Significance
Stochastic cell fate decisions are conserved and prominent processes during development, but the underlying molecular mechanisms are only partly understood. In the nematode Caenorhabditis elegans, the AWC sensory neuron pair asymmetrically differentiates into 2 distinct identities in a stochastic manner. Through identification of a unique transportin allele, we elucidate a mechanism by which a homeodomain-like factor couples voltage- and calcium-activated potassium channels to transactivation of a HMG-box transcription factor expression for the stochastic choice of AWC identities. We show that transportin drives nuclear import of the homeodomain-like factor to activate the expression of the HMG-box transcription factor for stochastic AWC identities. Our findings also provide structure-function insight into a conserved amino acid residue of transportins in cell type diversification.
Keywords: transportin 1, stochastic choice, sox-2, NSY-7, asymmetry
Abstract
Stochastic neuronal cell fate choice involving notch-independent mechanisms is a poorly understood biological process. The Caenorhabditis elegans AWC olfactory neuron pair asymmetrically differentiates into the default AWCOFF and induced AWCON subtypes in a stochastic manner. Stochastic choice of the AWCON subtype is established using gap junctions and SLO BK potassium channels to repress a calcium-activated protein kinase pathway. However, it is unknown how the potassium channel-repressed calcium signaling is translated into the induction of the AWCON subtype. Here, we identify a detailed working mechanism of how the homeodomain-like transcription factor NSY-7, previously described as a repressor in the maintenance of AWC asymmetry, couples SLO BK potassium channels to transactivation of sox-2 expression for the induction of the AWCON subtype through the identification of a unique imb-2 (transportin 1) allele. imb-2 loss-of-function mutants are not viable; however, we identify a viable imb-2 allele from an unbiased forward genetic screen that reveals a specific role of imb-2 in AWC olfactory neuron asymmetry. IMB-2 specifically drives nuclear import of NSY-7 within AWC neurons to transactivate the expression of the high mobility group (HMG)-box transcription factor SOX-2 for the specification of the AWCON subtype. This study provides mechanistic insight into how NSY-7 couples SLO BK potassium channels to transactivation of sox-2 expression for the induction of the AWCON subtype. Our findings also provide structure-function insight into a conserved amino acid residue of transportins in brain development and suggest its dysfunction may lead to human neurological disorders.
Stochastic cell fate decisions are conserved and prominent processes during development, but the underlying molecular mechanisms are only partly understood (1–4). The Caenorhabditis elegans (C. elegans) AWC pair of olfactory neurons acquires 2 mutually exclusive subtypes (AWCON and AWCOFF) and distinct functions through a stochastic coordinated cell signaling event (5–11), rendering it an excellent system to identify novel molecular mechanisms controlling stochastic cell fate specification.
Stochastic choice of AWC olfactory neuron subtypes is established during late embryogenesis and maintained throughout adulthood (12–15). The default AWCOFF neuron is specified via a calcium-activated protein kinase pathway downstream of voltage-gated calcium channels (15, 16). In this pathway, the TIR-1 (Sarm1) adaptor protein assembles a synaptic calcium-signaling complex that consists of UNC-43 calcium/calmodulin-dependent protein kinase (CaMKII) and NSY-1 MAP kinase kinase kinase (ASK1 MAPKKK) in a microtubule-dependent manner (11, 12, 17, 18). The NSY-5 gap junction protein innexin forms a transient gap junction neuronal network during embryogenesis to mediate intercellular calcium signaling to induce AWC asymmetry (19, 20). In addition, NSY-5 gap junction protein and NSY-4 claudin-like protein function in parallel to suppress calcium signaling in the AWCON neuron via voltage- and calcium-activated SLO BK potassium channels (19, 21, 22). It remains unknown what other mechanisms act downstream of the BK potassium channels to induce the AWCON identity.
Here, we identify a role of the karyopherin imb-2/transportin 1 downstream of the SLO BK potassium channels in promoting the AWCON subtype from an unbiased forward genetic screen. We show that asymmetrical expression of imb-2 in AWCON cells, which is dependent on nsy-5 (gap junction) and slo-1 (BK potassium channel), is necessary and sufficient for AWC asymmetry. In addition, IMB-2 localizes in close proximity to the homeodomain-like transcription factor NSY-7 and mediates nuclear transport of NSY-7 to specify the AWCON subtype. Furthermore, we reveal an activating function of NSY-7, which was previously described as a repressor in the maintenance of AWC asymmetry (14), in sox-2 expression by binding to its upstream regulatory sequence to induce the AWCON identity. Together, our study demonstrates that imb-2/transportin 1 functions to mediate nuclear transport of NSY-7 in AWC neurons, which, in turn, activates sox-2 expression to promote the AWCON subtype.
Results
The vy10 Mutation Causes a Defect in AWC Asymmetry.
Wild-type animals have 1 AWCON subtype, expressing the G protein-coupled receptor (GPCR) gene str-2, and one AWCOFF subtype, expressing the GPCR gene srsx-3 (15, 16) (Fig. 1 A, i and B). Gain-of-function mutations in the BK potassium channel gene slo-1 result in expression of str-2p::TagRFP in 2 AWC neurons (2AWCON phenotype) (15). We identified the vy10 allele from a forward genetic screen for mutants that suppress the 2AWCON phenotype in slo-1(ky399gf) mutants. The vy10 mutant lost expression of str-2p::TagRFP and, instead, expressed srsx-3p::GFP in both AWC neurons (2AWCOFF phenotype) from the first larval stage (L1) through adulthood (Fig. 1 A, ii and B). Since maintenance mutants display wild-type AWC asymmetry at the L1 stage but show a defect in AWC asymmetry later in development, these results suggest that the vy10 mutation affects the initial establishment of AWC asymmetry.
Fig. 1.
imb-2 is required and sufficient to promote the AWCON subtype. (A) Images of wild-type (i), imb-2(vy10) mutants (ii), and imb-2(OE) animals (iii) expressing the transgene str-2p::TagRFP (AWCON marker); srsx-3p::GFP (AWCOFF marker) in the adult stage. srsx-3p::GFP is also expressed in 2 AWB neurons. Asterisks indicate AWB cell bodies. (Scale bar, 10 μm.) (B) Expression of AWCON and AWCOFF markers in adults, unless otherwise indicated. n, total number of animals scored. (C) Quantification of the chemotaxis index. bu, butanone; pd, 2,3-pentanedione. Student’s t test was used to determine the P value. Error bars represent SEM.
