) Heatmap representing the analysis of splicing decisions altered in HSALR mice. The number in the boxes indicates the percentage of inclusion of the indicated exons obtained by semiquantitative RT-PCR. (B) Representative gels used to perform quantifications in A from quadriceps (qd) (Left) and gastrocnemius (gt) (Right) muscles. (C) RT-qPCR to analyze exon 9 inclusion of Limch1. Gadph values were used for normalization in the quantification of the percentage of exon inclusion. (D) Forelimb grip strength and (E) myotonia grade measured before injection (bi) and 7 d after the last dose (ai). All comparisons are relative to HSALR mice treated with the vehicle PBS (DM). (F) Quantification of the percentage of central nuclei in muscle fibers from qd and gt muscles. (G–J) Representative micrographs of muscle fibers stained with H&E and quantified in F. (Scale bar, 100 µm.) Experimental groups were control (CNT) (FVB; n = 8), DM (HSALR; n = 6), DM 25 mg/kg (HSALR; n = 3), and DM 50 mg/kg (HSALR; n = 5). All comparisons are relative to HSALR mice treated with the vehicle PBS (DM). *P < 0.05, **P < 0.01, ***P < 0.001 according to Student’s t test.
Functional and Histopathological Improvements in CQ-Treated HSALR Mice.
It has been proposed that muscular phenotypes in DM1 are partially caused by defects in AS, leading to developmentally inappropriate protein expression in adults (36, 37). Muscle weakness is widespread among patients, and HSALR (38) mice reproduce this phenotype in a variable, age- and sex-dependent manner. We measured the grip strength of the forelegs and observed that the HSALR mice had ∼25% less force than the reference mice. This phenotype was reevaluated 7 d after the end of treatment, observing an improvement of ∼50% in both groups of treated mice compared to controls (Fig. 7D). Another defining symptom of the disease is myotonia that was quantified by electromyography (EMG) using a 0 to 4 myotonic grade scale. EMG testing of the hindlegs showed that before treatment, HSALR mice had myotonic discharges in more than 80% of the electrode insertions. Four days after finishing the 7-d treatment with the drug, myotonia decreased from almost grade 3 to grade 2 or 1, indicating that myotonic discharges occurred in less than 50% of the insertions of the electrode (Fig. 7E). Interestingly, we found strong positive correlations between Clcn1 exon 7 inclusion and Clcn1 levels and also with myotonia grade (SI Appendix, Fig. S12). Finally, centrally located nuclei is one of the most characteristic histopathological changes in DM1 muscle (39) and is an indication of myopathic muscle attempting to regenerate. We observed that untreated HSALR muscle fibers had 35% and 33% of central nuclei in quadriceps and gastrocnemius, respectively. After CQ injections this phenotype improved significantly in both muscles in a dose-dependent manner (Fig. 7 F–J). Overall, these results confirm that molecular rescues brought about by CQ translate into an improvement of histological and functional phenotypes in HSALR animals.
Discussion
Several lines of evidence indicate that up-regulation of Muscleblind-like proteins is a valid therapeutic strategy against DM1 because MBNLs are reduced in DM1 models and patients, and transgenic overexpression did not have deleterious effects in murine models (15, 18, 40, 41). Three different epigenetic approaches have so far demonstrated the ability to up-regulate endogenous MBNL expression in mammalian models. Small molecule HDAC inhibitors ISOX and vorinostat increased MBNL1 expression in DM1 patient-derived fibroblasts (42) while phenylbutazone suppressed methylation of an enhancer region in Mbnl1 intron 1 (43). A third approach used antagomiRs to block miRNAs that inhibit MBNL1 and 2 expression in skeletal muscle tissue (19). This study provides evidence that CQ can increase Muscleblind levels in 3 models of disease and that this increase translates into molecular and functional improvements in fly and mouse models.
