Significance
To create an infectious HIV-1 particle, the envelope glycoprotein (Env) must find a budding virus assembly site on the surface of an infected cell. Assembly sites are composed of growing 2-dimensional lattices of the structural Gag protein anchored to the plasma membrane by the matrix domain. The cytoplasmic tail of Env and the matrix domain are physically proximal on the inner leaflet of the membrane, placing these elements in direct contact upon encounter. We provide single-molecule evidence that the Env cytoplasmic tail and a single residue in matrix are necessary for lattice retention of Env, with membrane budding being dispensable. This supports a model for direct interaction or corralling of Env by the matrix domain and suggests that a druggable interface exists.
Keywords: human immunodeficiency virus 1 (HIV-1), single-particle tracking, virus assembly
Abstract
The HIV-1 envelope glycoprotein (Env) is sparsely incorporated onto assembling virus particles on the host cell plasma membrane in order for the virus to balance infectivity and evade the immune response. Env becomes trapped in a nascent particle on encounter with the polymeric viral protein Gag, which forms a dense protein lattice on the inner leaflet of the plasma membrane. While Env incorporation efficiency is readily measured biochemically from released particles, very little is known about the spatiotemporal dynamics of Env trapping events. Herein, we demonstrate, via high-resolution single-molecule tracking, that retention of Env trimers within single virus assembly sites requires the Env cytoplasmic tail (CT) and the L12 residue in the matrix (MA) domain of Gag but does not require curvature of the viral lattice. We further demonstrate that Env trimers are confined to subviral regions of a budding Gag lattice, supporting a model where direct interactions and/or steric corralling between the Env-CT and a lattice of MA trimers promote Env trapping and infectious HIV-1 assembly.
HIV-1 utilizes a sparse number of viral envelope glycoproteins (Envs), up to 14 trimers per virus, to enter a naïve cell and spread infection (1). This paucity of displayed viral antigens on the surface of the viral particle is believed to contribute to the virus’ immune evasion strategy, leading to chronic disease (2, 3). To create an infectious particle, newly synthesized Env traffics through the secretory pathway as a heterodimeric trimer of gp120 and gp41 and is expressed on the plasma membrane. Env must then diffuse laterally on the plasma membrane and encounter an assembling viral lattice composed of the structural viral protein Gag. Gag is a soluble, multidomain protein synthesized in the cytoplasm and targets the host cell plasma membrane via interaction between the matrix (MA) domain of Gag and plasma membrane-enriched inositol phospholipids (4, 5). On anchoring to the inner leaflet of the plasma membrane, Gag laterally diffuses and begins to oligomerize, forming a large protein lattice on the plasma membrane (∼150 nm in diameter) (6–8). A spherical virus particle is then formed by tight packing of the capsid (CA) domain of Gag, leading to membrane bending above the viral lattice and budding of the nascent virus particle away from the cytoplasm.
Given the distinct biosynthetic pathways utilized by Env and Gag, the exact timing of encounter between these 2 species remains unclear. Env possesses several conserved endocytosis motifs within its cytoplasmic tail (CT), and upon presentation at the plasma membrane, these signals lead to cell surface down-regulation of Env. Our laboratory and others have suggested that the endocytic pathway acts to regulate cell surface levels of Env and limit antigen display to the host immune system but at the cost of reducing the amount of Env trimers per virion (9, 10). On recycling to the plasma membrane, Env trimers must then stochastically diffuse along the plasma membrane to encounter a single Gag lattice and complete infectious virus assembly. Evidence from our laboratory supports a model where intracellular retention of Env leads to late-stage incorporation of Env trimers into preformed Gag lattices (9); however, it is unclear what determinants in Env and Gag are required for confinement of Env at a nascent Gag lattice.
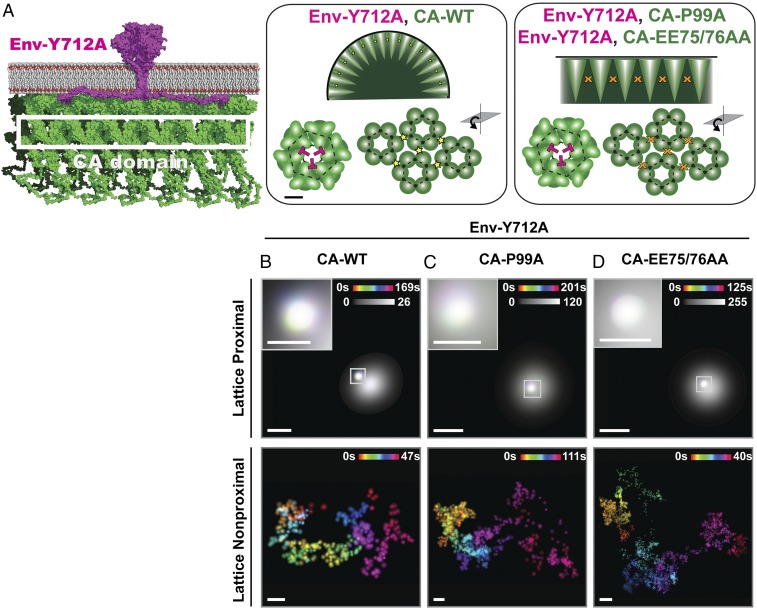
The Env-CT as well as the inner leaflet binding MA domain of Gag have been implicated in efficient virus incorporation (11–14). The putative proximity between the bulky Env-CT and the membrane-bound MA layer of the Gag lattice physically positions these 2 elements as master regulators of incorporation and retention of Env trimers at HIV-1 assembly sites (Fig. 1A). Strikingly, the bulky Env-CT has been previously shown to be required for efficient particle incorporation in T-cell lines but largely dispensable for particle incorporation in epithelial cell lines (15, 16). Paradoxically, we and others have shown that loss of the Env-CT (Env-∆CT) results in increased plasma membrane levels of Env due to loss of endocytosis motifs necessary for internalization (9, 17, 18). It remains unclear why deletion of the Env-CT reduces Env incorporation in T cells, despite increasing plasma membrane expression. These observations suggest that lattice trapping, through interaction between the Env-CT and the MA layer, is critical to efficiently incorporate Env trimers under physiologically relevant plasma membrane levels.
Fig. 1.
