Abstract
Background
Transcriptional profiling of the human immune response to malaria has been used to identify diagnostic markers, understand the pathogenicity of severe disease and dissect the mechanisms of naturally acquired immunity (NAI). However, interpreting this body of work is difficult given considerable variation in study design, definition of disease, patient selection and methodology employed. This work details a comprehensive review of gene expression profiling (GEP) of the human immune response to malaria to determine how this technology has been applied to date, instances where this has advanced understanding of NAI and the extent of variability in methodology between studies to allow informed comparison of data and interpretation of results.
Methods
Datasets from the gene expression omnibus (GEO) including the search terms; ‘plasmodium’ or ‘malaria’ or ‘sporozoite’ or ‘merozoite’ or ‘gametocyte’ and ‘Homo sapiens’ were identified and publications analysed. Datasets of gene expression changes in relation to malaria vaccines were excluded.
Results
Twenty-three GEO datasets and 25 related publications were included in the final review. All datasets related to Plasmodium falciparum infection, except two that related to Plasmodium vivax infection. The majority of datasets included samples from individuals infected with malaria ‘naturally’ in the field (n = 13, 57%), however some related to controlled human malaria infection (CHMI) studies (n = 6, 26%), or cells stimulated with Plasmodium in vitro (n = 6, 26%). The majority of studies examined gene expression changes relating to the blood stage of the parasite. Significant heterogeneity between datasets was identified in terms of study design, sample type, platform used and method of analysis. Seven datasets specifically investigated transcriptional changes associated with NAI to malaria, with evidence supporting suppression of the innate pro-inflammatory response as an important mechanism for this in the majority of these studies. However, further interpretation of this body of work was limited by heterogeneity between studies and small sample sizes.
Conclusions
GEP in malaria is a potentially powerful tool, but to date studies have been hypothesis generating with small sample sizes and widely varying methodology. As CHMI studies are increasingly performed in endemic settings, there will be growing opportunity to use GEP to understand detailed time-course changes in host response and understand in greater detail the mechanisms of NAI.
Keywords: Plasmodium falciparum, Gene expression, Malaria, Immunity
Background
Malaria, caused by infection with parasites of the genus Plasmodium, remains a significant public health concern [1]. Despite a vaccine in pilot implementation trials [2] and widespread application of control measures [3], the disease is still responsible for a huge burden of mortality and morbidity worldwide and a concerning increase in incidence has been seen in previously well-controlled areas [3].
With repeated exposure to infection, individuals in malaria-endemic regions develop naturally acquired immunity (NAI), first to the most severe clinical forms, such as cerebral malaria and then more slowly to infection itself [1]. Although the role of antibodies in controlling parasite density, symptomatology and severity of disease is well established [4, 5], less is known about mechanism in terms of the role of the innate and cellular immune responses [6]. Increased understanding of the immune response to malaria, in particular those that mediate NAI, could aid identification of diagnostic and prognostic markers, inform vaccine development and assist with the identification of treatment strategies to modify the immunological mechanisms mediating severe pathology [1].
Transcriptomics, which allows the expression of thousands of genes to be assessed in parallel for a single RNA sample, is an exciting, expanding area of research with vast potential application in the field of infection [7]. Facilitating a systems biology approach, gene expression data from high-throughput technologies (such as microarrays [8] and next generation sequencing enabling RNA sequencing for bulk cell populations and at single-cell resolution [9, 10]) can allow greater understanding of individuals’ response to infection. To date, expression data have been used to dissect mechanisms of vaccine immunogenicity [11], inform the design of new vaccines [12, 13], predict response to infection and outcome [14, 15], characterize and improve understanding of sepsis [16], and offer a novel approach to the diagnosis of infectious pathogens [17–19] together with RNA expression in the pathogen [20].
Given the limited understanding of the mechanisms of NAI to malaria from traditional immunological studies, a systems approach characterizing the gene expression patterns associated with infection could provide novel and valuable insights [21, 22]. Transcriptional profiling of the immune response to malaria in humans to date has sought to identify markers to aid diagnosis [23], to understand the pathogenicity of severe disease [24] and dissect the mechanisms of NAI [25, 26]. However, interpreting this body of work is difficult given considerable variation in study design, definition of disease, patient selection and methodology employed.
This review outlines a comprehensive analysis of all GEP studies of the human immune response to malaria with two aims: (i) to understand the application of this technology to date, in particular how these studies have informed understanding of NAI; and (ii) to determine the extent of variability in methodology between studies to allow informed comparison of data and interpretation of results.
Methods
A search of Gene Expression Omnibus (GEO) [27] for datasets including the search terms; ‘plasmodium’ or ‘malaria’ or ‘sporozoite’ or ‘merozoite’ or ‘gametocyte’ and ‘Homo sapiens’ was performed on 10th September 2019. Each of these datasets were examined and those not relating to the human immune response to malaria infection or using the Homo sapiens platform excluded. Of note, datasets of gene expression changes in relation to malaria vaccines were excluded.
Results
Studies identified
The search identified 30 GEO datasets. Seven of these datasets were excluded, as published analyses were unavailable. Twenty-three datasets and 25 related publications were therefore included in the final review (Table 1 and Additional file 1: Figure S1). All datasets related to Plasmodium falciparum infection except two that related to Plasmodium vivax infection (Table 1). The majority of datasets included samples from individuals infected with malaria ‘naturally’ in the field (n = 13, 57%), however some related to controlled human malaria infection (CHMI) studies (n = 6, 26%), or cells stimulated with Plasmodium in vitro (n = 6, 26%). Studies included samples from individuals with a wide range of ages (from 2 months—varying ages of adulthood) with differing degrees of prior exposure and, therefore, NAI to malaria. Samples were often collected as part of wider immuno-epidemiological studies or vaccine trials, leading to variation in study design and sampling intervals.
Table 1.
