Significance
Knowledge-based development of global warming-resilient crop varieties is hindered by limited understanding of warm-temperature signaling mechanisms. Using the Arabidopsis model, we show that the chromatin-modifying enzyme HISTONE DEACETYLASE 9 (HDA9) is essential for promoting an open plant architecture that allows for efficient mitigation of the impact of warm temperature. HDA9 does not affect hypocotyl elongation in response to different light conditions, setting it apart from the shade-avoidance response that phenotypically resembles acclimation to warmth. We demonstrate that HDA9 is required for transcriptional activation of YUCCA8, the rate-limiting enzyme in the biosynthesis of the phytohormone auxin, by facilitating net eviction of the H2A.Z histone variant from YUCCA8 nucleosomes at warm temperature.
Keywords: HDA9, thermomorphogenesis, Arabidopsis, shade avoidance, H2A.Z
Abstract
Many plant species respond to unfavorable high ambient temperatures by adjusting their vegetative body plan to facilitate cooling. This process is known as thermomorphogenesis and is induced by the phytohormone auxin. Here, we demonstrate that the chromatin-modifying enzyme HISTONE DEACETYLASE 9 (HDA9) mediates thermomorphogenesis but does not interfere with hypocotyl elongation during shade avoidance. HDA9 is stabilized in response to high temperature and mediates histone deacetylation at the YUCCA8 locus, a rate-limiting enzyme in auxin biosynthesis, at warm temperatures. We show that HDA9 permits net eviction of the H2A.Z histone variant from nucleosomes associated with YUCCA8, allowing binding and transcriptional activation by PHYTOCHROME INTERACTING FACTOR 4, followed by auxin accumulation and thermomorphogenesis.
Plants are immobile and lack homeostatic mechanisms to maintain body temperature. Therefore, projected global warming poses a serious threat to agricultural productivity, as each degree Celsius increase can lead up to 10% decrease in crop yields (1, 2). Several species, including the model plant Arabidopsis thaliana, can however mitigate warmth through adjustments of the vegetative body plan by a phytohormone-dependent process called thermomorphogenesis (3–5). The resulting “open” rosette architecture that is caused by traits, such as upward leaf movement and hypocotyl and petiole elongation, allows for more efficient cooling through improved evaporation and heat flux avoidance. As a result, thermomorphogenesis is essential for conferring optimized fitness under unfavorable high temperature conditions (3–5).
PHYTOCHROME INTERACTING FACTOR4 (PIF4) is an indispensable transcription factor for mediating thermomorphogenesis (6–8). PIF4 is transcriptionally induced by warm temperatures (6–8) and is tightly controlled by the evening complex component EARLY FLOWERING3 (ELF3) (9, 10). PIF4 directly binds and activates the expression of genes involved in biosynthesis of auxin, including the rate-limiting enzyme flavin monooxygenase YUCCA8 (YUC8) (7, 11).
Thermomorphogenesis phenotypically resembles the shade-avoidance response, which is a strategy exhibited by plants to outcompete neighbors under dense canopies (12). Accordingly, many of the proteins responsible for regulating thermomorphogenesis also play a role in light signaling and shade avoidance (4, 5). This includes PIF4 (6–8, 11) and the red-light photoreceptor phytochrome B (phyB), which was shown to also act as a thermosensor (13, 14).
Thermomorphogenesis is controlled by epigenetic mechanisms that regulate gene expression (11, 15–22). The histone variant H2A.Z is evicted from nucleosomes at thermoresponsive genes, in a heat-shock factor A1 (HSFA1a) family-dependent manner (16, 20). The ACTIN-RELATED PROTEIN6 (ARP6)-containing SWR1 histone-replacement complex is involved in the deposition of H2A.Z (16, 23, 24). In the context of high-temperature signaling, H2A.Z eviction is associated with greater chromatin accessibility and enhanced potential for transcriptional modulation by permitting the binding of transcriptional regulators, such as PIF4 (16, 20, 23). Furthermore, histone methylation contributes to regulation of temperature-responsive loci (15). More specifically, PICKLE (18) and SEUSS (19) stimulate thermomorphogenesis, whereas FLOWERING TIME CONTROL PROTEIN A (FCA)-mediated H3K4me2 removal from YUC8 attenuates thermomorphogenesis, preventing plant lodging (11). We (21) and others recently demonstrated that histone deacetylation mediated by the SANT domain-containing protein POWERDRESS (PWR) and the interacting REDUCED POTASSIUM DEPENDENCY 3 (RPD3)-like class I HISTONE DEACETYLASE 9 (HDA9) (25, 26), as well as HDA19 (22), are essential positive regulators of thermomorphogenesis, whereas HDA15 was identified as negative regulator of the response (22).
Here, we show that HDA9 defines a temperature signaling pathway that is uncoupled from shade avoidance. Under warm temperatures, HDA9 protein levels are high in young seedlings and mediate histone deacetylation at nucleosomes positioned at the transcriptional start-site and gene body of YUC8. This deacetylation event proposedly results in lower H2A.Z levels at warm temperatures, which allows for PIF4 binding to the G-box motif in the YUC8 promoter followed by conditional YUC8 transcriptional activation, resulting in auxin production and ultimately thermomorphogenesis.
Results
HDA9 Defines a Thermosignaling Pathway.
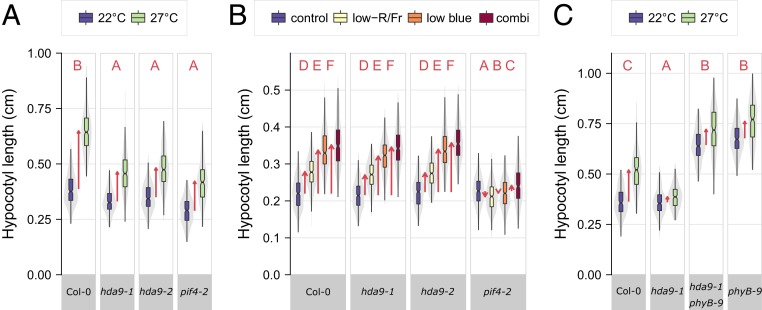
To investigate the role of HDA9 in thermomorphogenesis responses of vegetative organs [“type 3 thermomorphogenesis” (5)], we first examined the morphology of mutants of HDA9 in control (22 °C) and elevated (27 °C) ambient temperature. Arabidopsis hda9 mutants are compromised in thermomorphogenesis (21), as displayed by reduced hypocotyl elongation (Fig. 1A), petiole elongation, and hyponastic growth (SI Appendix, Fig. S1 A–D). Hypocotyl elongation of hda9 mutants was not affected in darkness (skotomorphogenesis) nor by spectral neutral shading (SI Appendix, Fig. S1E), indicating that elongation capacity is not mediated by HDA9. Furthermore, temperature-induced hypocotyl elongation was complemented in a line carrying functional HDA9 in the hda9-1 mutant background (27), confirming the requirement of HDA9 for thermomorphogenesis (SI Appendix, Fig. S1C). Some residual elongation was observed in hda9 mutant lines at high temperature (Fig. 1A and SI Appendix, Fig. S1 A–D), which suggests that while HDA9 is clearly essential, other factors contribute to thermomorphogenesis.
Fig. 1.
Mutations in HDA9 impair thermomorphogenesis independent of light-quality signaling and phyB. (A–C) Hypocotyl lengths of 8-d-old seedlings at (A and C) 22 °C or 27 °C and (B) different light-quality conditions and their combination. Boxes indicate boundaries of second and third quartiles of data distributions. Black bars indicate median and whiskers Q1 and Q4 values within 1.5 times the interquartile range. Violin plots designate phenotype distributions. Red arrows designate mean hypocotyl elongation responses. Red letters indicate statistical differences between hypocotyl responses (changes) in all panels (P < 0.05; 2-sided t test) (Dataset S1), with different letters indicating significantly different groups. (A–C) n = 208 to 295, 247 to 323, 131 to 236 seedlings per genotype and treatment, divided over 7, 12, 7, biological replicates, respectively.
Temperature-shift experiments, where seedlings were transferred from control to elevated temperature conditions and vice versa, indicated that hda9-1 and pif4-2 mutants exhibit reduced temperature sensitivity in hypocotyl elongation (SI Appendix, Fig. S1F). However, not all temperature-associated phenotypes are compromised in hda9 mutants. For example, high-temperature–induced expression of the HEAT SHOCK PROTEIN 70 (HSP70) marker gene (16), was not affected in the hda9-1 mutant (SI Appendix, Fig. S1G). Furthermore, the typical high-temperature–induced acceleration of the floral transition (28, 29) in hda9-1 was comparable to wild-type (SI Appendix, Fig. S1H), even though hda9 mutants exhibit a mild early-flowering phenotype in short-day conditions (27, 30). Notably, mutants in HDA9 also retained responsiveness to light-quality signals that induce shade avoidance, whereas shade avoidance was attenuated in the pif4-2 mutant, as expected (31) (Fig. 1B and SI Appendix, Fig. S1I). Moreover, the hda9-1 mutation could not suppress the constitutively elongated phenotype of the phyB-9 mutant (Fig. 1C and SI Appendix, Fig. S1J), suggesting that phyB effects do not depend on HDA9. In conclusion, HDA9 defines a thermosignaling pathway independent of temperature-induced flowering and is unlikely to have a role in hypocotyl elongation during shade avoidance.
HDA9 Promoter Activity, Expression, and Protein Dynamics.
