Significance
The oncoprotein transcription factor MYC is a validated but challenging anticancer target. In this work, we show that WDR5—a well-structured protein with druggable pockets—could be a focal point for effective anti-MYC therapies. We demonstrate that WDR5 recruits MYC to chromatin to control the expression of genes connected to protein synthesis, a process that is arguably deregulated in all cancers. We also show that disrupting the interaction between MYC and WDR5 causes existing tumors to regress. These findings raise the possibility that the MYC–WDR5 nexus could be targeted to treat cancer.
Keywords: cancer, chromatin, MYC, lymphoma, cancer therapy
Abstract
The oncoprotein transcription factor MYC is overexpressed in the majority of cancers. Key to its oncogenic activity is the ability of MYC to regulate gene expression patterns that drive and maintain the malignant state. MYC is also considered a validated anticancer target, but efforts to pharmacologically inhibit MYC have failed. The dependence of MYC on cofactors creates opportunities for therapeutic intervention, but for any cofactor this requires structural understanding of how the cofactor interacts with MYC, knowledge of the role it plays in MYC function, and demonstration that disrupting the cofactor interaction will cause existing cancers to regress. One cofactor for which structural information is available is WDR5, which interacts with MYC to facilitate its recruitment to chromatin. To explore whether disruption of the MYC–WDR5 interaction could potentially become a viable anticancer strategy, we developed a Burkitt's lymphoma system that allows replacement of wild-type MYC for mutants that are defective for WDR5 binding or all known nuclear MYC functions. Using this system, we show that WDR5 recruits MYC to chromatin to control the expression of genes linked to biomass accumulation. We further show that disrupting the MYC–WDR5 interaction within the context of an existing cancer promotes rapid and comprehensive tumor regression in vivo. These observations connect WDR5 to a core tumorigenic function of MYC and establish that, if a therapeutic window can be established, MYC–WDR5 inhibitors could be developed as anticancer agents.
The MYC oncogenes encode a set of 3 highly related proteins that feature prominently in cancer. Overexpression of at least one MYC protein is observed in more than half of all malignancies, and estimates suggest that up to one-third of all cancer deaths can be attributed to inappropriate MYC expression or activity (1). MYC proteins function by regulating gene expression, dimerizing with their obligate partner MAX (2) to form a DNA-binding transcription factor that controls the transcription of genes linked to protein synthesis, metabolism, proliferation, and genome instability—a near perfect suite of activities for the initiation, progression, and maintenance of the tumorigenic state (1).
The pervasive involvement of MYC in cancer has fueled interest in the notion that MYC can be targeted to treat malignancies. It is clear that inhibiting MYC in the context of an existing cancer promotes tumor regression in mice (3), even in cases where MYC is not the primary oncogenic lesion (4). A number of strategies have been developed to mitigate MYC overexpression (5) or to interfere with cellular processes hijacked by MYC (6), but in terms of blocking MYC function, the options are limited. Indeed, the most obvious route to MYC inhibition—disrupting target gene binding—is daunting, as the MYC:MAX interface is not readily amenable to pharmacological inhibition. Recently, however, it emerged that target gene recognition by MYC does not solely depend on interaction with MAX. Modeling reveals that ∼90% of MYC binding events in cells cannot be accounted for in terms of the affinity of MYC:MAX dimers for DNA (7), and a number of nuclear proteins have been shown to facilitate the recruitment of MYC to its target genes in the context of chromatin (8–11). If these chromatin cofactors can be understood, they could expose new avenues through which the ability of MYC to regulate tumorigenic transcriptional programs can be inhibited.
We identified WDR5, a protein that nucleates the assembly of histone modifier complexes (12), as a factor that facilitates the recruitment of MYC to chromatin (8). MYC binds directly to a shallow hydrophobic cleft on WDR5 that encompasses less than 800 Å2 of buried protein surface. Structure-guided mutations in MYC that disable interaction with WDR5 decrease binding of MYC to chromatin and attenuate its tumorigenic potential in mice. These observations led us to speculate that interaction of MYC with WDR5 stabilizes MYC:MAX dimers on DNA at key protumorigenic target genes and that as a result the MYC–WDR5 interface could be a focal point for pharmacological inhibition of MYC. But if the MYC–WDR5 connection is to be therapeutically pursued, we need to pinpoint target gene networks that are regulated by MYC and WDR5 in cancer cells and determine if these are relevant to the broad tumorigenic functions of MYC. Importantly, we also need to establish whether disrupting the MYC–WDR5 interaction in the context of a preexisting cancer will promote its regression. This latter criteria is of premier importance for cancer therapy, but is a standard that is rarely met for MYC cofactors.
Here, we describe a Burkitt’s lymphoma (BL) system that allows us to interrogate transcriptional programs that are controlled by the MYC–WDR5 connection and to evaluate its importance for tumor maintenance. We show that WDR5 recruits MYC to regulate a set of genes that are linked to protein synthesis and demonstrate that disrupting the MYC–WDR5 interaction is as effective as disabling all of the DNA-binding capabilities of MYC at promoting tumor regression. These findings solidify the concept that, if a therapeutic window can be established, inhibitors of the MYC–WDR5 connection could have antitumor activity.
Results
An Inducible Exon Swap System to Study the MYC–WDR5 Interaction.
To probe the importance of the MYC–WDR5 interaction in the context of a MYC-driven cancer, we built a system that allows exchange of wild-type (WT) for WDR5-interaction-defective (WBM) MYC in a BL cell line. We chose the Ramos BL line, because it carries the t(8:14) translocation that places c-MYC under the control of the IgH locus (Fig. 1A), and does not express the untranslocated c-MYC allele (13). This system enabled us to express the WBM mutant as the sole form of MYC in the cell and determine the impact of loss of interaction with WDR5 on primary transcriptional processes, as well as tumor engraftment and maintenance.
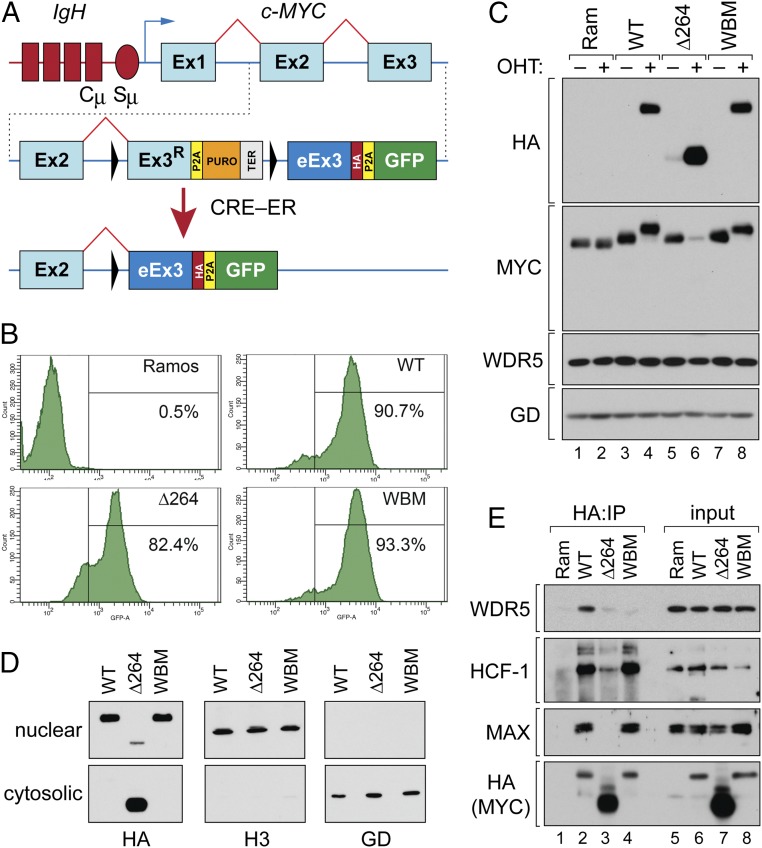
Fig. 1.