CQ is a potent inhibitor of autophagy that works by blocking autophagosome fusion with lysosomes (28). Autophagy is a prosurvival mechanism that performs a housekeeping function in removing exhausted, redundant, or unwanted cellular components, but that when abnormally activated, results in cell death (44). It has been demonstrated that autophagy is hyperactivated in different DM1 models (20, 26, 45) and could also contribute to muscular atrophy, one of the most serious symptoms of the disease (46). It has been shown in an inducible model of DM1 that overexpression of MblC, an isoform of the Drosophila Muscleblind protein, was sufficient to reduce autophagy (20). Low levels of Muscleblind proteins may, therefore, play a contributory role in hyperactivation of autophagy. Conversely, CQ prevents autophagosomes with cargo from fusing with lysosomes, so that the cargo is not degraded. Consequently, by blocking autophagy disposal of MBNLs with CQ treatment, Muscleblind levels build up, as we have demonstrated with colocalization studies of LysoTracker and MBNL proteins. Extra MBNL, in turn, may further contribute toward repressing excessive autophagy in a virtuous positive feedback loop, as demonstrated by iPDM treatment with mtf. We observed that overactivation of autophagy resulted in dramatic MBNL1 and 2 down-regulation that worsened splicing defects. This may also explain the observation that HSALR was the most refractory of the DM1 models in MBNL1 or 2 increase, which is in line with the lack of autophagy hyperactivation in this model compared to flies and iPDMs. In Drosophila Mbl activation also included small but significant up-regulation at the transcript level, which suggests that at least 2 pathways contribute to extra Mbl upon CQ treatment. It is important to note that CQ not only increased MBNL protein levels but also significantly, and dose-dependently, reduced colocalization with CUG RNA foci, as similarly reported for HSALR mice treated with phenylbutazone, which also increased endogenous levels of Mbnl1 (43). While the molecular basis for such an effect on the degree of colocalization remains unexplored, it indeed suggests an improved bioavailability of MBNL1 in the cell.
Mtf has been recently proposed as a potential anti-DM1 therapy (47, 48) and Bassez et al. (48) report that patients treated with mtf improved in the 6-min walk test. Because we detect down-regulation in MBNL levels upon treatment of iPDMs with mtf, we propose that this drug may work through alternative molecular pathways. In support of this notion, Laustriat et al. (47) studied splicing events dysregulated in human DM1 myoblasts and found a negligible effect by mtf (only inclusion of 4 out of 22 alternative exons were rescued). Remarkably, 1 of these splicing events was cTNT exon 5 inclusion, consistent with our own data. Furthermore, Laustriat et al. (47) did not study the effect of mtf on MBNL2 levels and regulated splicing events, in contrast with our data that found strong reduction of MBNL2 and worsening of MBNL2-dependent splicing. Therefore, the positive effects reported for mtf seem to stem from molecular rescues at levels other than MBNL protein amounts or activity. Taken together, the use of mtf, that reduces MBNL levels, CQ, that enhances them, and DM1 and healthy controls, establishes 4 data points that nicely correlate with the inclusion of a number of alternative exons (SI Appendix, Fig. S13). Muscleblind levels increased 4-fold in Drosophila muscles and 3-fold in gastrocnemius in HSALR mice. The increase was more limited in human cells and in other mouse tissues, but nevertheless sufficient to improve missplicing and expression of genes regulated by Muscleblind in the 3 models used (14, 16, 49). It was reported by in vitro studies that MBNL1 targets exhibit dose dependency and require different amounts of MBNL1 for the half-maximal response (29). Splicing analyses of CQ-treated HSALR muscles revealed no changes in the pattern of Atp2a1, that in vitro requires lower levels of Mbnl1 than Nfix, which was significantly restored with CQ. Similar results were obtained when HSALR mice were treated with an antagomiR that up-regulated Mbnl1 (19). These data, therefore, revealed differing in vivo and in vitro susceptibility of splicing regulation by MBNL1 levels.
