Significance
Hematopoietic stem cells (HSCs) are rare cells with the unique ability to self-renew and produce all blood cells throughout life. HSCs are functionally heterogeneous, and studies have shown that HSCs can differ in their contribution to major blood lineages. We found that the surface marker Neogenin-1, when combined with the reporter gene for long-term repopulating HSCs, Hoxb5, can stratify HSCs into 2 subpopulations—one that is more active and biased toward producing myeloid cells and another that is more dormant and capable of producing all blood lineages equally. Neogenin-1 reveals the diversity and hierarchical relationship of HSCs in the mouse bone marrow, enables the prospective isolation of myeloid-biased and balanced HSCs, and opens opportunities to do the same in humans.
Keywords: hematopoietic stem cell, transplantation, myeloid bias, aging, Neogenin-1
Abstract
Hematopoietic stem cells (HSCs) self-renew and generate all blood cells. Recent studies with single cell transplants and lineage tracing suggest that adult HSCs are diverse in their reconstitution and lineage potentials. However, prospective isolation of these subpopulations has remained challenging. Here, we identify Neogenin-1 (NEO1) as a unique surface marker on a fraction of mouse HSCs labeled with Hoxb5, a specific reporter of long-term HSCs (LT-HSCs). We show that NEO1+Hoxb5+ LT-HSCs expand with age and respond to myeloablative stress in young mice while NEO1−Hoxb5+ LT-HSCs exhibit no significant change in number. Furthermore, NEO1+Hoxb5+ LT-HSCs are more often in the G2/S cell cycle phase compared to NEO1−Hoxb5+ LT-HSCs in both young and old bone marrow. Upon serial transplantation, NEO1+Hoxb5+ LT-HSCs exhibit myeloid-biased differentiation and reduced reconstitution while NEO1−Hoxb5+ LT-HSCs are lineage-balanced and stably reconstitute recipients. Gene expression analysis reveals erythroid and myeloid priming in the NEO1+ fraction and association of quiescence and self-renewal–related transcription factors with NEO1− LT-HSCs. Finally, transplanted NEO1+Hoxb5+ LT-HSCs rarely generate NEO1−Hoxb5+ LT-HSCs while NEO1−Hoxb5+ LT-HSCs repopulate both LT-HSC fractions. This supports a model in which dormant, balanced NEO1−Hoxb5+ LT-HSCs can hierarchically precede active, myeloid-biased NEO1+Hoxb5+ LT-HSCs.
The hematopoietic system is hierarchically organized into distinct cell types and cellular states with unique functions and regenerative potentials (1). Residing at the apex of this hierarchy is the hematopoietic stem cell (HSC)—the cornerstone of all blood and immune development, maintenance, and regeneration. HSCs have the unique ability to self-renew and give rise to all major lineages of blood and immune cells throughout life. Over the years, combinations of surface markers (2–6), reporter genes (7–10), and other approaches (11, 12) have refined the definition of mouse HSCs and enabled the purification of long-term hematopoietic stem cells (LT-HSCs), a subset of HSCs capable of serially reconstituting irradiated recipients in a transplantation model. Recently, we identified Hoxb5 as a specific marker of LT-HSCs and generated a Hoxb5-mCherry reporter mouse strain for the prospective isolation of these cells (8). We demonstrated that the Hoxb5-mCherry reporter significantly improves the precise isolation of serially transplantable LT-HSCs compared to immunophenotypically defined HSCs (phenotypic HSCs [pHSCs]). As previously shown, only 7% to 35% of pHSCs are Hoxb5-mCherry+, and the potential for long-term multilineage reconstitution is restricted to this fraction (8).
Although mouse Hoxb5+ LT-HSCs are multilineage-contributing, self-renewing cells (8), the functional heterogeneity within this compartment has not yet been characterized. Understanding the composition of LT-HSCs may offer valuable insights into the mechanism of HSC expansion with age, as well as the competition of diverse HSCs for bone marrow niches (13, 14). On a per cell basis, HSCs from older mice exhibit biased differentiation toward myeloid lineages (15, 16). This trait, together with reduced stem cell activity, may be potentially ascribed to weaker responses to SDF-1 (14, 17) and poorer engraftability of HSCs in the G1 (18) and S/G2/M phases of the cell cycle (19). These aged HSCs may arise from either the cell-intrinsic transition of balanced to myeloid-biased LT-HSCs or the clonal expansion of preexisting fractions of myeloid-biased LT-HSCs (13, 14, 16, 20, 21). Several studies support the presence of preexisting myeloid-biased LT-HSCs by demonstrating that myeloid-biased subpopulations of LT-HSCs in young, healthy mice respond to environmental challenges, such as inflammation and infection (22, 23). Results from lineage tracing with genetic barcodes (24, 25) and single cell transplants of LT-HSCs also support the notion of inherent functional diversity among LT-HSCs (26–28). However, these studies did not identify markers to prospectively isolate distinct subpopulations of HSCs. Other groups have found protein and genetic reporters, such as CD150 (20), CD41 (29), vWF (30), and CD61 (22), that enrich for self-renewing lineage-biased subpopulations of HSCs. However, these markers were shown to segregate fractions of pHSCs, which contain both short-term (transiently self-renewing), Hoxb5−, and long-term (continuously self-renewing), Hoxb5+, HSCs. Our previous study showed that Hoxb5− pHSCs are homogenously lymphoid-biased (8), but the diversity of Hoxb5+ LT-HSCs has not yet been fully explored. Therefore, we sought to interrogate the heterogeneity among purified Hoxb5+ LT-HSCs and identify a strategy to prospectively isolate these cells with phenotypic markers.
Here, we find that Neogenin-1 (NEO1), a transmembrane receptor of the Ig family (31), is expressed on a fraction of mouse Hoxb5+ LT-HSCs and decreases with differentiation. Although NEO1 has been extensively investigated as a receptor for axon guidance (32, 33), neuronal survival (34), skeletal myofiber differentiation (35), intracellular iron homeostasis (36), mammary epithelial development (37), and endothelial migration (38), its expression in the bone marrow and association with LT-HSCs has not yet been explored, apart from a few unpublished reports (39–42). We find that NEO1+Hoxb5+ LT-HSCs represent a myeloid-biased subset of LT-HSCs that responds to myeloablative stress and expands with age. Contrastingly, NEO1−Hoxb5+ LT-HSCs exhibit greater reconstitution potential, balanced lineage output, and a more quiescent cell cycle status compared to NEO1+Hoxb5+ LT-HSCs. After transplant, NEO1−Hoxb5+ LT-HSCs give rise to NEO1+Hoxb5+ LT-HSCs, but the reverse transition is rarely observed. Therefore, we propose a model of early long-term hematopoiesis in which balanced, quiescent LT-HSCs self-renew and generate long-term myeloid-biased LT-HSCs in response to stress and during the course of aging.
Results
Neogenin-1 Marks a Subpopulation of Mouse Hoxb5+ LT-HSCs and Human HSCs.
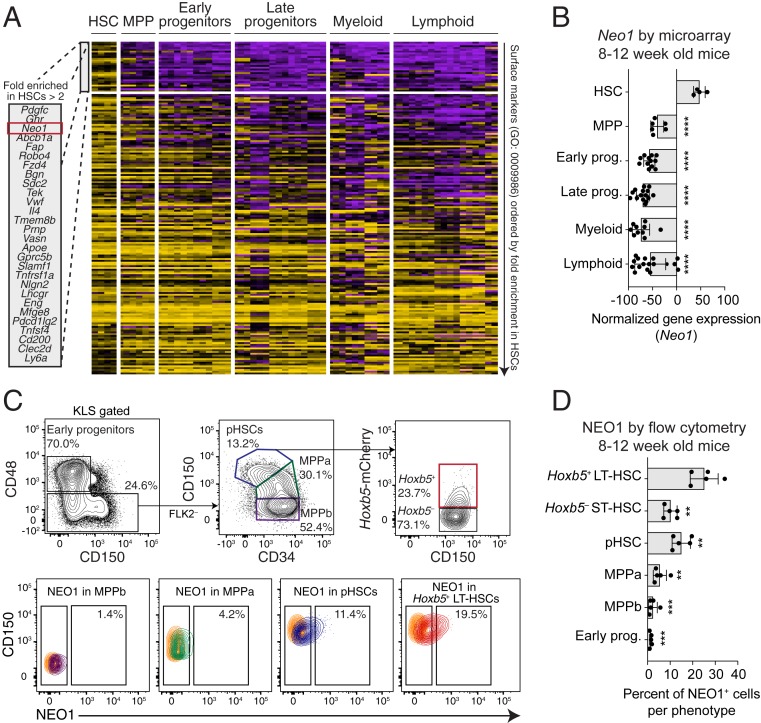
Functional heterogeneity within Hoxb5+ LT-HSCs is poorly understood. To identify surface candidates that fractionate Hoxb5+ LT-HSCs, we first pattern-searched 64 microarray expression profiles of 23 distinct mouse hematopoietic cell types (43) for 1) genes annotated to code for cell surface proteins (Gene Ontology [GO] Biological Process: 0009986) and 2) genes specifically expressed in HSCs compared to downstream cell types (Fig. 1A and Dataset S1). We found several known HSC-specific markers, including Robo4 (44), Slamf1 (5), Ly6a (2), Vwf (30), TEK (45), and a member of the Gpcr5 family (46), validating the utility of our approach. We also identified several markers of HSCs that have not been previously reported (Fig. 1A). Among the top 3 most enriched surface markers on HSCs, Neogenin-1 (Neo1) was more highly expressed on HSCs compared to the other 2 candidates (Fig. 1 A and B). Single-cell RNA sequencing (scRNA-seq) data of hematopoietic stem and progenitor cells validated the enriched expression of Neo1 in LT-HSCs compared to downstream short-term HSCs (ST-HSCs) and progenitors (SI Appendix, Fig. S1). Neo1 was also expressed on subsets of bone marrow stromal and endothelial cells and demarcated the contours of trabecular bone (SI Appendix, Fig. S2), suggesting its expression is not restricted to hematopoietic cells in the bone marrow.
Fig. 1.