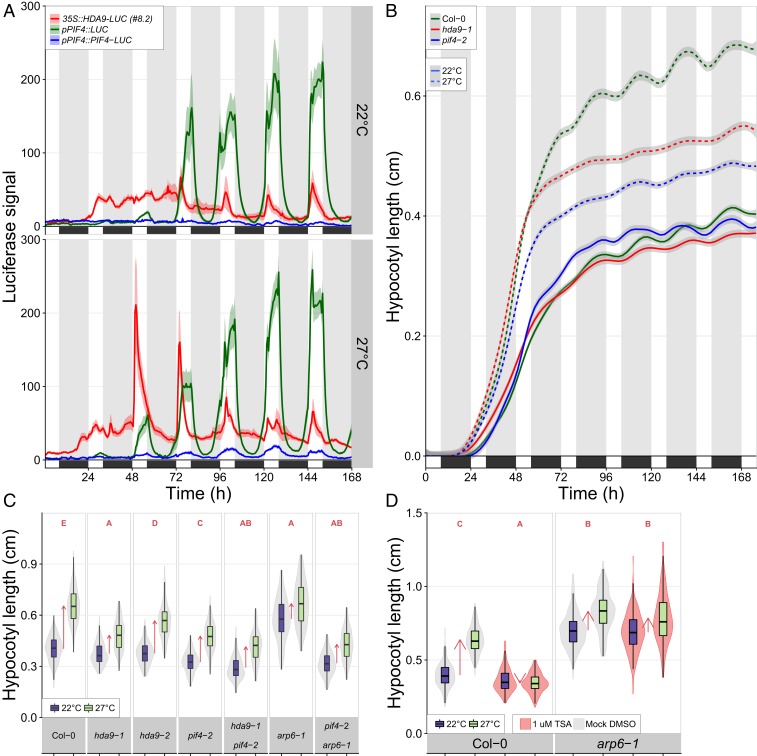
To examine if elevated temperature affects HDA9 promoter activity, we performed studies on transgenic lines carrying HDA9 promoter-reporter fusion constructs. Our study using pHDA9::GUS lines revealed that HDA9 promoter activity was largely, but not exclusively, restricted to roots, the root–shoot junction, and basal hypocotyl tissues of germinating seedlings and declined during seedling establishment (SI Appendix, Fig. S2 A and B). Testing pHDA9::GUS and pHDA9::LUC lines and qRT-PCR experiments demonstrated that high temperature had no effect on HDA9 transcript levels, nor promoter activity (SI Appendix, Figs. S2 A and C–F and S3A). Diurnal in planta luminescent profiling using HDA9 protein–reporter fusion constructs (35S::HDA9-LUC) lines revealed that HDA9 protein levels peak at dawn in response to warm-temperature conditions, starting from day 3 after initiation of germination (2-d-old seedlings) (Fig. 2A and SI Appendix, Fig. S3 A–C). HDA9 levels then gradually decreased during the day, with minimal levels displayed at dusk (Fig. 2A and SI Appendix, Fig. S3A). The peak amplitude of HDA9 protein levels at dawn faded during seedling establishment, reaching levels comparable to control temperature (22 °C) at approximately day 7 after initiation of germination. No apparent diurnal changes in HDA9 were observed at control temperature (22 °C). We therefore conclude that the elevated temperature signal is essential for observed increases in HDA9 protein levels.
Fig. 2.
HDA9, PIF4, and ARP6/H2A.Z present a thermosignaling module. (A) Dynamics of HDA9 and PIF4 protein and PIF4 transcript levels. n = 6 to 19 per genotype. See SI Appendix, Fig. S3 for details. (B) Progression of hypocotyl elongation. n = 110 to 212 seedlings per genotype, per treatment, divided over 32 replicates. Statistics (Tukey HSD per time point, genotype, and treatment) are presented in SI Appendix, Fig. S4A and Dataset S1. (A and B) Colored areas behind lines represent SEM. Black boxes and gray-shaded bands/white bands indicate darkness/daytime. (C and D) Hypocotyl lengths of 8-d-old seedlings in D, the presence of TSA and mock. (C) n = 157 to 324 and (D) n = 157 to 324 seedlings per genotype and treatment, divided over 7 (C) and 9 (D) replicates. Boxes indicate boundaries of second and third quartiles of data distributions. Black bars indicate median and whiskers Q1 and Q4 values within 1.5 times the interquartile range. Violin plots designate phenotype distributions. Red arrows indicate the mean hypocotyl response. Red letters in C and D indicate statistical differences between hypocotyl responses (changes) (P < 0.01; 2-sided t test), with different letters indicating significantly different groups.
Detected LUC signals of our 35S::HDA9-LUC lines (Fig. 2A and SI Appendix, Fig. S3A) were ∼50-fold lower than that of seedlings expressing 35S::LUC (SI Appendix, Fig. S3B). This difference likely can be attributed to suppression of the LUC signal by proteasomal degradation of HDA9 (32). Combined with the low expression from the native pHDA9 promoter (compared to the constitutive 35S promoter), this also explains why the diurnal peaks in LUC activity at warm temperature were not clearly detectable in seedlings expressing pHDA9::HDA9:LUC (SI Appendix, Fig. S3A). We therefore next analyzed whole-mount seedlings of several independent 35S::HDA9-LUC and pHDA9::HDA9-LUC lines (SI Appendix, Fig. S3 D and E) and normalized the detected LUC signals at warm temperatures to those at control temperature levels. This confirmed that temperature triggers HDA9 accumulation, also when expressed from the native promoter (SI Appendix, Fig. S3 F and G). PIF4 promoter activity and PIF4 protein levels followed a diurnal cycling pattern in response to high temperature starting at the dawn of day 3 (Fig. 2A and SI Appendix, Fig. S3H), as observed before (33–35).
To test whether the diurnal HDA9 and PIF4 protein abundance profiles match their requirement for thermomorphogenesis, we monitored the progression of hypocotyl elongation in pif4 and hda9 mutants in response to elevated temperature (Fig. 2B and SI Appendix, Fig. S4A). At control 22 °C, hda9-1 and pif4-2 exhibited overall wild-type rates of hypocotyl elongation during a 7-d exposure to short-day photoperiod conditions. An increase in hypocotyl elongation in response to elevated temperature (27 °C) started immediately after germination and initially occurred independent of PIF4 and HDA9 (Fig. 2B and SI Appendix, Fig. S4A). However, this high-temperature induction in hypocotyl elongation was largely impaired in hda9-1 and pif4-2 from day 3 onwards, whereas wild-type seedlings retained an increased elongation rate throughout the experimental period at 27 °C. This indicates that HDA9 and PIF4 promote thermomorphogenesis from day 3 (t = 48 h after initiation of germination), which correlates with the protein abundance profile of HDA9 and PIF4 (Fig. 2A). Of note, growth attenuation of hda9-1 and pif4-2 relative to wild-type occurred largely, but not exclusively during the night period (Fig. 2B and SI Appendix, Fig. S4A).
The HDA9, PIF4, ARP6/H2A.Z Module.
A possible genetic interaction between HDA9 and PIF4 was examined using a pif4-2 × hda9-1 double mutant. This double mutant showed impaired hypocotyl elongation in response to elevated temperature (Fig. 2C). Interestingly, temperature-induced hypocotyl elongation was suppressed by the specific histone deacetylase (HDAC)-inhibitor Trichostatin-A (TSA) (36) in a PIF4 overexpression line (35S::PIF4-HA), indicating that HDAC activity has a role downstream or parallel to PIF4 signaling during thermomorphogenesis (SI Appendix, Fig. S4B).
The next step was to assess the role of the ARP6-containing SWR1 complex, known to be required for the incorporation of H2A.Z (24) at nucleosomes of thermoresponsive genes (15, 16, 20, 23). Consequently, the arp6-1 mutant displays a warm-temperature transcriptome and elongated hypocotyls even at control temperatures (22 °C) (16). We observed that the arp6-1 mutant was resistant to TSA application (Fig. 2D). This observation suggests that ARP6 and H2A.Z genetically operate downstream of HDAC-mediated thermomorphogenesis. Attempts to obtain double homozygous hda9-1−/− × arp6-1−/− mutants were unsuccessful as independent progenies of hda9-1−/− × arp6-1−/+ lines segregated on average in a 56% (hda9-1−/− × arp6-1+/+)/44% (hda9-1−/− × arp6-1−/+) ± 5.3% ratio. Such a close to 50/50 non-Mendelian ratio implies that the double mutant is most likely lethal, indicating possible mutual dependency of HDA9 and ARP6/H2A.Z for viability. Examination of pif4-2 × arp6-1 double mutants revealed suppression of the arp6-1 phenotype (Fig. 2C), demonstrating that PIF4 is required for ARP6/H2A.Z-mediated thermomorphogenesis. Importantly, the transcriptional activation of PIF4 in response to warm temperature does not, however, require HDA9 and ARP6 (SI Appendix, Fig. S4 C and D).
We next tested the possibility of a direct interaction between HDA9 and PIF4 and between HDA9 and HSFA1a that allows for transcriptional induction of thermoresponsive genes by facilitating eviction of H2A.Z-nucleosomes during the day time (16, 20, 23), by bimolecular fluorescence complementation (BiFC) experiments (SI Appendix, Fig. S5 A–C) and yeast 2-hybrid assays (SI Appendix, Fig. S5 D and E). However, no direct interaction between HDA9 and PIF4 nor HSFA1a was observed with the approaches used and under the conditions tested, whereas the known (37) ZINC-FINGER HOMEODOMAIN 10 (ZFHD10) homodimerization (BiFC) and interaction between TANDEM ZINC-FINGER PLUS3 (TZP) and ZFHD10 (yeast 2-hybrid), here used as positive controls, were confirmed (SI Appendix, Fig. S5 D and E). Although, this does not exclude the possibility that HDA9, PIF4, and HSFA1a are part of a multiprotein complex. Together, these results indicate that HDA9, PIF4, ARP6, and H2A.Z present a genetic thermosignaling module that orchestrates thermomorphogenesis.
HDA9 Promotes Auxin Biosynthesis.
It is well-established that HDA9 is an epigenetic regulator of gene transcription. Therefore, we decided to investigate the effect of HDA9 on the transcriptional level to identify HDA9 targets during vegetative plant thermomorphogenesis. More specifically, to identify early and late differentially regulated genes, we performed RNA-sequencing and comparative analysis of the control (22 °C) and warm-temperature transcriptomes (27 °C) of hda9-1, pif4-2, and Col-0 (wild-type) seedlings at dawn of day 3 (2-d-old) and day 8 (7-d-old) (SI Appendix, Fig. S6A). Hierarchical clustering (SI Appendix, Fig. S6B) and principal component analyses (PCA) (SI Appendix, Fig. S6 C–E) indicated that expression differences were primarily explained by age of the plant and temperature. Two-day-old seedlings showed similar trends for high-temperature–regulated genes between Col-0 and pif4-2, whereas hda9-1 exhibited a distinct transcriptome (SI Appendix, Fig. S6 D and F). Seven-day-old seedlings exhibited apparent transcriptome differences between wild-type and pif4-2 and less so between wild-type and hda9-1 (SI Appendix, Fig. S6 E and G).