Lattice trapping of HIV-1 Env requires the Env-CT as well as the L12 residue in the Gag-MA layer. (A) Cartoon model highlighting essential domains of HIV-1 Gag (green) and Env (magenta) during viral assembly: the MA layer (highlighted with the white bar) anchored to the inner leaflet of the plasma membrane (gray) and the bulky Env-CT domain of a single Env trimer. Env-Y712A is putatively immobilized within a hexamer of trimers created by the MA layer (Upper Left); however, in the absence of the Env-CT, Env-∆CT is predicted to diffuse through the native MA layer (MA-WT; Upper Center), resulting in a reduction of Env incorporated into the budding virion. Altering the MA layer by introducing the MA-L12E mutation is predicted to relieve association with the Env-CT and/or alter the lattice hexamer structure to allow Env-Y712A to escape the viral lattice (Upper Right), resulting in potent reduction of Env trimer incorporation. (Scale bar: Upper, 10 nm.) (B–D) Representative examples of temporal projections of single Env trimer diffusion acquired by b12-QD605 labeling surface-exposed Env trimers on live infected COS7 cells. Env trimer trajectories proximal (Upper) and nonproximal (Lower) to individual Gag lattices (grayscale). Gag assembly site localizations are rendered at the measured diffraction limit. Color bars denote duration of single Env trimer tracks in seconds. Spatial confinement of Env yields an integrated white time projection. Grayscale bar denotes the number of localizations acquired for Gag assembly sites. Insets are magnified regions. (B) Env-Y712A becomes confined when proximal to the Gag-WT lattice but typically exhibits unconfined diffusion when nonproximal. (C) Env-∆CT shows unconfined diffusion whether proximal or nonproximal to the Gag WT lattice, suggesting that the Env-CT is necessary for Env trimer trapping by the Gag lattice. (D) Additionally, Env-Y712A trimers display unconfined diffusion when proximal to MA-L12E lattices, similar to the unconfined diffusion observed when nonproximal to the Gag lattice, suggesting that the MA-L12 residue plays a critical role in efficient Env trapping at assembly sites. WT = wild type. (Scale bars: B–D, 400 nm; Insets in B–D, 150 nm.)
To date, very little mechanistic evidence exists to support spatiotemporal models of the cellular encounter and potential interaction between the Env-CT and the MA layer of the assembled Gag lattice, yet new imaging technologies have begun to address this issue (9, 19–22). Herein, we directly visualize and statistically quantify the trapping events of single HIV-1 Env trimers within the Gag lattice on the surface of live infected cells. Using this methodology, we show that both the Env-CT and MA domain of Gag contribute to Env trapping at single virus assembly sites. Furthermore, we demonstrate that lattice curvature is dispensable for retention of Env trimers within single virus particles, suggesting that curvature-induced membrane remodeling does not contribute to Env trimer retention at virus assembly sites. Finally, we find that single Env trimers are highly confined to subviral regions of a budding particle, supporting a model for direct interaction or steric confinement between the Env-CT and the MA layer of the Gag lattice.
Results
Nanoscale Tracking of HIV-1 Assembly.
We utilized high-speed total internal reflection fluorescence microscopy and single-molecule tracking of HIV-1 Env combined with simultaneous tracking of single virus assembly sites on the surface of live infected cells to visualize lattice trapping events. Surface-exposed single Env trimers were labeled with a monovalent Fab fragment (b12) targeting the gp120 ectodomain of Env (23). Importantly, the b12 Fab was used to prevent bivalent antibody-mediated cross-linking of Env trimers as previously shown (24). The recombinantly produced b12 Fab was site specifically conjugated to a single quantum dot (QD605) using a p-azido-l-phenylalanine unnatural amino acid and copper-free click chemistry (25–27). This probe, b12-QD605, enabled single-molecule tracking of Env with a mean spatial localization uncertainty of 17.3 nm (SI Appendix, Fig. S1) and a temporal resolution of 100 Hz (SI Appendix, Supporting Discussion). Single b12-QD605 probes labeling a surface-exposed Env trimer periodically displayed intermittent photoblinking, which is diagnostic of single-molecule imaging (SI Appendix, Fig. S2). Additionally, we used the same methodology to conjugate QD605 to an anti-Env antibody Fab fragment, PGT145 (28, 29), recognizing trimeric Env gp120 only (SI Appendix, Fig. S3A). We optimized conditions to label single Env trimers at low labeling densities to prevent tracks from overlapping and ensure specific labeling. To further increase the sampling of Env trimer diffusion in single experiments, we introduced a single-point mutation into the Env-CT (Env-Y712A) that has been shown to reduce the rate of Env endocytosis (18). This resulted in a range of 2 to 5 b12-QD605–labeled Env trimers per cell (SI Appendix, Supporting Discussion).
To assess the proximity of Env trimer tracks to HIV-1 assembly sites, we labeled the Gag lattice with a genetically encoded GFP-tagged nanobody targeting the CA domain of Gag, previously shown to report on the spatial distribution of virus assembly sites without perturbing the rate of assembly (30). We show that the presence of this CA binding nanobody does not significantly perturb viral budding (SI Appendix, Fig. S4) and release (SI Appendix, Fig. S5). Furthermore, we utilized a release-incompetent mutant of Gag, PTAP(−), to ensure that observations of diffusion effects proximal to virus assembly sites were indeed cell-associated particles (SI Appendix, Fig. S6) (31). Only single-molecule tracks of Env and single Gag lattice assembly sites that were well resolved on the diffraction limit were selected for analysis. Using this approach, we observed single Env trimers freely diffusing in areas of the plasma membrane devoid of virus assembly sites (Fig. 1 B, Lower). Conversely, we observed drastic reductions in Env mobility when proximal to single virus assembly sites (Fig. 1 B, Upper and Movie S1). We then classified Env single-molecule tracks as inside or outside the diffraction-limited radius of single virus assembly sites. A maximum of 125 nm from a single track position and Gag lattice centroid was used as the criteria to assess proximity to the Gag lattice to compensate for the uncertainty of localizing a diffraction-limited object. To assess the diffusion rate of Env-Y712A trimers both inside and outside the radius of a single HIV-1 assembly site, we quantified the apparent diffusion coefficients (Dapparent) and slopes of the moment scaling spectrum (SMSS) for each track, which complementarily assess the modes of lateral motion of a diffusing single particle (SI Appendix, Fig. S7 has a detailed explanation of the SMSS parameter) (32–34). We observed that Env-Y712A trimers diffusing in proximity to virus assembly sites displayed a strong reduction in Dapparent and SMSS when compared with trimers diffusing outside of the radius of virus assembly sites (Fig. 2A) (lattice proximal: Dapparent = 0.0419 ± 0.0207 μm2 s−1, SMSS = 0.0155 ± 0.0723; lattice nonproximal: Dapparent = 0.0608 ± 0.0511 μm2 s−1, SMSS = 0.211 ± 0.277; P = 4.89 × 10−10). Similar qualitative diffusion behavior for Env-Y712A trimers was observed with the use of a quaternary-recognizing anti-gp120 probe, Fab PGT145-QD605 (SI Appendix, Fig. S3B), and using the b12-QD605 probe in the CEM-A T-cell line (SI Appendix, Fig. S8A).