Summary of gene expression datasets investigating the human immunological response to malaria infection
| GEO series | Title of dataset | Publication | Design | Infection/antigenic Stimulation | Species | Tissue | Age | Participant origin | Expression profiling | Subjects (samples)a | Controls | Platform name | Platform technology |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GSE2900 | Host response malaria | Griffiths et al. (2005) | Comparison of GEP in febrile children with convalescent samples 2 weeks post discharge | Field | P. falciparum | Whole blood: PAX gene | Children 2–126 months | Kenya | Array | 22 (28) | Subject paired samples: diagnosis and post treatment | LC-36 | Spotted DNA/cDNA |
| GSE5418 | Gene expression analysis in malaria infection | Ockenhouse et al. (2006) | Comparison of GEP in early, pre-symptomatic blood-stage infection post CHMI with symptomatic malaria-experienced adults with naturally acquired malaria | CHMI and Field | P. falciparum | PBMC | Adults; 19–49 years | USA and Cameroon | Array | 37 (74) | 22 un-infected malaria-naïve American adults | Affymetrix human genome U133A array | In situ oligonucleotide |
| GSE15221 | Malaria primes the innate immune response due to IFNγ induced enhancement of Toll-like receptor expression and function | Franklin et al. (2009) and Sharma et al. (2011) and Hirako et al. (2018) | Comparison GEP at malaria diagnosis and 28 days post treatment | Field | P. falciparum | PBMC | Adults 30 ± 10 years | Brazil; Porto Velho | Array | 21 (42) | Subject paired samples: diagnosis and post treatment | Illumina human-6 v2.0 | Oligonucleotide beads |
| GSE26876 | Time kinetics of gene expression in NK92 cells after P. falciparum-iRBC encounter | De Carvalho et al. (2011) | Comparison of GEP variation of NK92 cells after 6, 12, and 24 h of co-culture with either infected or uninfected RBC compared to time-point 0 | In vitro—iRBC | P. falciparum | NK92 cell line | N/A | N/A | Array | N/A (12) | Paired samples: pre and post exposure | Affymetrix human gene 1.0 ST array | In situ oligonucleotide |
| GSE33811 | Paired whole blood human transcription profiles from children with severe malaria and mild malaria | Krupka et al. (2012) | Comparison of GEP in severe malaria and subsequent mild malaria in same subjects 1 month later | Field | P. falciparum | Whole blood: tri-reagent BD | Children: 8–45 months | Malawi | Array | 5 (10) | Subject paired samples: severe and mild malaria | Affymetrix Human Gene 1.0 ST Array | In situ oligonucleotide |
| GSE34404 | The genomic architecture of host whole blood transcriptional response to malaria infection | Idaghdour et al. (2012) | Comparison of GEP in mild malaria with age matched un-infected controls | Field | P. falciparum | Whole blood: Tempus | Children; median age 3.7 years | Benin | Array | 94 subjects (94) and 64 controls (64) | Uninfected age matched | Illumina HumanHT-12 V4.0 expression bead chip | Oligonucleotide beads |
| GSE55843 | Loss and dysfunction of Vdelta2 + gamma delta-low T cells is associated with clinical tolerance to malaria | Jagannathan et al. (2014) | Comparison of GEP of Vδ2 + T cells from children with ‘high’ and ‘low’ episodes of malaria in the preceding year | In vitro—iRBC | P. falciparum | Vδ2 + T cells | Children: 4–5 years | Uganda | Array | 78 (156) | N/A | Agilent-039494 SurePrint G3 Human GE v2 8 × 60K Microarray 03938 | In situ oligonucleotide |
| GSE53292 | Transcriptomic analysis of Plasmodium PBANKA, PBSLTRiP-KO, PB268-KO parasite infected and uninfected host cell | Jaijyan et al. (2015) | Comparison of GEP of uninfected HepG2 with those infected with wild-type and knock out sporozoites | In vitro—sporozoites | P. falciparum | HepG2 cells | N/A | N/A | High throughput sequencing | NK | NK | Illumina Genome Analyzer IIx (Homo sapiens) | High-throughput sequencing |
| GSE50957 | Molecular hallmarks of experimentally acquired immunity to malaria [Pilot Study] | Tran et al. (2016) and Vallejo et al. (2018) | Comparison of GEP pre and post infection | CHMI | P. falciparum | Whole blood: PAX gene | Adults: 19–22 years | USA | High throughput sequencing | 5 (10) | Subject paired samples: Pre and post infection | Illumina HiSeq 2000 (Homo sapiens) | High-throughput sequencing |
| GSE52166 | Molecular hallmarks of naturally acquired immunity to malaria | Tran et al. (2016) | Comparison of GEP pre and post infection | Field | P. falciparum | Whole blood: Tempus | Adults and Children 13.5–23.3 years | Malawi | High throughput sequencing | 8 (16) | Paired same subject pre infection | Illumina HiSeq 2000 (Homo sapiens) | High-throughput sequencing |
| GSE64338 | Expression data from whole blood samples of Rwandan adults with mild malaria with matched sample 30 days later (convalescence) | Subramaniam et al. (2015) | Comparison of GEP in mild malaria and 30 days later | Field | P. falciparum | Whole blood: Tri-Reagent BD | Adults | Rwandan | Array | 19 (38) | Subject paired samples: diagnosis and post treatment | [HuGene-1_0-st] Affymetrix Human Gene 1.0 ST Array | In situ oligonucleotide |
| GSE64493 | FCRL5 delineates functionally impaired memory B cells associated with malaria exposure | Sullivan (2015) | Comparison of GEP between classical and atypical memory B cells in Uganda children | Field | P. falciparum | PBMC | Children 8–10 years | Uganda | Array | 12 | NK | Agilent-039494 SurePrint G3 Human GE v2 8 × 60K Microarray 039381 | In situ oligonucleotide |
| GSE67184 | Transcription profiling of malaria-naïve and semi-immune colombian volunteers in a Plasmodium vivax sporozoite challenge | Rojas-Penas (2015), Vallejo (2018) and Gardinassi (2018) | Comparison of GEP changes between malaria naïve and semi-immune adults pre-infection and at diagnosis | CHMI | P. vivax | Whole blood: Tempus | Adults | Columbia | High throughput sequencing | 12 (24) | Subject paired samples: pre-infection and diagnosis | Illumina HiSeq 2500 (Homo sapiens) | High-throughput sequencing |