Identification of Neogenin-1 (NEO1) as a unique surface marker on mouse hematopoietic stem cells (HSCs). (A and B) In silico screen to identify unique surface markers on HSCs from 64 microarray expression profiles of 23 distinct mouse hematopoietic cell types. (A) Heat map showing normalized gene expression of gene ontology-annotated (GO: 0009986) surface markers across different hematopoietic compartments; 186 surface markers expressed on HSCs and 23 phenotypes categorized as “HSC,” “MPP,” “Early progenitors,” “Late progenitors,” “Myeloid,” and “Lymphoid” are displayed (Dataset S1). Genes are ordered from top to bottom by log2 fold enrichment in HSCs compared to downstream cells, and the top most enriched genes (>2 log2 fold enrichment) are highlighted in a box. For further details, see Materials and Methods. (B) Bar plots showing normalized gene expression of Neo1 across the cell type categories shown in A. Statistical significance was calculated by unpaired, 2-tailed Student’s t test between “HSC” and each cell type. ****P < 0.0001. (C and D) Flow cytometry analysis of NEO1 surface expression in the 2-mo-old mouse bone marrow (n = 5 mice). (C) Contour plots with outliers showing the gating scheme for early progenitors, MPPb, MPPa, pHSCs, Hoxb5− ST-HSCs, and Hoxb5+ LT-HSCs (Top) and the corresponding surface expression of NEO1 for select populations (Bottom). Colors correspond to populations shown. Goat IgG isotype control for fluorescence staining with goat anti-mouse/human NEO1 antibody is highlighted in orange (Bottom). (D) Bar plots showing the percent of NEO1+ cells for each cell type gated in C. Statistical significance was calculated by a paired, 2-tailed Student’s t test between “Hoxb5+ LT-HSC” and each cell type. Bar plots in this figure indicate mean ± SD.
We next used flow cytometry to measure the relative protein levels of NEO1 on the surface of 2- to 3-mo-old early hematopoietic progenitors, multipotent progenitor subset A (MPPa), multipotent progenitor subset B (MPPb), phenotypic HSCs defined as Lin−c-KIT+SCA1+CD48−FLK2−CD150+CD34− (hereafter referred to as pHSCs), and 2 populations among pHSCs, including Hoxb5+ LT-HSCs and Hoxb5− ST-HSCs (Fig. 1C and SI Appendix, Fig. S3). Consistent with its gene expression, the relative protein levels of NEO1 and the frequency of NEO1+ cells progressively decreased with differentiation. NEO1+ cells comprised a significantly higher fraction of Hoxb5+ LT-HSCs compared to downstream cells (Fig. 1 C and D). NEO1 was also expressed on a fraction of long-term reconstituting Lin−CD34+CD38−CD45RA−CD90+ HSCs from human bone marrow (47), although NEO1 enrichment in human HSCs was diminished compared to that observed in mouse HSCs (SI Appendix, Fig. S4).
NEO1+Hoxb5+ LT-HSCs Selectively Expand with Age and Respond to Myeloablative Stress in Young Mice.
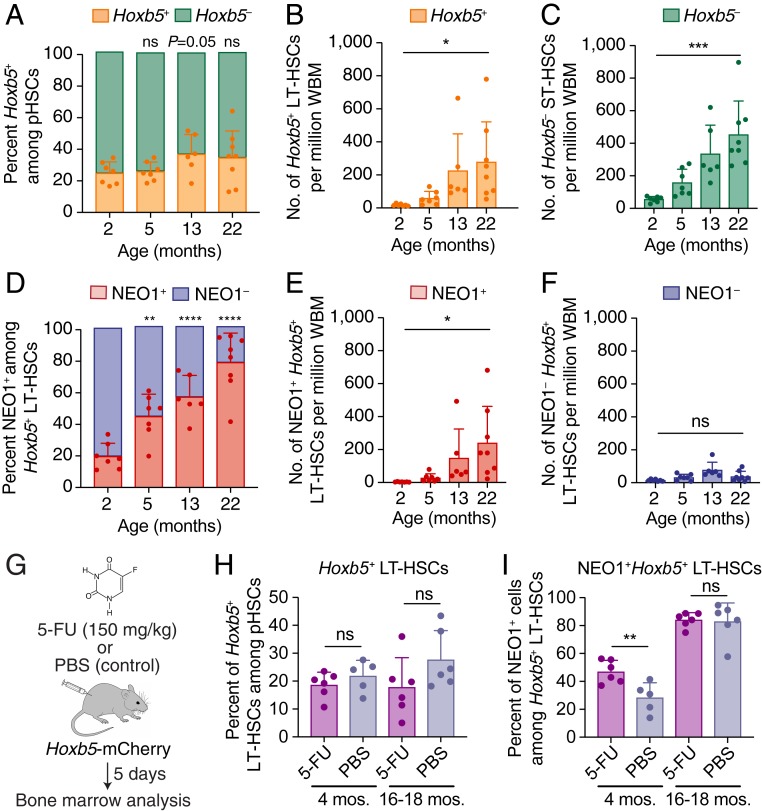
Previous studies have shown that a subpopulation of pHSCs expands with age (20, 27) and responds to environmental challenge (22, 23). However, the effect of aging and stress on Hoxb5+ LT-HSCs and their subpopulations has not yet been evaluated. To that end, we first measured the number and frequency of Hoxb5+ LT-HSCs (Fig. 2 A–C and SI Appendix, Fig. S5) and NEO1+ and NEO1− fractions (Fig. 2 D–F and SI Appendix, Fig. S5) in 2-, 5-, 13-, and 22-mo-old bone marrow. Consistent with the overall expansion of pHSCs (SI Appendix, Fig. S5 A and B), we observed that the total numbers of Hoxb5+ LT-HSCs and Hoxb5− ST-HSCs were significantly increased (Fig. 2 B and C and SI Appendix, Fig. S5 C–E). The frequency of Hoxb5+ LT-HSCs among pHSCs, although on average higher in bone marrow from older (13-mo-old and 22-mo-old) than younger (2-mo-old and 5-mo-old) mice, was highly variable in aged mice (Fig. 2A and SI Appendix, Fig. S5C).
Fig. 2.
NEO1+Hoxb5+ LT-HSCs selectively expand during aging and respond to myeloablative stress in young mice. (A–C) Frequency and number of Hoxb5+ LT-HSCs and Hoxb5− ST-HSCs at 2 (n = 7 mice), 5 (n = 7 mice), 13 (n = 6 mice), and 22 (n = 9 mice) months of age. (A) Percent of cells among pHSCs that are Hoxb5+ (orange) and Hoxb5− (green). Statistical significance was calculated using an unpaired, 2-tailed Student’s t test between 2 mo and each time point. ns, nonsignificant; P ≥ 0.05. (B) Number of Hoxb5+ LT-HSCs per million WBM cells. Statistical significance was calculated using an unpaired, 2-tailed Student’s t test between 2 mo and 22 mo of age. *P < 0.05. (C) Number of Hoxb5− ST-HSCs per million WBM cells. Statistical significance was calculated using an unpaired, 2-tailed Student’s t test between 2 mo and 22 mo of age. ***P < 0.001. (D–F) Frequency and number of NEO1+ and NEO1−Hoxb5+ LT-HSCs at 2 (n = 7 mice), 5 (n = 7 mice), 13 (n = 6 mice), and 22 (n = 9 mice) months of age. (D) Percent of cells among Hoxb5+ LT-HSCs that are NEO1+ (red) and NEO1− (blue). Statistical significance was calculated using an unpaired, 2-tailed Student’s t test between 2 mo and each time point. ***P < 0.001, ****P < 0.0001 (E) Number of NEO1+ Hoxb5+ LT-HSCs per million WBM cells. Statistical significance was calculated using an unpaired, 2-tailed Student’s t test between 2 mo and 22 mo of age. *P < 0.05. (F) Number of NEO1−Hoxb5+ LT-HSCs per million WBM cells. Statistical significance was calculated using an unpaired, 2-tailed Student’s t test between 2 mo and 22 mo of age. ns, nonsignificant; P ≥ 0.05. (G–I) Response of HSC subpopulations from 4-mo-old (n = 5 to 6 mice) and 16- to 18-mo-old (n = 6 mice) Hoxb5-mCherry mice 5 d after treatment with 5-fluoruracil (5-FU). (G) Experimental design of myeloablative stress with 5-FU or PBS control. (H) Frequency of Hoxb5+ LT-HSCs among all pHSCs after 5-FU or PBS injection at different ages. (I) Frequency of NEO1+Hoxb5+ LT-HSCs among all Hoxb5+ LT-HSCs after 5-FU or PBS injection at different ages. Statistical significance was calculated using an unpaired, 2-tailed Student’s t test. ns, nonsignificant or P ≥ 0.05; **P < 0.01. All bar plots in this figure indicate mean ± SD. mos., months.
Despite the variable expansion of Hoxb5+ LT-HSCs, the frequency of NEO1+ cells among Hoxb5+ LT-HSCs progressively increased with age in a consistent manner (Fig. 2D and SI Appendix, Fig. S5F). At all ages, NEO1 expression was significantly higher in Hoxb5+ LT-HSCs compared to Hoxb5− ST-HSCs, except at 22 mo, when NEO1 was similarly high in Hoxb5+ and Hoxb5− (SI Appendix, Fig. S5I). Fewer than 20% of Hoxb5+ LT-HSCs in 2-mo-old mice expressed surface NEO1 while more than 80% of 22-mo-old Hoxb5+ LT-HSCs were NEO1+ (Fig. 2D and SI Appendix, Fig. S5F). However, while the number of NEO1+ cells per million whole bone marrow cells increased with age (Fig. 2E and SI Appendix, Fig. S5G), the number of NEO1−Hoxb5+ LT-HSCs did not significantly change (Fig. 2F and SI Appendix, Fig. S5H). This suggests that the NEO1+ fraction selectively expands among Hoxb5+ LT-HSCs in the bone marrow while the number of NEO1−Hoxb5+ LT-HSCs remains stable with age.
We next evaluated the response of Hoxb5+ LT-HSCs and the NEO1+ and NEO1− subpopulations to myeloablative stress at different ages. The 4-mo-old and 16- to 18-mo-old mice were treated with 150 mg/kg 5-fluorouracil (5-FU), and their bone marrow was analyzed 5 d posttreatment when HSC proliferation is maximum (48) (Fig. 2G). While the frequency of Hoxb5+ and Hoxb5− cells among pHSCs did not change, irrespective of age (Fig. 2H), a significantly higher percentage of Hoxb5+ LT-HSCs were NEO1+ than NEO1− after treatment in 4-mo-old mice (Fig. 2I and SI Appendix, Fig. S6A). This selective response, however, was absent in 16- to 18-mo-old mice. As shown previously (48), the expression of c-KIT in Hoxb5+ LT-HSCs after 5-FU treatment was significantly decreased in both 4-mo-old and 16- to 18-mo-old mice (SI Appendix, Fig. S6B).