Inducible c-MYC exon swap in a BL cell line. (A) Structure of the c-MYC gene in Ramos cells (Top). Chromosome 14 is in red and shows part of the IgH locus including the constant (Cµ) and switch (Sµ) regions. Chromosome 8 is depicted in blue and shows exons 1 to 3 of c-MYC (Ex1–Ex3). Schematic of the modified locus in the unswitched state (Middle). Ex3R; recoded WT exon 3. P2A, “self-cleaving” peptide; PURO, puromycin resistance gene; TER, transcriptional terminator; eEx3, exchanged exon 3 (encoding WT, WBM, or ∆264 MYC sequences); HA, hemagglutinin epitope tag. LoxP sites are the black triangles. Configuration of the altered locus after activation of ER-linked CRE recombinase (Bottom). (B) Cells were treated with 20 nM OHT for 24 h, and GFP-positive cells scored by flow cytometry. The percentage of GFP-positive cells in each population is shown. (C) Western blot, performed on lysates from parental Ramos CRE–ER expressing cells (Ram), or modified cells expressing the WT, WBM, or ∆264 c-MYC proteins. Cells were untreated (−) or treated (+) with 20 nM OHT for 24 h prior to lysate preparation. Blots were probed with anti-HA, anti-MYC, anti-WDR5, and anti-GAPDH (GD) antibodies as indicated. (D) Engineered Ramos cells were switched to express the indicated MYC protein and subject to biochemical fractionation into nuclear and cytosolic extracts. Extracts were then probed with antibodies against HA (switched MYC), histone H3 (H3; nuclear), or GAPDH (GD; cytosolic). (E) Cell lines were treated with 20 nM OHT (24 h), lysates prepared, and HA-tagged MYC proteins immunoprecipitated (IP) by an anti-HA antibody. Immune complexes (lanes 1 through 4) were then probed for WDR5, HCF-1, MAX, or HA-tagged MYC by Western blotting. A sample of the lysates (input) were also probed by Western blot (lanes 5 through 8). This IP is representative of 3 independent experiments.
The WBM mutation (I262E/V264E/V265E) alters residues encoded within the third exon of c-MYC, permitting us to use an exon 3 swap strategy to express WBM mutant MYC (Fig. 1A). We also included a WT exon 3 swap as a control, and another exon 3 swap that truncates MYC at residue 264 (∆264), removing sequences required for nuclear localization and DNA binding. Ramos cells were engineered to express CRE recombinase linked to the estrogen receptor hormone binding domain (CRE–ER). We then used CRISPR-facilitated homologous recombination to introduce these modules into the translocated c-MYC locus (SI Appendix, Fig. S1A) and identified clones with appropriate integration by Southern blotting (SI Appendix, Fig. S1 B and C). This strategy replaces endogenous exon 3 of c-MYC with a LoxP-flanked cassette that carries a recoded wild-type exon 3 (Ex3R), a puromycin resistance gene (PURO), and a transcriptional terminator (TER). Activation of CRE–ER in these cells excises the Ex3R-PURO-TER cassette (Fig. 1A), bringing in place exchanged exon 3 (eEx3) encoding WT, WBM, or ∆264 MYC sequences. After swap, MYC proteins carry an HA-epitope tag, and swapped cells express green fluorescent protein (GFP). We refer to these lines as “WT,” “WBM,” and “∆264,” either “unswitched” or “switched.”
To trigger the switch, we treated cells with 4-hydroxytamoxifen (OHT) for 24 h, at which time ∼80 to 90% of cells were GFP positive (Fig. 1B). At that time, and in an OHT-dependent manner, we detected robust expression of all 3 HA-tagged proteins (Fig. 1C). Probing for total c-MYC levels (Fig. 1C), endogenous MYC was undetectable after switch and was replaced by the switched MYC species, which—for the WT and WBM MYC proteins—were expressed at levels similar to endogenous MYC. The ∆264 MYC protein is not detected by the α-MYC antibody we used, but is expressed at higher levels than WT MYC, as determined by α-HA blotting (Fig. 1C). The steady-state WDR5 levels are unaffected by the switch (Fig. 1C), and, as expected (8), the WBM MYC mutant: 1) localizes to the nuclear fraction (Fig. 1D), 2) retains the ability to associate with MAX and HCF-1 (14) (Fig. 1E), and 3) is defective for interaction with WDR5 (Fig. 1E). Also as expected, the ∆264 mutant is mostly localized to the cytosol (Fig. 1D) and is defective for interaction with WDR5, HCF-1, and MAX (Fig. 1E). We conclude that our system is appropriate for interrogating the consequences of disrupting the MYC–WDR5 interaction and for comparing these effects with those associated with a total loss in the ability of MYC to localize to the nucleus and bind chromatin (∆264).
MYC and WDR5 Colocalize on Chromatin at Genes Involved in Protein Synthesis.