| GSE67469 | Transcription profiling of malaria-naïve and semi-immune colombian volunteers in a Plasmodium vivax sporozoite challenge | Rojas-Penas (2015) | Comparison of GEP changes between malaria naïve and semi-immune adults over the time-course of malaria infection: pre-infection, day 5, day 7, day 9, diagnosis and month 4 | CHMI | P. vivax | Whole blood: Tempus | Adults | Columbia | RT-qPCR | 16 (85) | Subject paired samples: Pre infection and multiple time-points post infection | Fluidigm 96×96 nanofluidic arrays for 96 genes: blood informative transcripts | RT-PCR |
| GSE7586 | Genome wide analysis of placental malaria | Muehlenbachs (2007) | Comparison of GEP in women with placental malaria and those without | Field | P. falciparum | Placenta | Adults | Tanzania | Array | 20 (20) | NK | [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array | In situ oligonucleotide |
| GSE77122 | Involvement of β-defensin 130 (DEFB130) in the macrophage microbicidal mechanisms for killing Plasmodium falciparum | Terkawi (2017) | Human monocyte-derived macrophages were co-cultured with P. falciparum iRBCs, saponin-treated iRBCs, or non-infected RBCs | In vitro—iRBC | P. falciparum | Macrophages | NK | NK | Array | NK (8) | NK | Agilent-028004 SurePrint G3 Human GE 8 × 60K Microarray | In situ oligonucleotide |
| GSE93664 | Comparison of the transcriptomic profile of P. falciparum reactive polyfunctional and IFNγ monofunctional human CD4 T cells | Burel (2017) | Comparison of GEP in monofunctional and polyfunctional IFN producing T cells collected 21 days post CHMI infection | CHMI + in vitro—iRBC | P. falciparum | IFN producing T cells | 18–42 years | Australia | Array | 8 (2) | NK | [HuGene-2_0-st] Affymetrix Human Gene 2.0 ST Array | In situ oligonucleotide |
| GSE100562 | RNA-sequencing analysis of response to P. falciparum infection in Fulani and Mossi ethnic groups, Burkina Faso | Quin (2017) | Comparison of GEP in onocytes and CD14− cells in P. falciparum infected and uninfected malaria-exposed Fulani and Mossi sympatric ethnic groups | Field | P. falciparum | Monocytes (CD14+) and lymphocytes (CD14−) | 15–24 years | Burkino Faso | High throughput sequencing | 23 (23) | NK | Illumina HiSeq 2500 (Homo sapiens) | High-throughput sequencing |
| GSE1124 | Whole blood transcriptome of childhood malaria | Boldt (2019) | Comparison of GEP of children with asymptomatic parasitemia, uncomplicated malaria, malaria with severe anaemia and cerebral malaria | Field | P. falciparum | Whole blood: PAX gene | 0.5–6 years | Gabon | Array | NK | Healthy control children | [HG-U133A] Affymetrix Human Genome U133A Array | In situ oligonucleotide |
| GSE114076 | Differential gene expression profile of human neutrophils cultured with Plasmodium falciparum-parasitized erythrocytes | Terkawi (2018) | Comparison of GEP in neutrophils incubated with iRBC or non-infected RBC | In vitro—iRBC | P. falciparum | Neutrophils | NK | NK | Array | 1 (8) | Culture with non-infected RBC | Agilent-072363 SurePrint G3 Human GE v3 8 × 60K Microarray | In situ oligonucleotide |
| GSE97158 | Transcriptional responses induced by controlled human malaria infection (CHMI) | Rothan (2018) | Comparison of GEP in whole blood pre and post sporozoite CHMI in malaria exposed adults | CHMI | P. falciparum | Whole blood: PAX gene | Adults | Tanzania | High throughput sequencing | 10 (40) | Subject paired samples: pre and post CHMI | Illumina HiSeq 2000 (Homo sapiens) | High-throughput sequencing |
| GSE65928 | Malaria-associated atypical memory B cells exhibit markedly reduced B cell receptor signaling and effector function | Portugal (2015) | Comaprison of GEP of naïve B cells, classical and atypical memory B cells in immune adults | Field | P. falciparum | B cells | Adults: 18–37 years | Mali | Array | 20 (20) | US healthy adults | [HuGene-2_0-st] Affymetrix Human Gene 2.0 ST Array [transcript (gene) version] | In situ oligonucleotide |
| GSE72058 | Activated neutrophils are associated with pediatric cerebral malaria vasculopathy in Malawian children | Feintuch (2016) | Comparison of GEP in cerebral malaria between children with malaria retinopathy and those without | Field | P. falciparum | Whole blood: Tri-Reagent BD | Children 6 month–12 years | Mali | Array | 98 (98) | NK | [HuGene-1_0-st] Affymetrix Human Gene 1.0 ST Array [transcript (gene) version] | In situ oligonucleotide |
PBMC peripheral blood mononuclear cells, GEP gene expression profile, CHMI controlled human malaria infection, iRBCs infected red blood cells, N/A not applicable, NK not known
aSamples analysed for publication
Review of methodological approaches
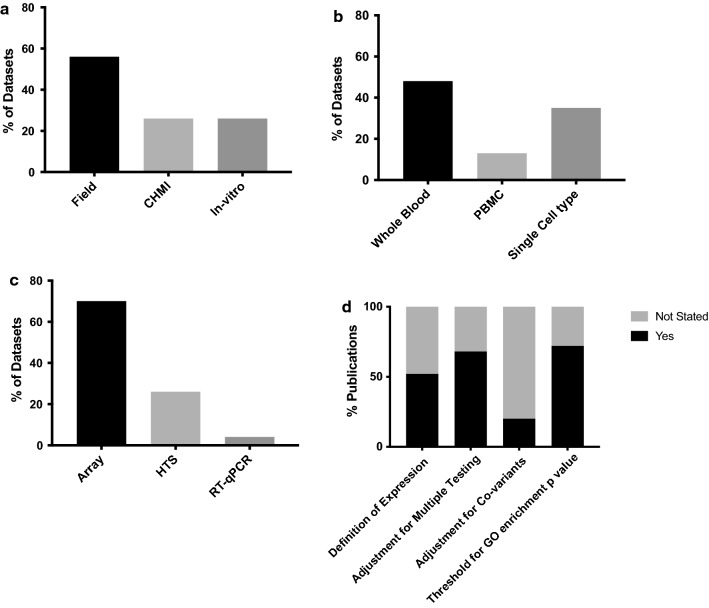
Significant heterogeneity in the datasets was found in terms of study design, sample type, platform used and method of analysis (Tables 1, 2 and Fig. 1), making direct comparison of results between studies difficult. Most datasets were generated from whole blood samples (n = 11, 48%), however some used PBMCs (n = 3, 13%) or individual tissue or cells types (n = 8, 35%) (Table 1). For the majority of studies, expression profiling was performed by array (n = 16, 70%), with others using high throughput sequencing (n = 6, 26%) or RT-qPCR [28] (n = 1, 4%) (Table 1). There was heterogeneity in data generation between studies with variation in methods used for normalization of data and adjustment for co-variables (Table 2). Thresholds for significance varied considerably and not all studies applied corrections for multiple testing. Choice of database used for gene ontology analysis also varied and there was variable, often incomplete reporting of analysis methods used (Table 2).
Table 2.