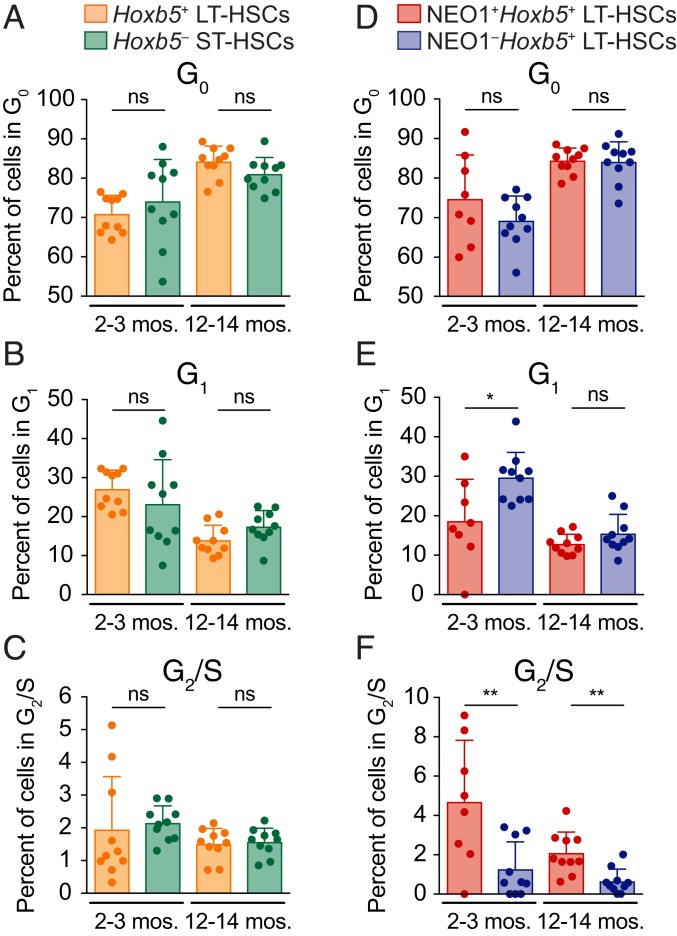
NEO1+Hoxb5+ LT-HSCs Are More Proliferative than NEO1−Hoxb5+ LT-HSCs in Young and Old Mice.
We next asked whether the difference in expansion of NEO1+ versus NEO1− Hoxb5+ LT-HSCs during aging and in response to myeloablative stress can be partially explained by differences in proliferation (49). To address this, we measured the percent of each cell population in different phases of the cell cycle, including G0, G1, and G2/S by KI-67 and DAPI staining at both 2 to 3 mo of age and 12 to 14 mo of age (Fig. 3 and SI Appendix, Fig. S7). Consistent with previous reports (50), 12- to 14-mo-old HSCs were more often in G0 compared to 2- to 3-mo-old HSCs, suggesting increased exhaustion and senescence with age in the absence of leukemia (Fig. 3 and SI Appendix, Fig. S7). In support of the limited expansion of Hoxb5+ LT-HSCs among pHSCs with age, we did not observe significant differences in G0, G1, or G2/S status between Hoxb5+ LT-HSCs and Hoxb5− ST-HSCs (Fig. 3 A–C and SI Appendix, Fig. S7). NEO1+ and NEO1− Hoxb5+ LT-HSCs also did not differ in the proportion of G0 cells in both 2- to 3-mo-old and 12- to 14-mo-old bone marrow (Fig. 3D and SI Appendix, Fig. S7)
Fig. 3.
NEO1 marks a more proliferative fraction of LT-HSCs. (A–F) Cell cycle analysis of 2- to 3-mo-old (n = 10 mice) and 12- to 14-mo-old (n = 10 mice) LT-HSC fractions with KI-67 and DAPI staining. mos., months. (A–C) Percent of Hoxb5+ LT-HSCs or Hoxb5− ST-HSCs in (A) G0, (B) G1, and (C) G2/S in 2- to 3-mo-old and 12- to 14-mo-old mouse bone marrow. Statistical significance was calculated using an unpaired, 2-tailed Student’s t test. ns, nonsignificant; P ≥ 0.05. (D–F) Percent of NEO1+ or NEO1− Hoxb5+ LT-HSCs in (D) G0, (E) G1, and (F) G2/S in 2- to 3-mo-old and 12- to 14-mo-old mouse bone marrow. Statistical significance was calculated using an unpaired, 2-tailed Student’s t test. ns, nonsignificant; P ≥ 0.05, *P < 0.05, **P < 0.01. All bar plots in this figure indicate mean ± SD.
However, NEO1+Hoxb5+ LT-HSCs were significantly more often in G2/S compared to NEO1−Hoxb5+ LT-HSCs in both young and old bone marrow (Fig. 3F and SI Appendix, Fig. S7). Moreover, in the young bone marrow, there was a significantly smaller percentage of NEO1+Hoxb5+ LT-HSCs in G1 compared to NEO1−Hoxb5+ LT-HSCs (Fig. 3E and SI Appendix, Fig. S7). Taken together, this suggests that NEO1+Hoxb5+ LT-HSCs are more proliferative, which may partially contribute to their selective expansion during aging and in response to myeloablative stress in the young.
NEO1 Marks a Less Regenerative, Myeloid-Biased Fraction of Hoxb5+ LT-HSCs.
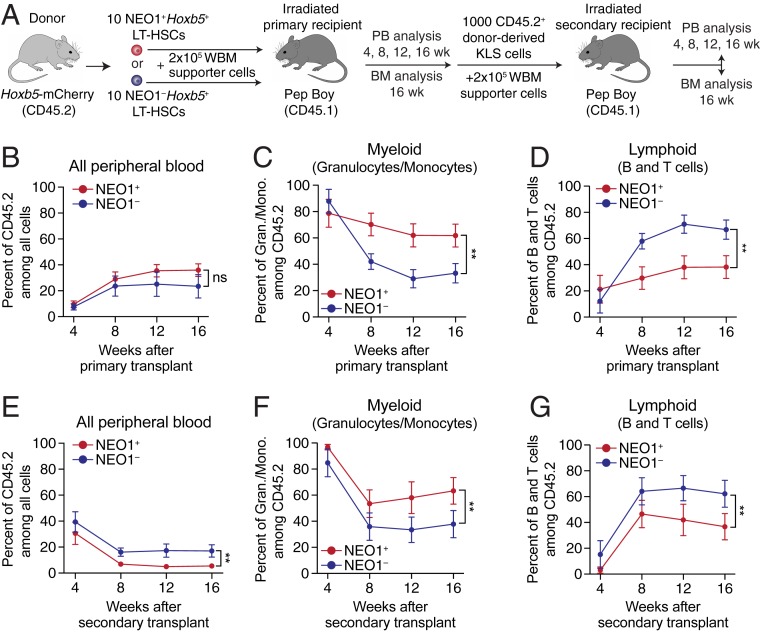
Given the aging and cell cycle differences between NEO1+ and NEO1− Hoxb5+ LT-HSCs, we next evaluated their reconstitution potential and lineage output by 10-cell transplants into congenic irradiated primary recipients (Fig. 4A). Over the course of 16 wk, the percent of total chimerism among peripheral blood that was donor-derived was similar between NEO1+ and NEO1− Hoxb5+ LT-HSC transplants (Fig. 4B). The frequency of Lin−c-KIT+SCA1+ (KLS) cells in the bone marrow 16 wk posttransplant was also comparable between the conditions (SI Appendix, Fig. S8). However, among all donor-derived peripheral blood, NEO1+Hoxb5+ LT-HSCs gave rise to a higher percentage of granulocytes and monocytes (myeloid) and a lower percentage of B and T cells (lymphoid) compared to NEO1−Hoxb5+ LT-HSCs (Fig. 4 C and D).
Fig. 4.
NEO1+Hoxb5+ LT-HSCs exhibit myeloid bias and reduced reconstitution potential upon serial transplantation. (A) Experimental design for primary and secondary transplantations of NEO1+ and NEO1− Hoxb5+ LT-HSCs. BM, bone marrow. (B–D) Measurements of reconstitution potential and lineage priming after primary transplantation (“NEO1+,” n = 14 mice; “NEO1−,” n = 11 mice) from 2 independent experiments. (B) Percent of donor-derived (CD45.2+) cells among all peripheral blood cells at 4, 8, 12, and 16 wk posttransplant. (C) Percent of myeloid cells (GR1+CD11B+ granulocytes and monocytes) among donor-derived (CD45.2+) cells at 4, 8, 12, and 16 wk posttransplant. (D) Percent of lymphoid cells (B220+ B cells and CD3+ T cells) among donor-derived (CD45.2+) cells at 4, 8, 12, and 16 wk posttransplant. (E–G) Same as in B–D but analyzing peripheral blood in secondary recipients transplanted with 1,000 donor-derived Lin−c-KIT+SCA1+ (KLS) cells from primary hosts (“NEO1+,” n = 8; “NEO1−,” n = 9) from 2 independent experiments. Statistical significance for B–G was calculated using 2-way ANOVA with time posttransplant and NEO1 status as factors. **P < 0.01; ns, nonsignificant. All line plots in this figure indicate mean ± SEM.
To evaluate the long-term reconstitution potential and stability of lineage bias, we serially transplanted 1,000 donor-derived KLS cells from primary recipients into congenic irradiated secondary hosts (Fig. 4A). Although KLS cells from both donors repopulated all major lineages in secondary hosts, NEO1−Hoxb5+-derived cells exhibited significantly higher reconstitution compared to NEO1+Hoxb5+-derived cells (Fig. 4E). Moreover, as during primary transplant, NEO1+Hoxb5+-derived cells maintained significant bias toward granulocytes and monocytes and away from B and T cells compared to NEO1−Hoxb5+-derived cells. (Fig. 4 F and G). This suggests that both myeloid-biased and balanced phenotypes are long-term maintained after secondary transplantation.
Transcriptional Programs Recapitulate Functional Differences between NEO1+ and NEO1−Hoxb5+ LT-HSCs.