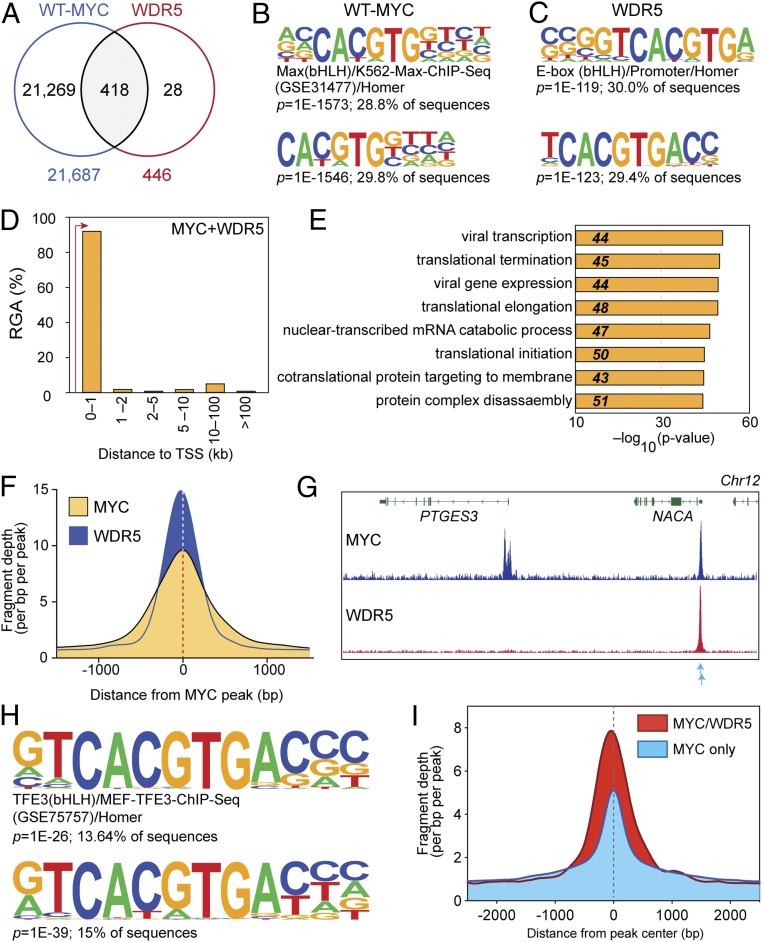
We first determined the location of MYC and WDR5 on chromatin in Ramos cells. Using chromatin immunoprecipitation coupled to next-generation sequencing (ChIP-Seq), we identified ∼22,000 MYC-binding sites (Fig. 2A), which were enriched in the “E-box” DNA motif (Fig. 2B), divided equally between transcription start site (TSS) proximal and distal (SI Appendix, Fig. S2A), and linked to genes connected to the core functions of MYC, including translation (SI Appendix, Fig. S2B). For WDR5, we tracked ∼450 binding sites (Fig. 2A)—30% of which contained an E-box (Fig. 2C), and 94% of which are also bound by MYC (Fig. 2A). As expected from the extensive overlap of MYC at WDR5-binding sites, the relevant properties of WDR5 sites and WDR5/MYC sites are similar; they are mostly promoter proximal (Fig. 2D and SI Appendix, Fig. S2C) and strongly enriched in genes connected to protein synthesis (Fig. 2E and SI Appendix, Fig. S2D), including ∼50 subunits of the ribosome, nucleolar RNAs, and translation initiation factors (Dataset S1). Looking closely at MYC/WDR5 cobound sites, several interesting features emerge. First, the ChIP-Seq profiles for the 2 proteins are very similar (Fig. 2 F and G and SI Appendix, Fig. S3) consistent with a close relationship between the sites bound by MYC and WDR5 at these locations. Second, although the relationship at cobound sites is close, there are many sites where MYC shows robust binding to chromatin in the absence of WDR5 (SI Appendix, Fig. S3). Third, E-boxes are strongly represented at cobound sites—60% (247) of these sites carry a perfect E-box, while 97% (408) carry either a perfect or an imperfect (CAN1N2TG; where N1 is not C and N2 is not G) E-box motif. Known and de novo motif analysis revealed that, compared to MYC-only bound sites, MYC/WDR5 cobound sites are enriched in an extended E-box element (GTCACGTGAC; Fig. 2H). Finally, looking specifically at sites with imperfect E-boxes, the ChIP-Seq signal for MYC is higher for MYC/WDR5 cobound than MYC-only sites (Fig. 2I), consistent with the notion that WDR5 facilitates loading of MYC to chromatin at locations where its DNA recognition sequence is imperfect. Based on these data, we conclude that most binding of MYC to chromatin in Ramos cells occurs independent of WDR5, but that there is a cohort of genes—connected to protein synthesis—where MYC and WDR5 share promoter-proximal binding sites.
Fig. 2.
MYC and WDR5 colocalize at cohort of protein synthesis genes in Ramos cells. (A) Venn diagram, showing overlap of HA–WT–MYC and WDR5-binding sites—as determined by ChIP-Seq (false discovery rate [FDR] = 0.01). HA–WT–MYC was monitored in switched WT cells; WDR5 was monitored in unswitched WBM cells that express endogenous, wild-type, MYC protein. (B) Motif analysis for HA-tagged MYC-binding sites. At Top is known-motif analysis; Bottom is de novo motif analysis. Significance values and the percentage of ChIP-Seq peaks containing the indicated motif are shown for each. (C) As in B, except for WDR5-binding sites. (D) Distribution of shared MYC- and WDR5-binding sites, binned according to distance from the nearest annotated TSS. RGA, region–gene association. (E) The top 8 GO enrichment categories for genes bound by MYC and WDR5 in Ramos cells. (F) Histogram of normalized MYC and WDR5 ChIP-Seq fragment counts at MYC–WDR5 cobound sites in a 1,500-bp window either side of the center of the MYC peaks. (G) IGV screenshot of representative ChIP-Seq data for HA–WT–MYC and WDR5 in Ramos cells. Shown is the PTGES3 gene, which is bound only by MYC, and NACA, which is bound by both MYC and WDR5. Blue arrows indicate the position of 2 imperfect E-boxes in the NACA locus. n = 3 independent ChIP-Seq experiments for MYC and WDR5. (H) Motif analysis for MYC-binding sites at WDR5 cobound genes. At Top is known-motif analysis; Bottom is de novo motif analysis. Other details are as in B. (I) Histogram of normalized MYC ChIP-Seq fragment counts at sites with imperfect E-boxes that are bound by both MYC and WDR5 (red) or MYC only (blue). A 2,500-bp window either side of the peak center is shown.
WDR5 Recruits MYC to Chromatin at Protein Synthesis Genes.
We next asked whether disrupting the MYC–WDR5 interaction impacts the binding of WDR5 or MYC to chromatin. First, we switched WT for WBM MYC and performed ChIP-Seq for WDR5. Here, we found that exchange of endogenous MYC for the WBM mutant modestly reduced the intensity of WDR5 ChIP-Seq peaks, but had no impact on the total number of WDR5 peaks detected or the location of WDR5 peaks across the genome (SI Appendix, Fig. S4 A and B). Because the WBM mutant is the sole form of MYC in these cells, we conclude that association of WDR5 with chromatin occurs largely independent of its ability to interact with MYC.