Comparison of methodological approaches for analysis of gene expression data
| Dataset | Data generation | Gene ontology analysis | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GEO series | Publication | RNA Quantification Platform | Normalization | Adjustment for covariates | Definition expression | Expressed genes | Threshold FC | Threshold P | Test | Multiple testing | GO analysis | Threshold GO enrichment p | Test | Multiple testing |
| GSE2900 | Griffiths (2005) | Stanford University cDNA lymphochip two color microarray | Scaled to geometric mean of sample:reference signal ratio from all array features | NS | Signal threshold | 9869 | 2.5 (from median in > 4 samples) | 0.1 | Permutation | FDR | NA | NA | NA | NA |
| GSE5418 | Ockenhouse (2006) | Affymetrix U133A GeneChips | RMA | NS | NS | NS | No | 0.01 | SAM, t-test | FDR | Onto Express and Pathway Architect | 0.05 | NS | FDR |
| GSE15221 | Franklin (2009) and Sharma (2011) | Illumina Human WG-6 v2.0 | Cubic spline | NS | Signal threshold | NS | 1.7 | 0.01 | Paired t-test | FDR | Onto Express | Varying | NS | NS |
| GSE15221 | Hirako (2018) | Illumina Human WG-6 v2.0 | Cubic spline | NS | Signal threshold | NS | 1.5 | 0.01 | Permutation and t-test | FDR | DAVID, GSEA | 0.05 | Multiple | FDR |
| GSE26876 | de Carvalho (2011) | Affymetrix Human Gene 1.0 ST Array | RMA | NS | NS | NS | 1.5 | 0.05 | Student t-test | No | Ingenuity pathway analysis | NS | NS | NS |
| GSE33811 | Krupka (2012) | Affymetrix Human Gene 1.0 ST Array | RMA and Quantile | NS | Signal and variation threshold | 3110 | 2 | 0.05 | Paired t-test | No | Gene set enrichment analysis on selected GO terms | 0.01 | Paired t-test | FDR |
| GSE34404 | Idaghdour (2012) | Illumina Human HT-12 BeadChips | Quantile | Location, Sex, Hb, total cell counts (RBCs and WBCs) and ancestry | Signal and normality threshold | NS | 2 (for comparison) | 0.01 | ANOVA, ANCOVA | FDR | Gene set enrichment analysis on customized MsigDB database | 0.05 | NS | Bonferroni |
| GSE55843 | Jagannathan (2014) | Agilent Sure Print G3 Human Gene Expression 8 × 60K v2 gene expression microarrays | Quantile | NS | Signal threshold | NS | 2 | 0.05 | SAM | FDR | NA | NA | NA | NA |
| GSE53292 | Jaijyan (2015) | Illumina Genome Analyzer Iix 72SE | NS | NS | NS | NS | NS | 0.05 | t-test | No | GeneCodis3, Bingo 2.3 plugin (Cytoscape 2.8.3) | 0.05 | NS | NS |
| GSE50957 GSE52166 | Tran (2016) | Illumina HiSeq 2000 2 × 100 PE | TAMM | Batch, Sex, Age, Pre-infection baseline | Signal and variation threshold, removal Y chromosomes | NS | 1.5 | 0.05 | Limma | FDR | Ingenuity pathway analysis | 0.05 | Fisher exact test | FDR |
| GSE50957 GSE67184 | Vallejo (2018) | Illumina HiSeq 2000 2 × 100 PE | CPM, TPM | NS | Signal threshold | NS | NS | 0.05 | EdgeR | FDR | WGSEA, ToppGene, STRING | 0.05 | Multiple | FDR |
| GSE64338 | Subramaniam (2015) | Affymetrix Human Gene 1.0 ST Array | Nonlinear normalization based on Li-Wong methods | NS | NS | NS | 1.2 | 0.001 | Paired t-test | FDR | Ingenuity Pathway Analysis | 0.05 | NS | FDR |
| GSE64493 | Sullivan (2015) | Agilent Sure Print G3 Human Gene Expression 8 × 60K v2 gene expression microarrays | Quantile | NS | Signal threshold | NS | 1.5 | 0.03 | Limma | FDR | DAVID | 0.05 | NS | FDR |
| GSE67184 | Rojas-Penas (2015) | Illumina HiSeq 2500 2 × 100 PE | SNM | Location/time-point, subject (random effect) | Signal threshold | 6154 | No | 0.05 | NS | FDR | NA | NA | NA | NA |
| GSE67184 | Gardinassi (2018) | Illumina HiSeq 2500 2 × 100 PE | NS | NS | NS | NS | No | 0.05 | Limma, repeated measures ANOVA | FDR | GSEA on blood transcriptome modules (BTM, Li et al.) | 0.05 | permutation | FDR |
| GSE7586 | Muehlenbachs (2007) | Affymetrix U133 Plus 2.0 GeneChip | GC RMA | NS | NS | NS | 2.5 | 0.01 | t-test | No | NA | NA | NA | NA |
| GSE77122 | Tarawa (2017) | Agilent Sure Print G3 Human Gene Expression 8 × 60K gene expression microarrays | Each gene expression array dataset was normalized to the in silicon pool for the macrophages cultured with RBCs | NS | NS | NS | No | 0.05 | Paired t-test | No | DAVID | 0.05 | Fisher exact test | No |
| GSE93664 | Burl (2017) | Affymetrix Human Gene ST 2.0 gene array | RMA | NS | NS | NS | 2 | 0.05 | NS | No | STRING | 0.01 | NS | Corrected unspecified |
| GSE100562 | Quin (2017) | Illumina HiSeq 2500 2 × 50 PE | NS | NS | NS | NS | No | 0.05 | Limma | FDR | NA | NA | NA | NA |
| GSE1124 | Boldt (2019) | Affymetrix U133A + B GeneChips | RMA | NS | Signal threshold | NS | 1.9 | 0.004 | SAM | FDR | DAVID and Ingenuity Pathway Analysis | 0.05 | NS | NS |
| GSE114076 | Terkawi (2018) | Agilent Sure Print G3 Human Gene Expression 8 × 60K gene expression microarrays | Each gene expression array dataset was normalized to the in silicon pool for the neutrophils cultured with RBCs | NS | NS | NS | 2 | 0.01 | Limma | No | Genomatix GeneRanker, DAVID, NET-GE and Enricher | 0.05 | NS | Corrected unspecified |
| GSE97158 | Rothan (2018) | Illumina HiSeq 2500 2 × 51 PE | TMM | Blocking by subject, in two separate models interaction with cell count and time of parasitemia was added | Signal threshold | 16,473 | 1.5 | 0.05 | Limma | FDR | GSEA (camera) on blood transcriptome modules (BTM, Li et al.) | 0.05 | Fisher exact test | FDR |
| GSE65928 | Portugal (2015) | Affymetrix Human Gene ST 2.0 gene array | RMA | NS | NS | NS | NS | 0.05 | ANOVA | FDR | Ingenuity pathway analysis | NS | NS | NS |
| GSE72058 | Feintuch (2016) | Affymetrix Human Gene 1.0 ST array | RMA and Quantile | Peripheral parasitemia | NS | NS | No | 0.05 | t-test | No | GSEA, CateGOrizer and ingenuity pathway analysis | 0.2 and 0.06 | NS | FDR |
FDR false discovery rate, Hb haemoglobin, NA not available, NS not specified in publication, RBCs red blood cells, RMA Robust Multichip average, SNM supervised normalization of microarray, TMM trimmed mean of M-values, GEO Gene Expression Omnibus, GE gene ontology
Fig. 1.