Fig. 2.
Diffusion analysis highlights Env mobility increases with loss of the Env-CT or altering of the Gag-MA layer (MA-L12E) when proximal to assembling HIV-1 lattices. (A–C) The 2D diffusion analyses statistically compare proximal (in) and nonproximal (out) Env trimer diffusion with respect to Gag lattices. Centroids of ellipses indicate the mean apparent diffusion coefficient (Dapparent) and slope of the moment scaling spectrum (SMSS) quantitatively describing single Env trimer displacement over time. Ellipses denote the SDs of the collective parameters. Inverted triangles in Upper Left and Lower of each plot represent the individual values of SMSS and D, respectively, for the genotype and spatial consideration. (A–D) Lighter shades are lattice proximal Env tracks (in), and (A–C) darker shades are lattice nonproximal Env tracks (out). (A) Env-Y712A diffusion proximal to an MA-WT lattice (Dapparent = 0.0419 ± 0.0207 μm2 s−1, SMSS = 0.0155 ± 0.0723) displays a statistically significant difference (P = 4.89 × 10−10) from that of nonproximal tracks (Dapparent = 0.0608 ± 0.0511 μm2 s−1, SMSS = 0.211 ± 0.277). (B) Env-∆CT trimers proximal to MA-WT lattices (Dapparent = 0.0848 ± 0.0680 μm2 s−1, SMSS = 0.426 ± 0.294) exhibit a slight but statistically significant reduction in mobility when compared with Env-∆CT diffusion nonproximal to the viral lattice (Dapparent = 0.181 ± 0.598 μm2 s−1, SMSS = 0.425 ± 0.294; P = 1.68 × 10−5), suggesting that reduced mobility, induced by membrane ordering, possibly occurs but that this force is insufficient to confine Env-∆CT trimers at assembly sites. (C) Diffusion of Env-Y712A proximal to lattices possessing an MA-L12E layer (Dapparent = 0.0685 ± 0.0787 μm2 s−1, SMSS = 0.126 ± 0.261) exhibits a statistically significant decrease in mobility in comparison with Env-Y712A diffusion away from MA-L12E lattices (Dapparent = 0.105 ± 0.196 μm2 s−1, SMSS = 0.325 ± 0.286; P = 3.25 × 10−13), indicating that a defective MA layer can still affect the diffusion of Env trimers despite the inability to incorporate Env into released particles (SI Appendix, Fig. S10) (12). (D) Comparison of the mobility of Env trimers when proximal to Gag lattices. Env trimers with an intact Env-CT statistically differ in mobility when proximal to MA-L12E vs. MA-WT lattices (P = 0.001), yet it is significant enough to allow Env-Y712A to diffuse through an altered MA layer. Interestingly, a larger statistical difference is observed when comparing the mobility of Env-Y712A with Env-∆CT trimers proximal to functional MA layers (P = 1.82 × 10−15), and a smaller reduction in mobility is observed when comparing Env-Y712A proximal to the MA-L12E lattice with Env-∆CT proximal to the MA-WT lattice (P = 6.38 × 10−9). These findings indicate that the Env-CT dominantly drives the observed reduction in mobility at virus assembly sites when compared with a mutated MA layer. Error estimates from the distribution of Dapparent and SMSS values are SD, and P values were obtained using the Peacock method and the 2D Kolmogorov–Smirnov test. The n values for lattice proximal and lattice nonproximal tracks per genotype are as follows: Env-Y712A/MA-WT: 51 and 628 tracks, respectively; Env-∆CT/MA-WT: 105 and 2,474 tracks, respectively; and Env-Y712A/MA-L12E: 90 and 2,692 tracks, respectively. WT = wild type.
Evidence for the Role of the Env-CT in Lattice Incorporation.
Given the role of the Env-CT in efficient particle incorporation, we sought to determine if the Env-CT is required for trapping Env trimers at sites of virus assembly. First, we confirmed that loss of the Env-CT in COS7 cells resulted in a bulk reduction in Env accumulation at single virus assembly sites (SI Appendix, Fig. S9) and a partial loss of incorporation into virus-like particles (SI Appendix, Fig. S10). We hypothesized that loss of the Env-CT, despite increasing surface levels of Env trimers, acts to reduce particle incorporation by preventing lattice trapping. To test this hypothesis, we measured the diffusion properties of Env trimers lacking the last 144 amino acid residues of the CT (Env-∆CT) both in proximity to Gag assembly sites and in areas of the plasma membrane devoid of detectable Gag signal (Fig. 1C and Movie S2). Interestingly, we observed a slight, but significant, reduction in the mobility of Env-∆CT when inside the Gag lattice compared with Env-∆CT outside the radius (Fig. 2B) (lattice proximal: Dapparent = 0.0848 ± 0.0680 μm2 s−1, SMSS = 0.426 ± 0.294; lattice nonproximal: Dapparent = 0.181 ± 0.598 μm2 s−1, SMSS = 0.425 ± 0.294; P = 1.68 × 10−5). Deletion of the Env-CT, however, led to a strong increase in the mobility of Env trimers inside the radius of virus assembly sites when compared with the mobility of Env-Y712A trimers (Fig. 2D) (lattice proximal: P = 1.82 × 10−15), demonstrating that the reduced mobility, induced by membrane ordering above the Gag lattice, is insufficient to trap Env-∆CT trimers. We observed similar behavior of Env-∆CT with the use of the quaternary binding PGT145-QD605 probe (SI Appendix, Fig. S3C). These results are consistent with previous findings of the diffusion behavior of Env-∆CT trimers on the surface of cells and released particles (9, 21) yet enable specific measurement of local diffusional effects on encounter with cell-associated assembly sites.
The MA Residue L12 of Gag Is Required for Env Confinement at Assembly Sites.