In model flies, CQ improved locomotor parameters and survival, phenotypes that had previously been shown to depend on Muscleblind levels (20, 41). The rescue was similar to previous results with an inducible Drosophila DM1 model where MblC overexpression was sufficient to increase the area of indirect flight muscles of model flies by 35% (24% by CQ) (20). In vitro, we observed that exposure of the cells to CQ improved the fusion index of iPDMs, which is characteristically reduced in patient cells. Myoblast fusion is critical for proper muscle growth and regeneration, and a defect in this process will therefore probably contribute to muscle atrophy. It has been reported in C2C12 cells that Mbnl1–3 levels vary significantly in the nucleus during myogenic differentiation and that MBNL family members are likely contributors to developmentally regulated myogenic AS (50). Analogously, HSALR mice showed partial recovery of muscle strength, decrease in the number of myotonic discharges in the skeletal muscles, and reduction in the number of central nuclei in the muscle fibers. These results are consistent with previous approaches in which MBNL1 was transgenically overexpressed in HSALR mice to correct myopathy (18) or where administration of different compounds generated similar results (19, 42, 43, 51). Importantly, these resulting improvements at the functional level make CQ a strong candidate for drug repurposing in DM1 either alone or as a systemic complement to the limited biodistribution of oligonucleotide-based therapies (19, 52, 53).
Materials and Methods
iCDFs and iPDFs (1,300 CTG) differentiable to myoblasts by MyoD expression were provided by D. Furling, Centre de Recherche en Myologie, Paris, France (27). Nonimmortalized CDFs and PDFs (333 and 1,000 CTG) differentiable to myoblasts by MyoD expression were generated in A.L.d.M.’s laboratory. Mouse handling and experimental procedures conformed to the European law regarding laboratory animal care and experimentation (2003/65/CE) and were approved by Conselleria de Agricultura, Generalitat Valenciana (reference number 2018/VSC/PEA/0182). Homozygous transgenic HSALR (line 20 b) mice were provided by C. Thornton (13) (University of Rochester Medical Center, Rochester, NY) and mice with the same genetic background (FVB) were used as controls. In all figures, graphs show mean ± SEM *P < 0.05, **P < 0.01, ***P < 0.001 according to Student’s t test. In all graphs error bars correspond to biological replicates. For additional details, see SI Appendix, Materials and Methods.
Data Availability.
All data discussed in the paper are available to readers.
Supplementary Material
Acknowledgments
This work was funded by a grant from the Ministerio de Economía y Competitividad (SAF2015-64500-R, including funds from the European Regional Development Fund) (awarded to R.A.). A.B. was supported by a postdoctoral fellowship (APOSTD2017/077), and M.S.-A. and J.E.-E. were supported by predoctoral fellowships (ACIF/2018/071 and GRISOLIAP/2018/098, respectively), all from the Conselleria d’Educació, Investigació, Cultura i Esport (Generalidad Valenciana). This work was cofinanced by the European Union through Programa Operativo del Fondo Europeo de Desarrollo Regional of Comunitat Valenciana 2014-2020.
Footnotes
Competing interest statement: The method described in this paper is the subject of a patent application (inventors: A.B., R.A.). M.S.-A., J.E.-E., M.Z., and A.L.d.M. declare no competing interests.
This article is a PNAS Direct Submission.
Data deposition: RNA-sequencing data have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE128844).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1820297116/-/DCSupplemental.