Comparison of key methodological variables between datasets or publications. a Antigenic stimulation; CHMI controlled human malaria infection, ‘field’ infection naturally by mosquito bite, ‘in-vitro’ in vitro stimulation by sporozoites or infected red blood cells. Some datasets employed more than one method of antigenic stimulation. b Tissue type analysed; PBMC peripheral blood mononuclear cells. c Expression profiling method: HTS high throughput sequencing. d Manipulation of data, go gene ontology
Transcriptional insights into the immune response to malaria infection
Seven datasets provided insight into the transcriptional changes associated with NAI to malaria (Table 3) [24–26, 28–31]. However, given the difficulty in defining or quantifying NAI for an individual, studies varied in their approach, choosing to examine GEPs in settings of varying history of prior exposure to malaria [25, 26, 28, 29], symptomatology during infection [25] or severity of disease [24, 32]. All studies examining NAI included small numbers of subjects and all deployed different experimental designs (Table 3).
Table 3.
Gene expression studies informing understanding of naturally acquired immunity to malaria infection
| Measure of NAI | Publication | Design | Sample | Species | Subjects for comparison | Key finding | Comment | |
|---|---|---|---|---|---|---|---|---|
| Prior exposure to malaria | Tran et al. (2016) | Comparison of GEP changes from paired infected and uninfected samples | Whole blood | P. falciparum | Malaria-naïve, symptomatic Dutch CHMI volunteers at diagnosis (n = 5) | Malaria experienced Malian children (> 13 years) and adults infected in the field (n = 8) | Graded activation of pathways of downstream proinflammatory cytokines with highest activation in malaria-naive subjects and significantly reduced activation in malaria experienced Malians | |
| Ockenhouse et al. (2006) | Comparison of GEP changes in infection-controls samples US malaria naïve subjects | PBMC | P. falciparum | US malaria-naïve CHMI volunteers with early, blood-stage infection (n = 22) | Malaria-experienced Cameroonian adults presenting with naturally acquired febrile malaria (n = 15) | Similar induction of pro-inflammatory cytokines seen between pre-symptomatic and symptomatic individuals regardless of prior malaria exposure | ||
| Rojas-Pena et al. (2015) and Vallejo et al. (2018) | Comparison of GEP changes from paired infected and uninfected samples | Whole blood | P. vivax | Columbian malaria-naïve (MN) CHMI volunteers at diagnosis (n = 7) | Columbian malaria-exposed (ME) CHMI volunteers at diagnosis (n = 9) | Little differentiation seen between MN and ME populations by Rojas-Penas et al. However network co-expression analysis by Vallejo et al. showed the inflammatory response was attenuated in ME volunteers with decreased class II antigen presentation in dendritic cells | No significant difference between groups for pre-patent period or parasitaemia at diagnosis suggesting there may have been no difference in functional immunity between groups | |
| Jagannathan et al. (2014) | Comparison of GEP between groups | Vδ2+ T cells | P. falciparum | Ugandan children with low prior malaria incidence (n = 4) | Ugandan children with low prior malaria incidence (n = 4) | Comparison of basal gene expression patterns of sorted, un-stimulated Vδ2+ T cells identified 48 differentially expressed genes, many with known roles in immunomodulation. For each of these genes, expression was higher among children with high prior exposure to malaria | Data suggest recurrent malaria infection causes up-regulation of immunoregulatory pathways that dampen the pro-inflammatory immune response to P. falciparum infection and help explain immunological tolerance to the parasite | |
| Symptoms at diagnosis | Tran et al. (2016) | Comparison of GEP changes from paired infected and uninfected samples | Whole blood | P. falciparum | Malaria experienced Malian children (> 13 years) and adults infected in the field and asymptomatic at diagnosis (EA, n = 5) | Malaria experienced Malian children (> 13 years) and adults infected in the field and symptomatic with fever at the time of diagnosis (EF, n = 3) | Only 70 differentially expressed genes (DEGs) were identified between these groups despite the apparent clinical differences | 2 of the 5 individuals in the EA group progressed to febrile malaria within 5 days of initial diagnosis by PCR |
| Disease severity | Krupka et al. (2012) | Comparison of GEP in same subjects at diagnosis with severe and subsequent mild malaria | Whole blood | P. falciparum | Malawian children who, after presenting with severe malaria (all had cerebral malaria), were found to have mild malaria one month later on screening by blood smear (n = 5) | Pathway analysis showed relative up regulation of Type I IFN signaling pathway, regulation of inflammation, regulation of leukocyte proliferation and T cell activation in episodes of mild malaria | ||
| Boldt et al. (2019) | Comparison of GEP between groups | Whole blood | P. falciparum | Healthy uninfected Gabonese children | Gabonese children with asymptomatic parasitaemia, mild malaria, malaria with severe anaemia and cerebral anaemia (0.5–6 years) | GEP of 22 genes significantly differed among groups. Immunoglobulin production, complement regulation and IFN beta signaling were most conspicuous | ||
PBMC peripheral blood mononuclear cells, GEP gene expression profile, CHMI controlled human malaria infection
The findings from a number of studies supported a dampening of the innate pro-inflammatory immune response as a mechanism underpinning NAI [24–26, 33] although this finding was not observed in all studies [28, 29, 31].
One study by Franklin et al. provided evidence of ‘pro-inflammatory priming’ of the innate immune system in acute malaria infection [34]. Comparison of GEP in Brazilian adults presenting with uncomplicated malaria with paired convalescent samples showed an increase in expression in genes involved in TLR signalling pathways supporting a role for TLR hyper responsiveness in the pathology of malaria infection [34, 35].
Quin et al. sought to use RNA sequencing to elucidate the mechanism driving lower infection rates, lower parasite densities and fewer symptomatic cases of P. falciparum in the population of Fulani compared to other sympatric ethnic groups [33]. Comparison of the GEP of monocytes from infected and uninfected Fulani and Mossi adults showed a marked difference, with a significantly greater number of differentially expressed (DE) genes in infected Fulani compared to infected Mossi participants (1239 versus 3 DE genes respectively). Pathway analysis showed that infected Fulani, but not infected Mossi, individuals demonstrated a marked reduction in expression of inflammasome pathway components, suggesting a blunting of the innate pro-inflammatory immune response post-infection could explain the differences in susceptibility.