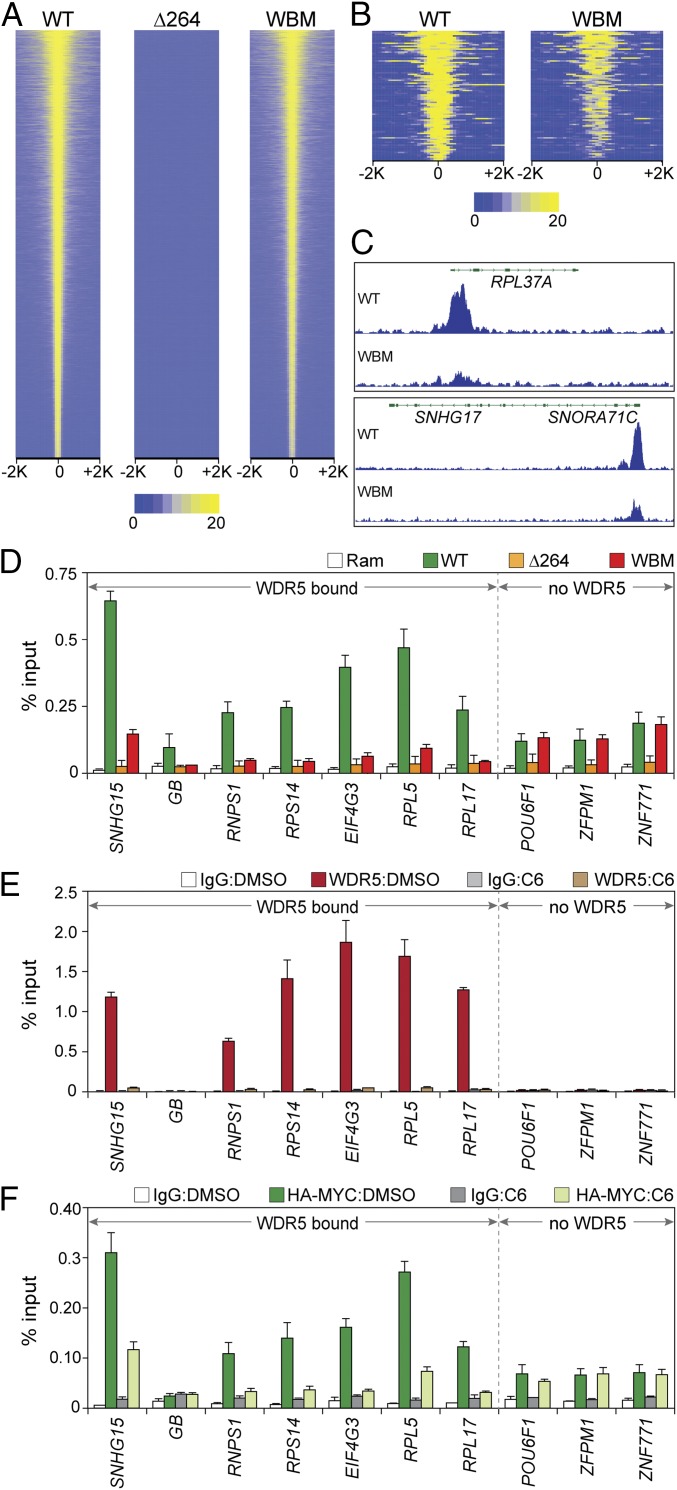
Next, we performed ChIP-Seq for HA-tagged MYC in both the ∆264 and WBM switched cells. The ∆264 MYC mutant failed to bind chromatin, with no ChIP-Seq peaks detected in these experiments (Fig. 3A). The WBM MYC, in contrast, displayed a modest reduction in the intensity of chromatin binding genome-wide (SI Appendix, Fig. S4 C and D), but the vast majority of MYC-binding events were preserved (Fig. 3A). Indeed, there are just 88 sites where MYC binding was significantly perturbed by the WBM mutation (Fig. 3 B and C). Interestingly, these 88 sites have fewer perfect E-boxes than the MYC/WDR5 cobound sites where MYC binding is insensitive to the WBM mutation—and they have more imperfect E-boxes (SI Appendix, Fig. S4E). Although the number of loci that are sensitive to the WBM mutation was small, the sites involved are strongly enriched in 2 important characteristics: 85% of them are bound by WDR5, and more than half are connected to protein synthesis (Dataset S2). We validated the ChIP-Seq results with ChIP-qPCR, probing 6 WDR5-bound loci and 3 genes where MYC binds, but WDR5 was not detected (Fig. 3D). Here, we observed identical results: the WBM mutation disrupts binding of MYC to WDR5-bound genes, but has little if any impact on genes that are not bound by WDR5 (Fig. 3D). These findings demonstrate that interaction with WDR5 is generally dispensable for chromatin binding by MYC, but that there is a rarefied cohort of genes—connected to protein synthesis and with reduced quality DNA-binding sequences—where WDR5 is important for MYC to associate with chromatin.
Fig. 3.
Interaction of MYC with protein synthesis genes is WDR5 dependent. (A) Heatmap, showing HA-tagged MYC peak intensity for each MYC protein (ChIP-Seq) after switching; the figure displays the combined average of normalized peak intensity (read density) in 100-bp bins ±2 kb around peak centers. Intensities are ranked based on WT–MYC. (B) As in A, except for the 88 loci where the WBM mutant was associated with a significant reduction in HA–MYC binding. (C) Two IGV screenshots of normalized representative ChIP-Seq data for HA-tagged MYC (WT or WBM) in the indicated cell lines after switching. Both RPL37 and SNROA71C/SNHG17 bind WDR5. n = 3 independent ChIP-Seq experiments for HA-tagged MYC proteins. (D) ChIP was performed (anti-HA antibody) in cells switched to the indicated MYC proteins (or parental Ramos cells). Coprecipitating DNAs were monitored (Q-PCR) with primer sets spanning ChIP-Seq peaks. Six WDR5-bound loci, and 3 non–WDR5-bound loci, were probed, as indicated. GB, corresponds to the SNHG15 gene body, where little MYC is detected. ChIP signal is represented as a percentage of input DNA. (E) WT engineered Ramos cells were selected after switching to permanently express HA-tagged WT MYC. These cells were then treated with either DMSO or 25 µM C6 for 4 h and WDR5 binding to the indicated loci probed via ChIP-qPCR. IgG is a negative control ChIP. Primer sets and abbreviations are as in D. (F) As in E, except that ChIP-qPCR was performed for MYC. The mean and SD of 3 experiments are shown.
Recently, we discovered a small molecule, C6, that binds tightly to the “WIN” site of WDR5 (12) and displaces it from chromatin (15). As an orthologous challenge to the concept that WDR5 is required for MYC recruitment at select loci, we asked if C6 also displaces MYC. We treated Ramos cells expressing HA-tagged WT MYC with C6 and performed ChIP for both WDR5 and MYC. Here, as we observed in MV4:11 cells (15), a 4-h treatment of Ramos cells with C6 displaced WDR5 from chromatin at the 6 sampled sites (Fig. 3E). Probing for MYC (Fig. 3F), we determined that C6 treatment disrupted MYC binding to these same 6 loci, but had little effect at sites where MYC binds without WDR5. Under these conditions, C6 did not alter MYC or WDR5 expression (SI Appendix, Fig. S4F) or the MYC–WDR5 interaction SI Appendix, Fig. S4G). Notably, the magnitude in reduction of MYC binding upon WIN site inhibitor treatment was very similar to that observed with the WBM mutation (Fig. 3D), consistent with the idea that either evicting WDR5 from chromatin, or disrupting the MYC–WDR5 interaction, is sufficient to attenuate MYC binding at these loci.
Interaction of MYC with WDR5 Is Important for Protein Synthesis Gene Transcription.
Next, we used precision run on (PRO)-Seq (16) to monitor the distribution and density of active RNA polymerases across the genome after switching. To establish a baseline, we first compared WT with ∆264 switched cells, 24 h after OHT treatment. Compared to the WT switch, switching to the ∆264 mutant decreased transcription at ∼3,400 genes and increased transcription at about 1,200 genes (Fig. 4A and SI Appendix, Fig. S5A). Although these transcriptional changes were modest, they were entirely consistent with MYC inactivation: there was a strong tendency of MYC-bound genes to be transcriptionally altered in the presence of the ∆264 mutant (SI Appendix, Fig. S5B), and we observed significant enrichment in gene signatures linked to MYC, including hallmark MYC gene sets, mTORC1 signaling, and glycolysis (SI Appendix, Fig. S5C). PRO-Seq can thus capture a broad range of transcriptional changes associated with loss of MYC function in this system.