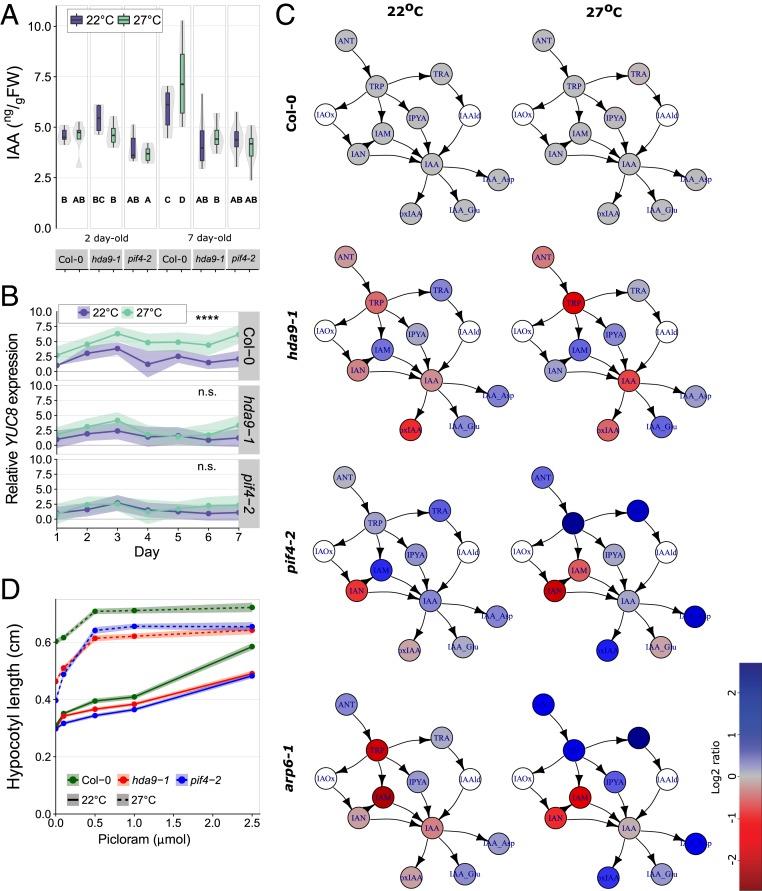
Gene ontology (GO) enrichment analysis revealed an auxin-responsive signature for the genes that were up-regulated in response to high temperature in 2-d-old wild-type, but not in hda9-1 seedlings (Table 1 and SI Appendix, Table S1). Given the requirement of auxin for thermomorphogenesis (6–8), the levels of bioactive indole-3-acetic acid (IAA) were quantified in hda9-1 and pif4-2 seedlings. These measurements showed that HDA9 is required for high-temperature–induced auxin accumulation in 7-d-old seedlings and confirmed the necessity of PIF4 (6–8) (Fig. 3A). However, no significant changes in IAA levels were observed in 2-d-old seedlings (Fig. 3A), suggesting that high-temperature–induced hypocotyl elongation during the first 2 d after germination (Fig. 2B) may not require de novo auxin biosynthesis. These data are consistent with our observation that temperature-dependent effects on HDA9 and PIF4 protein levels (Fig. 2A and SI Appendix, Fig. S3) and elongation growth (Fig. 2B) are only observed from dawn of day 3 after germination and onwards.
Table 1.
Top 5 enriched GO terms among genes up-regulated by 27 °C vs. control 22 °C in Col-0 but not hda9-1 (2-d-old seedlings)
| GO process | Total genes | Found genes | P value (nonadjusted) |
| Response to auxin | 279 | 39 | 1.11E-12 |
| Auxin-activated signaling pathway | 170 | 28 | 2.01E-11 |
| Regulation of organ growth | 10 | 7 | 2.16E-10 |
| Water transport | 19 | 8 | 1.29E-08 |
| Ion transmembrane transport | 26 | 8 | 3.42E-07 |
See Dataset S1 for full table.
Fig. 3.
HDA9 is required for YUCCA8-dependent auxin biosynthesis. (A) IAA levels. Boxes indicate boundaries of second and third quartiles of data distributions. Black bars indicate median and whiskers Q1 and Q4 values within 1.5 times the interquartile range. Violin plots designate phenotype distributions. Bold letters indicate statistical differences between IAA levels over all samples (P < 0.05; Tukey HSD), n = 4 to 10 replicas per genotype per treatment per time point, each of 100 mg (FW) seedlings. (B) Relative YUCCA8 expression, 3 to 4 replicas per genotype, per treatment, per timepoint, each of >25 seedlings. Asterisks indicate significant difference on the range of time points between 22 °C and 27 °C (****P < 0.001, n.s. indicates nonsignificant; 2-sided t test). For statistical comparisons of individual time points, see Dataset S1. (C) Auxin metabolite levels, normalized to Col-0 wild-type. White symbols indicate not detectable. See SI Appendix, Fig. S8 for details and abbreviations of metabolites. n = 4 replicates per genotype and treatment, each of 10 mg (FW) of 2-d-old seedlings. (D) Hypocotyl lengths of 8-d-old seedlings in the presence of different concentrations Picloram, n = 104 to 132 seedlings per genotype per treatment divided over 4 replicates. See Dataset S1 for comparative statistics. (B and D) Colored areas behind the lines represent SEM.
Detailed examination of auxin biosynthesis and signaling genes (SI Appendix, Table S1) revealed that high-temperature–induced up-regulation of, among others, the rate-limiting auxin biosynthesis gene YUC8, was suppressed in hda9-1, supporting our hypothesis that histone deacetylation is necessary for YUC8 induction (21). This requirement of HDA9 [and PIF4 (7)] for YUC8 induction was independently confirmed by qRT-PCR analysis (Fig. 3B) and luminometric assays using lines transiently expressing pYUC8-LUC (38) (SI Appendix, Fig. S7A). Assessment of a pYUC8-n3GFP transgenic line confirmed that YUC8 promoter activity is induced by high temperature in the root–hypocotyl junction (SI Appendix, Fig. S7B) in 2-d-old seedlings, the tissue where HDA9 promoter activity is prevalent (SI Appendix, Fig. S2 A and B). The application of the HDAC inhibitor TSA further confirmed that HDAC activity positively contributes to YUC8 induction (SI Appendix, Fig. S7B).
YUCCA enzymes catalyze the conversion of indole-3-pyruvic acid (IPyA) to bioactive IAA (39) and mutation of YUC8 alone is sufficient to impair thermomorphogenesis (7). Auxin metabolite profiling demonstrated an increase of IpyA relative levels in the hda9-1 and pif4-2 mutant backgrounds, consistent with the requirement of HDA9 and PIF4 (7, 8) for YUC8 induction (Fig. 3C and SI Appendix, Fig. S8 A and B). Accordingly, application of the synthetic auxin analog Picloram to hda9-1 and pif4-2 (7, 8) seedlings exposed to warm temperatures substantially attenuated the impairment of hypocotyl elongation (Fig. 3D and SI Appendix, Fig. S8C), whereas the response to IpyA application was less effective (SI Appendix, Fig. S8C). Strikingly, levels of various auxin metabolites and IAA were elevated in the arp6-1 mutant (Fig. 3C), suggesting that the constitutive elongated hypocotyls at control temperatures of arp6-1 (16) is auxin-dependent. Indeed, application of the polar auxin transport inhibitor 1-naphthylphthalamic acid (NPA) suppressed the arp6-1 hypocotyl elongation phenotype (SI Appendix, Fig. S8D), as in 35S::PIF4-HA and wild-type. We conclude that HDA9 mediates thermomorphogenesis by promoting YUC8-transcription and auxin accumulation and that this response is likely antagonized by ARP6/H2A.Z.
HDA9 Permits H2A.Z Depletion to Facilitate PIF4 Binding to the YUCCA8 Promoter.
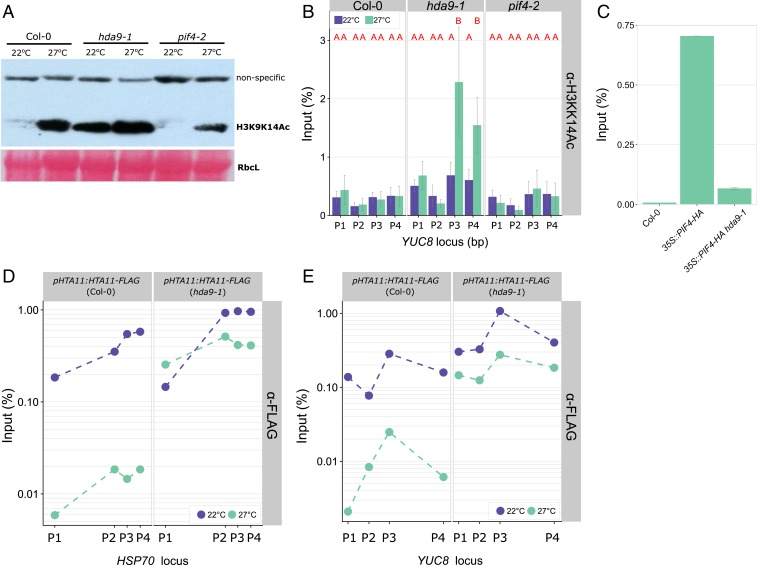
It has been reported that high ambient temperature (40) and heat stress (41) lead to increased levels of histone acetylation at specific loci across the genome. Moreover, the HDA9/PWR complex stimulates H3 deacetylation (25, 26) and we have shown that PIF4 and YUC8 +1 nucleosomes are hyperacetylated at elevated temperatures in 10-d-old pwr mutants (21). We also monitored H3K9K14 acetylation levels in 2-d-old seedlings by Western blot analysis and showed a genome-wide increase in this epigenetic mark in response to elevated temperature (Fig. 4A). Acetylation levels in the pif4-2 mutant were similar to wild-type, suggesting that PIF4 does not have a role in mediating global acetylation levels (Fig. 4A). Furthermore, the data confirmed that hda9-1 seedlings exhibit constitutive H3K9K14 hyperacetylation at both control and high-temperature (26, 30, 42) conditions (Fig. 4A).
Fig. 4.
HDA9 permits H2A.Z eviction. (A) Western blot analysis of H3K9K14ac levels of 2-d-old seedlings at 22 °C or 27 °C (50-mg pooled seedlings per genotype and treatment). Ponceau staining of RIBULOSE BIPHOSPHATE CARBOXYLASE LARGE CHAIN (RbcL) is shown as loading control. (B–E) ChIP-qPCR analysis of (B) H3K9K14Ac levels, (C) PIF4 binding to the G-Box motif and (D and E) H2A.Z (HTA11) enrichment at (B, C, and E) YUCCA8 and (D) HSP70 loci, in 2-d-old seedlings of indicated genotypes. Tested chromatin regions are (B and E) P1 (−1,374 bp), P2 (−657 bp), P3 (4 bp) and P4 (1,813 bp) and (D) P1 (−359 bp), P2 (4 bp), P3 (80 bp), and P4 (159 bp) relative to the transcriptional start site and are from refs. 11 and 16. (B) n = 4 and (E) n = 2, independent replicates of pooled seedlings (SI Appendix, Fig. S9B), error bars represent SEM. (B) Red letters indicate statistical differences on input fraction per tested position (P < 0.05; Tukey HSD).
To assess the effect of HDA9 on histone acetylation levels at the YUC8 locus, we performed chromatin immunoprecipitation (ChIP) assays in 2-d-old seedlings, since our data (Fig. 2) showed that thermomorphogenesis is initiated at this time point. This revealed a warm-temperature–specific increase in H3K9K14 acetylation at the YUC8 transcriptional start site and gene body in hda9-1 seedlings (Fig. 4B and SI Appendix, Fig. S9 A and B) compared to wild-type and pif4-2 mutant plants. In 10-d-old wild-type seedlings we also observed H3K9K14 deacetylation at the transcriptional start site, as described previously (21) (SI Appendix, Fig. S9C). HDAC activity has been primarily associated with transcriptional repression (43–45); however, these results, together with our earlier findings (21), indicate that HDA9-mediated H3K9K14 deacetylation permits thermal induction of YUC8 transcription (Figs. 3B and 4B and SI Appendix, Figs. S7 and S9 A and B). Accordingly, PIF4 binding to the G-box motif (CACGTG) in the YUCCA8 promoter (Fig. 4C and SI Appendix, Fig. S9D), but not the PIF4 promoter (SI Appendix, Fig. S9E), was abolished in the hda9-1 mutant. Although PIF4 protein levels in the hda9-1 35S::PIF4 line background were slightly lower than in the 35S::PIF4 background (SI Appendix, Fig. S9F), likely due to (partial) silencing of the 35S::PIF4 transgenic construct by the hda9-1 SALK T-DNA insert, an effect that is not uncommon for T-DNA insertion lines (46).