Another study sought to examine the genetic basis of gene expression variation in malaria [36]. Idaghdour et al. compared GEP in children diagnosed with uncomplicated malaria (n = 94) in Benin with age matched controls (n = 64) [36] and performed a genome wide association test of transcript abundance. Testing for genotype-by-infection interactions demonstrated the existence of genome wide significant interactions and other genes subject to interaction effects beneath genome-wide significance but still likely to have important roles in modulating the course of infection. These interactions affected the complement system, antigen processing and presentation and T cell activation [36].
In work to identify a transcriptional signature to distinguish acute malaria from other febrile illnesses, Griffiths et al. compared the GEP of twenty-two Kenyan children admitted with febrile illnesses (fifteen of which had malaria infection alone) with six convalescent samples collected 2 weeks post discharge [23]. Two main GEPs relating to neutrophil and erythroid activity were shown to differentiate acutely ill and convalescent children, with significantly higher expression of genes in the neutrophil-related gene region in subjects with bacterial infections and significantly higher expression of genes related to lymphocyte and T cell activation in subjects with malaria. The authors also identified two gene profiles whose expression intensity correlated with host parasitaemia.
Only two datasets included gene expression changes following P. vivax infection [28, 30, 37]. Rojas-Penas et al. interrogated GEP changes in malaria naïve (MN) and malaria-exposed (ME) Columbian volunteers following infection with P. vivax in a CHMI setting [28]. Significant GEP changes were consistent with time-point rather than prior malaria exposure, with a decline in innate immune signalling and neutrophil number (in contrast to strong up regulation of the same genes reported by Igadour et al. [36]) and an increase in interferon induction seen at diagnosis. No significant GEP changes were noted at other time points, including those relating to the liver stage of infection. Further analysis of this dataset by Vallejo et al. using network co-expression analysis showed that while P. vivax infection induced strong inflammatory responses in all participants, the inflammatory response was attenuated with pathways associated with antigen processing and presentation less enriched in those with prior exposure to P. vivax, suggesting a more ‘tolerogenic’ immune response in these individuals [30].
In contrast to this work, Rothen et al. found that transcriptional changes post-CHMI via intradermal injection of cryopreserved P. falciparum sporozoites were most pronounced on day 5 after inoculation, during the clinically silent liver stage rather than during the blood-stage of infection [38].
Transcriptomic studies in specific cell types
Whilst the majority of studies examined the immune response from whole blood or PBMCs, some examined transcriptomic changes in other cell types or tissues [26, 33, 39–45]. For example, the work of Muehlenbachs et al. with placental tissue highlighted a previously unappreciated role for B cells in chronic placental malaria [39]; whilst Sullivan et al. compared GEPs of classical and ‘atypical’ memory B cells obtained from Ugandan children showing the latter demonstrated down-regulation of B cell receptor signalling and apoptosis [43].
Discussion
GEP is a powerful tool to analyse the immune response to infection. As this review demonstrates, the application of these studies for malaria are wide-ranging, from attempts to dissect the mechanisms of NAI to improving understanding of the interaction between host genotype and infection outcome. However, as a field in its relative infancy, studies are often hypothesis generating with extremely small sample sizes. There is a lack of standardization ranging from methodological (such as sample type, RNA extraction, platform and analysis) to phenotype (including precision in disease context and immune status). This variation means interpreting published data and comparison between studies is challenging. Some of this is unavoidable, however, much could be addressed, for example by implementing standardization in blood sampling, methodological protocols for data generation and analysis with robust significance testing and approaches to confounders, use of ontologies (for example human phenotype and gene ontologies) and expert curation and annotation of data on deposition [46–49].
GEP studies are well placed to examine the mechanisms of NAI and have already helped highlight the role of the innate and early adaptive immune responses [24–26]. However, work has been limited by the lack of an in vitro correlate or universally accepted definition of NAI, meaning identifying the immune status of individuals or quantification of immunity is problematic [6, 50]. In field studies where the timing of infection and parasite burden and dynamics are unknown, and potentially hugely variable between individuals, only limited information can be reliably extrapolated from any GEP changes seen. Most studies assess gene expression from peripheral blood or its components, which does not provide reliable information regarding the transcriptional changes in key organs such as spleen, liver, and bone marrow. In addition, when subjects are recruited at presentation with disease, no baseline comparator data are available to use as a control. Even if a clear difference in GEP were to be reported between individuals with and without NAI, it would be near impossible to distinguish GEP changes associated with parasitaemia from those mediating immunity.
However, there is much potential for the future use of GEP studies, particularly in CHMI studies [51, 52] where the parasite burden can be pre-defined and dynamics of infection closely monitored using highly sensitive qPCR. As these studies are increasingly performed in endemic settings [53–55], there will be growing opportunity to use GEP to understand detailed time-course changes in immune response, particularly at the skin, liver and pre-symptomatic blood-stage, which to date have been difficult to study in human subjects infected in the field.
Conclusion
GEP in malaria is a potentially powerful tool, but to date studies have been hypothesis generating with small sample sizes and widely varying methodology. As CHMI studies are increasingly performed in endemic settings, there will be growing opportunity to use GEP to understand detailed time-course changes in host response and understand in greater detail the mechanisms of NAI.
Supplementary information
Additional file 1: Figure S1. Flowchart summarizing identification of GEO datasets and publications.
Acknowledgements
Not applicable.
Abbreviations
- CHMI
controlled human malaria infection
- GEO
gene expression omnibus
- GEP
gene expression profile
- NAI
naturally acquired immunity
- PBMC
peripheral blood mononuclear cells
Authors’ contributions
SH conceived the work, analysed the datasets and wrote the manuscript. JM conducted the methodological review of the datasets. HEL, SJD, AVSH, JCK and KM made significant contributions to the conception of the work. JCK substantially revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by The Wellcome Trust [Grant Number 097940/Z/11/Z to SHH and Wellcome Trust Core Award Grant Number 090532/Z/09/Z]. SHH is a NIHR Academic Clinical Lecturer in Infectious Diseases & Microbiology at the University of Oxford and Research Fellow at St. Peter’s College, University of Oxford. SJD and AVSH are Jenner Investigators, and SJD is also a Lister Institute Research Prize Fellow and a Wellcome Trust Senior Fellow [106917/Z/15/Z]. JCK is a Wellcome Trust Investigator. The funders had no role in the design, collection, analysis, or interpretation of data; in the writing of the manuscript; or in the decision to submit the manuscript for publication.
Availability of data and materials
Data sharing is not applicable to this article as no datasets were generated or analysed during the current study
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12936-019-3035-0.