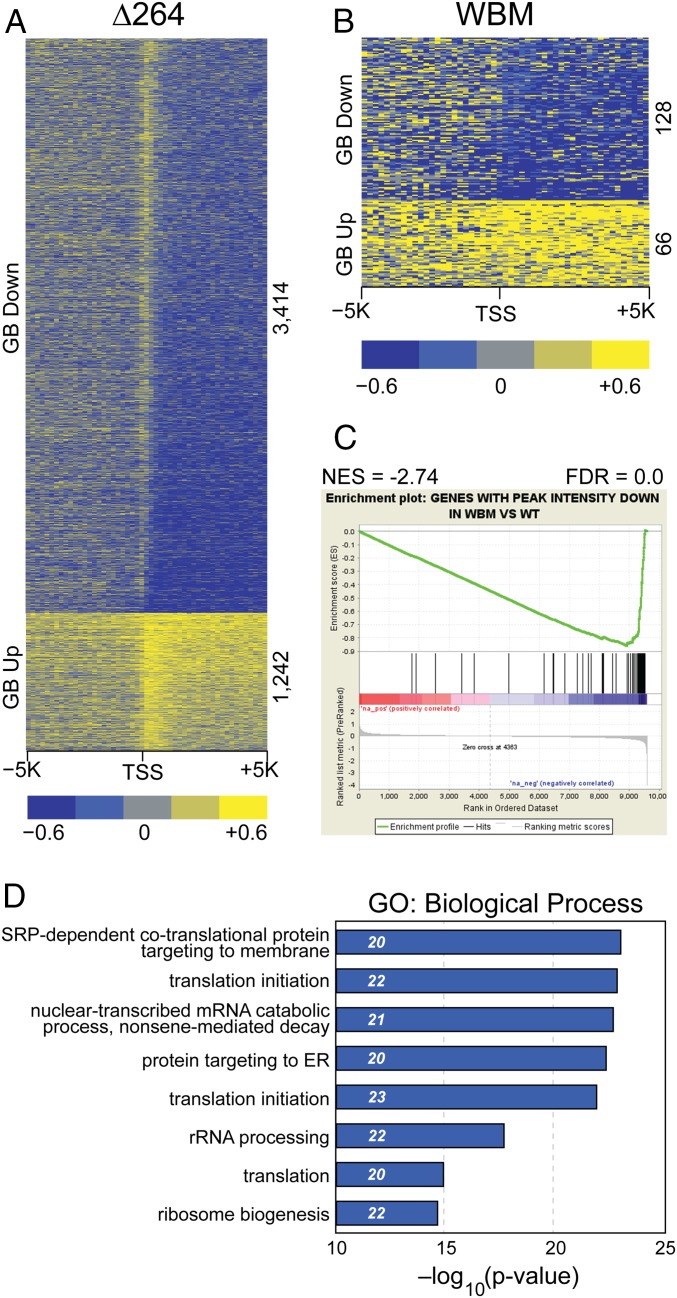
Fig. 4.
The MYC–WDR5 interaction is important for transcription of genes connected to protein synthesis. (A) Heatmap, displaying log2-fold change of active polymerases in ∆264 switched cells (compared to WT switched cells) in the promoter-proximal region and ±5 kb around the TSS (200-bp bins), as determined by PRO-Seq. (24 h after switch). The Top of the figure shows genes where transcription in the GB decreased (GB down); the Lower part shows genes where gene body transcription increased (GB up). (B) As in A but for WBM switched cells. (C) Gene set enrichment analysis (GSEA) comparing genes with a reduction in MYC binding in WBM mutation determined by ChIP-Seq against a gene list ranked by alteration in the density of gene body-associated transcribing polymerases in the WBM mutant vs. WT, as determined by PRO-Seq. (D) The top 8 GO enrichment categories for genes displaying a decrease in gene body-associated RNA polymerases in WBM switched cells. Numbers in italics represent the number of genes in each category. n = 2 independent PRO-Seq experiments.
Next, we compared the WT switch to that of the WBM mutant. Here, transcriptional changes were of the same magnitude as the ∆264 MYC protein, but affected fewer genes (Fig. 4B and SI Appendix, Fig. S5A). In total, there were 128 genes where transcription decreased, and 66 genes where it increased, in the presence of WBM MYC. Although the number of genes showing decreased transcription in the presence of WBM mutant MYC was small, the genes themselves are strongly clustered—they are significantly enriched in those where MYC is displaced from chromatin by the WBM mutation (Fig. 4C), and as expected contain multiple protein synthesis genes (Fig. 4D), including ribosome subunits, nucleolar RNAs, and translation factors (Dataset S3). These findings demonstrate that WDR5 has a functional consequence on MYC-driven transcriptional events in BL cells and highlight biomass accumulation as the overwhelming process controlled by the MYC–WDR5 interaction in this setting.
Interaction with WDR5 Is Essential for Tumor Maintenance by MYC.
The ability of MYC to stimulate protein synthesis has long been recognized as an important part of its tumorigenic repertoire (1). The biological clustering of gene expression changes we observe with the WBM MYC mutant, therefore, predicts that it should have impaired tumorigenic potential. But how will it compare to the ∆264 mutant, which globally eliminates chromatin binding by MYC? To answer this question, we switched WT, ∆264, or WBM cells in culture, injected these cells into the flanks of nude mice, and assayed tumor size and mouse survival over time (Fig. 5A). Consistent with a published report (17), mice receiving WT switched Ramos cells developed tumors rapidly (Fig. 5B and SI Appendix, Fig. S6A) and with complete penetrance, and all mice had to be killed by 19 d after injection (Fig. 5C). In comparison, both the ∆264 and WBM switched cells displayed delayed tumor growth (Fig. 5B) and a lag in mouse morbidity (Fig. 5C and SI Appendix, Fig. S6A). Although mice receiving the ∆264 and WBM switched cells all eventually developed tumors, these appear to arise from contaminating unswitched cells in the injected populations, as the ∆264 and WBM tumors had much higher levels of cells with the unswitched exon 3 cassette than WT tumors (Fig. 5D). Based on these data, we conclude that the ∆264 and WBM forms of MYC are both compromised in their ability to drive tumorigenesis in vivo.
Fig. 5.