The expression of 2 general AWC cell identity markers odr-1p::TagRFP and ceh-36p:;TagRFP was not affected in vy10 mutants (SI Appendix, Fig. S1A). These results suggest that the vy10 mutation does not affect general AWC identity; rather it results in a defect in the induction of AWC asymmetry.
vy10 mutants chemotaxed normally to the AWCOFF-sensed odor 2,3-pentanedione but had a significant reduction in their ability to chemotax toward the AWCON-sensed odor butanone (P < 0.001) (Fig. 1C). Taken together, our results show that the vy10 mutation results in a loss of the AWCON cell identity at both molecular and functional levels.
vy10 Is a Missense Mutation in imb-2/Transportin 1.
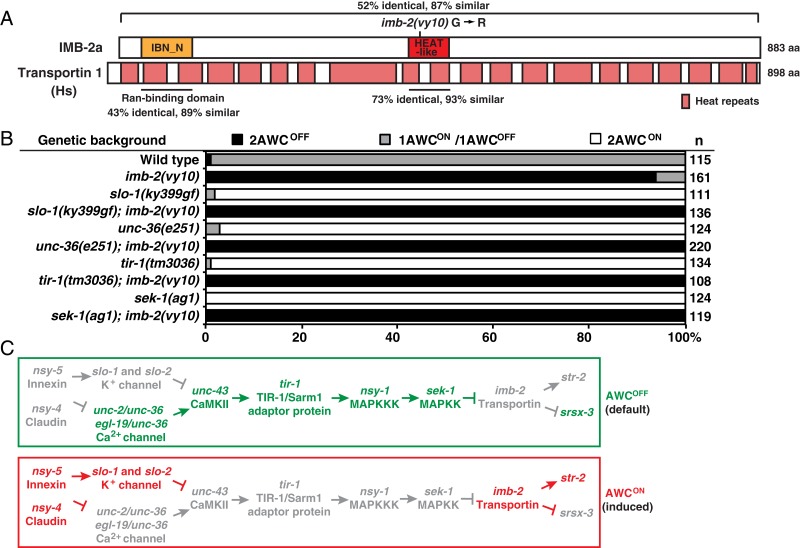
We identified the molecular lesion in vy10 mutants with 1-step whole-genome sequencing and single-nucleotide polymorphism mapping (23). The vy10 lesion was identified as a G to A mutation, resulting in a glycine to arginine change, in the second exon of both R06A4.4a/imb-2a and R06A4.4c/imb-2c isoforms (SI Appendix, Fig. S1B). imb-2 encodes an importin β-type nuclear transport receptor that mediates import of cargo proteins into the nucleus (24). There are 3 predicted importin β genes in C. elegans (wormbase.org), 12 in Drosophila melanogaster (flybase.org), at least 10 in mice (Mouse Genome Informatics), and more than 20 in humans (24, 25). C. elegans IMB-2 and human transportin 1 are 52% identical and 87% similar throughout the entire proteins (Fig. 2A and SI Appendix, Fig. S2). Human transportin 1 is a superhelical protein, consisting of 20 HEAT repeats with adjacent repeats connected by a linker (UniProt) (26–28). Similar to other IMB-2 and transportin proteins, C. elegans IMB-2 protein has a predicted IBN_N Ran-bonding domain and a HEAT-like repeat [Pfam (29)]. The predicted HEAT-like repeat of C. elegans IMB-2 and human transportin 1 are 73% identical and 93% similar. The glycine residue affected by the vy10 mutation is conserved in the predicted HEAT-like repeat of IMB-2 and transportin proteins (SI Appendix, Fig. S1C). This conserved glycine residue is located in the linker between HEAT repeats 9 and 10 of human transportin 1. imb-2 has 3 alternatively spliced isoforms (wormbase.org) (SI Appendix, Fig. S1B). The predicted HEAT-like repeat is present in IMB-2a and IMB-2c but is absent in IMB-2b.
Fig. 2.
imb-2/transportin 1 acts downstream of the calcium-activated MAPK pathway in promoting AWCON. (A) Structure of C. elegans IMB-2a and human transportin 1 proteins. Hs, Homo sapiens. (B) Double mutant analysis of imb-2(vy10) with 2AWCON mutants. Animals were scored at the adult stage. n, total number of animals scored. (C) The AWC asymmetry genetic pathway that demonstrates imb-2/transportin 1 acting downstream of the calcium-activated MAPK pathway to promote AWCON. Genes in green represent AWCOFF promoting; genes in red represent AWCON promoting; and those in gray represent less active or inactive genes.
Both a PCR-amplified imb-2p::imb-2 genomic DNA fragment and an imb-2 fosmid clone almost completely rescued the 2AWCOFF mutant phenotype in vy10 mutants (Fig. 1B and SI Appendix, Fig. S1B). Together, these results support that the vy10 2AWCOFF mutant phenotype was caused by the identified missense mutation in imb-2.
Both odr-3p::imb-2a and odr-3p::imb-2c, expressing imb-2 cDNA isoforms from an AWC odr-3 promoter (30) (SI Appendix, Fig. S1B), rescued the vy10 2AWCOFF phenotype to a degree similar to imb-2 genomic clones (Fig. 1B). However, odr-3p::imb-2b only partially rescued the vy10 2AWCOFF phenotype. These results suggest that imb-2 mainly acts in AWC cells to regulate AWC asymmetry and suggest an important role of the HEAT-like repeat of IMB-2 in AWC asymmetry. In addition, the transgene expressing human transportin 1 cDNA from the AWC odr-3 promoter rescued the vy10 mutant asymmetry phenotype, similar to the rescuing ability of odr-3p::imb-2a and imb-2c (Fig. 1B and SI Appendix, Fig. S1B). This result suggests conservation of imb-2/transportin 1 gene function in C. elegans and humans. Furthermore, overexpression of odr-3p::imb-2a, odr-3p::imb-2b, and odr-3p::imb-2c in wild-type animals resulted in a high penetrance of the 2AWCON phenotype, opposite to the 2AWCOFF phenotype of the vy10 mutants (Fig. 1 A, iii and B). In addition to the 2AWCON phenotype, overexpression of odr-3p::imb-2c also caused a high penetrance of the 2AWCOFF phenotype (Fig. 1B). Taken together, these results suggest that imb-2 is essential and sufficient to promote the AWCON cell identity and that the vy10 mutation causes loss of or reduction of imb-2 function in AWC asymmetry. Heterozygous imb-2(vy10/+) animals displayed wild-type AWC asymmetry (Fig. 1B), indicating that the imb-2(vy10) allele is recessive.