References
- 1.Thornton C. A., Myotonic dystrophy. Neurol. Clin. 32, 705–719, viii (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Overby S. J., Cerro-Herreros E., Llamusi B., Artero R., RNA-mediated therapies in myotonic dystrophy. Drug Discov. Today 23, 2013–2022 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Timchenko L. T., et al. , Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 24, 4407–4414 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barreau C., Paillard L., Méreau A., Osborne H. B., Mammalian CELF/Bruno-like RNA-binding proteins: Molecular characteristics and biological functions. Biochimie 88, 515–525 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Cardani R., et al. , Overexpression of CUGBP1 in skeletal muscle from adult classic myotonic dystrophy type 1 but not from myotonic dystrophy type 2. PLoS One 8, e83777 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Savkur R. S., Philips A. V., Cooper T. A., Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat. Genet. 29, 40–47 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Charlet-B N., et al. , Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol. Cell 10, 45–53 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Fugier C., et al. , Misregulated alternative splicing of BIN1 is associated with T tubule alterations and muscle weakness in myotonic dystrophy. Nat. Med. 17, 720–725 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Batra R., et al. , Loss of MBNL leads to disruption of developmentally regulated alternative polyadenylation in RNA-mediated disease. Mol. Cell 56, 311–322 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masuda A., et al. , CUGBP1 and MBNL1 preferentially bind to 3′ UTRs and facilitate mRNA decay. Sci. Rep. 2, 209 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rau F., et al. , Misregulation of miR-1 processing is associated with heart defects in myotonic dystrophy. Nat. Struct. Mol. Biol. 18, 840–845 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Wang E. T., et al. , Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell 150, 710–724 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mankodi A., et al. , Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science 289, 1769–1773 (2000). [DOI] [PubMed] [Google Scholar]
- 14.Du H., et al. , Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat. Struct. Mol. Biol. 17, 187–193 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanadia R. N., et al. , Reversal of RNA missplicing and myotonia after muscleblind overexpression in a mouse poly(CUG) model for myotonic dystrophy. Proc. Natl. Acad. Sci. U.S.A. 103, 11748–11753 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Charizanis K., et al. , Muscleblind-like 2-mediated alternative splicing in the developing brain and dysregulation in myotonic dystrophy. Neuron 75, 437–450 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang P. Y., et al. , Reduced cytoplasmic MBNL1 is an early event in a brain-specific mouse model of myotonic dystrophy. Hum. Mol. Genet. 26, 2247–2257 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Chamberlain C. M., Ranum L. P., Mouse model of muscleblind-like 1 overexpression: Skeletal muscle effects and therapeutic promise. Hum. Mol. Genet. 21, 4645–4654 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cerro-Herreros E., et al. , miR-23b and miR-218 silencing increase Muscleblind-like expression and alleviate myotonic dystrophy phenotypes in mammalian models. Nat. Commun. 9, 2482 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bargiela A., et al. , Increased autophagy and apoptosis contribute to muscle atrophy in a myotonic dystrophy type 1 Drosophila model. Dis. Model. Mech. 8, 679–690 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Llamusi B., et al. , Muscleblind, BSF and TBPH are mislocalized in the muscle sarcomere of a Drosophila myotonic dystrophy model. Dis. Model. Mech. 6, 184–196 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yum K., Wang E. T., Kalsotra A., Myotonic dystrophy: Disease repeat range, penetrance, age of onset, and relationship between repeat size and phenotypes. Curr. Opin. Genet. Dev. 44, 30–37 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swank D. M., et al. , Alternative exon-encoded regions of Drosophila myosin heavy chain modulate ATPase rates and actin sliding velocity. J. Biol. Chem. 276, 15117–15124 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Silvers R., Keller H., Schwalbe H., Hengesbach M., Differential scanning fluorimetry for monitoring RNA stability. ChemBioChem 16, 1109–1114 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Bargiela A., Llamusi B., Cerro-Herreros E., Artero R., Two enhancers control transcription of Drosophila muscleblind in the embryonic somatic musculature and in the central nervous system. PLoS One 9, e93125 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loro E., et al. , Normal myogenesis and increased apoptosis in myotonic dystrophy type-1 muscle cells. Cell Death Differ. 17, 1315–1324 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Arandel L., et al. , Immortalized human myotonic dystrophy muscle cell lines to assess therapeutic compounds. Dis. Model. Mech. 10, 487–497 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mauthe M., et al. , Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy 14, 1435–1455 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wagner S. D., et al. , Dose-dependent regulation of alternative splicing by MBNL proteins reveals biomarkers for myotonic dystrophy. PLoS Genet. 