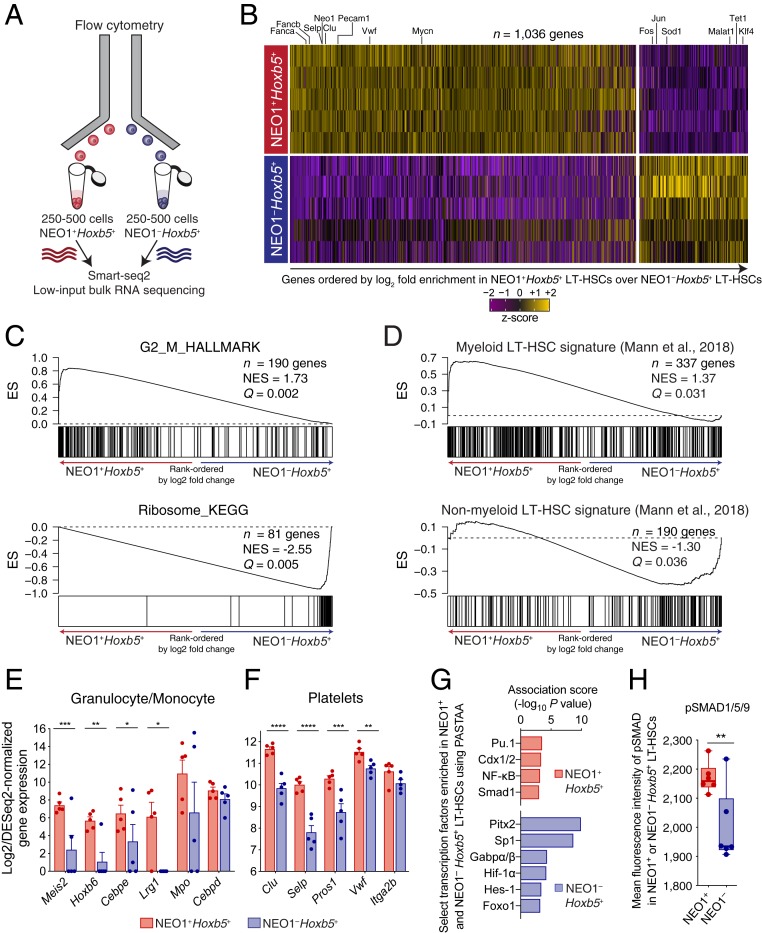
We next sought to understand the transcriptional programs that drive the observed functional differences between NEO1+ and NEO1− Hoxb5+ LT-HSCs. We isolated 250 to 500 NEO1+Hoxb5+ and NEO1−Hoxb5+ cells from female, 8- to 12-wk-old Hoxb5-mCherry mice and performed low-input full-length RNA sequencing using the Smart-seq2 protocol (51, 52) (Fig. 5A). Paired gene expression comparison of the 2 populations identified 1,036 differentially expressed genes (false discovery rate [FDR] P-adjusted < 0.1) (Fig. 5B, SI Appendix, Fig. S9A, and Dataset S2) (53). Genes implicated in activation, cell cycle, and differentiation, such as Fanca, Fancb, and Mycn (54, 55), were enriched in NEO1+Hoxb5+ LT-HSCs while genes involved in anti-redox (e.g., Sod1) (56) and regulation of stem cell potency (e.g., Klf4 and Malat1) (57, 58) were enriched in NEO1−Hoxb5+ LT-HSCs (Fig. 5B). Gene set enrichment analysis (GSEA) (59) and a hypergeometric test with gene ontology (GO) biological processes (60, 61) revealed that the driving differences (FDR < 0.05, P value < 0.05) between NEO1+ and NEO1− cells were cell cycle and ribosomal RNA expression (Fig. 5C and SI Appendix, Fig. S9B).
Fig. 5.
Distinct molecular signatures of NEO1+ and NEO1− Hoxb5+ LT-HSCs. (A) Experimental design for bulk RNA sequencing of NEO1+ and NEO1− Hoxb5+ LT-HSCs. (B) Heat map of differentially expressed genes (n = 1,036 genes; FDR < 0.1) after pairwise comparison of NEO1+ (n = 5 samples) and NEO1− (n = 5 samples) Hoxb5+ LT-HSC transcriptomes using DESeq2. Select genes are highlighted. Genes are ordered from left to right by log2 fold enrichment in NEO1+ over NEO1− Hoxb5+ LT-HSCs. (C and D) Gene set enrichment analysis (GSEA) plots of molecular signatures significantly enriched (Q value < 0.05) over a gene list ordered by log2 fold change, including (C) “G2_M_HALLMARK” (Top), “Ribosome_KEGG” (Bottom), (D) Myeloid LT-HSC signature (Top), and Non-myeloid LT-HSC signature (Bottom) from Mann et al. (22). ES, enrichment score; NES, normalized enrichment score. (E and F) Bar plots showing log2 and DESeq2-normalized gene expression for select genes associated with (E) granulocyte or monocyte and (F) platelet programs. Statistical significance was calculated using a paired, 2-tailed Student’s t test adjusted for multiple hypothesis testing with Benjamini–Hochberg procedure. *P-adjusted < 0.05, **P-adjusted < 0.01, ***P-adjusted < 0.001, ****P-adjusted < 0.0001. Bars and error bars indicate mean ± SEM. (G) Transcription factor analysis of NEO1+ and NEO1− Hoxb5+ LT-HSCs with PASTAA. Bar plots showing select transcription factors (TFs) significantly associated with NEO1+Hoxb5+ LT-HSCs (red) and NEO1−Hoxb5+ LT-HSCs (blue). Bars indicate the association score, or the −log of the most significant hypergeometric P value, as calculated by PASTAA. An extended list of all significant TFs and all TFs identified by PASTAA can be found in SI Appendix, Fig. S11 and Dataset S3, respectively. (H) Box plot showing mean fluorescent intensity of pSMAD1/5/9 in NEO1+ and NEO1− Hoxb5+ LT-HSCs by intracellular FACS. Statistical significance was calculated using a paired, 2-tailed Student’s t test. **P < 0.01.
We next searched for the expression of lineage-specific transcripts that may indicate signs of early myeloid and lymphoid priming in LT-HSCs. Among the genes significantly enriched in NEO1+ compared to NEO1− Hoxb5+ LT-HSCs, we found several myeloid genes, such as Lrg1, lineage-related transcription factors, such as Meis2, Hoxb6, and Cebpe (Fig. 5E), and platelet genes, such as Vwf, Clu, and Selp (Fig. 5F). We also found that NEO1+ LT-HSCs were significantly enriched (Q < 0.05) for previously reported gene signatures of megakaryocyte progenitors (MkPs) and preerythrocyte colony-forming units (preCFU-E) (SI Appendix, Fig. S9C) (30). Moreover, the gene expression signature of NEO1+Hoxb5+ LT-HSCs significantly aligned with expression profiles of previously reported myeloid-biased LT-HSCs (Fig. 5 D, Top) while NEO1− LT-HSCs were enriched for the balanced LT-HSC signature (Fig. 5 D, Bottom). Altogether, these data suggest that LT-HSCs may sample regions of the transcriptome associated with their lineage fate decisions.
We also compared NEO1+ and NEO1− Hoxb5+ LT-HSC gene expression with respect to known stemness-associated genes. Overall, NEO1+Hoxb5+ LT-HSCs had higher expression of known stem-related genes, including Ctnnal1 (9), Fgd5 (7), Bmi1 (62), Gprc5c (46), and Slamf1 (CD150) (5) (SI Appendix, Fig. S9D), the latter of which was confirmed by flow cytometry (SI Appendix, Fig. S10). This suggests that, although previously identified stemness genes enrich for a self-renewing phenotype, their expression may also be associated with myeloid bias.
Finally, we searched for transcription factors (TFs) associated with NEO1+ and NEO1− Hoxb5+ LT-HSCs using predicting associated transcription factors from annotated affinities (PASTAA). Among the 38 TFs significantly enriched in NEO1+Hoxb5+ LT-HSCs, we found TFs associated with leukemogenesis, such as CDX1/2 (63, 64), aging and inflammation, such as NF-κB (65), and regulators of myeloid differentiation, such as PU.1 and SMAD1 (66, 67) (Fig. 5G, SI Appendix, Fig. S11, and Dataset S3). Several gene sets of BMP, TGF-β, and SMAD signaling were enriched in NEO1+, and this was also confirmed by intracellular fluorescence-activated cell sorting (FACS) for pSMAD1/5/9 (Fig. 5H and SI Appendix, Fig. S11B). On the other hand, NEO1−Hoxb5+ LT-HSCs were enriched in TFs associated with maintenance of HSCs, such as GABPα/β (68), primitive hematopoiesis, such as PITX2 (69), resistance to oxidative stress, such as FOXO1 (70), and regulators of quiescence, such as HES1 and HIF1α (71, 72) (Fig. 5G, SI Appendix, Fig. S11A, and Dataset S3). Interestingly, SP1, a TF upstream of CDX genes (63, 73)—the TFs enriched in NEO1+Hoxb5+ LT-HSCs—was found to be uniquely associated with NEO1−Hoxb5+ LT-HSCs (Fig. 5G, SI Appendix, Fig. S11A, and Dataset S3). Notably, these TFs were mutually exclusive between NEO1+ and NEO1− Hoxb5+ LT-HSCs.
Lineage-Balanced NEO1−Hoxb5+ LT-HSCs Outcompete NEO1+Hoxb5+ LT-HSCs in Reconstitution and Reside at the Apex of Hematopoiesis.
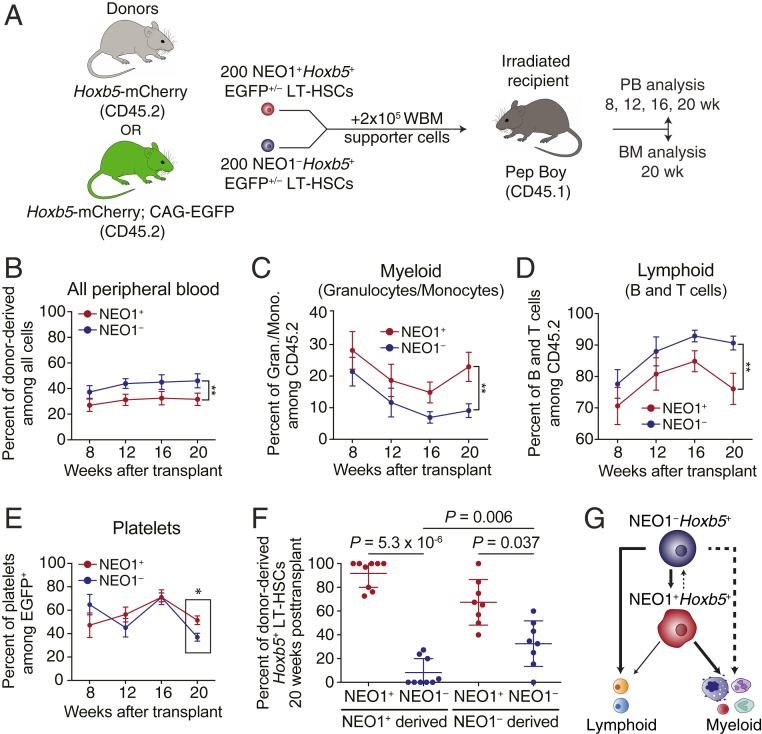
To directly compare the relative fitness of NEO1+ and NEO1− Hoxb5+ LT-HSCs, we cotransplanted 200 cells from each fraction with host supporter cells into irradiated 2- to 3-mo-old congenic recipients (Fig. 6A). Donor origin was distinguished by CD45.2 and EGFP expression using Hoxb5-mCherry and CAG-EGFP;Hoxb5-mCherry mice (Fig. 6A). In order to eliminate any strain-specific biases, NEO1+ and NEO1− Hoxb5+ LT-HSCs were isolated from both strains and reciprocally transplanted into recipient mice (Fig. 6A). Unlike the primary transplants above, NEO1−Hoxb5+ LT-HSCs exhibited significantly higher reconstitution potential compared to NEO1+Hoxb5+ LT-HSCs in the cotransplantation setting (Fig. 6B). This suggests that NEO1−Hoxb5+ LT-HSCs are more fit to reconstitute young recipients compared to NEO1+Hoxb5+ LT-HSCs.