Both imb-2(tm6328) and imb-2(tm6405) alleles in which partial exon and intron regions of all 3 imb-2 isoforms are deleted (SI Appendix, Fig. S1B) caused early embryonic arrest. This prevented us from scoring AWC phenotypes in the potential imb-2 null mutants. We also observed a high penetrance of embryonic lethal phenotype in RNA interference (RNAi) knockdown of imb-2, and we did not observe any defective AWC phenotypes in the survivors of imb-2(RNAi) animals. Unlike imb-2 deletion mutants and severe imb-2(RNAi) animals, imb-2(vy10) mutants are viable and fertile, suggesting that imb-2(vy10) is likely not a null allele. To circumvent the embryonic lethal phenotypes of imb-2 null alleles, we generated conditional imb-2 knockdown or knockout insertion and deletion mutations in AWC neurons using CRISPR-Cas9 technology (31). Transgenic animals of AWC imb-2 CRISPR-Cas9, expressing Cas9 under the AWC odr-3 promoter and an imb-2 single guide RNA (target site in exon 2 of imb-2a and imb-2c) under a U6 small nuclear RNA promoter, recapitulated the 2AWCOFF phenotype observed in imb-2(vy10) mutants (Fig. 1B and SI Appendix, Fig. S1B). Together, these results further support that the vy10 mutation is a partial loss-of-function allele of imb-2 in AWC asymmetry.
imb-2 Acts Downstream of a Calcium-Activated MAP Kinase Cascade to Promote AWCON.
imb-2(vy10) was identified as a suppressor of slo-1(ky399gf) mutants, suggesting imb-2 acts downstream of slo-1 to promote AWCON (Fig. 2 B and C). The position of imb-2/transportin 1 in the AWC asymmetry genetic pathway was further determined by double mutant analysis of imb-2(vy10) with 2AWCON mutants. The 2AWCON phenotype of unc-36(e251) (calcium channel subunit), tir-1(tm3036) (Sarm 1 adaptor protein), and sek-1(ag1) (MAPKK) mutants was suppressed by the 2AWCOFF phenotype of imb-2(vy10) mutants. These results suggest that imb-2/transportin 1 acts downstream of the MAP kinase cascade to promote the AWCON subtype (Fig. 2C).
imb-2/Transportin 1 Is Asymmetrically Expressed in AWCON Neurons.
The expression pattern of imb-2 was analyzed in imb-2::mNG knock-in (where [mNG] represents mNeonGreen) animals in which we tagged the C-terminal end of endogenous IMB-2 with the fluorescent reporter mNG using Cas9-triggered homologous recombination (31–33) (SI Appendix, Fig. S1B). imb-2::mNG knock-in animals displayed wild-type AWC asymmetry (Fig. 1B), indicating that the tagged IMB-2::mNG fusion protein is functional. IMB-2::mNG was detected in the nucleus and around the nuclear envelope of numerous cells in the head and body during embryogenesis, larval stages, and adulthood (Fig. 3A). The broad expression pattern of imb-2 in the head is consistent with expression of vertebrate transportin 1 in multiple regions of the brain, including the olfactory bulb (34).
Fig. 3.
imb-2/transportin 1 acts cell autonomously to promote the AWCON subtype. (A) Images of imb-2::mNG knock-in expression in a 3-fold stage embryo (i), a first-stage larva (ii), and an adult (iii). AWC neurons were labeled with odr-1p::TagRFP. AWC cell bodies are outlined with dashed lines. (Scale bars, 5 μm [i and ii] and 50 μm [iii].) Asterisks indicate nonadult animals. (B) Images of imb-2::mNG knock-in expression at a higher level in the AWCL neuron than in AWCR at the L1 stage (ventral view). Both AWCL and AWCR were marked by odr-1p::TagRFP. (Scale bar, 5 μm.) (C) Quantification of asymmetric imb-2::mNG knock-in expression in AWCL and AWCR neurons. No significant difference was observed between AWCL > AWCR and AWCL < AWCR in wild-type animals. ns, not significant. n, total number of animals scored. P values were calculated using Fisher’s exact test. Error bars represent SE of proportion. (D) Images of imb-2::mNG knock-in expression at a higher level in AWCON than in AWCOFF in a L1 animal (dorsal view). AWCON was marked by str-2p::TagRFP and ceh-36p::myrTagRFP, while AWCOFF was only marked by ceh-36p::myrTagRFP. (Scale bar, 5 μm.) (E) Quantification of asymmetric expression of imb-2::mNG knock-in in AWCON and AWCOFF. n, total number of animals scored. P values were determined using a Z test. Error bars represent SE of proportion. (F) Quantification of AWC asymmetry phenotypes in wild-type, imb-2(vy10), and imb-2(vy10) mutants containing the extrachromosomal transgene imb-2 fosmid(OE); odr-1p::DsRed. (G) Quantification of AWC phenotypes in imb-2(vy10) mosaic animals containing the extrachromosomal transgene imb-2 fosmid(OE) in only 1 AWC neuron, inferred by the presence of the coinjected odr-1p::DsRed AWC marker. The data were obtained from a subset of animals scored in F.
IMB-2::mNG was detected in both AWC neurons in late embryogenesis during which AWC asymmetry was established and was maintained until adulthood (Fig. 3A). IMB-2::mNG was asymmetrically expressed in the left AWC neuron (AWCL) or the right AWC neuron (AWCR) in a stochastic manner (Fig. 3 B and C). Stochastic asymmetry of imb-2 expression in AWC neurons is consistent with the random nature of AWC asymmetry. The percentage of animals with equivalent imb-2 expression in AWCL and AWCR was significantly higher in nsy-5(ky634) (gap junction) and slo-1(ky399gf) (BK potassium channel) mutants (P < 0.001 and P = 0.04, respectively) (Fig. 3C). However, asymmetric imb-2 expression was not significantly affected in nsy-4(ky627) (claudin), slo-1(eg142lf);slo-2(ok2214lf) (BK potassium channels), or unc-43(n498gf) (CaMKII) mutants. These results suggest that NSY-5 gap junctions and SLO-1 BK potassium channels regulate asymmetric expression of imb-2 in AWC neurons.
IMB-2::mNG was expressed at a significantly higher expression level in the AWCON neuron than the AWCOFF neuron in the majority of animals (Fig. 3 D and E). Asymmetric expression of imb-2 in AWCON was also observed in our initial assessment of the imb-2 expression pattern using imb-2 fosmid::GFP in which we fused GFP with the C terminus of IMB-2 by homologous recombination (35), that rescued imb-2(vy10) mutants (Fig. 1B and SI Appendix, Figs. S1B and S3 A–C). These results are consistent with the hypothesis that imb-2 acts cell autonomously to promote the AWCON cell identity.
imb-2/Transportin 1 Acts Cell Autonomously in AWCON Neurons.