12, e1006316 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y., et al. , Metformin induces autophagy and G0/G1 phase cell cycle arrest in myeloma by targeting the AMPK/mTORC1 and mTORC2 pathways. J. Exp. Clin. Cancer Res. 37, 63 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bargiela Sabater-Arcis A., et al. , Increased Muscleblind levels by chloroquine treatment improves myotonic dystrophy type 1 phenotypes in in vitro and in vivo models. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?&acc=gse128844. Deposited 26 March 2019. [DOI] [PMC free article] [PubMed]
- 32.Wei C., et al. , Correction of GSK3β at young age prevents muscle pathology in mice with myotonic dystrophy type 1. FASEB J. 32, 2073–2085 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koshelev M., Sarma S., Price R. E., Wehrens X. H., Cooper T. A., Heart-specific overexpression of CUGBP1 reproduces functional and molecular abnormalities of myotonic dystrophy type 1. Hum. Mol. Genet. 19, 1066–1075 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalsotra A., et al. , A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc. Natl. Acad. Sci. U.S.A. 105, 20333–20338 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ebralidze A., Wang Y., Petkova V., Ebralidse K., Junghans R. P., RNA leaching of transcription factors disrupts transcription in myotonic dystrophy. Science 303, 383–387 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Lin X., et al. , Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum. Mol. Genet. 15, 2087–2097 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Nakamori M., et al. , Splicing biomarkers of disease severity in myotonic dystrophy. Ann. Neurol. 74, 862–872 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones K., et al. , GSK3β mediates muscle pathology in myotonic dystrophy. J. Clin. Invest. 122, 4461–4472 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vihola A., et al. , Histopathological differences of myotonic dystrophy type 1 (DM1) and PROMM/DM2. Neurology 60, 1854–1857 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Kim H. J., et al. , Therapeutic modulation of eIF2α phosphorylation rescues TDP-43 toxicity in amyotrophic lateral sclerosis disease models. Nat. Genet. 46, 152–160 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cerro-Herreros E., Fernandez-Costa J. M., Sabater-Arcis M., Llamusi B., Artero R., Derepressing muscleblind expression by miRNA sponges ameliorates myotonic dystrophy-like phenotypes in Drosophila. Sci. Rep. 6, 36230 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang F., et al. , A flow cytometry-based screen identifies MBNL1 modulators that rescue splicing defects in myotonic dystrophy type I. Hum. Mol. Genet. 26, 3056–3068 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen G., et al. , Phenylbutazone induces expression of MBNL1 and suppresses formation of MBNL1-CUG RNA foci in a mouse model of myotonic dystrophy. Sci. Rep. 6, 25317 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Madeo F., Zimmermann A., Maiuri M. C., Kroemer G., Essential role for autophagy in life span extension. J. Clin. Invest. 125, 85–93 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brockhoff M., et al. , Targeting deregulated AMPK/mTORC1 pathways improves muscle function in myotonic dystrophy type I. J. Clin. Invest. 127, 549–563 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Foff E. P., Mahadevan M. S., Therapeutics development in myotonic dystrophy type 1. Muscle Nerve 44, 160–169 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laustriat D., et al. , In vitro and in vivo modulation of alternative splicing by the biguanide metformin. Mol. Ther. Nucleic Acids 4, e262 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bassez G., et al. , Improved mobility with metformin in patients with myotonic dystrophy type 1: A randomized controlled trial. Brain 141, 2855–2865 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Picchio L., Plantie E., Renaud Y., Poovthumkadavil P., Jagla K., Novel Drosophila model of myotonic dystrophy type 1: Phenotypic characterization and genome-wide view of altered gene expression. Hum. Mol. Genet. 22, 2795–2810 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Bland C. S., et al. , Global regulation of alternative splicing during myogenic differentiation. Nucleic Acids Res. 38, 7651–7664 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Herrendorff R., et al. , Identification of plant-derived alkaloids with therapeutic potential for myotonic dystrophy type I. J. Biol. Chem. 291, 17165–17177 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sobczak K., Wheeler T. M., Wang W., Thornton C. A., RNA interference targeting CUG repeats in a mouse model of myotonic dystrophy. Mol. Ther. 21, 380–387 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Horsham J. L., et al. , Clinical potential of microRNA-7 in cancer. J. Clin. Med. 4, 1668–1687 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data discussed in the paper are available to readers.