Mutations in the MA domain of Gag have been shown to cause a drastic reduction in the levels of Env incorporated into released virus particles (11, 12, 35, 36). As the MA layer resides proximal to the Env-CT and inner leaflet of the plasma membrane, we hypothesized that specific MA mutations can cause a reduction in the trapping of Env trimers at virus assembly sites as has been previously suggested (12) but never microscopically observed. We found that introduction of the mutation L12E into the MA domain of Gag (MA-L12E), as previously shown (11, 37), led to reduced Env accumulation at virus assembly sites (SI Appendix, Fig. S9) and incorporation into released virus (SI Appendix, Fig. S10), and strikingly, it created assembly sites incapable of confining single Env trimers (Fig. 1 D, Upper vs. Lower and Movie S3). We did observe, however, that the MA-L12E layer caused a reduction in diffusion of Env trimers within Gag lattices when compared with freely diffusing trimers (Fig. 2C) (lattice proximal: Dapparent = 0.0685 ± 0.0787 μm2 s−1, SMSS = 0.126 ± 0.261 vs. lattice nonproximal: Dapparent = 0.105 ± 0.196 μm2 s−1, SMSS = 0.325 ± 0.286; P = 3.25 × 10−13). Reduction in the mobility of Env-Y712A within the radius of MA-L12E lattices was, however, less pronounced than the effect of Env-CT removal at MA wild type (MA-WT) assembly sites (Fig. 2D) (Env-Y712A proximal to the MA-L12E lattice vs. Env-∆CT proximal to the MA-WT lattice; P = 6.38 × 10−9). Similar qualitative observations of Env-Y712A diffusion in the presence of MA-L12E lattices were observed in the T-cell line CEM-A (SI Appendix, Fig. S8B).
Curvature of the Gag Lattice Is Not Required for Confining Env Diffusion.
The plasma membrane is the site at which Env interfaces with the Gag lattice. As membrane curvature has been shown to have an impact on the lipid composition of biological membranes (38, 39), we hypothesized that membrane curvature could also contribute to the confinement of single Env trimers at virus assembly sites (Fig. 3A). Previous studies have identified 2 mutations in the Gag CA domain (CA-P99A and CA-EE75/76AA) that inhibit membrane budding and curvature at virus assembly sites yet still form electron dense flat lattices (40–42). To address if curvature contributes to Env trapping at virus assembly sites, we performed single-molecule tracking of Env-Y712A trimers on the surface of infected cells expressing Gag containing the CA-P99A curvature mutation (Fig. 3C). We demonstrate that the mobility of Env trimers is drastically reduced when proximal to flat viral lattices containing the CA-P99A mutation (Figs. 3C and 4A) (lattice proximal: Dapparent = 0.0351 ± 0.0237 μm2 s−1, SMSS = 0.156 ± 0.273 vs. lattice nonproximal: Dapparent = 0.0930 ± 0.0791 μm2 s−1, SMSS = 0.312 ± 0.297; P = 5.89 × 10−14), showing indistinguishable diffusion when compared with CA wild type (CA-WT) assembly sites (Figs. 3 B and C and 4C) (P = 0.007; expectation value α < 0.001). To confirm that curvature indeed was not required for confining Env at virus assembly sites, we introduced a separate curvature-inhibiting mutation into the Gag CA domain, CA-EE75/76AA. Under these conditions, the CA-EE75/76AA lattice was still able to strongly confine the diffusion of Env trimers at flat lattice assembly sites (Figs. 3D and 4B and Movie S4) (lattice proximal: Dapparent = 0.0452 ± 0.0409 μm2 s−1, SMSS = 0.0787 ± 0.247 vs. lattice nonproximal: Dapparent = 0.120 ± 0.209 μm2 s−1, SMSS = 0.322 ± 0.307; P = 5.18 × 10−22), supporting our finding that inhibition of curvature does not significantly affect lattice confinement of Env in comparison with the CA-WT lattice (Fig. 4C) (CA-P99A vs. CA-EE75/76AA lattice proximal: P = 0.0390; CA-P99A vs. CA-WT lattice proximal: P = 0.007; and CA-EE75/76AA vs. CA-WT lattice proximal: P = 7.93 × 10−4). Similar qualitative diffusion behavior was observed both in the CEM-A T-cell line (SI Appendix, Fig. S8C) and when using the trimer-specific Fab PGT145-QD605 in COS7 cells (SI Appendix, Fig. S3D). Next, we sought to determine the role of the Env-CT in flat lattice trapping by introducing the Env-CT deletion (Env-∆CT) into the CA-EE75/76AA proviral genetic background. We found that the diffusivity of Env-∆CT trimers significantly increases within the radius of single CA-EE75/76AA lattice assembly sites when compared with Env-Y712A (SI Appendix, Fig. S11) (lattice proximal: Dapparent = 0.193 ± 0.0814, SMSS = 0.525 ± 0.195; lattice nonproximal: Dapparent = 0.136 ± 0.124, SMSS = 0.391 ± 0.300; P = 0.0139). Furthermore, the diffusion rates of Env-∆CT in the presence or absence of lattice curvature were found to be highly similar (CA-WT vs. CA-EE75/76AA with Env-∆CT; P = 3.67 × 10−4) (SI Appendix, Supporting Discussion). Together, our results demonstrate that the Env-CT and MA layer play a dominant role in reducing the mobility of Env trimers at virus assembly sites, with membrane curvature being dispensable for Env confinement within the lattice.
Fig. 3.
Membrane curvature is not required for Env trimer confinement within HIV-1 lattices. (A) Cartoon representation of an Env trimer (magenta) proximal to the flat Gag lattice (green). During assembly, the CA domain of Gag oligomerizes to form the hexameric lattice (highlighted by the white box). Further packing leads to membrane bending to create the budding virion (Middle). Introduction of mutations into the CA domain (CA-P99A and CA-EE75/76AA) prevents membrane curvature but still results in formation of dense flat lattices on the plasma membrane (Right). (Scale bar: Middle, 10 nm.) (B–D) Representative tracking of single Env trimers proximal (Upper) and nonproximal (Lower) to individual Gag lattices (grayscale) imaged in COS7 cells. Env (multicolor) and Gag (gray) assembly site localization centroids are rendered as before. Grayscale bars denote the number of localizations acquired for Gag. Insets are magnified regions. (B) Confinement of Env-Y712A trimer diffusion is observed when proximal to a CA-WT lattice, but diffusion is unconfined when nonproximal to virus assembly sites. The curvature mutations (C) CA-P99A and (D) CA-EE75/76AA produce flat Gag lattices capable of confining the diffusion of Env-Y712A trimers, similar to CA-WT lattices, indicating that Env trapping by Gag lattices does not require membrane curvature or curvature-induced changes in lipid composition. WT = wild type. (Scale bars: B–D, 400 nm; Insets in B–D, 150 nm.)
Fig. 4.