Expression of imb-2 fosmid(OE), odr-1p::DsRed or odr-3p::imb-2a(OE), odr-1p::DsRed transgenes in both AWC cells significantly rescued the 2AWCOFF phenotype of imb-2(vy10) mutants (Fig. 3F and SI Appendix, Fig. S3D). Spontaneous loss of the extrachromosomal array resulted in mosaic animals in which only 1 of the AWC neurons retained the imb-2(OE) rescuing transgene (inferred by the AWC marker odr-1p::DsRed) and the other AWC cell remained imb-2(vy10). In the majority of the mosaic animals that exhibited rescue of the mutant phenotype (1 AWCON/1 AWCOFF), the AWC cell expressing imb-2 fosmid or odr-3p::imb-2a became AWCON, and the imb-2(vy10) AWC cell became AWCOFF (Fig. 3G and SI Appendix, Fig. S3E). These results suggest that imb-2 acts largely cell autonomously to induce AWCON.
Mosaic analysis was also performed in transgenic lines in which odr-3p::imb-2a was overexpressed in a wild-type background, resulting in a 2AWCON phenotype (SI Appendix, Fig. S3F). When the odr-3p::imb-2a array was present in only a single AWC neuron, the imb-2(OE) cell became AWCON, and the wild-type cell became AWCOFF in the majority of mosaic animals (SI Appendix, Fig. S3G). These data further support the notion that imb-2 specifies the AWCON fate in a largely cell autonomous fashion.
imb-2/Transportin 1 Is Required for Nuclear Localization of NSY-7 Homeodomain-like Transcription Factor to Specify the AWCON Subtype.
To identify the cargo of IMB-2/transportin in promoting AWCON, we examined fluorescent reporters translationally fused with 5 candidate transcription factors that are implicated in AWC asymmetry (14, 36–38) (SI Appendix, Fig. S4A). Nuclear localization of SOX-2, CEH-36 (OTX/OTD), MLS-2 (HMX/NKX), and DIE-1 (zinc finger) was not affected in imb-2(vy10) mutants (SI Appendix, Fig. S4B). However, NSY-7 was detected in the cytosol and largely excluded from the nucleus of AWC neurons in imb-2(vy10) mutants (Fig. 4A). These results suggest that imb-2 is required for the transport of NSY-7 into the nucleus of AWC neurons.
Fig. 4.
imb-2 is required for nuclear localization of NSY-7 homeodomain-like transcription factor in the specification of AWCON identity. (A) Images of NSY-7::GFP and NSY-7::2xnlsGFP expressed from single copy insertion transgenes odr-3p::nsy-7::GFP and odr-3p::nsy-7::2xnlsGFP, respectively, in AWC at the L1 stage. AWC neurons were labeled with odr-1p::TagRFP. (Scale bar, 5 μm.) (B) Quantification of AWC asymmetry phenotypes in L1 or adults. (C) Images of transgenic animals for bimolecular fluorescence complementation (BiFC) assays between different forms of IMB-2 and NSY-7 proteins, each fused to nonfluorescent fragments of Venus, at the L1 stage. (Scale bar, 5 μm.) VN, VN173 (Venus 1–172); VC, VC155 (Venus 155–238).
To directly determine the requirement of IMB-2 for nuclear localization of NSY-7 in promoting the AWCON subtype, we generated Mos1-mediated single copy insertion transgenes of odr-3p::nsy-7::GFP or odr-3p::nsy-7::2xnlsGFP (containing 2 copies of the nuclear localization signal [NLS] derived from SV40) on the same locus of a chromosome. NSY-7::2xnlsGFP was localized to the AWC nucleus in imb-2(vy10) mutants (Fig. 4A), suggesting that the SV40 NLS bypassed the requirement of IMB-2 for the transport of NSY-7::2xnlsGFP to the nucleus. The imb-2(vy10) 2AWCOFF mutant phenotype was not rescued by cytoplasmic localized NSY-7::GFP. However, nuclear localized NSY-7::2xnlsGFP not only rescued the imb-2(vy10) 2AWCOFF mutant phenotype, but also resulted in a 2AWCON phenotype. These results suggest that imb-2 is required for the transport of NSY-7 into the nucleus of AWC neurons to induce the AWCON cell identity.
IMB-2/Transportin 1 Is in Close Proximity to NSY-7 in AWC Neurons.
To determine whether IMB-2/transportin 1 interacts with NSY-7 in AWC neurons, we used a bimolecular fluorescence complementation (BiFC) assay (39). Two nonfluorescent Venus fragments, VN173 and VC155, were fused to IMB-2 and NSY-7, respectively, and the odr-3 promoter was used to drive their expression in AWC neurons (Fig. 4C). Coexpression of odr-3p::imb-2::VN173 and odr-3p::nsy-7::VC155 transgenes resulted in punctate Venus fluorescence in the nucleus of AWC neurons (Fig. 4C and SI Appendix, Fig. S5A). Similarly, coexpression of odr-3p::imb-2::VC155 and odr-3p::nsy-7::VN173 transgenes in which VC155 and VN173 were fused to IMB-2 and NSY-7, respectively, also showed a punctate pattern of Venus fluorescence in the AWC nucleus (SI Appendix, Fig. S5B). These BiFC results suggest that IMB-2 and NSY-7 proteins may be close enough to interact. The interaction of IMB-2 with MLS-2 was tested as a negative control of this assay since the nuclear localization of MLS-2 was not affected in imb-2(vy10) mutants (SI Appendix, Fig. S4B). As expected, we did not observe Venus expression from the control assays (SI Appendix, Fig. S5C).
The nsy-7(ky630) allele is a missense mutation leading to exclusively cytoplasmic localization of the mutant NSY-7 protein (14) (SI Appendix, Fig. S5D). Venus expression was observed between IMB-2G417R with the vy10 mutation and NSY-7; however, the localization pattern was mostly excluded from the nucleus (Fig. 4 C, Middle). Similarly, Venus expression of IMB-2 and NSY-7H179Y with the ky630 mutation also resulted in localization outside of the AWC nucleus (Fig. 4 C, Bottom). These results suggest that imb-2(vy10) and nsy-7(ky630) mutations do not abolish the interaction between IMB-2 and NSY-7 but rather prevent the protein complex from entering the nucleus.