Fig. 6.
NEO1−Hoxb5+ LT-HSCs outcompete NEO1+Hoxb5+ LT-HSCs in reconstitution potential and reside at the apex of the hematopoietic hierarchy. (A) Experimental design for cotransplantation of NEO1+ and NEO1− Hoxb5+ LT-HSCs (n = 12 mice). BM, bone marrow. (B–D) Measurements of reconstitution potential and lineage priming after cotransplantation. (B) Percent of donor-derived cells among all peripheral blood cells at 8, 12, 16, and 20 wk posttransplant. (C) Percent of myeloid cells (GR1+CD11B+ granulocytes and monocytes) among donor-derived cells at 8, 12, 16, and 20 wk posttransplant. (D) Percent of lymphoid cells (B220+ B cells and CD3+ T cells) among donor-derived cells at 8, 12, 16, and 20 wk posttransplant. Statistical significance for B–D was calculated using 2-way ANOVA with time posttransplant and NEO1 status as factors. **P < 0.01. (E) Percent of platelets (EGFP+CD41+) among donor-derived cells at 8, 12, 16, and 20 wk posttransplant. Statistical significance at 20 wk posttransplant was calculated using an unpaired, 2-tailed Student’s t test. *P < 0.05. (F) Percent of NEO1+ and NEO1− Hoxb5+ LT-HSCs derived from donor NEO1+ and NEO1− Hoxb5+ LT-HSCs in the mouse bone marrow 20 wk posttransplant. Only samples for which Hoxb5+ LT-HSCs were present are shown (“NEO1+ derived,” n = 9; “NEO1− derived,” n = 8). Statistical significance was calculated using a paired, 2-tailed Student’s t test between the percent of NEO1+ and NEO1− Hoxb5+ LT-HSCs derived from the same donor and an unpaired, 2-tailed Student’s t test between the percent of NEO1−Hoxb5+ LT-HSCs between NEO1+ and NEO1− donors. P values are indicated on the graph. (G). Schema depicting a revised model of long-term hematopoiesis with a lineage-balanced, quiescent NEO1−Hoxb5+ LT-HSC residing above a downstream myeloid-biased NEO1+Hoxb5+ LT-HSC. The thickness of the lines indicates the degree of contribution, and dashed lines mark putative differentiation paths. All line plots in this figure indicate mean ± SEM. Scatter dot plot in F indicates mean ± SD.
Cotransplantation also confirmed that NEO1+Hoxb5+ LT-HSCs contribute significantly more to granulocytes and monocytes (myeloid) and less to B and T cells (lymphoid) compared to NEO1−Hoxb5+ LT-HSCs (Fig. 6 C and D). We also quantified platelet fractions among EGFP+ donors. Relative platelet contribution from NEO1+Hoxb5+ was not significantly different from NEO1−Hoxb5+ donors during the first 16 wk posttransplant but significantly increased among NEO1+Hoxb5+-derived peripheral blood (PB) at 20 wk posttransplant (Fig. 6E).
Finally, we also measured the composition of LT-HSCs between NEO1+ and NEO1−-derived bone marrow in the cotransplantation setting. Both NEO1+ and NEO1− Hoxb5+ cells produced on average equal numbers of Hoxb5+ LT-HSCs per million bone marrow cells (SI Appendix, Fig. S12) and, among Hoxb5+ LT-HSCs, more NEO1+ than NEO1− Hoxb5+ cells (Fig. 6F). However, NEO1+Hoxb5+ cells produced significantly fewer NEO1−Hoxb5+ cells compared to NEO1−Hoxb5+ cells (P = 0.006). This suggests limited transition from NEO1+ to NEO1− while NEO1−Hoxb5+ cells are capable of giving rise to high percentages of both populations (Fig. 6F). Therefore, NEO1−Hoxb5+ LT-HSCs likely precede NEO1+Hoxb5+ LT-HSCs in the differentiation hierarchy (Fig. 6G).
Discussion
Previously, we showed that phenotypic HSCs (pHSCs) are variable in their reconstitution potential and that Hoxb5 expression distinguishes long-term from short-term repopulating HSCs (8). While Hoxb5− pHSCs were unable to repopulate secondary recipients and were homogeneously lymphoid-biased, Hoxb5+ pHSCs serially reconstituted recipients and exhibited variable contribution to hematopoietic lineages. Therefore, we sought to understand the diversity of self-renewing HSCs in the mouse bone marrow using Hoxb5 as a reporter to mark long-term HSCs (LT-HSCs).
To accomplish this, we screened gene expression profiles for candidate surface markers that are strictly enriched in HSCs and stratify Hoxb5+ LT-HSCs into subpopulations. We identified Neogenin-1 (Neo1; NEO1) as a transmembrane receptor specifically expressed on a subfraction of Hoxb5+ LT-HSCs. We found that, while both NEO1+ and NEO1− Hoxb5+ LT-HSCs are long-term repopulating and multilineage-contributing LT-HSCs, NEO1+Hoxb5+ LT-HSCs are more myeloid-biased and less productive in secondary transplants compared to NEO1−Hoxb5+ LT-HSCs. We show that NEO1+ cells comprise a minor fraction of Hoxb5+ LT-HSCs in young mice that progressively expands with age and represents >80% of 22-mo-old Hoxb5+ LT-HSCs. This expansion can be partially explained by the higher frequency of NEO1+Hoxb5+ in the G2/S cell cycle phase compared to NEO1−Hoxb5+ LT-HSCs. We also observed increased response of NEO1+Hoxb5+ LT-HSCs to myeloablative stress, although only in young mice. Given that several candidate ligands of NEO1 (e.g., BMPs, Netrin-1, Rgma, Rgmb, Rgmc, and Cdon) (31, 74) are also expressed by mesenchymal and endothelial cells in the bone marrow, we speculate that both aging and response to myeloablative stress in young may be regulated by the relative availability of these key factors. Relatedly, the absence of NEO1 response in aged may be due to various cell-intrinsic or niche-related changes with age (13–16) that diminish the differentiation potential of NEO1− to NEO1+ or expansion of NEO1+ cells.
Although the function of NEO1 in LT-HSCs remains elusive, our analysis of lineage-specific transcripts and TFs provides some important clues to the molecular drivers of these different cell states. Firstly, from gene expression analysis (Fig. 5 and SI Appendix, Figs. S9 and S11), we find that the NEO1+Hoxb5+ LT-HSCs are associated with myeloid-related TFs (e.g., PU.1, SMAD1, and NF-κB) (65–67) and several lineage-specific transcripts (e.g., Meis2, Hoxb6, and Cebp). These lineage-related programs are reduced or absent in the transcriptomes of NEO1−Hoxb5+ LT-HSCs. This suggests that, while NEO1−Hoxb5+ LT-HSCs remain undecided, downstream NEO1+Hoxb5+ LT-HSCs start exploring myeloid fate decisions by expressing lineage-related TFs and transcripts. On the other hand, many of the TFs found to be associated with NEO1−Hoxb5+ LT-HSCs (e.g., PITX2, FOXO1, GABPα/β, HES1, and HIF1α) (68–72) are involved in early development, antioxidation, quiescence, self-renewal, or maintenance of HSCs. This is in line with a model in which NEO1−Hoxb5+ LT-HSCs precede NEO1+Hoxb5+ LT-HSCs. Interestingly, we found that the NEO1−Hoxb5+ LT-HSCs are associated with SP1, an early TF that targets and activates CDX genes (63, 73)—the same CDX genes that are associated with NEO1+Hoxb5+ LT-HSCs in our PASTAA analysis. Therefore, one may hypothesize that SP1 activation of CDX genes may be involved in directing the differentiation of NEO1− to NEO1+ Hoxb5+ LT-HSCs. Given Neogenin-1 is expressed in various cell types in the niche and multiple tissues of the body, cell type-specific and conditional knockout of NEO1 in HSCs will be crucial to determine the function of this protein in hematopoiesis.
Several previous studies have used single cell transplants to describe HSC heterogeneity and the existence of different lineage-primed states. Single cell HSC transplants by Dykstra and colleagues described 2 fractions of long-term self-renewing HSCs, “α cells” and “β cells,” which were myeloid-biased and lineage-balanced, respectively (26). Yamamoto and colleagues also have demonstrated the presence of myeloid-restricted progenitors with long-term repopulating activity (MyRPs) that are derived directly from balanced HSCs and expand with age (27, 28). While single HSC transplants evidenced the presence of balanced and myeloid-biased LT-HSCs, these studies did not identify surface or transcriptional markers to distinguish these populations. We have demonstrated a strategy for the prospective isolation of balanced and myeloid-biased LT-HSCs using Neogenin-1 and Hoxb5. The functional potential of NEO1+ and NEO1− Hoxb5+ LT-HSCs matches the characteristics of the myeloid-biased and balanced LT-HSC fractions previously predicted by single cell transplants (26–28).