The resistance of the arp6-1 mutant to TSA application (Fig. 2D) indicated that ARP6/H2A.Z operates genetically downstream of HDA9. Furthermore, studies in rat (47) and yeast (48) demonstrated that histone acetylation levels positively correlate with H2A.Z occupancy. Taking this information into consideration, we hypothesized that HDA9-mediated H3K9K14 deacetylation at warm temperature conditions could modulate H2A.Z occupancy in Arabidopsis as well, allowing for PIF4 binding followed by thermal induction of YUC8 expression. We tested this by ChIP experiments using transgenic lines expressing epitope-tagged H2A.Z (HTA11-FLAG) (16) in the Col-0 wild-type and hda9-1 mutant backgrounds (SI Appendix, Fig. S10 A and B). Our results showed that in wild-type seedlings YUC8 nucleosomes are indeed depleted from H2A.Z in response to warm temperature, in a similar manner to the temperature-regulated locus HSP70 (16) used here as a positive control (Fig. 4D and SI Appendix, Fig. S10C). However, H2A.Z eviction from YUC8 and HSP70 chromatin was abolished in the hda9-1 mutant background (Fig. 4E and SI Appendix, Fig. S10D). As expected, the GYPSY retrotransposon used as a negative control that is not regulated by temperature or H2A.Z (16) showed no signs of depletion (SI Appendix, Fig. S10E). Thus, HDA9 activity is indeed required for the net eviction of H2A.Z from YUC8 nucleosomes at warm temperatures during thermomorphogenesis, which proposedly allows for PIF4 binding to the G-box of YUCCA8 followed by auxin biosynthesis and thermomorphogenesis.
Discussion
Plant thermomorphogenesis depends on accumulation of the phytohormone auxin (4, 7, 8, 49). We show that HDA9 allows for PIF4 binding and transcriptional activation of YUC8 to stimulate auxin biosynthesis during warm days (Fig. 5). In agreement, a recent study showed that hda9-1 silique valve cells exhibit attenuated auxin signaling and likely have reduced auxin levels (50).
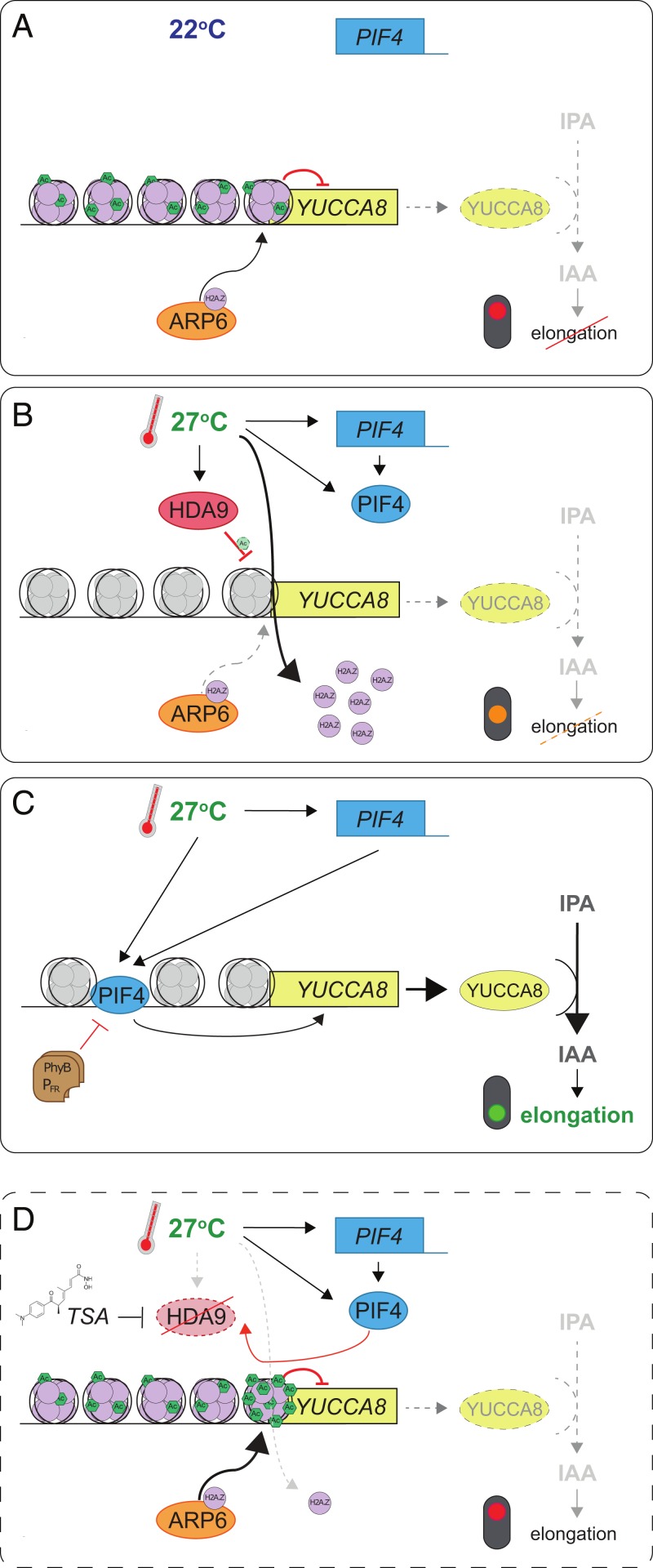
Fig. 5.
Schematic model of proposed HDA9-mediated thermomorphogenesis regulation in 2 d-old seedlings. (A) At control temperatures, PIF4 expression (blue box) is limited and nucleosomes associated with YUCCA8 (yellow box) contain high levels of H2A.Z (purple circles), deposited by ARP6 (orange ellipse). Therefore, auxin (IAA) levels are low and elongation growth is repressed (dashed lines; red traffic light). (B) When temperatures rise, HDA9 protein (red ellipse) levels are high during day time, resulting in maintenance of a low acetylation level at high temperature, comparable to control temperature levels at the YUCCA8 locus. This facilitates the warm-temperature–induced eviction of H2A.Z from nucleosomes (gray nucleosomes) over ARP6-mediated deposition, providing a net permissive chromatin environment (orange traffic light). At the same time, PIF4 levels rise independently of HDA9 and (C) subsequently bind the G-Box motif in the YUCCA8 promoter. This triggers YUC8 accumulation, followed by turnover of IPyA to IAA, which induces elongation growth (green traffic light). Thermosensing by photo-activated Phytochrome B (PhyB-PFR; brown rectangles) inhibits PIF4 activity by a parallel mechanism. (D) When HDA9 is inactivated by mutation or TSA application, YUCCA8 is hyperacetylated at warm temperatures. This shifts the balance from net H2A.Z eviction to net deposition. As a result, PIF4 cannot efficiently bind the YUCCA8 promoter and IAA accumulation is prohibited, resulting in attenuation of thermomorphogenesis, despite a permissive warm-temperature environment. Possible secondary effects of HDA9 on other regulators of YUC8 expression (e.g., transcriptional regulation of repressors) or HDA9-mediated deacetylation of nonhistone proteins are not illustrated in this model.
Recent work demonstrated that high-temperature–induced hypocotyl elongation requires a cotyledon-derived mobile auxin signal and a permissive hypocotyl-localized thermosensor in established 4- to 8-d-old seedlings (49). In apparent contrast, HDA9 promoter activity was mainly detected in the hypocotyl/root junction and roots of germinating seedlings (Fig. 2A and SI Appendix, Fig. S2). Moreover, HDAC-dependent induction of YUC8 promoter activity at warm-temperature conditions (SI Appendix, Fig. S7) spatially and temporally colocalizes with HDA9 promoter activity (SI Appendix, Fig. S2A). Furthermore, HDA9 promoter activity and HDA9 protein levels declined during seedling establishment (Fig. 2 and SI Appendix, Fig. S2). Therefore, we propose that HDA9 defines an early temperature signaling pathway in the hypocotyl that is substituted by cotyledon-mediated temperature signaling during the course of seedling establishment (49). In agreement, although YUC8 expression remains detectable in the hypocotyl, YUC8 expression is more pronounced in the cotyledons of 4-d-old (51) and 7-d-old seedlings (49).
We propose, that due to HDA9-mediated deacetylation, eviction of H2A.Z at warm temperature can supersede SWR1/ARP6-mediated incorporation at the YUC8 locus in wild-type plants. This results in net eviction of H2A.Z, allowing for accessible YUC8 chromatin to which PIF4 can bind and activate transcription (Fig. 5). It is worth noting that acetylation changes in hda9-1 mutant seedlings were apparent only at the transcription start site and gene body of YUC8 at 27 °C (Fig. 4B), whereas H2A.Z is evicted upstream of the gene as well (Fig. 4E). This suggests that HDA9-mediated histone deacetylation is not directly causal for H2A.Z eviction but has a facilitating role.
Our model (Fig. 5) is supported by the observation that the SWR1 complex preferentially binds to acetylated nucleosomes and acetylation enhances the exchange of H2A for H2A.Z in yeast (48, 52). Moreover, H2A and H4 acetylation can stimulate SWR1 activity (52). Nevertheless, we cannot exclude alternative possibilities by which HDA9 might influence YUC8 transcription, for example, by silencing of a repressor or deacetylation of a nonhistone transcriptional regulator by HDA9. Whether HDA9 dependency for H2A.Z eviction extends beyond the YUC8 (and HSP70) locus is a subject for further study.
We demonstrate that HDA9 is not required for hypocotyl elongation in response to different light quality signals and by the red (R)/far-red (FR) photoreceptor and thermosensor phytochrome B. The thermosensory event triggering the HDA9-dependent thermosignaling pathway is still unknown. Since the HDA9 protein accumulates at high temperatures and recent data indicate that HDA9 is prone to proteasomal regulation (32), studying HDA9 proteasomal degradation and posttranslational modifications could provide hints toward a possible thermosensory mechanism.
HDA19 was recently identified as a positive regulator of thermomorphogenesis, whereas HDA15 negatively regulates thermomorphogenesis by directly suppressing high-temperature–responsive genes at control temperature (22). Whether HDA19 and HDA15, like HDA9, control thermomorphogenesis by mediating H2A.Z remains to be established. However, the limited overlap in differentially regulated genes in response to warm temperatures suggests that different RPD3-like HDACs affect separate branches of the thermomorphogenesis regulatory network (22). In accordance, we show here that HDA9 operates at least partly independent of the phyB thermosignaling and shade avoidance pathway, whereas HDA15 can be considered an integrator of light and temperature signaling as it interacts with HFR1 (LONG HYPOCOTYL IN FAR RED1), a positive regulator of photomorphogenesis (22), and with PIF3, which guides HDA15 to its target genes (53).