Importin α proteins function as adaptor proteins in the classical nuclear transport process by binding to importin β and the NLS of cargoes (40, 41). In nonclassical nuclear import pathways, importin β proteins can directly bind certain cargoes, such as transcription factors and ribosomal proteins, independent of importin α (42). There are 3 importin α genes, ima-1, ima-2, and ima-3 in the C. elegans genome (wormbase.org). All of ima-1, ima-2, and ima-3 deletion mutants analyzed displayed wild-type AWC asymmetry (SI Appendix, Fig. S6), suggesting that these importin α genes are not required for AWC asymmetry. Consistent with these results, RNAi knockdown of ima-1, ima-2, and ima-3 individually or together did not cause abnormal AWC asymmetry phenotypes. Together with the BiFC assay result that implies directly binding of IMB-2 to its cargo NSY-7, these results suggest that IMB-2 may mediate a nonclassical nuclear import of NSY-7 independent of importin α to establish AWC asymmetry.
nsy-7 Is Required for sox-2 Expression to Promote the AWCON Subtype.
Previous studies have shown that nsy-7 functions to maintain AWC asymmetry by studying nsy-7(ky630) mutants (14). We found that nsy-7(tm3080) deletion mutants displayed a complete penetrance of the 2AWCOFF phenotype at both L1 and L4 stages (Fig. 4B and SI Appendix, Fig. S5D), revealing an additional role of nsy-7 in the establishment of AWC asymmetry besides maintenance. Overexpression of nsy-7 in AWC caused a strong 2AWCON phenotype (Fig. 4B), suggesting that nsy-7 is sufficient to induce the AWCON identity.
The HMG-box transcription factor sox-2 is a candidate target of NSY-7 since sox-2 has been implicated in promoting the AWCON identity (37) and contains a potential NSY-7 binding site, previously identified by protein-binding microarrays (14), in the upstream regulatory sequence. An integrated sox-2 transcriptional reporter transgene, sox-2ps::2xnlsGFP (37), was expressed in both AWC neurons at the L1 stage in the majority of wild-type animals (Fig. 5 A and B). The number of AWC neurons expressing sox-2ps::2xnlsGFP was significantly reduced in nsy-7(tm3080) and imb-2(vy10) mutants, indicating that nsy-7 and imb-2 are required for sox-2 expression in AWC at the L1 stage. However, sox-2 expression was lost in both AWC neurons of wild-type, nsy-7(tm3080), and imb-2(vy10) mutants at the L4 stage (Fig. 5B). Together, these results suggest that IMB-2 transports NSY-7 into the nucleus of AWC neurons to activate sox-2 expression at the L1 stage but not at the L4 stage.
Fig. 5.
nsy-7 is required for sox-2 expression in promoting the AWCON subtype. (A) Images of sox-2ps::2xnlsGFP expression at the L1 stage. (Scale bar, 5 μm.) Asterisks indicate AWB cell body. (B) Quantification of the percentage of animals expressing sox-2ps::2xnlsGFP in AWC at the L1 and L4 stages. (C) Quantification of AWC asymmetry phenotypes at the L1 and L4 stages. (D) Quantification of sox-2ps::2xnlsGFP and sox-2ps(NSY-7m)::2xnlsGFP expression in AWC at the L1 stage. NSY-7m, mutated NSY-7-binding site within the sox-2 upstream regulatory sequence. ***P < 0.0001. Statistic comparison was performed by Fisher’s exact test. Quantification of the transgene expression in each of the independent lines is included in SI Appendix, Fig. S7. (E) A representative gel image of electrophoretic mobility shift assays (EMSA) with 6xHis-tagged NSY-7 protein and an IRDye-labeled DNA probe containing the NSY-7 consensus-binding site in the sox-2 promoter. Unlabeled wild-type or mutant competitor probes were added to lanes 3–6 and 7–10, respectively at increasing concentrations (20×, 40×, 60×, and 80×). Competitive-binding assays were performed 9 independent times, and these independent assays showed the same trend that the mutant competitor probe was not as efficient at competing away the NSY-7-DNA complex as the wild-type competitor probe. Nucleotides in gray are sequences of Caenorhabditis remani, Caenorhabditis briggsae, and Caenorhabditis brenneri that differ from the C. elegans sequence. Sequence alignment between species was performed on http://genome.ucsc.edu. (F) Relative band intensities of the NSY-7-DNA complex were plotted against the concentration of the competitor probe. Images and respective band intensity plots from 2 other independent EMSA assays are presented in SI Appendix, Fig. S8.
Overexpression of sox-2 fosmid::mCherry almost completely rescued the 2AWCOFF phenotype of imb-2(vy10) and nsy-7(tm3080) mutants at the L1 stage (Fig. 5C, row d compared with row c and row f compared with row e, respectively). However, the rescue ability of sox-2 fosmid::mCherry was greatly reduced in imb-2(vy10) mutants and completely lost in nsy-7(tm3080) mutants at the L4 stage (Fig. 5C, rows i and j and rows k and l). These results are consistent with sox-2 expression in AWC at L1 but not in L4 (Fig. 5B). The data also suggest that nsy-7 activates sox-2 expression for the establishment of AWC asymmetry during early development and regulates other factors for the maintenance of AWC asymmetry later in development. NSY-7 was previously described as a repressor of the AWCOFF-specific marker srsx-3 expression to maintain the AWCON subtype (14). Our results reveal a role of NSY-7 as a transcriptional activator of sox-2 expression in the specification of the AWCON identity.
NSY-7 Transcription Factor Binds to sox-2 Upstream Regulatory Sequence.
The NSY-7 target site CCTTAAC, identified by protein-binding microarrays (14), is located in the upstream regulatory sequence of sox-2 (Fig. 5 D and E). The extrachromosomal transgene sox-2ps::2xnlsGFP, containing the putative NSY-7-binding site, was expressed in, at least, 1 AWC neuron in the majority of animals (Fig. 5D and SI Appendix, Fig. S7, 10 lines examined). The number of AWC neurons expressing sox-2ps(NSY-7m)::2xnlsGFP, containing a mutated NSY-7-binding site, was significantly decreased (Fig. 5D and SI Appendix, Fig. S7, 6 lines examined) compared to that of AWC expressing sox-2ps::2xnlsGFP. These results suggest that the NSY-7-binding site is required for appropriate expression of sox-2 in AWC neurons.