Other markers and approaches have also been proposed to enrich for myeloid-biased HSCs. For example, our group and others previously showed that high CD150 surface expression enriches for myeloid-biased HSCs (20, 75). KLS cells with the capacity to rapidly efflux Hoechst dye (i.e., lower side-population, or lower-SPKLS cells) also exhibit biased differentiation to myeloid lineages (11, 12). However, the association of CD150 and rapid dye efflux capacity with myeloid bias was not evaluated in highly purified long-term repopulating cells. In fact, these previous studies found that the CD150high compartment, although uniformly self-renewing, contains various distinct HSC cell types with unique lineage potentials. We found that, among CD150high cells, NEO1+Hoxb5+ LT-HSCs are akin to the previously described “myeloid-predominant” long-term reconstituting cells (LTRCs) while NEO1−Hoxb5+ LT-HSCs resemble the “balanced” LTRCs (75). Consistently, we found that NEO1+Hoxb5+ LT-HSCs indeed express higher CD150 (SI Appendix, Fig. S10), validating our initial attempts to prospectively isolate myeloid-biased HSCs (20). Additional work is needed to evaluate whether differences in dye efflux capacity can also distinguish NEO1+ and NEO1− Hoxb5+ LT-HSCs.
CD41 has also been suggested to mark myelo-erythroid HSCs (29) although it likely separates different fractions. CD41− HSCs are lymphoid-biased and proliferative while NEO1−Hoxb5+ LT-HSCs are balanced and quiescent. The vWF reporter mouse is another system used to isolate platelet-biased and myeloid-biased HSCs (30). However, like the CD41− HSCs, vWF− HSCs are lymphoid-biased HSCs that are phenotypically distinct from the balanced, quiescent NEO1−Hoxb5+ cells identified in this study. Finally, CD61 was recently described as a surface marker on myeloid-biased LT-HSCs that respond to inflammatory stress and expand with age (22). The CD61+ and CD61− LT-HSCs are transcriptionally similar to NEO1+ and NEO1− Hoxb5+ LT-HSCs, suggesting that these markers may capture similar cell types (Fig. 5D). Leveraging combinations of both surface markers will likely improve the purification of balanced LT-HSCs from lineage-biased LT-HSCs.
Our results bring to question the hierarchical relationship between lineage-primed and balanced LT-HSCs. Previous transplantation studies suggest some degree of plasticity between LT-HSC fractions with different differentiation potentials (11, 26). Our cotransplantation experiment indicates that NEO1−Hoxb5+ LT-HSCs are likely upstream of NEO1+Hoxb5+ LT-HSCs, as NEO1−Hoxb5+ LT-HSCs produced NEO1+Hoxb5+ LT-HSCs, while conversion of NEO1+Hoxb5+ LT-HSCs to NEO1−Hoxb5+ HSCs was rare. The few instances of NEO1+Hoxb5+ donors producing NEO1−Hoxb5+ LT-HSCs may be attributable to sorting impurity among the 200 cells that were injected. This is also consistent with our gene expression and cell cycle analysis demonstrating that NEO1+Hoxb5+ LT-HSCs are more often cycling compared to NEO1−Hoxb5+ cells (76). Moreover, NEO1−Hoxb5+ LT-HSCs contributed more to total hematopoiesis in secondary transplants and cotransplants compared to NEO1+Hoxb5+ LT-HSCs. Therefore, our data suggest that balanced quiescent LT-HSCs precede myeloid-biased LT-HSCs during differentiation, corroborating a recent study showing that myeloid-biased MyRPs are derived from balanced HSCs (27, 28). These results are also in agreement with the hierarchical model proposed with CD150 expression (20, 75). Both NEO1+ and NEO1− Hoxb5+ LTHSCs are contained within CD150high cells that give rise to downstream CD150med/low cells. However, initial studies did not evaluate the heterogeneity within CD150high cells. We find that, among CD150high cells, NEO1 expression is highly correlated with CD150 expression (SI Appendix, Fig. S10). These findings contrast with the view that vWF+ and CD41+ platelet/myeloid-biased cells reside at the apex of the hematopoietic hierarchy (29, 30). Moreover, our transplantation data support a hybrid between the previously proposed “clonal succession” and “clonal diversity” models (11), in that there likely exist stable lineage-biased LT-HSCs that succeed highly potent, lineage-balanced LT-HSCs.
Additional work will also be required to not only verify the hierarchical relationship between NEO1−Hoxb5+ and NEO1+Hoxb5+ LT-HSCs but also to delineate the differentiation path those distinct LT-HSC fractions follow to generate various blood cells. Our studies suggest that NEO1−Hoxb5+ LT-HSCs likely precede NEO1+Hoxb5+ LT-HSCs hierarchically (Fig. 6G); nevertheless, experiments like a single cell transplant are necessary to confirm that notion. Also, we suspect that balanced NEO1−Hoxb5+ LT-HSCs contribute to myeloid lineage through a NEO1+Hoxb5+ LT-HSC intermediate. However, it is unclear whether NEO1−Hoxb5+ LT-HSCs require the NEO1+Hoxb5+ intermediate state or can independently produce lineage progenitors through alternative routes. Moreover, it remains to be answered whether all NEO1+Hoxb5+ LT-HSCs are derived from NEO1−Hoxb5+ LT-HSCs. To fully reveal the hierarchical order and sequence of differentiation events, single cell tracking experiments will be required. Critically, our experimental results are based on the behavior of cells upon transplantation into young, irradiated mice. A recent study using individually barcoded HSCs showed that lineage biases are more pronounced after transplantation into lethally irradiated mice compared to unirradiated or anti-c-KIT–depleted syngeneic mice (24). This suggests that posttransplant lineage bias may be either due to plasticity in lineage output or the selective engraftment of preexisting HSC subsets. Therefore, it will be important to evaluate the potential and hierarchical relationship between NEO1+ and NEO1− Hoxb5+ LT-HSCs during in situ, unperturbed hematopoiesis with in vivo lineage tracing. This will likely require the identification of a surrogate positive marker for NEO1−Hoxb5+ LT-HSCs or a dual reporter system that labels both Hoxb5 and Neo1 expressing cells.
Furthermore, we note that comparing NEO1+ and NEO1− fractions within pHSCs without gating Hoxb5+ LT-HSCs may mislead the significance of NEO1 to separate myeloid-biased from balanced cells. The vast majority of NEO1− cells among pHSCs are short-term, lymphoid-biased Hoxb5− ST-HSCs (8) that far outnumber the long-term, balanced NEO1−Hoxb5+ LT-HSCs we find in this study. NEO1 is also rarely expressed in Hoxb5− downstream cells and bone marrow niche cells, and its functional role in these populations has not been evaluated in this study.
The functional differences between NEO1+ and NEO1− Hoxb5+ LT-HSCs may be influenced by intrinsic programs, external cues, or both. As NEO1 is a receptor to many known ligands, ongoing studies are evaluating the role of the bone marrow niche and particular ligands to NEO1 in influencing lineage bias and stem cell maintenance.
Finally, our antibody against NEO1 also marked subpopulations of human HSCs, MPPs, and LMPPs, suggesting that NEO1 may also associate with functionally distinct cell types in human HSC biology. Further evaluation of NEO1 function in mouse and human bone marrow cells may offer insights into evolutionarily conserved mechanisms of lineage bias during long-term hematopoiesis.
Taken together, we have identified a marker on the surface of Hoxb5+ LT-HSCs, Neogenin-1, that enables the separation of myeloid-biased LT-HSCs from quiescent, balanced LT-HSCs with the highest long-term repopulation potential. Our findings reveal a previously undescribed layer of functional heterogeneity among strictly defined functional LT-HSCs and enable the precise and prospective study of LT-HSCs and their fractions.
Materials and Methods
A full description of experimental materials and methods is provided in SI Appendix, Supplementary Materials and Methods.
Mice.
The 2- to 3-mo-old female Hoxb5-mCherry mice (MGI:5911679; available through RIKEN BioResource Research Center: RBRC09733) were used as donors (CD45.2) for transplant experiments, bulk RNA sequencing, and intracellular FACS analysis of pSMAD. Additionally, 2- to 3-mo-old CAG-EGFP;Hoxb5-mCherry mice (in-house colony) were used for cotransplant assays. The 2- to 3-mo-old female B6.SJL-Ptprca Pepcb/BoyJ mice (The Jackson Laboratory) were used as recipients (CD45.1) for transplant experiments and for supporter bone marrow. The 4-mo-old and 16- to 18-mo-old female Hoxb5-mCherry mice were used for experiments with 5-fluorouracil. Five-, 13-, and 22-mo-old female Hoxb5-mCherry mice were used for aging analysis. The 2- to 3-mo-old and 12- to 14-mo-old female Hoxb5-mCherry mice were used for cell cycle analysis. The 2- to 3-mo-old C57BL/6J female mice (The Jackson Laboratory) were used for fluorescent-minus-one (FMO) controls for Hoxb5-mCherry expression.
Gene Expression Profiles of Mouse Hematopoietic Cells.
All microarray data used in this study are accessible through the Gene Expression Commons platform (https://gexc.riken.jp) and the Gene Expression Omnibus (GEO) accession number, GSE34723. We analyzed 64 microarray gene expression profiles (GEPs) of 23 distinct mouse hematopoietic cell types (Dataset S1) for surface markers enriched in HSCs compared to downstream progeny. GEPs were normalized against a large common reference of >11,939 Affymetrix Mouse Genome 430 2.0 microarrays as described before (43). For each gene, the probeset with the largest dynamic range was selected and transformed to percentile ranks (range: –100% to +100%) based on its relative expression to the reference. Genes were further subset based on 2 main criteria: 1) positive percentile expression in HSCs and 2) annotation as a cell surface protein based on GO:0009986, leaving 186 gene candidates. Fold-change enrichment in HSCs was calculated as the average percentile rank for each gene among HSCs divided by the average percentile rank for that gene across all other cells.
Mouse Hematopoietic Stem Cell Isolation by Flow Cytometry.
For bone marrow isolation, tibia, femur, and pelvis were dissected, crushed with mortar and pestle in FACS buffer (2% fetal bovine serum [FBS] in phosphate-buffered saline [PBS] with 100 U/mL DNase), and the supernatant was collected. For whole bone marrow (WBM) isolation, red blood cells were depleted with ACK (ammonium-chloride-potassium) lysis buffer by incubating at room temperature for 10 min and Fc-blocked by incubating with rat IgG (LifeSpan BioSciences) for 10 min. For c-KIT+ cell isolation, samples were Fc-blocked with rat IgG for 10 min, incubated in c-KIT magnetic beads (Miltenyi) with 100 U/mL DNase, and MACS-isolated using LS magnetic columns (Miltenyi) as per the manufacturer’s protocol.