The ability of MYC to interact with WDR5 is required for tumor growth and maintenance. (A) Schematic of tumor formation assay. Unswitched WT, ∆264, or WBM cells were grown in culture and pulsed 3 times with OHT to induce the switch. Cells were then injected into a flank of nude mice, and tumor growth was monitored. (B) Average tumor volumes in mice injected with the indicated switched Ramos cells over days 5 to 17 of the experiment. Tumor volumes for ∆264 and WBM cells were significantly different (*) from WT (P < 4.84E-3; 2-tailed t test). (C) Kaplan–Meier survival curves of mice injected with switched WT, ∆264, or WBM cells (n = 9 per group); P < 0.0001 (log-rank test). (D) PCR assay for presence of unswitched cells in tumor samples taken at time of sacrifice. Each dot represents a single mouse. The horizontal cross represents the mean for each group. Note that we were unable to extract usable DNA from one of the tumors in the ∆264 group. (E) Schematic of tumor maintenance assay. Unswitched WT, ∆264, or WBM cells were injected into a flank of nude mice, and tumor growth was monitored. At day 15, mice received 3 consecutive daily injections of tamoxifen to induce the switch, and tumor development was monitored. (F) Average tumor volumes in mice injected with the indicated unswitched Ramos cells before and after switching were induced, beginning at day 15 (red arrow “Tam”). After switch, tumor volumes for ∆264 and WBM cells were significantly different (*) from WT (P < 7.0E-9; 2-tailed t test). (G) Kaplan–Meier survival curves of mice (in F) (n = 9 per group); P < 0.0001 (log-rank test). (H) For the indicated cell types, tumors were harvested (n = 4 per group) 48 and 96 h following tamoxifen administration. Apoptosis was evaluated in the isolated lymphoma cells by staining for Annexin V positivity (Annexin V+) and analysis by flow cytometry. The extent of Annexin V positivity in ∆264 and WBM tumor cells was significantly different (*) from WT tumors (P < 1.65E-5; 2-tailed t test).
Finally, we asked whether switching cells in the context of an established tumor would impact tumor growth (Fig. 5E). We injected cells in their unswitched state into the flanks of nude mice, allowed tumors to grow to ∼200 mm3, and starting at day 15 administered tamoxifen treatment for 3 consecutive days. In WT MYC cells, switching did not affect the rapid rate of tumor growth (Fig. 5F and SI Appendix, Fig. S6B), and mouse survival time was analogous to that observed with preswitched WT cells (Fig. 5G). In contrast, for both the ∆264 and WBM cells, tumors regressed rapidly and completely within a week after tamoxifen treatment (Fig. 5F), and these mice survived the entire 60-d duration of the experiment (Fig. 5G and SI Appendix, Fig. S6B). Tumor regression in the ∆264 and WBM cases was accompanied by widespread apoptosis, with most of the ∆264 and WBM tumor cells reporting as Annexin V positive 96 h after switch induction (Fig. 5H and SI Appendix, Fig. S6 C and D). Consistent with overlapping phenotypic response of WBM and ∆264 tumors to switching, RNA-Seq of tumors (SI Appendix, Fig. S7A) revealed widespread and highly similar gene expression changes in both cases (SI Appendix, Fig. S7 B and C), with enrichment in gene expression signatures connected to RAS and NOTCH signaling, p53, apoptosis, MYC, the cell cycle, and mTORC1 (Dataset S4). Importantly, despite the overall similarities in the tumor regression transcriptional profiles, comparison of differentially expressed genes between the WBM and ∆264 tumors demonstrated that genes selectively down-regulated in the WBM samples were enriched in genes that are decreased in the WBM mutant for MYC binding (SI Appendix, Fig. S7D) and for gene expression, as determined by PRO-Seq (SI Appendix, Fig. S7E). Thus the transcriptional changes we measured for the WBM mutant in cell culture occurred after switching in vivo.
Together, these results establish that the ability of MYC to interact with WDR5 is critical for tumor maintenance in mice. Moreover, the near identical behavior of the ∆264 and WBM MYC mutants in both the tumor engraftment and maintenance assays demonstrates that disrupting the MYC–WDR5 interactions is as effective at promoting tumor regression as disabling all of the chromatin-binding capabilities of MYC.
Discussion
There is overwhelming evidence that strategies to inhibit MYC could form the basis of broadly effective anticancer therapies (3, 18). But the absence of well-defined and druggable pockets within MYC makes the prospect of direct pharmacological inhibition almost impossible. Here, we present evidence that WDR5—a small, well-structured, protein with pockets suitable for drug discovery (12)—could be a viable route for MYC inhibition. Within the context of a canonical MYC-driven cancer, we show that interaction with WDR5 is essential for MYC to bind to and regulate a set of genes manifestly linked to biomass accumulation and demonstrate that disrupting the MYC–WDR5 interaction within an existing cancer promotes widespread apoptosis and tumor collapse. These findings raise the possibility that the MYC–WDR5 nexus could be an actionable venue for the discovery of new ways to inhibit MYC.
Mechanistically, our findings provide insights into the MYC–WDR5 connection. First, because we replace WT for WBM MYC in the switched cells, and see little impact on WDR5 binding by CHIP-Seq, we conclude that interaction with MYC does not serve to recruit WDR5 to chromatin. This finding supports our earlier contention (8) that recruitment of MYC to chromatin at select sites depends on recognition of a prebound and proximal WDR5 protein. What localizes WDR5 to these sites is, however, unknown. Recruitment of WDR5 is WIN site dependent (15), and so it is likely that WDR5 is tethered to these locations by engaging an arginine-containing WIN motif (consensus “ARA”; ref. 12) in a chromatin-resident protein. Motif analysis revealed that MYC/WDR5 cobound genes are enriched in an extended E-box consensus (Fig. 2H) that matches the “CLEAR element” present in lysosomal biogenesis genes (19) where it is bound by the micropthalmia-transcription factor E (MiT/TFE) subfamily of basic helix–loop–helix (bHLH) transcription factors (20); whether members of the MiT/TFE family of transcription factors play a role in the initial recruitment of WDR5 to these sites awaits further investigation. Second, we show that not all sites of MYC/WDR5 colocalization on chromatin are sites of facilitated recruitment; the WBM mutation reduces binding of MYC to just 88 of the 418 MYC/WDR5-binding sites in Ramos cells (Fig. 4B), and it is within these 88 “WBM sensitive” genes that we observe transcriptional decreases by PRO-Seq (Fig. 4C). Importantly, we find that the 88 WBM sensitive sites differ from other MYC/WDR5 cobound sites in that they have significantly fewer perfect—and more imperfect—E-box elements. Thus the dependency of MYC on interaction with WDR5 correlates with the lower quality of DNA elements in these targets, providing a simple rationale for the importance of the MYC–WDR5 interaction in stabilizing MYC on chromatin at these sites.
Identification of protein synthesis as the biological context in which MYC and WDR5 cooperate is notable, as connections between the tumorigenic actions of MYC and the protein synthesis machinery are extensive (1). MYC regulates transcription by all 3 nuclear RNA polymerases (21), and has widespread control of RNA polymerase II-transcribed genes linked to protein synthesis (22). There is growing support for the concept that targeting ribosome biogenesis could be an effective anti-MYC strategy (23) and an established precedent that a 2-fold reduction in the level of a single ribosome protein subunit can suppress tumorigenesis by MYC in vivo (24). Disrupting the interaction of MYC with WDR5 decreases transcription of more than 20 ribosome protein subunits, as well as nucleolar RNAs and factors required for translation initiation. The commonality of enhanced protein synthesis as a tumorigenic MYC mechanism, together with the finding that all 3 MYC family members interact with WDR5 (8, 25), suggests that strategies to inhibit MYC through WDR5 could be pursued to treat a range of malignancies.