Diffusion analysis establishes that Env-Y712A incorporation into virus assembly sites does not rely on Gag-induced membrane curvature. (A and B) Diffusion analyses comparing proximal (in) and nonproximal (out) Env tracking with respect to Gag assembly sites. The mean Dapparent and slope of the moment scaling spectrum (SMSS) are indicated by closed circles. Boundaries of each ellipse denote the combined SDs for the diffusion parameters. Inverted triangles in Left and Lower of each plot represent the individual values of SMSS and Dapparent, respectively, for each CA genotype and proximity condition. (A–C) Lighter shades are lattice proximal Env-Y712A diffusion properties, while (A and B) darker shades are lattice nonproximal. (A) When proximal to flat lattices produced by the curvature mutant CA-P99A, Env-Y712A diffusivity (Dapparent = 0.0351 ± 0.0237 μm2 s−1, SMSS = 0.156 ± 0.273) varies significantly from Env-Y712A diffusivity nonproximal to these flat lattices (Dapparent = 0.0930 ± 0.0791 μm2 s−1, SMSS = 0.312 ± 0.297; P = 5.89 × 10−14). (B) Env-Y712A diffusivity proximal to flat CA-EE75/76-AA lattices (Dapparent = 0.0452 ± 0.0409 μm2 s−1, SMSS = 0.0787 ± 0.247) similarly exhibits a significant reduction in mobility when compared with diffusion of Env-Y712A nonproximal to these flat lattices (Dapparent = 0.120 ± 0.209 μm2 s−1, SMSS = 0.322 ± 0.307; P = 5.18 × 10−22). (C) Diffusion and confinement of Env-Y712A trimers at sites of flat lattice assembly are indistinguishable between CA-P99A and CA-EE75/76AA mutations (P = 0.0389). Similarly, Env-Y712A trimer diffusion and confinement at CA-P99A vs. CA-WT lattices were indistinguishable (P = 0.007); however, there was a minor, but statistically significant, difference in the diffusion of Env-Y712A in proximity to CA-EE75/76AA vs. CA-WT lattices (P = 7.93 × 10−4), suggesting that Env trapping at HIV-1 assembly sites does not require a curved membrane. Errors reported for Dapparent and SMSS are SD, and P values were obtained using the Peacock method and the 2D Kolmogorov–Smirnov statistical test. The n values for lattice proximal and lattice nonproximal tracks per genotype are as follows: Env-Y712A/CA-WT: 51 and 628 tracks, respectively; Env-Y712A/CA-P99A: 87 and 829 tracks, respectively; and Env-Y712A/CA-EE75/76AA: 121 and 2,306 tracks, respectively. WT = wild type.
Subviral Confinement of Env Trimers in the Gag Lattice.
We have shown that the Env-CT and a single residue in the Gag-MA layer can influence the ability of a viral lattice to ensnare Env trimers. It remains unclear, however, if the Env-CT and MA layer cooperate on a local, subviral scale or if Env trimers exhibiting viral confinement are capable of diffusing within the Gag lattice. To address whether the Gag lattice acts to trap Env trimers locally, on a subviral scale, or on a single assembly site scale (Fig. 5A), we analyzed the displacements of highly confined Env-Y712A trimers (mean squared displacement parameter α < 0.3) present at single virus assembly sites for both CA-WT and CA curvature-mutant (CA-P99A, CA-EE75/76AA) lattices (Fig. 5B). Our results show that measured displacements greater than our mean localization precision (SI Appendix, Fig. S1) (∼17.3 nm), over minutes of time, only occur in as many as 21 and 24% of the total displacements for averaged Env-Y712A tracks found at assembly sites with CA-WT and both curvature mutants (CA-P99A, CA-EE75/76AA), respectively (SI Appendix, Fig. S12). These results, when compared with b12-QD605 probes immobilized directly to a coverslip surface (Fig. 5B) (20% of displacements larger than the mean localization precision of ∼17.3 nm), demonstrate that Env trimers are highly confined to local regions of the lattice, and this underlying diffusion barrier is smaller than our localization precision limit.
Fig. 5.
Quantification of the displacements of lattice proximal Env-Y712A trimers at native HIV-1 assembly sites. (A) Cartoon model demonstrating subviral (Left) and viral (Right) confinement of Env in a budding lattice. A 2D projection of Env trimer position measurements for subviral confinement (Left Inset) vs. confinement to the entire budding lattice (Right Inset) will produce distinct spatial distribution patterns depending on the mechanism of Env confinement. A direct lattice interaction or steric corralling within a hexamer of MA trimers would confine Env trimer motion to subviral regions of the viral membrane. Passive incorporation of Env trimers would allow lateral diffusion on the surface of the bud over extended periods of observation. (B) Integrated single-molecule localizations of lattice associated tracks exhibiting extreme confinement (mean squared displacement parameter α < 0.3 and total variance in position, σ2 < 58.5 nm) were aligned for CA-WT assembly sites (n = 12 tracks), CA-P99A and CA-EE75/76AA (compiled) assembly sites (n = 27 tracks), and the anti-Env probe (b12-QD605) immobilized on a bare coverslip surface (n = 15 tracks). Stars represent mean positions, and dotted lines represent the 2D 68% confidence intervals calculated by principal component analysis (PCA). The average SD from the mean position over the perimeter of the ellipses was σCA-WT = 30.1 nm (n = 7,050 localizations), σCA-EE75/76AA, CA-P99A = 37.2 nm (n = 21,282 localizations), and σimmobilized = 28.0 nm (n = 22,699 localizations). The SD from the centroid for the point clouds (σCA-WT = 19.9 nm, σCA-EE75/76AA, CA-P99A = 24.6 nm, and σimmobilized = 18.5 nm) was found to be smaller than those found from PCA. These results provide upper and lower bounds to the potential for Env trimer displacement in confined regions of a single viral lattice.
Discussion
Here, we demonstrate methodology to observe the acquisition of a single envelope glycoprotein trimer into a virus assembly site with subviral displacement sensitivity. Our imaging approach using a balance of speed and precision (100 Hz and less than 20 nm, respectively) has enabled the quantized measurement of the diffusion of single HIV-1 Env trimers with respect to Gag lattice assembly sites. In this study, we utilized this approach to directly uncover the consequences of perturbing both the Env-CT as well as the MA layer. Furthermore, we introduced key Gag mutations to directly show that membrane curvature is not required for Env retention within the viral lattice.