References
- 1.White NJ, Pukrittayakamee S, Hien TT, Faiz MA, Mokuolu OA, Dondorp AM. Malaria. Lancet. 2014;383:723–735. doi: 10.1016/S0140-6736(13)60024-0. [DOI] [PubMed] [Google Scholar]
- 2.Cockburn IA, Seder RA. Malaria prevention: from immunological concepts to effective vaccines and protective antibodies. Nat Immunol. 2018;19:1199–1211. doi: 10.1038/s41590-018-0228-6. [DOI] [PubMed] [Google Scholar]
- 3.WHO . World malaria report. Geneva: World Health Organization; 2018. [Google Scholar]
- 4.Cohen S, Mc GI, Carrington S. Gamma-globulin and acquired immunity to human malaria. Nature. 1961;192:733–737. doi: 10.1038/192733a0. [DOI] [PubMed] [Google Scholar]
- 5.Sabchareon A, Burnouf T, Ouattara D, et al. Parasitologic and clinical human response to immunoglobulin administration in falciparum malaria. Am J Trop Med Hyg. 1991;45:297–308. doi: 10.4269/ajtmh.1991.45.297. [DOI] [PubMed] [Google Scholar]
- 6.Doolan DL, Dobano C, Baird JK. Acquired immunity to malaria. Clin Microbiol Rev. 2009;22:13–36. doi: 10.1128/CMR.00025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conesa A, Mortazavi A. The common ground of genomics and systems biology. BMC Syst Biol. 2014;8(Suppl 2):S1. doi: 10.1186/1752-0509-8-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schulze A, Downward J. Navigating gene expression using microarrays—a technology review. Nat Cell Biol. 2001;3:E190–E195. doi: 10.1038/35087138. [DOI] [PubMed] [Google Scholar]
- 9.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papalexi E, Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol. 2018;18:35–45. doi: 10.1038/nri.2017.76. [DOI] [PubMed] [Google Scholar]
- 11.Querec TD, Akondy RS, Lee EK, Cao W, Nakaya HI, Teuwen D, et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol. 2009;10:116–125. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Six A, Bellier B, Thomas-Vaslin V, Klatzmann D. Systems biology in vaccine design. Microb Biotechnol. 2012;5:295–304. doi: 10.1111/j.1751-7915.2011.00321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rappuoli R, Aderem A. A 2020 vision for vaccines against HIV, tuberculosis and malaria. Nature. 2011;473:463–469. doi: 10.1038/nature10124. [DOI] [PubMed] [Google Scholar]
- 14.Hogan CM, Hammer SM. Host determinants in HIV infection and disease. Part 1: cellular and humoral immune responses. Ann Intern Med. 2001;134:761–776. doi: 10.7326/0003-4819-134-9_Part_1-200105010-00013. [DOI] [PubMed] [Google Scholar]
- 15.Blohmke CJ, Darton TC, Jones C, Suarez NM, Waddington CS, Angus B, et al. Interferon-driven alterations of the host’s amino acid metabolism in the pathogenesis of typhoid fever. J Exp Med. 2016;213:1061–1077. doi: 10.1084/jem.20151025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong HR. Clinical review: sepsis and septic shock—the potential of gene arrays. Crit Care. 2012;16:204. doi: 10.1186/cc10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anderson ST, Kaforou M, Brent AJ, Wright VJ, Banwell CM, Chagaluka G, et al. Diagnosis of childhood tuberculosis and host RNA expression in Africa. N Engl J Med. 2014;370:1712–1723. doi: 10.1056/NEJMoa1303657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mejias A, Ramilo O. Transcriptional profiling in infectious diseases: ready for prime time? J Infect. 2014;68(Suppl 1):S94–S99. doi: 10.1016/j.jinf.2013.09.018. [DOI] [PubMed] [Google Scholar]
- 19.Berry MP, Graham CM, McNab FW, Xu Z, Bloch SA, Oni T, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466:973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Westermann AJ, Forstner KU, Amman F, Barquist L, Chao Y, Schulte LN, et al. Dual RNA-seq unveils noncoding RNA functions in host–pathogen interactions. Nature. 2016;529:496–501. doi: 10.1038/nature16547. [DOI] [PubMed] [Google Scholar]
- 21.Cummings CA, Relman DA. Using DNA microarrays to study host–microbe interactions. Emerg Infect Dis. 2000;6:513–525. doi: 10.3201/eid0605.000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunachie S, Hill AV, Fletcher HA. Profiling the host response to malaria vaccination and malaria challenge. Vaccine. 2015;33:5316–5320. doi: 10.1016/j.vaccine.2015.07.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griffiths MJ, Shafi MJ, Popper SJ, Hemingway CA, Kortok MM, Wathen A, et al. Genomewide analysis of the host response to malaria in Kenyan children. J Infect Dis. 2005;191:1599–1611. doi: 10.1086/429297. [DOI] [PubMed] [Google Scholar]
- 24.Krupka M, Seydel K, Feintuch CM, Yee K, Kim R, Lin CY, et al. Mild Plasmodium falciparum malaria following an episode of severe malaria is associated with induction of the interferon pathway in Malawian children. Infect Immun. 2012;80:1150–1155. doi: 10.1128/IAI.06008-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tran TM, Jones MB, Ongoiba A, Bijker EM, Schats R, Venepally P, et al. Transcriptomic evidence for modulation of host inflammatory responses during febrile Plasmodium falciparum malaria. Sci Rep. 2016;6:31291. doi: 10.1038/srep31291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jagannathan P, Kim CC, Greenhouse B, Nankya F, Bowen K, Eccles-James I, et al. Loss and dysfunction of Vdelta2(+) gammadelta T cells are associated with clinical tolerance to malaria. Sci Transl Med. 2014;6:251ra117. doi: 10.1126/scitranslmed.3009793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets-update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rojas-Pena ML, Vallejo A, Herrera S, Gibson G, Arevalo-Herrera M. Transcription profiling of malaria-naive and semi-immune Colombian volunteers in a Plasmodium vivax sporozoite challenge. PLoS Negl Trop Dis. 2015;9:e0003978. doi: 10.1371/journal.pntd.0003978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ockenhouse CF, Hu WC, Kester KE, Cummings JF, Stewart A, Heppner DG, et al. Common and divergent immune response signaling pathways discovered in peripheral blood mononuclear cell gene expression patterns in presymptomatic and clinically apparent malaria. Infect Immun. 2006;74:5561–5573. doi: 10.1128/IAI.00408-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vallejo AF, Read RC, Arevalo-Herrera M, Herrera S, Elliott T, Polak ME. Malaria systems immunology: Plasmodium vivax induces tolerance during primary infection through dysregulation of neutrophils and dendritic cells. J Infect. 2018;77:440–447. doi: 10.1016/j.jinf.2018.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boldt ABW, van Tong H, Grobusch MP, Kalmbach Y, Dzeing Ella A, Kombila M, et al. The blood transcriptome of childhood malaria. EBioMedicine. 