How HDA9 is recruited to its target loci at warm temperatures remains elusive, as HDA9 lacks DNA binding capacity. Obvious candidates to target HDA9 would be PIF4, which is required for YUC8 activation at high temperatures (6–8), and HSFA1a, which is required for daytime H2A.Z eviction at warm temperatures (20). However, we did not detect a direct interaction between HDA9 and HSFA1 nor between HDA9 and PIF4 (SI Appendix, Fig. S5). Future studies using in planta IP followed by mass spectrometry would be essential for identifying the multiprotein complex in which HDA9 operates.
HDA9 does interact with the SANT (SWI3/DAD2/N-CoR/TFIII-B) domain protein PWR (21, 25, 26, 54), which confers substrate binding specificity to HDA9, but does not bind chromatin directly. Both HDA9 (this work) and PWR (21) are required for YUC8 transcriptional induction and thermomorphogenesis. However, the role of PWR is more pleiotropic than that of HDA9, as the majority of genes misregulated in the pwr mutant are not affected in hda9-1 (32). In contrast, almost all genes misregulated in hda9-1 are also affected by the pwr mutation (32). Moreover, in the context of thermomorphogenesis, PWR is involved in the thermal induction of HSP70, PIF4, and YUC8 (21) transcription, whereas HDA9 appears to be essential primarily for the thermal induction of YUC8.
Recently, it was shown that HDA9 interacts with the Evening Complex (EC) component EARLY FLOWERING 3 (ELF3) to attenuate TIMING OF CAB EXPRESSION 1 (TOC1) core clock gene transcription in the early night (55). Furthermore, HDA9 interacts with HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES15 (HOS15) to control growth (32, 53, 56) and development and—together with HOS15, ELF3, ELF4, and LUX—functions in the EC to repress transcription of the floral activator GIGANTEA (56). Both ELF3 (9, 10) and TOC1 (57) are negative regulators of thermomorphogenesis during the night period. Our findings indicate that HDA9 is a positive regulator of thermomorphogenesis that operates during the light period, as the protein is stabilized at dawn and subsequently allows for YUC8-mediated thermomorphogenesis. These apparent contrasting roles of HDA9 in growth regulation during high-temperature signaling on one hand and as a member of the EC complex on the other, suggests that HDA9-mediated thermomorphogenesis likely occurs independent of its role in the EC, at least in young seedlings. This notion is supported by the observation that in the hda9-1 mutant the diurnal rhythmicity of elongation growth is attenuated during daytime and nighttime in both high-temperature and control temperature conditions (SI Appendix, Fig. S2A), whereas EC-complex mutants typically display enhanced elongation growth during the night period under high-temperature conditions (9, 10).
Other HDA9 interacting factors include the DNA-binding AHL22 protein (58) and the HDAC complex components SAP30 FUNCTION-RELATED 1 (AFR1) and AFR2 (59). Whether and how HDA9 interacting factors play a role in “type 3” vegetative plant thermomorphogenesis remains to be investigated further. Yet the finding of a HDA9-specific thermosignaling pathway provides a very promising target for knowledge-based breeding of robust thermotolerant crops that maintain productivity under the regime of projected global climate warming, without compromising for important light-responsiveness cues.
Materials and Methods
Plant Materials and Growth Conditions.
Arabidopsis seeds were obtained from the Nottingham Arabidopsis stock center (http://arabidopsis.info/; stock number between brackets) or were kind gifts of colleagues. The following genotypes were used: Col-0 wild type (N1092), hda9-1 (SALK_007123) (42), hda9-2 (GABI_305G03) (42), 35S::HDA9-8 (42), pHDA9-HDA9-HA (27), pif4-2 (N66043), 35S::PIF4-HA (31), phyB-9 (60) (N6217), pPIF4::LUC and pPIF4::PIF4-LUC (gift of Salomé Prat, Spanish National Research Council, Spain), arp6-1 (16), pHSP70::LUC (16), p35S:LUC (N9966), pHTA11:HTA11-FLAG (16). The pYUC8-n3GFP line contains a 2-kb fragment upstream of the YUC8 start codon coupled to a nuclear triple GFP reporter and was generated as described (61) (gift of Dolf Weijers, Wageningen University, Wageningen, The Netherlands). Double-mutant combinations were generated by crossing and (homozygous) progeny was selected by genotyping PCRs and/or testing for antibiotic resistance (SI Appendix, Table S2).
Plants were cultivated on sterile 0.8% agar, full strength Murashige-Skoog (MS including MES Buffer and vitamins, Duchefa) medium without sucrose on Petri dishes (plates), unless stated otherwise. Where applicable, TSA, Picloram, IAA, IPyA, or NPA (all Sigma-Aldrich) were dissolved in DMSO and supplemented to the MS-agar medium. DMSO solvent lacking added compounds was used as mock control (0.1% final concentration in all cases).
Before sowing, seeds were surface-sterilized by incubation in a solution of 0.8% bleach (Glorix, 4.5% [Cl]) in ethanol for 10 min, followed by twice washing with ethanol for 10 min, or seeds were gas-sterilized by chlorine gas for 3 h. Seeds were stratified for 2 to 3 d in darkness at 4 °C to synchronize germination. Thereafter, the plates were cultivated in identical climate-controlled growth cabinets (Snijders, Microclima 1,000) at either 22 °C (control) or 27 °C (high ambient temperature), in a short-day photoperiod (8-h light/16-h darkness), 100 to 125 µmol m−2 s−1 photosynthetic active radiation (PAR) white-light conditions and 70% relative humidity, unless stated otherwise. Long-day conditions consisted of 16-h light/8-h darkness, at otherwise identical conditions.
For skotomorphogenesis experiments, germination was induced by exposure to 4 h of white light. After this the plates were packed light-tight in aluminum foil and incubated in the growth cabinets as described above. Low R/FR light conditions (654 to 664 nm/724 to 734 nm: 0.15 in low R/FR compared to 1.8 in control white light) were obtained by supplementing the white growth chamber lights (120 µmol m−2 s−1 PAR) with FR LEDs (730 nm, Philips), without affecting the PAR. Low-blue light (B, 300 to 400 nm: 4.3 µmol m−2 s−1 in low blue compared to 58.4 µmol m−2 s−1 in control white light) was achieved by shading plants using a Lee Medium Yellow 100 filter, without affecting the PAR. Spectral neutral low light conditions were achieved by reducing the PAR by 90% to ∼10 to 15 µmol m−2 s−1 using spectral neutral shade cloth. Spectra were obtained by a Jaz Spectrofotometer and SpectraSuite analysis software (Ocean Optics).
Phenotyping.
Seedlings used for hypocotyl elongation quantifications on MS-plates were scanned using a flatbed scanner and hypocotyl lengths were measured using ImageJ image-analysis software (https://imagej.nih.gov/ij/).
For shift experiments, plants were sown on several MS-agar plates and put in 22 °C or 27 °C conditions. At dawn (lights on) and dusk (lights off) of each photoperiod, 1 plate was shifted to the other temperature condition (22 °C to 27 °C and vice versa) and left there for the remaining experimental period. At dusk of day 4 after initiation of germination the plates were scanned, and hypocotyl lengths were measured as described above.
Plants containing 10 to 12 true leaves, grown on standard potting soil (Primasta), were used for petiole length, leaf blade length, and leaf angle measurements. The plants were first photographed from the side for leaf angle measurements and subsequently harvested, flattened, and photographed from the top. Leaf angles (hyponasty) were determined from the photos by measuring the average elevation of 2 opposing leaves per plant relative to the horizontal (62), by ImageJ. Petiole and leaf blade lengths of all leaves of the plant were measured from the top photos using ImageJ, starting from the youngest leaf where a petiole was visible. The petiole and leaf blade length per plant was defined as the average of the lengths of the 3rd to 6th youngest leaves.
Flowering time was determined of plants grown on standard potting soil (Primasta) by counting the number of rosette leaves and the number of days after germination at floral bud appearance.
Construction of Transgenic HDA9 β-GLUCURONIDASE and LUCIFERASE Lines.
The 1,035-bp intergenic region upstream of the HDA9 (At3g44680) start codon (pHDA9), starting from the stop codon of the upstream gene At3g44690 and the HDA9 coding sequence (CDS), excluding the stop codon (CDS HDA9-stop) were PCR-amplified (primers in SI Appendix, Table S2), using the proofreading Phusion DNA polymerase (ThermoScientific) and cloned by GATEWAY BP recombination (Invitrogen) in entry vector pDONRP4-P1 (Box1; attL4 & attR1) or pDONR-221 (Box2 attL1 and attL2). To generate a pHDA9:GUS/GFP vector pDONR-221_pHDA9 (Box2) was recombined into destination vector pFAST-GO4 (63) by a LR recombination. pHDA9:fLUC-nosT (pHDA9:LUC) was constructed by multisite Gateway (Three-Fragment Vector Construction Kit; Invitrogen; Thermo Fisher Scientific) by recombining pDONRP4-P1-pHDA9 (Box1), pDONR221-fLUC-F7 (64) (Box2), and pEN-R2-6-L3_nosT (attR2 and attL3; Box3) (65) into destination vector pGreenII0125, with its resistance cassette replaced by a Norflurazon resistance marker (gift from Renze Heidstra, Wageningen University, Wageningen, The Netherlands). pHDA9:HDA9-fLUC was constructed by combining pDONRP4-P1-pHDA9 (Box1), pDONR-221_CDS HDA9-stop (Box2), and pDONR-L2rL3_fLUC (Box3) (64) into destination vector pGreenII0125. 35S::HDA9-fLUC was constructed by combining pDONR_L4L1r_35S (Box1), pDONR221-HDA9-stop (Box2) and pDONR-L2rL3_fLUC (Box3) (64) into destination vector pGreenII0125. All (multisite) GATEWAY reactions were performed according to manufacturer protocols. Primers used for cloning are listed in SI Appendix, Table S2.
Constructs were transformed to Escherichia coli DH5α cells, selected for by colony PCR and confirmed by restriction analysis and sequencing (Macrogen) before further usage. Expression vectors for plant transformation were transformed to electro-competent Agrobacterium tumefaciens strain C58 and plants were transformed by floral-dip transformation (66). Seeds carrying pHDA9-GUS/GFP were selected on their GFP fluorescence using a Zeiss fluorescence microscope and pHDA9:LUC, pHDA9:HDA9-LUC, and 35S:HDA9-LUC seedlings were selected and propagated on MS-agar plates containing 10 μM norflurazon, until homozygous lines were obtained.
Luciferase Assays.