The spatiotemporal dynamics of single Env trimers on the surface of infected cells showed marked diffusive differences between those trimers residing proximal to Gag lattices compared with trimers diffusing in regions of the plasma membrane lacking appreciable Gag signal. Our 2-dimensional (2D) tracking approach suggests that Env trimers diffuse on the plasma membrane relatively freely; however, on encounter of a Gag lattice, these trimers are effectively trapped, and this motion is now largely confined to an extent below our localization precision limit (SI Appendix, Supporting Discussion has 2D tracking considerations; SI Appendix, Fig. S13). Furthermore, the confinement of a single Env trimer to a subviral region of the Gag lattice (less than 30 nm) over minutes of observation supports a model where direct interaction and/or steric corralling between the bulky Env-CT and a lattice of MA trimers lead to irreversible Env incorporation (13, 43, 44).
Removal of the Env-CT (Env-∆CT) leads to a potent reduction in Env incorporation in most T-cell lines but has less detrimental effects in the HeLa and MT-4 cell lines (15). While it remains unclear how infected HeLa or MT-4 lines can overcome the Env incorporation defect when lacking CT sorting signals, one consequence of removal of the Env-CT, shown by us and others, is that surface-exposed Env levels are drastically increased (9, 18). Increasing Env-∆CT levels creates a high steady-state density of trimers on the plasma membrane that can promote stochastic or passive incorporation onto releasing particles. In this study, we demonstrate that Env-∆CT does not become sufficiently trapped by the underlying Gag lattice on encounter in the membrane and proceeds to diffuse through individual lattices, albeit at an apparent 2-fold slower rate when compared with Env-∆CT diffusing outside of viral lattices. In contrast, we observed that flat lattices, produced by the curvature mutant CA-EE75/76AA, show a marginal increase in the apparent diffusion coefficient at sites of virus assembly. These results suggest that the curved lattice may impart specific properties to the membrane that reduce Env trimer mobility. The apparent reduction in Env-∆CT diffusion is also consistent with spectroscopic and lipidomic studies suggesting a strong ordering of membrane lipids by the underlying viral lattice in released particles (21, 45, 46). Lipid ordering at lattice assembly sites is likely not sufficient to completely confine the diffusion of Env trimers at assembly sites. Therefore, Env-CT–independent cell lines likely make use of higher steady-state plasma membrane levels to increase passive incorporation. Taken together, our results support a model where the Env-CT is a dominant driver of lattice retention.
The membrane-proximal MA domain has been shown to create an ordered hexamer of trimers array in vitro (43). This MA layer can potentially serve as a gated diffusion barrier to the bulky Env-CT and prevent Env trimer escape from assembling lattices. We demonstrate that a previously reported MA mutation, MA-L12E (11), prevents confinement of Env trimers at Gag MA-L12E assembly sites. This result suggests that the L12E mutation acts to significantly permeabilize the MA layer and enables Env diffusion through assembly sites. While Env diffusion at MA-L12E assembly sites was increased relative to that of MA-WT lattices, we observe an intermediate mobility of Env trimers when compared with the dynamics of Env-∆CT at native Gag lattices. This result demonstrates that the MA-L12E lattice is unable to potently confine the diffusion of Env trimers; however, the physical proximity of the bulky Env-CT and the MA-L12E layer at the inner leaflet of the plasma membrane can still create a force that acts to significantly reduce Env trimer mobility. Previous biochemical measurements (11, 36, 37) and results from this study (SI Appendix, Fig. S10) have shown that the MA-L12E mutation markedly reduces Env incorporation efficiency, suggesting, despite our detection of reduced mobility of Env-Y712A at MA-L12E assembly sites, that stronger forces are required for confinement and lattice incorporation of Env trimers (SI Appendix, Supporting Discussion has an interpretation of Env-Y712A diffusion nonproximal to MA-L12E lattices). This result is significant, as it suggests that the MA-L12E layer does not reject Env-Y712A due to noncomplementarity, but instead allows Env trimers to sample the lattice less encumbered by the MA-layer topography.
The HIV-1 Gag lattice creates a large ordered protein domain anchored to the plasma membrane, which contributes to the enrichment of cholesterol and sphingolipids observed in released particles (42, 46). The induction of membrane curvature has been shown to consequently reorganize certain lipid and host cell proteins at virus assembly sites (42, 47). Gag-induced membrane remodeling could, therefore, contribute to Env trapping at assembly sites. We demonstrate, however, that membrane curvature is not an important determinant for confining Env trimers within viral lattices. Two separate and well-characterized CA mutations (40–42), known to inhibit membrane curvature and virus budding but still capable of forming flat densely packed lattices, were able to strongly confine the diffusion of Env-Y712A trimers to a subviral region of an assembly site. These results suggest that curvature-induced membrane reorganization, while still producing significant effects on other HIV-1 assembly and restriction processes (47–49), does not contribute significantly to Env confinement within assembling particles. This suggests that a direct interaction or steric corralling between the Env-CT and the underlying MA layer is responsible for Env incorporation (13, 36, 43, 44). Furthermore, this suggests that viral confinement of Env trimers is not due to curvature-induced diffusion barriers at the neck of the bud, but rather that physical proximity to the lattice is a more important determinant of particle incorporation.
Previous studies have suggested that a direct biochemical interaction exists between the Env-CT and Gag-MA (13, 44), but it is also possible that the MA layer acts to sterically corral the bulky Env-CT between adjacent MA trimers (43, 50). Our measurements of Env displacements within the viral lattice suggest that Env trimers infrequently display mobility beyond our mean localization precision of 17.3 nm, if at all (Fig. 5 and SI Appendix, Figs. S1 and S12). The potential displacements would correspond to only 2 hexameric MA protomers, estimated at ∼10 nm in diameter (43). The lack of appreciable Env trimer displacement over significant observation periods (minutes), relative to the overall size of the viral lattice, suggests that Env is unable to efficiently move laterally beyond a few hexameric protomers of the MA layer during the lifetime of virus assembly, typically 7 to 14 min (51). If Env displacements were larger than our measurements, escape events would become more probable, and this would alter the steady-state levels of Env trimers per virus particle. This consideration is intriguing, because it suggests that the Gag lattice could also act as a diffusion barrier for Env trimers attempting to enter the lattice, providing an additional layer of regulation to limit the number of trimers per particle. Further studies will be required to understand how the levels of Env trimers are regulated when frequent encounters between viral lattices and Env trimers must occur. Recent work from our laboratory suggests that regulation of the surface levels of Env, through intracellular sequestration, contributes to Env sparsity on released particles (9), but additional mechanisms must exist to achieve a precise number of trimers per particle to balance infectivity and escape from immune surveillance.