Samples for mouse HSC isolation were stained with a mixture of antibodies against lineage markers: i.e., CD3, Gr-1, CD11B, B220, and TER119 (AF700), c-KIT (APC-Cy7), SCA-1 (PE-Cy7), CD48 (BV711), FLK2 (PerCP-Cy5.5), CD150 (BV421 or APC), CD34 (primary: biotin; secondary: Strep-BUV737), and NEO1 (primary: goat anti-mouse/human [cat. no. AF1079; R&D]; secondary: donkey anti-goat IgG [H+L] cross-absorbed AF488-conjugated; negative control: normal goat IgG). Although the antibody to NEO1 is polyclonal, results were consistent across multiple reagent lots and experiments. Antibody-labeled cells were shown to express higher NEO1 mRNA compared to unlabeled cells (SI Appendix, Fig. S9A). Primary and secondary antibody incubations were 20 to 30 min each with a 5-min wash step in between. Catalog number, concentrations, and clone information are provided in Dataset S4.
Flow cytometry and cell sorting were performed on the BD FACSAria and BD LSRFortessa. Gating strategy for the different populations is shown in SI Appendix, Fig. S3. Either 7-AAD or DAPI was used as a viability dye for dead cell exclusion, depending on the assay. All cells were suspended in FACS buffer (2% FBS in PBS) on ice unless otherwise indicated.
Methods for cell cycle and pSMAD analysis are described in detail in SI Appendix, Supplementary Materials and Methods.
Myeloablative Stress with 5-Fluorouracil.
The 4-mo-old and 16- to 18-mo-old female Hoxb5-mCherry mice were injected once with 150 mg of 5-FU (Sigma-Aldrich) per kg body weight from a stock solution of 10 mg/mL in PBS (48). Bone marrow populations were isolated and analyzed 5 d after treatment as described above. Notably, given the up-regulation of CD11B in HSCs posttreatment with 5-FU (48), the antibody to CD11B was omitted from the lineage staining panel.
Transplantation Assays.
The 2- to 3-mo-old female B6.SJL-Ptprca Pepcb/BoyJ (CD45.1) recipient mice were lethally irradiated at a single dose of 9 Gy. For reconstitution assays, 10 NEO1+Hoxb5+ or NEO1−Hoxb5+ LT-HSCs were isolated from donor CD45.2+ Hoxb5-mCherry mice (MGI:5911679) as described in Mouse Hematopoietic Stem Cell Isolation by Flow Cytometry and coinjected with 2 × 105 recipient whole bone marrow cells in 200 μL of PBS with 2% FBS into the retroorbital venous plexus. For secondary transplants, 1,000 CD45.2+ Lin−cKIT+SCA1+ (KLS) cells were isolated by flow cytometry and transplanted together with 2 × 105 recipient (CD45.1) whole bone marrow cells into lethally irradiated recipient CD45.1+ mice as described above. For cotransplantation assays, 200 NEO1+ and NEO1− Hoxb5+ LT-HSCs were isolated from either CD45.2+ Hoxb5-mCherry mice (MGI:5911679) or an in-house strain of CD45.2+ CAG-EGFP;Hoxb5-mCherry mice and transplanted into lethally irradiated recipient CD45.1+ mice at a split dose of 9 Gy with a 4-h interval. Any strain-specific bias was controlled for by transplanting the same condition from both strains (e.g., in one experiment, NEO1+ Hoxb5+ LT-HSCs were isolated from Hoxb5-mCherry mice and in another from CAG-EGFP;Hoxb5-mCherry mice). In all cases, recipients with lower than 1% total chimerism were considered failed transplantations and excluded from analysis.
Peripheral Blood Analysis for Chimerism.
Peripheral blood collections for assessment of donor chimerism were performed at 4, 8, 12, and 16 wk after primary and secondary transplantations and 8, 12, 16, and 20 wk after cotransplantations. At each time point, 50 to 100 μL of blood was collected from the retroorbital venous plexus using heparinized capillary tubes (Fisher Scientific) and added to K2/EDTA-coated MiniCollect tubes (Greiner Bio-One). Red blood cells were depleted with 2 rounds of ACK lysis buffer by incubating at room temperature for 5 min each. Cells were then washed with cold PBS. Cells were Fc-blocked with rat IgG (LifeSpan BioSciences) and stained with 5 μg/mL rat anti-mouse antibodies (catalog nos., concentrations, and clones are provided in Dataset S4) to CD45.1, CD45.2, CD11B, GR1, B220, CD3, and, only in the cotransplantation assay, CD41. Then 7-aminoactinomycin D (7-AAD; BD Bioscience) was added for live and dead cell discrimination.
Additional information on how chimerism was calculated for reconstitution and cotransplantation experiments is provided in SI Appendix, Supplementary Materials and Methods.
RNA Sequencing.
For RNA sequencing experiments, 250 to 500 cells from 2 pooled mice per sample were sorted directly into 100 μL of lysis buffer (Buffer RL), and RNA was isolated with the Single Cell RNA Purification Kit (Norgen Biotek Corp.) according to the manufacturer’s protocol. RNA quality was measured by capillary electrophoresis using the Agilent 2100 Bioanalyzer with Nano mRNA assay at the Stanford Protein and Nucleic Acid (PAN) Facility.
Libraries were prepared using the Smart-seq2 protocol by Picelli et al. (51) with minor modifications (see SI Appendix, Supplementary Materials and Methods for more details). Ten samples were sequenced with 151 bp paired-end reads on a single lane of NextSeq 500 (Illumina, San Diego, CA) at the Stanford Functional Genomics Facility.
After sequencing, bcl2fastq2 v2.18 (Illumina) was used to extract the data and generate FASTQ files for each sample by using unique barcode combinations from the Nextera preparation. Raw reads were trimmed for base call quality (PHRED score ≥21) and for adapter sequences using Skewer v0.2.2 (77). Trimmed reads were then aligned to the mouse genome assembly (mm10) from the University of California, Santa Cruz (http://genome.ucsc.edu) using STAR v2.4 with default setting (78).
Methods for RNA-seq data analysis are described in detail in SI Appendix, Supplementary Materials and Methods, and raw and processed data are available in the GEO database under accession no. GSE130504 (52).
Statistics.
Statistical significance between 2 groups was determined using a paired or unpaired Student’s t test, as appropriate. For comparison of 2 groups across multiple time points, statistical significance was determined using a 2-way ANOVA using the groups and time points as factors. Multiple hypothesis correction was applied to gene expression comparisons using the Benjamini–Hochberg procedure. Results with P-adjusted < 0.1 were considered significant. Data analyses were performed with R 3.5.1, Prism v7 (GraphPad Software, Inc.) and FlowJo v10 (FlowJo, LLC). The investigators were not blinded to allocation during experiments and outcome assessment. No sample-size estimates were performed to ensure adequate power to detect a prespecified effect size.
Supplementary Material
Acknowledgments
We thank R. Yamamoto, C. K. F. Chan, J. Xiang, V. Mascetti, V. G. Alvarado, A. Chandra, and A. Manjunath for technical assistance and discussion. We thank T. Naik for laboratory management, A. McCarty and C. Wang for mouse breeding and management, P. Lovelace and S. Weber for their support and assistance with FACS, and the Stanford Functional Genomic Facility (SFGF) for assistance with sequencing scRNA-seq libraries. We thank K. Kao, T. Sakamaki, and M. Miyanishi for assistance with breeding the Hoxb5-mCherry and CAG-EGFP;Hoxb5-mCherry mouse strains. This study was supported by California Institute for Regenerative Medicine Grant RT3-07683 and the Virginia and D. K. Ludwig Fund for Cancer Research (to I.L.W.); the Thomas and Stacey Siebel Foundation (Siebel Stem Cell Institute), the National Cancer Institute, Department of Health and Human Services (Public Health Service Grant CA09302), National Heart, Lung, and Blood Institute (NHLBI) Ruth L. Kirschstein National Research Service Award F30HL147460, and the Stanford Medical Science Training Program (to G.S.G.); Foundation For Polish Science Grant HOMING POIR.04.04.00-00-5F16/18-00 and a MOBILITY PLUS Fellowship from the Polish Ministry of Science and Higher Education (to K.S.); National Science Centre Poland HARMONIA UMO-2015/18/M/NZ3/00387 to Alicja Jozkowicz and the Leading National Research Center (KNOW) supported by the Polish Ministry of Science and Higher Education to the Faculty of Biochemistry, Biophysics, and Biotechnology of Jagiellonian University (supporting M.Z.); and a Stanford Vice President for Undergraduate Research office (VPUE) grant and Bio-X summer funding (to J.J.N.).
Footnotes
The authors declare no competing interest.
Data deposition: The bulk RNA sequencing data generated in this study have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE130504).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1911024116/-/DCSupplemental.