For quantitative luciferase (LUC) assays, protein extracts were prepared from ∼25 mg harvested seedlings that were snap-frozen in liquid nitrogen. Tissue was homogenized using a mortar and a pestle in 100 µL 1× passive lysis buffer (PLB, Promega E1941), followed by 10-min incubation at room temperature. Col-0 wild-type seedlings lacking LUC activity were included with each replica as negative control. Debris was pelleted by centrifugation and subsequently 20 µL of supernatant was transferred to a 96-well Lumitrac plate (Promega). Unless stated otherwise, LUC activity was assayed using the LUC Assay System detection kit (Promega, #E1500) in a Glomax 96 microplate luminometer (Promega, #E6521), with the “LUC Assay System with Injector” protocol (2-s delay between injection and measurement, 10-s integration time). Thereafter, protein concentrations were determined of each sample using Bradford reagent (Sigma-Aldrich, #B6916). Absorbance was measured using a Biotech synergy HT-plate reader. A BSA (Sigma-Aldrich, #A7906) standard curve was included to calculate protein concentrations, to normalize the LUC signal to the protein concentration of each sample.
Visualization of LUC signals was done using a Hamamatsu ImaEM-X2 camera, with electron-multiplying gain.
For pYUC8-Luciferase experiments, seedlings were transiently transformed as previously described (67) with the dual-reporter construct carrying the YUC8 promoter fused to firefly-luciferase (fLUC) and the constitutive 35S:CaMV promoter fused to Renilla-luciferase (pYUC8:fLUC-35S:rLUC) (38). Next, 1.0-µm gold microcarriers (Bio-Rad, #1652263) were coated with the vector and transformed to 7-d-old seedlings that were pregrown on MS-agar plates under control 22 °C, SD conditions, using a Bio-Rad PDS-1000 He Biolistic particle delivery system, with 600-psi rupture discs (Bio-Rad, #1652327). After transformation, plants were transferred back to either control (22 °C) conditions or were placed in high-temperature (27 °C) conditions in a short-day regime. At dawn of the next morning (18- to 20-h incubation), the whole seedlings were snap-frozen in liquid N2. To determine fLUC activity, protein extracts were made as described above and activity was measured using the “Dual-Luciferase Reporter Assay System” (Promega, #E1960) with the “LUC Assay System with Injector” protocol (2-s delay between injection and measurement, 10-s integration time). The fLUC signal was normalized to the rLUC internal control.
Diurnal Luciferase Profiling in Germinating Seeds and Seedlings.
Seeds were sown on felt, drenched in 100 mL 0.5 mM D-luciferin (Promega) in 0.5× nutrient solution (Hyponex). For positioning of seeds, the felt was covered by an aluminum plate with 200 holes, each hole containing a single seed. After sowing the imbibed seeds were stratified for 3 d at 5 °C. Subsequently, seeds were placed in a custom-made climate cabinet with a high-performance PIXIS 1024 CCD camera (Princeton Instruments) fitted with a 35-mm f/1.4 Nikon SLR lens (Nikon). Temperature was kept constant 22 °C or 27 °C and relative humidity at 40%. The germinating seeds were grown for 7 d at a short-day photoperiod (8-h light/16-h dark cycles). Light was provided from the top by LEDs emitting light in B (420 to 500 nm), R (590 to 660 nm), and FR (680 to 760 nm) spectra. To mimic the low-light intensities and altered spectrum at the start of the day and end of the day, during the first and last hour of the daytime period the light intensity was 30 µmol m−2 s−1 and the B:R:FR ratio was 1:2:1. During the remaining hours of the daytime, light intensity was 100 µmol m−2 s−1 and B:R:FR ratio was 3:6:1. Every morning the seedlings were provided with fresh luciferin and nutrient solution by injecting 50 mL 0.5 mM D-luciferin (Promega) in 0.5× nutrient solution (Hyponex) into the felt on which the seedlings were growing. Top-view images were taken every 30 min with an exposure time of 7 min. The camera (Princeton Instruments) was fitted with a ZBPB074 Bandpass Filter (Asahi Spectra) to block chlorophyll fluorescence and LED lights were turned off 30 s before imaging to allow for chlorophyll fluorescence decay. LUC signal values were quantified from the images using ImageJ software by determining mean gray values of each single grid-compartment holding 1 seed/seedling. Effectiveness of thermomorphogenesis in these experimental conditions was determined by measuring hypocotyl length of 7-d-old seedlings shortly after finishing the LUC measurements (SI Appendix, Fig. S3C).
Measurements of Free IAA and Auxin Metabolite Profiling.
IAA, was extracted, purified, and analyzed as previously described by liquid chromatography-tandem mass spectrometry analysis, with minor modifications (68). IAA was extracted from ∼10 mg (fresh weight, FW) seedling tissue (2-d-old or 7-d-old, grown at either 22 °C or 27 °C) at 4 °C overnight in 1 mL methanol containing [phenyl 13C6]-IAA (0.02 nmol/mL) as internal standard. The methanol fraction was purified by anion-exchange column (Grace Extra Clean Amino 100 mg/1.5 mL solid-phase extraction; Grace Davison Discovery Sciences). The volume of the wash and elution solvent was scaled down to 1 mL each to compensate for the reduced column size.
Quantification of auxin metabolites and IAA were performed according to the method described by Pencík et al. (69). Samples (10-mg FW) were homogenized and extracted in 1.0 mL of ice-cold Na-phosphate buffer (50 mM, pH 7.0, 4 °C) containing 0.1% diethyldithiocarbamic acid sodium salt together with a mixture of stable isotope-labeled internal standards (5 pmol of [13C6]ANT, [13C6]IAA, [13C6]IAAsp, [13C6]IAGlu, [13C6]IAM, [2H5]IAOx, [13C6]oxIAA, and [2H2]TRA, 5 pmol of [13C6]IAN and [2H4]IPyA, and 50 pmol of [2H5]Trp per sample added). The extracts were purified using in-tip microSPE based on the StageTips technology (70). Briefly, a volume of 250 μL of each plant extract was acidified to pH 2.7 with 0.1 M hydrochloric acid (∼100 μL). Combined multi-StageTips (containing C18/SDB-XC layers) were activated sequentially, with 50 μL each of acetone, methanol, and water (by centrifugation at 434 × g, 15 min, 4 °C). After application of aliquots of the acidified sample (678 × g, 30 min, 4 °C), the microcolumns were washed with 50 μL of 0.1% acetic acid (525 × g, 20 min, 4 °C), and elution of samples was performed with 50 μL of 80% (vol/vol) methanol (525 × g, 20 min, 4 °C). Another 250 μL of the extract was derivatized by adding 100 μL of 0.75 M cysteamine (pH 8.2) to convert the labile compound IPyA to its respective thiazolidine derivatives (71). After 15-min incubation, the sample was adjusted to pH 2.7 and purified as described above. Both eluates were then evaporated to dryness in vacuum and stored at −20 °C. The levels of IAA, its precursors, conjugates and catabolites were determined using ultrahigh-performance liquid chromatography-electrospray tandem mass spectrometry (a 1290 Infinity LC system and a 6490 Triple Quadrupole LC/MS system, Agilent Technologies) using stable isotope-labeled internal standards as a reference (72). Four independent biological replicates were performed.
Time-Lapse Growth Assays.
A custom digital time-lapse camera system was developed in house to quantify hypocotyl growth throughout the photo- and dark period. A modified Canon EOS 350D DSLR camera was used, in which the standard internal IR and UV rejection filters were replaced by a 715-nm long-pass filter, allowing detection of wavelengths beyond 715 nm (73). The camera was placed in front of vertical-positioned MS-agar plates containing the seedlings. Photos were taken every 2 h, from imbibed stratified seed to 8-d-old seedlings, using an Aputure AP-R1C LCD Timer Remote controller. Hypocotyl lengths of individual plants were measured manually using ImageJ Image analysis software. A LED spotlight (940 nm ± 50 nm; Kingbright, #BL0106-15-28) was used to illuminate seedlings during the night. The emitted light by the LED was very weak and the emitted light was barely detectable above background levels during the day time (SI Appendix, Fig. S11 A–F), as determined using a Jaz Spectrofotometer and SpectraSuite analysis software (Ocean Optics). The emitted light did not interfere with plant development, as no de-etiolation and germination was observed in LED-exposed dark-grown seedlings, respectively imbibed stratified seeds, in otherwise complete darkness (SI Appendix, Fig. S11G).
qRT-PCR.
Seedlings were harvested at dawn and snap-frozen in liquid N2. Each sample for qPCRs contained at least 20 seedlings. RNA was isolated as described previously (74) with minor modifications. Genomic DNA was removed using DNaseI (Thermo Scientific) and cDNA was synthesized using RevertAid H Minus Reverse Transcriptase (Thermo Scientific) using a mix of odT18VN primer and oligo dT primers. qRT-PCR reactions were performed using SYBR green mastermix (Life Technologies) on a ViiA7 Real Time PCR system and ViiA7 software was used to analyze the data. Relative expression levels were calculated using the ΔΔCt method (75), normalized to the expression of the reference genes: At1G57870 and At5g08290. See SI Appendix, Table S2 for primers used.
β-Glucuronidase Staining and Microscopy.
Seedlings were harvested at dawn and vacuum-infiltrated in β-glucuronidase (GUS) staining buffer containing 0.2 mM X-gluc (5-bromo-4-chloro-3-indolyl glucuronide) in a sodium phosphate buffer (pH = 7.2, supplemented with Triton X-100, potassium ferrocyanide and potassium ferricyanide) on ice. Samples were incubated at room temperature for 30 min and subsequently bleached in ethanol baths with increasing concentration (20%, 30%, 50%, and 70%). The tissues were fixed in FAA fixative (50% ethanol, 5% [vol/vol] acetic acid, 3.7% [vol/vol] formaldehyde) and stored in 70% ethanol. Intensity-scoring of GUS staining per organ was performed visually, using a binocular on a scale from 0 (GUS staining absent) to 4 (GUS staining saturated).
GFP signals were captured using a Zeiss Axio Zoom.V16 Fluorescence Stereo Zoom Microscope with Axiocam 506 Color and HXP 200C Fluorescent Illuminator and ZEN Image acquiring software.
Transcriptomics.
For transcriptomic experiments, 12 batches, each containing >50 individual Col-0 wild-type, hda9-1, or pif4-2 mutant seedlings, were grown on MS-agar plates under control (22 °C) or high temperature (27 °C) conditions, with each batch sown and harvested on different dates. At dawn of day 3 (2 d-old) and day 8 (7 d-old), the plates were photographed and seedlings snap-frozen in liquid N2. Effectiveness of the treatments was confirmed by measuring the hypocotyl lengths of the replicates using ImageJ (SI Appendix, Fig. S6A). Next, RNA was isolated using the Sigma Spectrum Plant Total RNA isolation kit and gDNA was removed by on-column DNase treatment (Sigma-Aldrich). RNA integrity and concentration were checked using RNA 6000 Nano Chips on a Bioanalyzer (Agilent-2100). For RNA-seq library preparation, in total 3 samples were prepared for each genotype, treatment, and time-point, by combining isolated RNA of 3 individually harvested batches per sample. Illumina TruSeq RNA Library preparation and Illumina HiSeq2500 (high-throughput) single-end 50-bp sequencing was outsourced to Macrogen, Korea.