Our approach for visualizing single-molecule events that contribute to infectious particle biogenesis identifies the mechanistic role of viral determinants in choreographing this process. Importantly, this methodology balances spatial and temporal resolution to visualize dynamic virus assembly events on the single virus scale. Our results show that, outside of virus assembly sites, Env trimers are highly diffusive and can sample significant regions of the infected cell plasma membrane. However, when Env encounters a virus assembly site, the Gag lattice potently confines Env trimers. We directly show that the Env-CT and MA layer of the Gag lattice significantly contribute to reducing the lateral mobility of Env trimers at virus assembly sites, with perturbation to either of these determinants unencumbering Env diffusion within the lattice. Furthermore, we demonstrate that membrane curvature is not required for Env retention at virus assembly sites, suggesting that interaction, either direct or steric, between the Env-CT and MA layer drives the creation of new infectious virus particles. Future studies should focus on the role of additional motifs in mediating lattice trapping of Env, with mechanistic studies aimed at differentiating between models of lattice interaction and corralling of Env diffusion.
Materials and Methods
Monovalent Fab anti-Env gp120 probes b12-QD605 and PGT145-QD605 were generated by recombinant bacterial expression and incorporation of a p-azido-l-phenylalanine unnatural amino acid (23, 26). Fab probes were conjugated to single fluorescent quantum dots (Thermo Fisher) using copper-free click chemistry (25, 27). Gag and Env were expressed from the HIV-1 NL4-3 reference genome via single-round infection of COS7 or CEM-A cells and imaged at 40 to 44 h postinfection. The p6 motif of Gag was mutated to remove the PTAP motif and ensure that virus assembly sites were cell associated. To detect single virus assembly sites, Gag was indirectly labeled by cytoplasmic expression of a capsid N-terminal domain-specific nanobody fused to the enhanced green fluorescent protein gene (CANTD-eGFP) (30). CANTD-eGFP was expressed through the nef splice acceptor site in the NL4-3 genome. Surface-exposed Env was labeled with b12-QD605 or PGT145-QD605 probes and imaged under physiological conditions (37 °C and 5% CO2) using a custom-built total internal reflection fluorescence microscope. The fluorescence emission from both the eGFP and QD605 probes was split into 2 channels and simultaneously imaged onto 2 halves of an sCMOS camera operating at 100 Hz (Hamamatsu). Raw images were corrected for pixel nonuniformity, split into respective channels, and corrected for chromatic aberration using fiduciary reference. Image channels were separately processed using custom peak finding software (IDL; Harris Geospatial), and the resulting subpixel coordinates were segmented for individual regions of interest using custom Matlab code (Mathworks). Sparse QD605 localizations were linked to create particle trajectories using custom single-particle tracking analysis software written in Matlab. Those trajectories for QD605 residing within 125 nm of an eGFP-labeled virus assembly site, the estimated diffraction-limited resolution of the microscope, were classified as proximal (Rayleigh criteria). SI Appendix, SI Text has further detail of the methods performed in this study.
Data and Materials Availability.
Data and reagents will be made available on request.
Supplementary Material
Acknowledgments
We thank Dr. Eric Freed and Dr. Melissa Victoria Fernandez for comments and careful reading of the manuscript. We also thank Courtney Ozzello for preparing electron microscopy (EM) samples and diligently imaging EM sections. Proviral clones (NL4-3 reference genome) for MA-L12E and Env-∆CT (CTdel-144) were a gift from Dr. Eric Freed. pEVOL-pAzF was a gift from Dr. Peter Schultz (Addgene; plasmid 31186). pCOMB3H-b12 was a gift from Dr. Dennis Burton. pCANTD-eGFP was a gift from Dr. Heinrich Leonhardt. The pSPAX2 packaging vector was provided by Dr. Didier Trono (Addgene; plasmid 12260). The following reagents were provided through the NIH AIDS Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases (NIAID), NIH: anti-p24 IgG from Dr. Bruce Chesebro and Kathy Wehrly, anti-gp41 IgG 10E8v4 from Dr. Peter Kwong, anti-gp120 IgG 2G12 from Polymun Scientific, and anti-gp120 IgG b12 from Dr. Dennis Burton and Dr. Carlos Barbas. This research was supported by NIAID Grant R01AI138625 and the Boettcher Foundation Webb-Waring Biomedical Research Award.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1910008116/-/DCSupplemental.
References
- 1.Zhu P., et al. , Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature 441, 847–852 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Hogan M. J., et al. , Increased surface expression of HIV-1 envelope is associated with improved antibody response in vaccinia prime/protein boost immunization. Virology 514, 106–117 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stano A., et al. , Dense array of spikes on HIV-1 virion particles. J. Virol. 91, e00415–e00417 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ono A., Ablan S. D., Lockett S. J., Nagashima K., Freed E. O., Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc. Natl. Acad. Sci. U.S.A. 101, 14889–14894 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saad J. S., et al. , Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc. Natl. Acad. Sci. U.S.A. 103, 11364–11369 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manley S., et al. , High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat. Methods 5, 155–157 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Floderer C., et al. , Single molecule localisation microscopy reveals how HIV-1 Gag proteins sense membrane virus assembly sites in living host CD4 T cells. Sci. Rep. 8, 16283 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Briggs J. A., Wilk T., Welker R., Kräusslich H. G., Fuller S. D., Structural organization of authentic, mature HIV-1 virions and cores. EMBO J. 22, 1707–1715 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buttler C. A., et al. , Single molecule fate of HIV-1 envelope reveals late-stage viral lattice incorporation. Nat. Commun. 9, 1861 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egan M. A., Carruth L. M., Rowell J. F., Yu X., Siliciano R. F., Human immunodeficiency virus type 1 envelope protein endocytosis mediated by a highly conserved intrinsic internalization signal in the cytoplasmic domain of gp41 is suppressed in the presence of the Pr55gag precursor protein. J. Virol. 70, 6547–6556 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freed E. O., Martin M. A., Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J. Virol. 70, 341–351 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freed E. O., Martin M. A., Virion incorporation of envelope glycoproteins with long but not short cytoplasmic tails is blocked by specific, single amino acid substitutions in the human immunodeficiency virus type 1 matrix. J. Virol. 69, 1984–1989 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wyma D. J., Kotov A., Aiken C., Evidence for a stable interaction of gp41 with Pr55(Gag) in immature human immunodeficiency virus type 1 particles. J. Virol. 74, 9381–9387 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roy N. H., Chan J., Lambelé M., Thali M., Clustering and mobility of HIV-1 Env at viral assembly sites predict its propensity to induce cell-cell fusion. J. Virol. 