In the context of a preformed tumor, switching cells to express the WBM mutant MYC causes widespread induction of apoptosis, associated with rapid tumor loss. This mechanism of tumor regression is similar to what has been described for other models of MYC inactivation (26–28), and supports the idea that MYC can be therapeutically targeted through WDR5 to reverse malignancy. The MYC–WDR5 interface is small and well structured and should be amenable to pharmacological inhibition (8). But one of the surprising aspects of this work is the realization that targeting the MYC–WDR5 interface may not be absolutely required to inhibit chromatin binding of MYC through WDR5. Our recent discovery that inhibitors of the WIN site of WDR5—which is on the opposite side of WDR5 to the MYC interface—displace WDR5 from chromatin (15), together with evidence here that the small molecule C6 also displaces MYC, hints that WIN site inhibitors could be developed to block MYC. The WIN site of WDR5 is the subject of intense drug discovery efforts from several groups (15, 29–33) and a number of distinct chemotypes have been reported. Ultimately, whether this avenue can be pursued depends on whether a therapeutic window can be established for WDR5 inhibitors between normal and cancer cells, which likely both make use of the MYC–WDR5 interaction to regulate protein synthesis gene expression. It has, however, been possible to establish a therapeutic window with a protein-based inhibitor of the MYC–MAX interaction (34), demonstrating that essential MYC cofactors can be targeted to selectively kill cancer cells. Development of potent inhibitors with suitable in vivo pharmacological properties is needed to definitely address this issue.
Finally, a particularly provocative aspect of this work is our finding that disrupting the MYC–WDR5 interaction has the same antitumor effect as eliminating the entire carboxy terminus of MYC (∆264)—in terms of the mechanism, extent, and permanency of tumor loss we observe. The carboxy terminus of MYC carries sequences required for nuclear localization (35), and for interaction with cofactors including WDR5 (8), HCF-1 (14), MIZ-1 (36), and MAX (2); yet from an antitumor perspective, disrupting the interaction with WDR5 is sufficient to recapitulate loss of all of these critical processes in this model. The importance of this result is that it demonstrates that challenges associated with pharmacological inhibition of the MYC:MAX interaction (18) do not need to be overcome to discover and implement MYC inhibitors, and that additional MYC cofactors can serve as a viable focal point for new drug discovery initiatives. Our understanding of the MYC interactome has blossomed in recent years (37–40), and if some of these interactors can be understood at the level of structural and biological detail now available for WDR5, the prospect of multimodal approaches to effectively block MYC in the clinic could become reality.
Materials and Methods
Reagents.
Details on plasmid constructions, Ramos cell engineering and validation, and antibodies used in this study are provided in SI Appendix, Materials and Methods.
Immunoprecipitations.
For each immunoprecipitation, 12 × 106 cells were treated for 24 h with OHT to activate CRE–ER and 1.5 h with 25 µM MG132 (VWR). Cell lysates were prepared in Kischkel buffer (50 mM Tris⋅HCl pH 8.0, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, and Complete Protease Inhibitor mixture [Sigma-Aldrich]), briefly sonicated, and subject to immunoprecipitation overnight with the 12CA5 antibody. Immune complexes were captured with Protein-G agarose (Cell Signaling Technologies), washed in Kischkel buffer, and resolved by SDS/PAGE followed by immunoblotting.
Flow Cytometry.
Flow cytometry experiments were performed in the Vanderbilt University (VU) and Thomas Jefferson University Flow Cytometry Shared Resources on an LSRII flow cytometer and data analyzed with FACSDiva software (BD Biosciences). For apoptosis assays, cells were stained with Annexin-V-PE (1:100, Thermo Fisher BDB556422) and 7-AAD prior to analysis.
Chromatin Immunoprecipitation.
Ramos cells were plated at 7.5 × 105 per milliliter ± 20 nM 4-OHT for 24 h for ChIP using switched MYC alleles. To displace WDR5 with C6, cells were plated at 8 × 106 per milliliter and treated with 25 µM C6 (or DMSO) for 4 h. ChIP experiments were performed as described (8) using 12 × 106 cells per reaction. Decrosslinked DNA was diluted to 500 µL with water and 7.5 µL was used for each qPCR reaction with amplicons centered on ChIP-Seq peaks. Percent input was calculated by comparison to a 30-fold dilution of decrosslinked chromatin. For ChIP-Seq, 3 replicates of ChIP DNA were converted into Illumina sequencing libraries using the NEBNext Ultra II DNA Library Prep Kit (New England Biolabs). Sequencing was performed at the VU VANTAGE Shared Resource on an Illumina HiSeq 2500 (single-end 75-bp reads, ∼40 M reads per sample). Methods for bioinformatic analyses are described in SI Appendix, Materials and Methods.
PRO-Seq.
For PRO-Seq analysis, 3 × 107 cells were switched for 24 h prior to analysis and nuclei prepared according to published protocols (41, 42). Further details, including methods for bioinformatic analyses, are provided in SI Appendix, Materials and Methods.
In Vivo Studies.
Methods for tumor engraftment and maintenance assays, as well as tumor switch quantification and tumor RNA-Sequencing are described in SI Appendix, Materials and Methods. All studies complied with state and federal guidelines and were approved by the Institutional Animal Care and Use Committee at Thomas Jefferson University.
Data Availability.
All genomic data have been deposited at GEO (GSE126207).
Supplementary Material
Acknowledgments
For reagents we thank C. Bautista, C. Cepko, and F. Zhang. The VU Flow Cytometry Shared Resource is supported by the Vanderbilt Ingram Cancer Center (P30CA68485) and the Vanderbilt Digestive Disease Research Center (DK058404). The Thomas Jefferson Flow Cytometry and Research Animals shared resource cores are supported by the Sidney Kimmel Cancer Center National Cancer Institute (NCI) P30CA056036 grant. VANTAGE is supported by the Vanderbilt Ingram Cancer Center, the Vanderbilt Vision Center (P30EY08126), and NIH/National Center for Research Resources (G20RR030956). This project has been funded in part with federal funds from the NCI, under Chemical Biology Consortium Contract No. HHSN261200800001E. This work was also supported by grants from the Robert J. Kleberg, Jr. and Helen C. Kleberg Foundation (to W.P.T. and S.W.F.), the TJ Martell Foundation (to W.P.T. and S.W.F.), St. Baldrick’s Foundation (W.P.T.), Alex’s Lemonade Stand Foundation (W.P.T.), Edward P. Evans Foundation (W.P.T.), the NCI/NIH (CA200709; W.P.T.), the NCI/NIH (CA211305; L.R.T.), the NCI/NIH (CA148950; C.M.E.), the Integrated Biological Systems Training in Oncology Training Program (T32 CA119925; A.M.W. and W.P.T.), the Rally Foundation for Childhood Cancer Research Fellowship (A.M.W.), Open Hands Overflowing Hearts cofunded research fellowship (A.M.W.), the American Association for Cancer Research Basic Cancer Research Fellowship (A.M.W.), and the Sidney Kimmel Cancer Center/Thomas Jefferson University.