In EMSA, 6xHis-tagged NSY-7 was able to bind an IRDye-labeled DNA probe containing 39 bp of the sox-2 upstream regulatory sequence, which includes the NSY-7 consensus site (Fig. 5E and SI Appendix, Fig. S8 A and C, lane 2). A wild-type unlabeled probe (sox-2p-wt) was able to compete away NSY-7 from the IRDye-labeled probe, whereas the mutated unlabeled probe (sox-2p-m), containing the same mutated NSY-7 consensus site as in sox-2ps(NSY-7m)::2xnlsGFP, was not as efficient as a competitor (Fig. 5 E and F and SI Appendix, Fig. S8 A–D, lanes 3–6 compared to lanes 7–10). These results suggest that NSY-7 binding was specific to the upstream regulatory sequence of sox-2. Together with the requirement of nsy-7 for expression of sox-2, these results further support the role of NSY-7 as a transcriptional activator, whereas it was previously reported as a repressor (14).
nsy-7 and sox-2 Are Asymmetrically Expressed in the AWCON Neuron.
The expression pattern of nsy-7 in AWC neurons was further analyzed in transgenic animals expressing a nsy-7p::GFP extrachromosomal transgene in which GFP was driven by a 21 kb nsy-7 promoter (14) (SI Appendix, Fig. S9A). Consistent with previous findings (14), only 1 of the AWC neurons expressed nsy-7p::GFP in those transgenic animals that displayed GFP expression in AWC cells (SI Appendix, Fig. S9B). Asymmetric expression of nsy-7p::GFP in AWCL or AWCR was stochastic (SI Appendix, Fig. S9C). Neither stochastic asymmetry of nsy-7 expression in AWC neurons or the number of AWC expressing nsy-7p::GFP was affected in imb-2(vy10) mutants (SI Appendix, Fig. S9 C and D). nsy-7 was exclusively expressed in AWCON neurons, while no expression was observed in AWCOFF cells (SI Appendix, Fig. S9 E and F). This result further suggests that the NSY-7 transcription factor promotes the AWCON identity in a cell-autonomous manner.
The expression pattern of sox-2 was examined in sox-2::mNG knock-in animals in which we tagged the C-terminal end of endogenous SOX-2 with mNG using Cas9-triggered homologous recombination (31–33) (SI Appendix, Fig. S9A). Like imb-2 and nsy-7, sox-2 was asymmetrically expressed at a higher level in AWCON neurons than in AWCOFF neurons in a stochastic manner (SI Appendix, Fig. S9 G–J). Together, these results support the role of nsy-7 in activating sox-2 expression to promote the AWCON identity.
Discussion
Here, we identify an essential role of imb-2/transportin 1 in a stochastic choice of asymmetric olfactory neuron subtypes in C. elegans from an unbiased forward genetic screen. We show that imb-2/transportin 1 mediates transport of the NSY-7 homeodomain-like transcription factor into the nucleus of AWC olfactory neurons, which, in turn, activates sox-2 expression to promote the AWCON subtype. This study implicates karyopherins in the establishment of stochastic cell identity choice and left–right patterning. As C. elegans imb-2 is highly conserved with human transportin 1, this process may prove to be conserved in establishing stochastic cell identity and left–right asymmetry in mammals.
Our results suggest a mechanistic model for imb-2 function in a stochastic choice of AWC olfactory neuron subtypes (Fig. 6). In wild type, imb-2, nsy-7, and sox-2 are asymmetrically expressed in AWCON in a stochastic manner (Fig. 6A). IMB-2 binds to NSY-7 and mediates nuclear transport of NSY-7 in AWCON neurons. Nuclear-localized NSY-7 directly activates expression of sox-2 to promote the AWCON identity. In imb-2(vy10) mutants, as in wild type, nsy-7 is asymmetrically expressed in the AWC neurons in a stochastic manner (Fig. 6B). However, NSY-7 is largely excluded from the nucleus of AWC neurons in imb-2(vy10) mutants. Although IMB-2G417R (containing the vy10 mutation) and NSY-7 are still in close proximity, the imb-2(vy10) mutation abolishes the ability of the IMB-2-NSY-7 complex to enter the nucleus. Thus, sox-2 expression is subsequently lost, resulting in the inability to promote the AWCON identity and a 2AWCOFF mutant phenotype in imb-2(vy10) mutants.
Fig. 6.
Model of imb-2 function in AWC asymmetry. (A) In wild-type animals, imb-2 and nsy-7 are asymmetrically expressed in the AWC neuron that becomes AWCON. IMB-2 binds to NSY-7 and mediates the transport of NSY-7 into the nucleus to activate sox-2 expression thereby inducing the AWCON identity. (B) In imb-2(vy10) mutants, NSY-7 fails to enter the nucleus, leading to loss of sox-2 expression and a 2AWCOFF phenotype. Gray, less active or inactive molecules or expression.
It was previously shown that NSY-7 responds to transient embryonic signaling of the NSY-5 gap junction neuronal network by acting as a transcriptional repressor (repressing AWCOFF genes in AWCON neurons) in the maintenance of AWC asymmetry (14). Our results reveal a role of NSY-7 as a transcriptional activator of sox-2 expression in establishing the stochastic choice of the AWCON subtype. Our study extends the previous model of AWC asymmetry by identifying a role of imb-2/transportin 1 in the coupling transient NSY-5 gap junction signaling from the cell membrane and the cytosol to the NSY-7-SOX-2 transcriptional cascade in the nucleus.
In nuclear import pathways, importin β interacts with the nuclear pore complex to promote selective and efficient transport of importin-cargo complexes across the nuclear envelope into the nucleus (43, 44). Once importin-cargo complexes enter the nucleus, binding of GTP-bound Ran GTPase to importin β induces structural changes in importin β, leading to the release of cargoes (24). Our study suggests that the conserved glycine in the HEAT-like repeat mutated in the vy10 mutant may not be required for IMB-2 to recognize and bind to the cargo NSY-7 in the cytoplasm but is important for transport of the IMB-2-NSY-7 complex across the nuclear envelope into the nucleus. It is possible that the glycine to arginine change in the vy10 mutants may affect the binding affinity of IMB-2 to the nuclear pore complex and, thus, abolishes the ability of IMB-2-NSY-7 to enter into the AWC nucleus.