87, 7516–7525 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murakami T., Freed E. O., The long cytoplasmic tail of gp41 is required in a cell type-dependent manner for HIV-1 envelope glycoprotein incorporation into virions. Proc. Natl. Acad. Sci. U.S.A. 97, 343–348 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akari H., Fukumori T., Adachi A., Cell-dependent requirement of human immunodeficiency virus type 1 gp41 cytoplasmic tail for Env incorporation into virions. J. Virol. 74, 4891–4893 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhakta S. J., Shang L., Prince J. L., Claiborne D. T., Hunter E., Mutagenesis of tyrosine and di-leucine motifs in the HIV-1 envelope cytoplasmic domain results in a loss of Env-mediated fusion and infectivity. Retrovirology 8, 37 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Byland R., Vance P. J., Hoxie J. A., Marsh M., A conserved dileucine motif mediates clathrin and AP-2-dependent endocytosis of the HIV-1 envelope protein. Mol. Biol. Cell 18, 414–425 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muranyi W., Malkusch S., Müller B., Heilemann M., Kräusslich H. G., Super-resolution microscopy reveals specific recruitment of HIV-1 envelope proteins to viral assembly sites dependent on the envelope C-terminal tail. PLoS Pathog. 9, e1003198 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakin V., et al. , A versatile tool for live-cell imaging and super-resolution nanoscopy studies of HIV-1 Env distribution and mobility. Cell Chem. Biol. 24, 635–645.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Chojnacki J., et al. , Envelope glycoprotein mobility on HIV-1 particles depends on the virus maturation state. Nat. Commun. 8, 545 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chojnacki J., et al. , Maturation-dependent HIV-1 surface protein redistribution revealed by fluorescence nanoscopy. Science 338, 524–528 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Barbas C. F., 3rd, et al. , In vitro evolution of a neutralizing human antibody to human immunodeficiency virus type 1 to enhance affinity and broaden strain cross-reactivity. Proc. Natl. Acad. Sci. U.S.A. 91, 3809–3813 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anand S. P., et al. , Antibody-induced internalization of HIV-1 Env proteins limits surface expression of the closed conformation of Env. J. Virol. 93, e00293-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baskin J. M., et al. , Copper-free click chemistry for dynamic in vivo imaging. Proc. Natl. Acad. Sci. U.S.A. 104, 16793–16797 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chin J. W., et al. , Addition of p-azido-L-phenylalanine to the genetic code of Escherichia coli. J. Am. Chem. Soc. 124, 9026–9027 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Ning X., Guo J., Wolfert M. A., Boons G. J., Visualizing metabolically labeled glycoconjugates of living cells by copper-free and fast huisgen cycloadditions. Angew. Chem. Int. Ed. Engl. 47, 2253–2255 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walker L. M., et al. ; Protocol G Principal Investigators , Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477, 466–470 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee J. H., et al. , A broadly neutralizing antibody targets the dynamic HIV envelope trimer apex via a long, rigidified, and anionic β-Hairpin structure. Immunity 46, 690–702 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Helma J., et al. , Direct and dynamic detection of HIV-1 in living cells. PLoS One 7, e50026 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garrus J. E., et al. , Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107, 55–65 (2001). [DOI] [PubMed] [Google Scholar]
- 32.Ewers H., et al. , Single-particle tracking of murine polyoma virus-like particles on live cells and artificial membranes. Proc. Natl. Acad. Sci. U.S.A. 102, 15110–15115 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferrari R., Manfroi A., Young W. R., Strongly and weakly self-similar diffusion. Physica D 154, 111–137 (2001). [Google Scholar]
- 34.Sbalzarini I. F., Moments of displacement and their spectrum. https://www.researchgate.net/publication/251535342_Moments_of_displacement_and_their_spectrum (January 2005).
- 35.Tedbury P. R., Freed E. O., The role of matrix in HIV-1 envelope glycoprotein incorporation. Trends Microbiol. 22, 372–378 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murakami T., Freed E. O., Genetic evidence for an interaction between human immunodeficiency virus type 1 matrix and alpha-helix 2 of the gp41 cytoplasmic tail. J. Virol. 74, 3548–3554 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tedbury P. R., Ablan S. D., Freed E. O., Global rescue of defects in HIV-1 envelope glycoprotein incorporation: Implications for matrix structure. PLoS Pathog. 9, e1003739 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McMahon H. T., Boucrot E., Membrane curvature at a glance. J. Cell Sci. 128, 1065–1070 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larsen J. B., et al. , Membrane curvature and lipid composition synergize to regulate N-ras anchor recruitment. Biophys. J. 113, 1269–1279 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kong L. B., et al. , Cryoelectron microscopic examination of human immunodeficiency virus type 1 virions with mutations in the cyclophilin A binding loop. J. Virol. 72, 4403–4407 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.von Schwedler U. K., Stray K. M., Garrus J. E., Sundquist W. I., Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J. Virol. 77, 5439–5450 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hogue I. B., Grover J. R., Soheilian F., Nagashima K., Ono A., Gag induces the coalescence of clustered lipid rafts and tetraspanin-enriched microdomains at HIV-1 assembly sites on the plasma membrane. J. Virol. 85, 9749–9766 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alfadhli A., Barklis R. L., Barklis E., HIV-1 matrix organizes as a hexamer of trimers on membranes containing phosphatidylinositol-(4,5)-bisphosphate. Virology 387, 466–472 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cosson P., Direct interaction between the envelope and matrix proteins of HIV-1. EMBO J. 15, 5783–5788 (1996). [PMC free article] [PubMed] [Google Scholar]
- 45.Huarte N., et al. , Functional organization of the HIV lipid envelope. Sci. Rep. 6, 34190 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lorizate M., et al. , Comparative lipidomics analysis of HIV-1 particles and their producer cell membrane in different cell lines. Cell Microbiol. 15, 292–304 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Sengupta P., et al. , A lipid-based partitioning mechanism for selective incorporation of proteins into membranes of HIV particles. Nat. Cell Biol. 21, 452–461 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Perez-Caballero D., et al. , Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell 139, 499–511 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grover J. R., et al. , Roles played by capsid-dependent induction of membrane curvature and Gag-ESCRT interactions in tetherin recruitment to HIV-1 assembly sites. J. Virol. 87, 4650–4664 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tedbury P. R., Novikova M., Ablan S. D., Freed E. O., Biochemical evidence of a role for matrix trimerization in HIV-1 envelope glycoprotein incorporation. Proc. Natl. Acad. Sci. U.S.A. 113, E182–E190 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jouvenet N., Bieniasz P. D., Simon S. M., Imaging the biogenesis of individual HIV-1 virions in live cells. Nature 454, 236–240 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.