Quality control (QC) was performed in house on the raw sequencing reads prior to analysis using FastQC (www.bioinformatics.babraham.ac.uk/projects/fastqc). Subsequently, the RAW reads were aligned to the Arabidopsis genome (TAIR10) using TopHat v2.0.131 with the parameter settings: “bowtie” (76), “no-novel-juncs,” “p 6,” “G,” “min-intron-length 40,” “max-intron-length 2000.” On average, 91.6% (54.4 to 97.9%) of the RAW reads could be aligned to the genome per biological replicate. This represents an average of 45.1 (23.2 to 71.3) million mapped reads per sample. Aligned reads were summarized over annotated gene models using HTSeq-count (77) v0.6.12 with settings: “-stranded no,” “-i gene_id.” From the TAIR10 GTF file all ORFs of which the annotation starts with “CPuORF” were manually removed prior summarization to avoid not counting all double annotated bZIP TF family members. Sample counts were depth-adjusted and differential expression was determined using the DESeq package (78) with default settings. Gene-expression differences were determined by a full factorial ANOVA (genotype × temperature) per time point, genes with a P < 0.001 and an absolute log2 ratio difference >0.5 were taken as significant. Tukey honest significant difference (HSD) was used to pairwise compare samples. Genes with a temperature effect are show in the Venn diagrams in SI Appendix, Fig. S6 F and G.
All statistics associated with testing for differential gene expression were performed with R (www.r-project.org). Gene-expression profiles were hierarchically clustered using the Euclidean distance measure with average linkage using the Bioinformatics package of Matlab release 2014a. PCA analysis (SI Appendix, Fig. S6 C–E) was done on the whole dataset obtained by taking the log2 ratio of each sample with the mean for each gene by using the “prcomp” function from R v3.4.1 and visualization using package “ggplot2”; GO analysis (Table 1 and SI Appendix, Table S1) was done by using the hypergeometric test function “phyper” from R.
Protein Extraction and Western Blot Analysis.
Seedlings were snap-frozen in liquid N2 at dawn of day 3, postgermination (2-d-old seedlings). Total protein was extracted by grinding 50 mg of tissue in 4× Laemmli sample buffer (79). SDS/PAGE analysis was performed using 4 to 12% precast, Bis⋅Tris gradient gels in MES buffer (Life Technologies). The Bio-Rad transfer system and nitrocellulose membrane were used for Western blot transfer. The following antibodies were used for Western blot analysis: anti-H3K9K14Ac (Diagenode), anti–HA-Peroxidase, High Affinity Roche; anti-Flag (Sigma) (all 1:1,000 dilution); anti-UGPase (Agrisera) (1:5,000); anti-mouse HRP (Bio-Rad), anti-rabbit HRP (Bio-Rad) (1:10000 dilution).
ChIP-qPCR.
ChIP assays were performed on 2 g of plant tissue. Chromatin extraction was carried out as previously described (80, 81). DNA was sheared using a Bioruptor sonicator (Diagenode, B01020001) using the following settings: 20 cycles 30-s ON, 30-s OFF at high power. Anti-H3K9K14Ac (Diagenode), anti-FLAG (Sigma), and anti-HA tag-ChIP Grade (ab9110; Abcam) were used to IP the chromatin according to the manufacturer’s instructions. ChIP-qPCR was performed using the following cycles: 95 °C × 2 min, 95 °C × 3 s, 59.5 °C × 30 s for 50 cycles, 95 °C × 1 min and 60 °C × 30 s to calculate the melting curve. Oligonucleotides for YUC8, HSP70, and GYPSY genomic regions were derived from Lee et al. (11) and Kumar and Wigge (16), respectively, and for the PIF4 promoter from Zhu et al. (57). Relative enrichment was calculated as described previously (80). Effectiveness of the high-temperature treatment was confirmed by measurement of hypocotyl lengths of 7-d-old seedlings, as described above.
BiFC.
BiFC experiments on transiently transformed Nicotiana benthamiana epidermal cells and protein extraction was performed as previously described (37). For protein detection, ∼100 mg of tissue was homogenized in 4× Laemmli protein sample buffer, boiled at 100 °C, and 25 µL of the protein extract was separated on a 4 to 12% Bolt SDS/PAGE (ThermoFisher). Western blot analysis was subsequently performed using a Bio-Rad system on nitrocellulose membrane probed with an anti-HA- HRP [(1:1,000 dilution (Roche)] and an anti-cMYC antibody (1:2,000 [Santa Cruz]). Confocal microscopy was performed using a Leica SP8 inverted microscope. All BiFC constructs were cloned first in pDONR207 (ThermoFisher) followed by LR recombination into Gateway-compatible BiFC plasmids (82). All inserts in pDONR, spYNe, and spYCe were sequenced prior to BiFC experiments. Primers used for cloning are listed in SI Appendix, Table S2.
Yeast 2-Hybrid.
Assays were performed as previously described (37). The yeast strain MaV203 was used for the auxotrophy and GUS assays according to the manufacturer’s instructions (Invitrogen). HSFA1 and PIF4 were cloned in pDEST32 (GAL4 DB) and HDA9 in pDEST22 (GAL4 AD) by LR recombination from pDONR221. All constructs were cloned in pDONR221 using the same strategy as for pDONR207 according to the manufacturer’s instructions.
Statistical Analyses.
Statistical analyses per experiment are described in the figure legends and results and full test models are shown in Dataset S1. Two-sided t test using the means and SDs were used to determine statistical significance for the difference in responses. These significance tests were done by calculating the mean response from the means of the 2 treatments and the SD from the 2 treatments, then testing was done on these mean responses (change) and SD, taking into account the number of observations in each case. Full factorial ANOVA’s were done to determine significance of involved variables. Subsequent pairwise ANOVA’s were done to further determine the significance of the observed differences. Tukey HSD tests were used for all pairwise statistical differences and effects.
Data Availability.
RNA Sequencing data generated in this study have been deposited in the GEO repository (https://www.ncbi.nlm.nih.gov/geo/) under accession code GSE121383 (83). All data supporting the findings of this study are available within the paper, its SI Appendix, and in Dataset S1. Computational codes used for the analysis of the data are available upon request.
Supplementary Material
Acknowledgments
We thank Phil Wigge (The Sainsbury Laboratory at the University of Cambridge), Kerry Franklin (University of Bristol), Yoh-Sun Noh (Seoul National University), Salomé Prat (Spanish National Research Council), Hongtao Liu (National Key Laboratory, China), Renze Heidstra, and Barbara Möller and Dolf Weijers (Wageningen University) for sharing materials; Ronald Pierik and Lot Gommers (Utrecht University) for sharing equipment; Wiebe Nijland and Steven de Jong (Utrecht University) for advising on the development of the time-lapse camera set-up and for sharing their camera; and Machiel Beijaert, Kim Nijhof, Bram Janssen, Koen van Oostrom, Rebecca Lippmann, and Molly Harper for technical assistance. This work was supported by VENI Grants 863.11.008 (to M.v.Z.) and 863.15.010 (to W.K.), and Graduate School Starting Materials Grant ALWGSU 831.130.02 (to L.C.v.d.W. and M.v.Z.) and Graduate School Green Top Sectors GSGT.2018.007 (to M.P. and M.v.Z.), of the Netherlands Organization for Scientific Research. K.L. acknowledges support from the Swedish Foundation for Strategic Research and the Swedish Research Council, and the Swedish Metabolomics Centre for the use of instrumentation. O.N. was financially supported by the Ministry of Education, Youth, and Sports of the Czech Republic (European Regional Development Fund-Project “Plants as a tool for sustainable global development” no. CZ.02.1.01/0.0/0.0/16_019/0000827). E.K. is grateful to the John Grieve Bequest and the Biotechnology and Biological Sciences Research Council for New Investigator Grant Award BB/M023079/1. E.V. is supported by a Medical, Veterinary, and Life Sciences Doctoral Studentship from the University of Glasgow. M.v.H., U.S., and A.R.v.d.K. acknowledge the research programme Compact Plants (13149), which is (partly) financed by the Netherlands Organisation for Scientific Research. U.S. was supported by Erasmus, Mundus Action 2 project TIMUR (Training of Individuals through Mobility from Uzbek Republic to European Union). S.B. acknowledges the Australian Research Council Discovery Project DP190101818.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE121383).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1911694116/-/DCSupplemental.