In contrast with embryonic lethality of imb-2 deletion mutants and RNAi knockdown animals, imb-2(vy10) mutants are viable and fertile. Our approach revealed a role of imb-2 in postmitotic diversification of olfactory neuron subtypes, which would have been masked by candidate gene approaches of analyzing the phenotypes in imb-2 deletion mutants or imb-2 RNAi knockdown animals. In humans, importin β proteins have been shown to carry a diverse set of cargoes into the nucleus. Crystal structures of human importin β-1 bound with cargoes show that cargoes bind at different sites on importin β (45, 46). We propose that broadly expressed IMB-2 mediate nuclear transport of selective cargoes for different functions and that functional specificity of IMB-2 is conferred by different amino acid residues, which mediate nuclear transport of specific cargoes for distinct functions. It is possible that the glycine to arginine change within the HEAT-like repeat in vy10 mutants specifically affects nuclear import of the NSY-7 homeodomain-like transcription factor for a subset of imb-2 functions including establishing stochastic choice of AWC subtypes.
Although karyopherins have been implicated in various cell biological and developmental processes, the majority of implications derive from RNAi knockdown of karyopherins, expression of karyopherins during particular cell biological events or in particular tissues, or mutational analyses of candidate cargo proteins. For example, RNAi knockdown of imb-2 prevents redox-dependent nuclear import of the transcription factor FOXO/DAF-16 (47) and suppresses the life span of long-lived mitochondrial mutants with increased expression of FOXO/DAF-16 target genes (48). Defective nuclear transport of key cellular regulator molecules has been reported in a variety of diseases and cancers; however, the majority of pathogenic mutations are identified in the cargo proteins rather than the karyopherins that transport them (49–51). Although deregulation and/or mutations of karyopherins have been reported in some cancers, the molecular mechanisms of these karyopherin mutations and how the mutations lead to cancers remain to be elucidated (49). Our study sets a precedent by mechanistically implicating a widely expressed karyopherin protein in a specific function of a multicellular organism directly through the analysis of missense karyopherin mutants identified from an unbiased forward genetic screen. Our findings reveal structure-function insight into a conserved amino acid residue of karyopherins in a stochastic choice of cell identity and left–right patterning.
Materials and Methods
Strains and Transgenes.
The wild-type C. elegans strain is N2, Bristol variety. Strains were maintained by standard methods (52). A list of strains and transgenes is included in SI Appendix, Supplemental Materials and Methods.
Isolation of imb-2(vy10) Mutants.
A forward genetic screen was performed as previously described (52). kyIs140I; slo-1(ky399gf) P0 mutants were treated with EMS. Ten F1 progenies were picked onto single plates, and F2 were screened for suppression of the slo-1(ky399gf) 2AWCON phenotype using a Zeiss fluorescence dissecting microscope. The vy10 mutation was identified from a screen of 5,700 genomes.
Whole-Genome Sequencing.
The 1-step whole-genome-sequencing and SNP mapping strategy (23) was used to identify the vy10 mutation with an Illumina GAIIX sequencing platform and 100-nucleotide reads. Analysis of sequencing results was performed using CloudMap software as previously described (53).
BiFC Assays.
BiFC assays were performed as previously described (39). Nonfluorescent VN173 and VC155 fragments of Venus were fused to IMB-2 and NSY-7, respectively. The fusion protein constructs, driven by the AWC odr-3 promoter, were coinjected into animals. Expression of Venus was imaged at the L1 stage.
Supplementary Material
Acknowledgments
We thank Cori Bargmann, Alex Boyanov, Oliver Hobert, Dan Dickinson, Bob Goldstein, Jim Wells, David Crowe, Shohei Mitani, WormBase, and C. elegans Genetic Center (funded by the NIH Office of Research Infrastructure Programs P40 OD010440) for assistance, strains, reagents, and/or protocols. This work was supported by the National Science Foundation Grant (IOS-1455758 to C.C.), a Whitehall Foundation Research Award (to C.-F.C.), an Alfred P. Sloan Research Fellowship (to C.-F.C.), and the National Institutes of Health Grants (5R01GM098026-05 to C.-F.C., R01GM111320 to C.C.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1908168116/-/DCSupplemental.
References
- 1.Johnston R. J. Jr, Desplan C., Stochastic neuronal cell fate choices. Curr. Opin. Neurobiol. 18, 20–27 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnston R. J. Jr, Desplan C., Stochastic mechanisms of cell fate specification that yield random or robust outcomes. Annu. Rev. Cell Dev. Biol. 26, 689–719 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Losick R., Desplan C., Stochasticity and cell fate. Science 320, 65–68 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reiner S. L., Adams W. C., Lymphocyte fate specification as a deterministic but highly plastic process. Nat. Rev. Immunol. 14, 699–704 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Alqadah A., Hsieh Y. W., Chuang C. F., microRNA function in left-right neuronal asymmetry: Perspectives from C. elegans. Front. Cell. Neurosci. 7, 158 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alqadah A., Hsieh Y. W., Morrissey Z. D., Chuang C. F., Asymmetric development of the nervous system. Dev. Dyn. 247, 124–137 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alqadah A., Hsieh Y. W., Xiong R., Chuang C. F., Stochastic left-right neuronal asymmetry in Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371, 20150407 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsieh Y. W., Alqadah A., Chuang C. F., Asymmetric neural development in the Caenorhabditis elegans olfactory system. Genesis 52, 544–554 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsieh Y. W., Alqadah A., Chuang C. F., Mechanisms controlling diversification of olfactory sensory neuron classes. Cell. Mol. Life Sci. 74, 3263–3274 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor R. W., Hsieh Y. W., Gamse J. T., Chuang C. F., Making a difference together: Reciprocal interactions in C. elegans and zebrafish asymmetric neural development. Development 137, 681–691 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Troemel E. R., Kimmel B. E., Bargmann C. I., Reprogramming chemotaxis responses: Sensory neurons define olfactory preferences in C. elegans. Cell 91, 161–169 (1997). [DOI] [PubMed] [Google Scholar]
- 12.Chuang C. F., Bargmann C. I., A Toll-interleukin 1 repeat protein at the synapse specifies asymmetric odorant receptor expression via ASK1 MAPKKK signaling. Genes Dev. 19, 270–281 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lesch B. J., Bargmann C. I., The homeodomain protein hmbx-1 maintains asymmetric gene expression in adult C. elegans olfactory neurons. Genes Dev. 24, 1802–1815 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lesch B. J., Gehrke A. R., Bulyk M. L., Bargmann C. I., Transcriptional regulation and stabilization of left-right neuronal identity in C. elegans. Genes Dev. 23, 345–358 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Troemel E. R., Sagasti A., Bargmann C. I., Lateral signaling mediated by axon contact and calcium entry regulates asymmetric odorant receptor expression in C. elegans. Cell 99, 387–398 (1999). [DOI] [PubMed] [Google Scholar]