Footnotes
Competing interest statement: S.W.F., S. R. Stauffer, W.P.T., E. T. Olejniczak, J. Phan, F. Wang, K. Jeon, and R. D. Gogliotti were granted US Patent 10,160,763, “WDR5 Inhibitors and Modulators,” on December 25, 2018.
This article is a PNAS Direct Submission.
Data deposition: The genomic data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE126207).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1910391116/-/DCSupplemental.
References
- 1.Tansey W. P., Mammalian MYC proteins and cancer. New J. Sci. 2013, 1–27 (2014). [Google Scholar]
- 2.Blackwood E. M., Eisenman R. N., Max: A helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science 251, 1211–1217 (1991). [DOI] [PubMed] [Google Scholar]
- 3.Chen H., Liu H., Qing G., Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduct. Target. Ther. 3, 5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soucek L., et al. , Inhibition of Myc family proteins eradicates KRas-driven lung cancer in mice. Genes Dev. 27, 504–513 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delmore J. E., et al. , BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146, 904–917 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dang C. V., Therapeutic targeting of Myc-reprogrammed cancer cell metabolism. Cold Spring Harb. Symp. Quant. Biol. 76, 369–374 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Lorenzin F., et al. , Different promoter affinities account for specificity in MYC-dependent gene regulation. eLife 5, e15161 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas L. R., et al. , Interaction with WDR5 promotes target gene recognition and tumorigenesis by MYC. Mol. Cell 58, 440–452 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carugo A., et al. , In vivo functional platform targeting patient-derived xenografts identifies WDR5-myc association as a critical determinant of pancreatic cancer. Cell Rep. 16, 133–147 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Richart L., et al. , BPTF is required for c-MYC transcriptional activity and in vivo tumorigenesis. Nat. Commun. 7, 10153 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerlach J. M., et al. , PAF1 complex component Leo1 helps recruit Drosophila Myc to promoters. Proc. Natl. Acad. Sci. U.S.A. 114, E9224–E9232 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guarnaccia A. D., Tansey W. P., Moonlighting with WDR5: A cellular multitasker. J. Clin. Med. 7, E21 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bemark M., Neuberger M. S., The c-MYC allele that is translocated into the IgH locus undergoes constitutive hypermutation in a Burkitt’s lymphoma line. Oncogene 19, 3404–3410 (2000). [DOI] [PubMed] [Google Scholar]
- 14.Thomas L. R., et al. , Interaction of MYC with host cell factor-1 is mediated by the evolutionarily conserved Myc box IV motif. Oncogene 35, 3613–3618 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aho E. R., et al. , Displacement of WDR5 from chromatin by a WIN site inhibitor with picomolar affinity. Cell Rep. 26, 2916–2928.e13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwak H., Fuda N. J., Core L. J., Lis J. T., Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science 339, 950–953 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pagel J. M., et al. , Comparison of a tetravalent single-chain antibody-streptavidin fusion protein and an antibody-streptavidin chemical conjugate for pretargeted anti-CD20 radioimmunotherapy of B-cell lymphomas. Blood 108, 328–336 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whitfield J. R., Beaulieu M. E., Soucek L., Strategies to inhibit myc and their clinical applicability. Front. Cell Dev. Biol. 5, 10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sardiello M., et al. , A gene network regulating lysosomal biogenesis and function. Science 325, 473–477 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Meadows N. A., et al. , The expression of Clcn7 and Ostm1 in osteoclasts is coregulated by microphthalmia transcription factor. J. Biol. Chem. 282, 1891–1904 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Campbell K. J., White R. J., MYC regulation of cell growth through control of transcription by RNA polymerases I and III. Cold Spring Harb. Perspect. Med. 4, a018408 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsieh A. L., Dang C. V., MYC, metabolic synthetic lethality, and cancer. Recent Results Cancer Res. 207, 73–91 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Poortinga G., Quinn L. M., Hannan R. D., Targeting RNA polymerase I to treat MYC-driven cancer. Oncogene 34, 403–412 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Barna M., et al. , Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature 456, 971–975 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun Y., et al. , WDR5 supports an N-Myc transcriptional complex that drives a pro-tumorigenic gene expression signature in neuroblastoma. Cancer Res. 75, 5143–5154 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Jain M., et al. , Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science 297, 102–104 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Marinkovic D., Marinkovic T., Mahr B., Hess J., Wirth T., Reversible lymphomagenesis in conditionally c-MYC expressing mice. Int. J. Cancer 110, 336–342 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Giuriato S., et al. , Sustained regression of tumors upon MYC inactivation requires p53 or thrombospondin-1 to reverse the angiogenic switch. Proc. Natl. Acad. Sci. U.S.A. 103, 16266–16271 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bolshan Y., et al. , Synthesis, optimization, and evaluation of novel small molecules as antagonists of WDR5-MLL interaction. ACS Med. Chem. Lett. 4, 353–357 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Senisterra G., et al. , Small-molecule inhibition of MLL activity by disruption of its interaction with WDR5. Biochem. J. 449, 151–159 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao F., et al. , Targeting MLL1 H3K4 methyltransferase activity in mixed-lineage leukemia. Mol. Cell 53, 247–261 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grebien F., et al. , Pharmacological targeting of the Wdr5-MLL interaction in C/EBPα N-terminal leukemia. Nat. Chem. Biol. 11, 571–578 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang F., et al. , Discovery of potent 2-Aryl-6,7-dihydro-5 H-pyrrolo[1,2- a]imidazoles as WDR5-WIN-site inhibitors using fragment-based methods and structure-based design. J. Med. Chem. 61, 5623–5642 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beaulieu M. E., et al. , Intrinsic cell-penetrating activity propels Omomyc from proof of concept to viable anti-MYC therapy. Sci. Transl. Med. 11, eaar5012 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dang C. V., Lee W. M., Identification of the human c-myc protein nuclear translocation signal. Mol. Cell. Biol. 8, 4048–4054 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Riggelen J., et al. , The interaction between Myc and Miz1 is required to antagonize TGFbeta-dependent autocrine signaling during lymphoma formation and maintenance. Genes Dev. 24, 1281–1294 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kalkat M., Wasylishen A. R., Kim S. S., Penn L., More than MAX: Discovering the Myc interactome. Cell Cycle 10, 374–375 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Dingar D., et al. , BioID identifies novel c-MYC interacting partners in cultured cells and xenograft tumors. J. Proteomics 118, 95–111 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Kalkat M., et al. , MYC protein interactome profiling reveals functionally distinct regions that cooperate to drive tumorigenesis. Mol. Cell 72, 836–848.e7 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Baluapuri A., et al. , MYC recruits SPT5 to RNA polymerase II to promote processive transcription elongation. Mol. Cell 74, 674–687.e11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mahat D. B., et al. , Base-pair-resolution genome-wide mapping of active RNA polymerases using precision nuclear run-on (PRO-seq). Nat. Protoc. 11, 1455–1476 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weissmiller A. M., et al. , Inhibition of MYC by the SMARCB1 tumor suppressor. Nat. Commun. 10, 2014 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All genomic data have been deposited at GEO (GSE126207).