References
- 1.Challinor A. J., et al. , A meta-analysis of crop yield under climate change and adaptation. Nat. Clim. Chang. 4, 287–291 (2014). [Google Scholar]
- 2.Zhao C., et al. , Temperature increase reduces global yields of major crops in four independent estimates. Proc. Natl. Acad. Sci. U.S.A. 114, 9326–9331 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crawford A. J., McLachlan D. H., Hetherington A. M., Franklin K. A., High temperature exposure increases plant cooling capacity. Curr. Biol. 22, R396–R397 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Quint M., et al. , Molecular and genetic control of plant thermomorphogenesis. Nat. Plants 2, 15190 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Casal J. J., Balasubramanian S., Thermomorphogenesis. Annu. Rev. Plant Biol. 70, 321–346 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Koini M. A., et al. , High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr. Biol. 19, 408–413 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Sun J., Qi L., Li Y., Chu J., Li C., PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating arabidopsis hypocotyl growth. PLoS Genet. 8, e1002594 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franklin K. A., et al. , Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl. Acad. Sci. U.S.A. 108, 20231–20235 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Box M. S., et al. , ELF3 controls thermoresponsive growth in Arabidopsis. Curr. Biol. 25, 194–199 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Raschke A., et al. , Natural variants of ELF3 affect thermomorphogenesis by transcriptionally modulating PIF4-dependent auxin response genes. BMC Plant Biol. 15, 197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee H.-J., et al. , FCA mediates thermal adaptation of stem growth by attenuating auxin action in Arabidopsis. Nat. Commun. 5, 5473 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Ballaré C. L., Pierik R., The shade-avoidance syndrome: Multiple signals and ecological consequences. Plant Cell Environ. 40, 2530–2543 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Legris M., et al. , Phytochrome B integrates light and temperature signals in Arabidopsis. Science 354, 897–900 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Jung J.-H., et al. , Phytochromes function as thermosensors in Arabidopsis. Science 354, 886–889 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Sidaway-Lee K., Costa M. J., Rand D. A., Finkenstadt B., Penfield S., Direct measurement of transcription rates reveals multiple mechanisms for configuration of the Arabidopsis ambient temperature response. Genome Biol. 15, R45 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar S. V., Wigge P. A., H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140, 136–147 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Boden S. A., Kavanová M., Finnegan E. J., Wigge P. A., Thermal stress effects on grain yield in Brachypodium distachyon occur via H2A.Z-nucleosomes. Genome Biol. 14, R65 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zha P., Jing Y., Xu G., Lin R., PICKLE chromatin-remodeling factor controls thermosensory hypocotyl growth of Arabidopsis. Plant Cell Environ. 40, 2426–2436 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Huai J., et al. , SEUSS and PIF4 coordinately regulate light and temperature signaling pathways to control plant growth. Mol. Plant 11, 928–942 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Cortijo S., et al. , Transcriptional regulation of the ambient temperature response by H2A.Z-nucleosomes and HSF1 transcription factors in Arabidopsis. Mol. Plant 10, 1258–1273 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tasset C., et al. , POWERDRESS-mediated histone deacetylation is essential for thermomorphogenesis in Arabidopsis thaliana. PLoS Genet. 14, e1007280 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen Y., et al. , Arabidopsis histone deacetylase HDA15 directly represses plant response to elevated ambient temperature. Plant J., 10.1111/tpj.14492 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Kumar S. V., H2A.Z at the core of transcriptional regulation in plants. Mol. Plant 11, 1112–1114 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Mizuguchi G., et al. , ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303, 343–348 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Chen X., et al. , POWERDRESS interacts with HISTONE DEACETYLASE 9 to promote aging in Arabidopsis. eLife 5, e17214 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim Y. J., et al. , POWERDRESS and HDA9 interact and promote histone H3 deacetylation at specific genomic sites in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 113, 14858–14863 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang M.-J., Jin H.-S., Noh Y.-S., Noh B., Repression of flowering under a noninductive photoperiod by the HDA9-AGL19-FT module in Arabidopsis. New Phytol. 206, 281–294 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Capovilla G., Schmid M., Posé D., Control of flowering by ambient temperature. J. Exp. Bot. 66, 59–69 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Verhage L., Angenent G. C., Immink R. G. H., Research on floral timing by ambient temperature comes into blossom. Trends Plant Sci. 19, 583–591 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Kim W., Latrasse D., Servet C., Zhou D.-X., Arabidopsis histone deacetylase HDA9 regulates flowering time through repression of AGL19. Biochem. Biophys. Res. Commun. 432, 394–398 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Lorrain S., Allen T., Duek P. D., Whitelam G. C., Fankhauser C., Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53, 312–323 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Mayer K. S., et al. , HDA9-PWR-HOS15 is a core histone deacetylase complex regulating transcription and development. Plant Physiol. 180, 342–355 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Foreman J., et al. , Light receptor action is critical for maintaining plant biomass at warm ambient temperatures. Plant J. 65, 441–452 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Yamashino T., et al. , A Link between circadian-controlled bHLH factors and the APRR1/TOC1 quintet in Arabidopsis thaliana. Plant Cell Physiol. 44, 619–629 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Nusinow D. A., et al. , The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475, 398–402 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshida M., Horinouchi S., Beppu T., Trichostatin A and trapoxin: Novel chemical probes for the role of histone acetylation in chromatin structure and function. BioEssays 17, 423–430 (1995). [DOI] [PubMed] [Google Scholar]
- 37.Perrella G., et al. , ZINC-FINGER interactions mediate transcriptional regulation of hypocotyl growth in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 115, E4503–E4511 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma D., et al. , Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc. Natl. Acad. Sci. U.S.A. 113, 224–229 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao Y., et al. , A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291, 306–309 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Ha J.-H., Lee H.-J., Jung J.-H., Park C.-M., Thermo-induced maintenance of photo-oxidoreductases underlies plant autotrophic development. Dev. Cell 41, 170–179.e4 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Hu Z., et al. , Histone acetyltransferase GCN5 is essential for heat stress-responsive gene activation and thermotolerance in Arabidopsis. Plant J. 84, 1178–1191 (2015). [DOI] [PubMed] [Google Scholar]
- 42.van Zanten M., et al. , HISTONE DEACETYLASE 9 represses seedling traits in Arabidopsis thaliana dry seeds. Plant J. 80, 475–488 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Perrella G., et al. , Histone deacetylase complex1 expression level titrates plant growth and abscisic acid sensitivity in Arabidopsis. Plant Cell 25, 3491–3505 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li J., Lin Q., Wang W., Wade P., Wong J., Specific targeting and constitutive association of histone deacetylase complexes during transcriptional repression. Genes Dev. 16, 687–692 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tian L., et al. , Reversible histone acetylation and deacetylation mediate genome-wide, promoter-dependent and locus-specific changes in gene expression during plant development. Genetics 169, 337–345 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Daxinger L., et al. , Unexpected silencing effects from T-DNA tags in Arabidopsis. Trends Plant Sci. 13, 4–6 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Zerzaihi O., Chriett S., Vidal H., Pirola L., Insulin-dependent transcriptional control in L6 rat myotubes is associated with modulation of histone acetylation and accumulation of the histone variant H2A.Z in the proximity of the transcriptional start site. Biochem. Cell Biol. 92, 61–67 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Ranjan A., et al. , Nucleosome-free region dominates histone acetylation in targeting SWR1 to promoters for H2A.Z replacement. Cell 154, 1232–1245 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bellstaedt J., et al. , A mobile auxin signal connects temperature sensing in cotyledons with growth responses in hypocotyls. Plant Physiol. 180, 757–766 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yuan L., Chen X., Chen H., Wu K., Huang S., Histone deacetylases HDA6 and HDA9 coordinately regulate valve cell elongation through affecting auxin signaling in Arabidopsis. Biochem. Biophys. Res. Commun. 508, 695–700 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Stavang J. A., et al. , Hormonal regulation of temperature-induced growth in Arabidopsis. Plant J. 60, 589–601 (2009). [DOI] [PubMed] [Google Scholar]
- 52.Altaf M., et al. , NuA4-dependent acetylation of nucleosomal histones H4 and H2A directly stimulates incorporation of H2A.Z by the SWR1 complex. J. Biol. Chem. 285, 15966–15977 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu X., et al. , PHYTOCHROME INTERACTING FACTOR3 associates with the histone deacetylase HDA15 in repression of chlorophyll biosynthesis and photosynthesis in etiolated Arabidopsis seedlings. Plant Cell 25, 1258–1273 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suzuki M., et al. , OLIGOCELLULA1/HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES15 promotes cell proliferation with HISTONE DEACETYLASE9 and POWERDRESS during leaf development in Arabidopsis thaliana. Front. Plant Sci. 9, 580 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee K., Mas P., Seo P. J., The EC-HDA9 complex rhythmically regulates histone acetylation at the TOC1 promoter in Arabidopsis. Commun. Biol. 2, 143 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park H. J., et al. , HOS15 interacts with the histone deacetylase HDA9 and the evening complex to epigenetically regulate the floral activator GIGANTEA. Plant Cell 31, 37–51 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu J.-Y., Oh E., Wang T., Wang Z.-Y., TOC1-PIF4 interaction mediates the circadian gating of thermoresponsive growth in Arabidopsis. Nat. Commun. 7, 13692 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yun J., Kim Y.-S., Jung J.-H., Seo P. J., Park C.-M., The AT-hook motif-containing protein AHL22 regulates flowering initiation by modifying FLOWERING LOCUS T chromatin in Arabidopsis. J. Biol. Chem. 287, 15307–15316 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gu X., Wang Y., He Y., Photoperiodic regulation of flowering time through periodic histone deacetylation of the florigen gene FT. PLoS Biol. 11, e1001649 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reed J. W., Nagpal P., Poole D. S., Furuya M., Chory J., Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5, 147–157 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Möller B. K., et al. , Auxin response cell-autonomously controls ground tissue initiation in the early Arabidopsis embryo. Proc. Natl. Acad. Sci. U.S.A. 114, E2533–E2539 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Millenaar F. F., et al. , Differential petiole growth in Arabidopsis thaliana: Photocontrol and hormonal regulation. New Phytol. 184, 141–152 (2009). [DOI] [PubMed] [Google Scholar]
- 63.Shimada T. L., Shimada T., Hara-Nishimura I., A rapid and non-destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana. Plant J. 61, 519–528 (2010). [DOI] [PubMed] [Google Scholar]
- 64.Peviani A., Lastdrager J., Hanson J., Snel B., The phylogeny of C/S1 bZIP transcription factors reveals a shared algal ancestry and the pre-angiosperm translational regulation of S1 transcripts. Sci. Rep. 6, 30444 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Karimi M., Bleys A., Vanderhaeghen R., Hilson P., Building blocks for plant gene assembly. Plant Physiol. 145, 1183–1191 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Clough S. J., Bent A. F., Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998). [DOI] [PubMed] [Google Scholar]
- 67.Rahmani F., et al. , Sucrose control of translation mediated by an upstream open reading frame-encoded peptide. Plant Physiol. 150, 1356–1367 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ruyter-Spira C., et al. , Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: Another belowground role for strigolactones? Plant Physiol. 155, 721–734 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pencík A., et al. , Ultra-rapid auxin metabolite profiling for high-throughput mutant screening in Arabidopsis. J. Exp. Bot. 69, 2569–2579 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rappsilber J., Ishihama Y., Mann M., Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 (2003). [DOI] [PubMed] [Google Scholar]
- 71.Novák O., et al. , Tissue-specific profiling of the Arabidopsis thaliana auxin metabolome. Plant J. 72, 523–536 (2012). [DOI] [PubMed] [Google Scholar]
- 72.Rittenberg D., Foster G. L., A new procedure for quantitative analysis by isotope dilution, with application to the determination of amino acids and fatty Acids. J. Biol. Chem. 133, 737–744 (1940). [Google Scholar]
- 73.Nijland W., et al. , Monitoring plant condition and phenology using infrared sensitive consumer grade digital cameras. Agric. For. Meteorol. 184, 98–106 (2014). [Google Scholar]
- 74.Oñate-Sánchez L., Vicente-Carbajosa J., DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Res. Notes 1, 93 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Livak K. J., Schmittgen T. D., Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) Method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- 76.Trapnell C., Pachter L., Salzberg S. L., TopHat: Discovering splice junctions with RNA-seq. Bioinformatics 25, 1105–1111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Anders S., Pyl P. T., Huber W., HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Anders S., Huber W., Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brown B. A., et al. , A UV-B-specific signaling component orchestrates plant UV protection. Proc. Natl. Acad. Sci. U.S.A. 102, 18225–18230 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kaiserli E., et al. , Integration of light and photoperiodic signaling in transcriptional nuclear foci. Dev. Cell 35, 311–321 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bowler C., et al. , Chromatin techniques for plant cells. Plant J. 39, 776–789 (2004). [DOI] [PubMed] [Google Scholar]
- 82.Walter M., et al. , Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40, 428–438 (2004). [DOI] [PubMed] [Google Scholar]
- 83.Woude L. C. v. d., Snoek L. B., Verk M. C. v., Zanten M. v., High temperature transcriptomes of mutants in HDA9, PIF4 and Col-0 wild type of young Arabidopsis seedlings. Gene Expression Omnibus (GEO). https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE121383. Deposited 17 October 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA Sequencing data generated in this study have been deposited in the GEO repository (https://www.ncbi.nlm.nih.gov/geo/) under accession code GSE121383 (83). All data supporting the findings of this study are available within the paper, its SI Appendix, and in Dataset S1. Computational codes used for the analysis of the data are available upon request.