- 16.Bauer Huang S. L., et al. , Left-right olfactory asymmetry results from antagonistic functions of voltage-activated calcium channels and the Raw repeat protein OLRN-1 in C. elegans. Neural Dev. 2, 24 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang C., Hsieh Y. W., Lesch B. J., Bargmann C. I., Chuang C. F., Microtubule-based localization of a synaptic calcium-signaling complex is required for left-right neuronal asymmetry in C. elegans. Development 138, 3509–3518 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sagasti A., et al. , The CaMKII UNC-43 activates the MAPKKK NSY-1 to execute a lateral signaling decision required for asymmetric olfactory neuron fates. Cell 105, 221–232 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Chuang C. F., Vanhoven M. K., Fetter R. D., Verselis V. K., Bargmann C. I., An innexin-dependent cell network establishes left-right neuronal asymmetry in C. elegans. Cell 129, 787–799 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Schumacher J. A., et al. , Intercellular calcium signaling in a gap junction-coupled cell network establishes asymmetric neuronal fates in C. elegans. Development 139, 4191–4201 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alqadah A., et al. , SLO BK potassium channels couple gap junctions to inhibition of calcium signaling in olfactory neuron diversification. PLoS Genet. 12, e1005654 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vanhoven M. K., Bauer Huang S. L., Albin S. D., Bargmann C. I., The claudin superfamily protein nsy-4 biases lateral signaling to generate left-right asymmetry in C. elegans olfactory neurons. Neuron 51, 291–302 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Doitsidou M., Poole R. J., Sarin S., Bigelow H., Hobert O., C. elegans mutant identification with a one-step whole-genome-sequencing and SNP mapping strategy. PLoS One 5, e15435 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harel A., Forbes D. J., Importin beta: Conducting a much larger cellular symphony. Mol. Cell 16, 319–330 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Mosammaparast N., Pemberton L. F., Karyopherins: From nuclear-transport mediators to nuclear-function regulators. Trends Cell Biol. 14, 547–556 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Chook Y. M., Blobel G., Structure of the nuclear transport complex karyopherin-beta2-Ran x GppNHp. Nature 399, 230–237 (1999). [DOI] [PubMed] [Google Scholar]
- 27.Chook Y. M., Jung A., Rosen M. K., Blobel G., Uncoupling Kapbeta2 substrate dissociation and ran binding. Biochemistry 41, 6955–6966 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Lee B. J., et al. , Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell 126, 543–558 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finn R. D., et al. , The pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 44, D279–D285 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roayaie K., Crump J. G., Sagasti A., Bargmann C. I., The G alpha protein ODR-3 mediates olfactory and nociceptive function and controls cilium morphogenesis in C. elegans olfactory neurons. Neuron 20, 55–67 (1998). [DOI] [PubMed] [Google Scholar]
- 31.Dickinson D. J., Ward J. D., Reiner D. J., Goldstein B., Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10, 1028–1034 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dickinson D. J., Goldstein B., CRISPR-based methods for Caenorhabditis elegans genome engineering. Genetics 202, 885–901 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dickinson D. J., Pani A. M., Heppert J. K., Higgins C. D., Goldstein B., Streamlined genome engineering with a self-excising drug selection cassette. Genetics 200, 1035–1049 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sato M., et al. , Transportin 1 in the mouse brain: Appearance in regions of neurogenesis, cerebrospinal fluid production/sensing, and circadian clock. J. Comp. Neurol. 519, 1770–1780 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Tursun B., Cochella L., Carrera I., Hobert O., A toolkit and robust pipeline for the generation of fosmid-based reporter genes in C. elegans. PLoS One 4, e4625 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alqadah A., Hsieh Y. W., Chuang C. F., A molecular link between distinct neuronal asymmetries. Cell Cycle 13, 1515–1516 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alqadah A., et al. , Postmitotic diversification of olfactory neuron types is mediated by differential activities of the HMG-box transcription factor SOX-2. EMBO J. 34, 2574–2589 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cochella L., et al. , Two distinct types of neuronal asymmetries are controlled by the Caenorhabditis elegans zinc finger transcription factor die-1. Genes Dev. 28, 34–43 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shyu Y. J., et al. , Visualization of protein interactions in living Caenorhabditis elegans using bimolecular fluorescence complementation analysis. Nat. Protoc. 3, 588–596 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Conti E., Uy M., Leighton L., Blobel G., Kuriyan J., Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell 94, 193–204 (1998). [DOI] [PubMed] [Google Scholar]
- 41.Goldfarb D. S., Corbett A. H., Mason D. A., Harreman M. T., Adam S. A., Importin alpha: A multipurpose nuclear-transport receptor. Trends Cell Biol. 14, 505–514 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Palmeri D., Malim M. H., Importin beta can mediate the nuclear import of an arginine-rich nuclear localization signal in the absence of importin alpha. Mol. Cell. Biol. 19, 1218–1225 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aramburu I. V., Lemke E. A., Floppy but not sloppy: Interaction mechanism of FG-nucleoporins and nuclear transport receptors. Semin. Cell Dev. Biol. 68, 34–41 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bayliss R., Littlewood T., Stewart M., Structural basis for the interaction between FxFG nucleoporin repeats and importin-beta in nuclear trafficking. Cell 102, 99–108 (2000). [DOI] [PubMed] [Google Scholar]
- 45.Cingolani G., Bednenko J., Gillespie M. T., Gerace L., Molecular basis for the recognition of a nonclassical nuclear localization signal by importin beta. Mol. Cell 10, 1345–1353 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Lee S. J., et al. , The structure of importin-beta bound to SREBP-2: Nuclear import of a transcription factor. Science 302, 1571–1575 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Putker M., et al. , Redox-dependent control of FOXO/DAF-16 by transportin-1. Mol. Cell 49, 730–742 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Senchuk M. M., et al. , Activation of DAF-16/FOXO by reactive oxygen species contributes to longevity in long-lived mitochondrial mutants in Caenorhabditis elegans. PLoS Genet. 14, e1007268 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Çağatay T., Chook Y. M., Karyopherins in cancer. Curr. Opin. Cell Biol. 52, 30–42 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dormann D., et al. , ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J. 29, 2841–2857 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harley V. R., et al. , Defective importin beta recognition and nuclear import of the sex-determining factor SRY are associated with XY sex-reversing mutations. Proc. Natl. Acad. Sci. U.S.A. 100, 7045–7050 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brenner S., The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Minevich G., Park D. S., Blankenberg D., Poole R. J., Hobert O., CloudMap: A cloud-based pipeline for analysis of mutant genome sequences. Genetics 192, 1249–1269 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.