References
- 1.Seita J., Weissman I. L., Hematopoietic stem cell: Self-renewal versus differentiation. Wiley Interdiscip. Rev. Syst. Biol. Med. 2, 640–653 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spangrude G. J., Heimfeld S., Weissman I. L., Purification and characterization of mouse hematopoietic stem cells. Science 241, 58–62 (1988). [DOI] [PubMed] [Google Scholar]
- 3.Ikuta K., Weissman I. L., Evidence that hematopoietic stem cells express mouse c-kit but do not depend on steel factor for their generation. Proc. Natl. Acad. Sci. U.S.A. 89, 1502–1506 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Osawa M., Hanada K., Hamada H., Nakauchi H., Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science 273, 242–245 (1996). [DOI] [PubMed] [Google Scholar]
- 5.Kiel M. J., et al. , SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell 121, 1109–1121 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Christensen J. L., Weissman I. L., Flk-2 is a marker in hematopoietic stem cell differentiation: A simple method to isolate long-term stem cells. Proc. Natl. Acad. Sci. U.S.A. 98, 14541–14546 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gazit R., et al. , Fgd5 identifies hematopoietic stem cells in the murine bone marrow. J. Exp. Med. 211, 1315–1331 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J. Y., et al. , Hoxb5 marks long-term haematopoietic stem cells and reveals a homogenous perivascular niche. Nature 530, 223–227 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Acar M., et al. , Deep imaging of bone marrow shows non-dividing stem cells are mainly perisinusoidal. Nature 526, 126–130 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sawai C. M., et al. , Hematopoietic stem cells are the major source of multilineage hematopoiesis in adult animals. Immunity 45, 597–609 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Challen G. A., Boles N. C., Chambers S. M., Goodell M. A., Distinct hematopoietic stem cell subtypes are differentially regulated by TGF-beta1. Cell Stem Cell 6, 265–278 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uchida N., Dykstra B., Lyons K. J., Leung F. Y., Eaves C. J., Different in vivo repopulating activities of purified hematopoietic stem cells before and after being stimulated to divide in vitro with the same kinetics. Exp. Hematol. 31, 1338–1347 (2003). [DOI] [PubMed] [Google Scholar]
- 13.de Haan G., Lazare S. S., Aging of hematopoietic stem cells. Blood 131, 479–487 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Morrison S. J., Wandycz A. M., Akashi K., Globerson A., Weissman I. L., The aging of hematopoietic stem cells. Nat. Med. 2, 1011–1016 (1996). [DOI] [PubMed] [Google Scholar]
- 15.Sudo K., Ema H., Morita Y., Nakauchi H., Age-associated characteristics of murine hematopoietic stem cells. J. Exp. Med. 192, 1273–1280 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rossi D. J., et al. , Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc. Natl. Acad. Sci. U.S.A. 102, 9194–9199 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wright D. E., Bowman E. P., Wagers A. J., Butcher E. C., Weissman I. L., Hematopoietic stem cells are uniquely selective in their migratory response to chemokines. J. Exp. Med. 195, 1145–1154 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Passegué E., Wagers A. J., Giuriato S., Anderson W. C., Weissman I. L., Global analysis of proliferation and cell cycle gene expression in the regulation of hematopoietic stem and progenitor cell fates. J. Exp. Med. 202, 1599–1611 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fleming W. H., et al. , Functional heterogeneity is associated with the cell cycle status of murine hematopoietic stem cells. J. Cell Biol. 122, 897–902 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beerman I., et al. , Functionally distinct hematopoietic stem cells modulate hematopoietic lineage potential during aging by a mechanism of clonal expansion. Proc. Natl. Acad. Sci. U.S.A. 107, 5465–5470 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rossi D. J., et al. , Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature 447, 725–729 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Mann M., et al. , Heterogeneous responses of hematopoietic stem cells to inflammatory stimuli are altered with age. Cell Rep. 25, 2992–3005.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacLean A. L., et al. , Single cell phenotyping reveals heterogeneity among hematopoietic stem cells following infection. Stem Cells 35, 2292–2304 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Lu R., Czechowicz A., Seita J., Jiang D., Weissman I. L., Clonal-level lineage commitment pathways of hematopoietic stem cells in vivo. Proc. Natl. Acad. Sci. U.S.A. 116, 1447–1456 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodriguez-Fraticelli A. E., et al. , Clonal analysis of lineage fate in native haematopoiesis. Nature 553, 212–216 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dykstra B., et al. , Long-term propagation of distinct hematopoietic differentiation programs in vivo. Cell Stem Cell 1, 218–229 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Yamamoto R., et al. , Large-scale clonal analysis resolves aging of the mouse hematopoietic stem cell compartment. Cell Stem Cell 22, 600–607.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamamoto R., et al. , Clonal analysis unveils self-renewing lineage-restricted progenitors generated directly from hematopoietic stem cells. Cell 154, 1112–1126 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Gekas C., Graf T., CD41 expression marks myeloid-biased adult hematopoietic stem cells and increases with age. Blood 121, 4463–4472 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Sanjuan-Pla A., et al. , Platelet-biased stem cells reside at the apex of the haematopoietic stem-cell hierarchy. Nature 502, 232–236 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Wilson N. H., Key B., Neogenin: One receptor, many functions. Int. J. Biochem. Cell Biol. 39, 874–878 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Wilson N. H., Key B., Neogenin interacts with RGMa and netrin-1 to guide axons within the embryonic vertebrate forebrain. Dev. Biol. 296, 485–498 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Xu K., et al. , Neural migration. Structures of netrin-1 bound to two receptors provide insight into its axon guidance mechanism. Science 344, 1275–1279 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsunaga E., et al. , RGM and its receptor neogenin regulate neuronal survival. Nat. Cell Biol. 6, 749–755 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Bae G. U., et al. , Neogenin regulates skeletal myofiber size and focal adhesion kinase and extracellular signal-regulated kinase activities in vivo and in vitro. Mol. Biol. Cell 20, 4920–4931 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang A. S., West A. P. Jr, Wyman A. E., Bjorkman P. J., Enns C. A., Interaction of hemojuvelin with neogenin results in iron accumulation in human embryonic kidney 293 cells. J. Biol. Chem. 280, 33885–33894 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Srinivasan K., Strickland P., Valdes A., Shin G. C., Hinck L., Netrin-1/neogenin interaction stabilizes multipotent progenitor cap cells during mammary gland morphogenesis. Dev. Cell 4, 371–382 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Park K. W., et al. , The axonal attractant Netrin-1 is an angiogenic factor. Proc. Natl. Acad. Sci. U.S.A. 101, 16210–16215 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Renders S., et al. , The netrin-1–Neogenin axis regulates hematopoietic stem cell dormancy and function with implications for stem cell ageing. Blood 132, 637 (2018). [Google Scholar]
- 40.Renders S., et al. , Neogenin regulates hematopoietic stem cell quiescence and maintenance. Exp. Hematol. 53, S49 (2017). [Google Scholar]
- 41.Svendsen A., et al. , Neogenin-1: Identification of a novel receptor associated with hematopoietic stem cell aging. Exp. Hematol. 64, S106 (2018). [Google Scholar]
- 42.de Haan G., et al. , Neogenin-1: A new receptor critical for hematopoietic stem cell function. Exp. Hematol. 53, S47 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Seita J., et al. , Gene expression commons: An open platform for absolute gene expression profiling. PLoS One 7, e40321 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith-Berdan S., et al. , Robo4 cooperates with CXCR4 to specify hematopoietic stem cell localization to bone marrow niches. Cell Stem Cell 8, 72–83 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yano M., et al. , Expression and function of murine receptor tyrosine kinases, TIE and TEK, in hematopoietic stem cells. Blood 89, 4317–4326 (1997). [PubMed] [Google Scholar]
- 46.Cabezas-Wallscheid N., et al. , Vitamin A-retinoic acid signaling regulates hematopoietic stem cell dormancy. Cell 169, 807–823.e19 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Majeti R., Park C. Y., Weissman I. L., Identification of a hierarchy of multipotent hematopoietic progenitors in human cord blood. Cell Stem Cell 1, 635–645 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Randall T. D., Weissman I. L., Phenotypic and functional changes induced at the clonal level in hematopoietic stem cells after 5-fluorouracil treatment. Blood 89, 3596–3606 (1997). [PubMed] [Google Scholar]
- 49.Bernitz J. M., Kim H. S., MacArthur B., Sieburg H., Moore K., Hematopoietic stem cells count and remember self-renewal divisions. Cell 167, 1296–1309.e10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pietras E. M., Warr M. R., Passegué E., Cell cycle regulation in hematopoietic stem cells. J. Cell Biol. 195, 709–720 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Picelli S., et al. , Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Gulati G. S., et al. , Bulk RNA-sequencing profiles of NEO1+Hoxb5+ and NEO1−Hoxb5+ LT-HSCs from the bone marrow of 2- to 3-month-old Hoxb5-mCherry mice. National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO). https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE130504. Deposited 30 April 2019.
- 53.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Du W., et al. , Inflammation-mediated notch signaling skews fanconi anemia hematopoietic stem cell differentiation. J. Immunol. 191, 2806–2817 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Laurenti E., et al. , Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell 3, 611–624 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pervaiz S., Taneja R., Ghaffari S., Oxidative stress regulation of stem and progenitor cells. Antioxid. Redox Signal. 11, 2777–2789 (2009). [DOI] [PubMed] [Google Scholar]
- 57.Park C. S., Shen Y., Lewis A., Lacorazza H. D., Role of the reprogramming factor KLF4 in blood formation. J. Leukoc. Biol. 99, 673–685 (2016). [DOI] [PubMed] [Google Scholar]
- 58.Ma X. Y., et al. , Malat1 as an evolutionarily conserved lncRNA, plays a positive role in regulating proliferation and maintaining undifferentiated status of early-stage hematopoietic cells. BMC Genomics 16, 676 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Subramanian A., et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu G., Wang L. G., Han Y., He Q. Y., clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ashburner M., et al. ; The Gene Ontology Consortium , Gene ontology: Tool for the unification of biology. Nat. Genet. 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park I. K., et al. , Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 423, 302–305 (2003). [DOI] [PubMed] [Google Scholar]
- 63.Scholl C., et al. , The homeobox gene CDX2 is aberrantly expressed in most cases of acute myeloid leukemia and promotes leukemogenesis. J. Clin. Invest. 117, 1037–1048 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Naudin C., et al. , PUMILIO/FOXP1 signaling drives expansion of hematopoietic stem/progenitor and leukemia cells. Blood 129, 2493–2506 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen Z., et al. , Cohesin-mediated NF-κB signaling limits hematopoietic stem cell self-renewal in aging and inflammation. J. Exp. Med. 216, 152–175 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nerlov C., Graf T., PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes Dev. 12, 2403–2412 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Trompouki E., et al. , Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell 147, 577–589 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu S., et al. , GABP controls a critical transcription regulatory module that is essential for maintenance and differentiation of hematopoietic stem/progenitor cells. Blood 117, 2166–2178 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Degar B. A., et al. , The homeodomain gene Pitx2 is expressed in primitive hematopoietic stem/progenitor cells but not in their differentiated progeny. Exp. Hematol. 29, 894–902 (2001). [DOI] [PubMed] [Google Scholar]
- 70.Tothova Z., et al. , FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell 128, 325–339 (2007). [DOI] [PubMed] [Google Scholar]
- 71.Yu X., et al. , HES1 inhibits cycling of hematopoietic progenitor cells via DNA binding. Stem Cells 24, 876–888 (2006). [DOI] [PubMed] [Google Scholar]
- 72.Takubo K., et al. , Regulation of the HIF-1alpha level is essential for hematopoietic stem cells. Cell Stem Cell 7, 391–402 (2010). [DOI] [PubMed] [Google Scholar]
- 73.Gilmour J., et al. , A crucial role for the ubiquitously expressed transcription factor Sp1 at early stages of hematopoietic specification. Development 141, 2391–2401 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Healey E. G., et al. , Repulsive guidance molecule is a structural bridge between neogenin and bone morphogenetic protein. Nat. Struct. Mol. Biol. 22, 458–465 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Morita Y., Ema H., Nakauchi H., Heterogeneity and hierarchy within the most primitive hematopoietic stem cell compartment. J. Exp. Med. 207, 1173–1182 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Morrison S. J., Weissman I. L., The long-term repopulating subset of hematopoietic stem cells is deterministic and isolatable by phenotype. Immunity 1, 661–673 (1994). [DOI] [PubMed] [Google Scholar]
- 77.Jiang H., Lei R., Ding S. W., Zhu S., Skewer: A fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics 15, 182 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dobin A., et al. , STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.