2019;40:614–625. doi: 10.1016/j.ebiom.2018.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Subramaniam KS, Spaulding E, Ivan E, Mutimura E, Kim RS, Liu X, et al. The T-cell inhibitory molecule Butyrophilin-Like 2 is up-regulated in mild Plasmodium falciparum infection and is protective during experimental cerebral malaria. J Infect Dis. 2015;212:1322–1331. doi: 10.1093/infdis/jiv217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quin JE, Bujila I, Cherif M, Sanou GS, Qu Y, Vafa Homann M, et al. Major transcriptional changes observed in the Fulani, an ethnic group less susceptible to malaria. Elife. 2017;6:e29156. doi: 10.7554/eLife.29156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Franklin BS, Parroche P, Ataide MA, Lauw F, Ropert C, de Oliveira RB, et al. Malaria primes the innate immune response due to interferon-gamma induced enhancement of toll-like receptor expression and function. Proc Natl Acad Sci USA. 2009;106:5789–5794. doi: 10.1073/pnas.0809742106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma S, DeOliveira RB, Kalantari P, Parroche P, Goutagny N, Jiang Z, et al. Innate immune recognition of an AT-rich stem-loop DNA motif in the Plasmodium falciparum genome. Immunity. 2011;35:194–207. doi: 10.1016/j.immuni.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Idaghdour Y, Quinlan J, Goulet JP, Berghout J, Gbeha E, Bruat V, et al. Evidence for additive and interaction effects of host genotype and infection in malaria. Proc Natl Acad Sci USA. 2012;109:16786–16793. doi: 10.1073/pnas.1204945109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gardinassi LG, Arevalo-Herrera M, Herrera S, Cordy RJ, Tran V, Smith MR, et al. Integrative metabolomics and transcriptomics signatures of clinical tolerance to Plasmodium vivax reveal activation of innate cell immunity and T cell signaling. Redox Biol. 2018;17:158–170. doi: 10.1016/j.redox.2018.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rothen J, Murie C, Carnes J, Anupama A, Abdulla S, Chemba M, et al. Whole blood transcriptome changes following controlled human malaria infection in malaria pre-exposed volunteers correlate with parasite prepatent period. PLoS ONE. 2018;13:e0199392. doi: 10.1371/journal.pone.0199392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muehlenbachs A, Fried M, Lachowitzer J, Mutabingwa TK, Duffy PE. Genome-wide expression analysis of placental malaria reveals features of lymphoid neogenesis during chronic infection. J Immunol. 2007;179:557–565. doi: 10.4049/jimmunol.179.1.557. [DOI] [PubMed] [Google Scholar]
- 40.Burel JG, Apte SH, Groves PL, McCarthy JS, Doolan DL. Polyfunctional and IFN-gamma monofunctional human CD4+ T cell populations are molecularly distinct. JCI Insight. 2017;2:e87499. doi: 10.1172/jci.insight.87499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jaijyan DK, Singh H, Singh AP. A sporozoite- and liver stage-expressed tryptophan-rich protein plays an auxiliary role in Plasmodium liver stage development and is a potential vaccine candidate. J Biol Chem. 2015;290:19496–19511. doi: 10.1074/jbc.M114.588129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Terkawi MA, Takano R, Furukawa A, Murakoshi F, Kato K. Involvement of beta-defensin 130 (DEFB130) in the macrophage microbicidal mechanisms for killing Plasmodium falciparum. Sci Rep. 2017;7:41772. doi: 10.1038/srep41772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sullivan RT, Kim CC, Fontana MF, Feeney ME, Jagannathan P, Boyle MJ, et al. FCRL5 delineates functionally impaired memory B cells associated with Plasmodium falciparum exposure. PLoS Pathog. 2015;11:e1004894. doi: 10.1371/journal.ppat.1004894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Terkawi MA, Takano R, Kato K. Differential gene expression profile of human neutrophils cultured with Plasmodium falciparum-parasitized erythrocytes. J Immunol Res. 2018;2018:6709424. doi: 10.1155/2018/6709424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Portugal S, Tipton CM, Sohn H, Kone Y, Wang J, Li S, et al. Malaria-associated atypical memory B cells exhibit markedly reduced B cell receptor signaling and effector function. Elife. 2015;4:e07218. doi: 10.7554/eLife.07218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bammler T, Beyer RP, Bhattacharya S, Boorman GA, Boyles A, Bradford BU, et al. Standardizing global gene expression analysis between laboratories and across platforms. Nat Methods. 2005;2:351–356. doi: 10.1038/nmeth0605-477a. [DOI] [PubMed] [Google Scholar]
- 47.Conesa A, Madrigal P, Tarazona S, Gomez-Cabrero D, Cervera A, McPherson A, et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016;17:13. doi: 10.1186/s13059-016-0881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chowdhury HA, Bhattacharyya DK, Kalita JK. (Differential) co-expression analysis of gene expression: a survey of best practices. IEEE/ACM Trans Comput Biol Bioinform. 2019 doi: 10.1109/TCBB.2019.2893170. [DOI] [PubMed] [Google Scholar]
- 49.Rung J, Brazma A. Reuse of public genome-wide gene expression data. Nat Rev Genet. 2013;14:89–99. doi: 10.1038/nrg3394. [DOI] [PubMed] [Google Scholar]
- 50.Langhorne J, Ndungu FM, Sponaas AM, Marsh K. Immunity to malaria: more questions than answers. Nat Immunol. 2008;9:725–732. doi: 10.1038/ni.f.205. [DOI] [PubMed] [Google Scholar]
- 51.Barton AJ, Hill J, Pollard AJ, Blohmke CJ. Transcriptomics in human challenge models. Front Immunol. 2017;8:1839. doi: 10.3389/fimmu.2017.01839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stanisic DI, McCarthy JS, Good MF. Controlled human malaria infection: applications, advances, and challenges. Infect Immun. 2018;86:e00479. doi: 10.1128/IAI.00479-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lell B, Mordmuller B, Dejon Agobe JC, Honkpehedji J, Zinsou J, Mengue JB, et al. Impact of sickle cell trait and naturally acquired immunity on uncomplicated malaria after controlled human malaria infection in adults in Gabon. Am J Trop Med Hyg. 2018;98:508–515. doi: 10.4269/ajtmh.17-0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hodgson SH, Juma E, Salim A, Magiri C, Kimani D, Njenga D, et al. Evaluating controlled human malaria infection in Kenyan adults with varying degrees of prior exposure to Plasmodium falciparum using sporozoites administered by intramuscular injection. Front Microbiol. 2014;5:686. doi: 10.3389/fmicb.2014.00686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shekalaghe S, Rutaihwa M, Billingsley PF, Chemba M, Daubenberger CA, James ER, et al. Controlled human malaria infection of Tanzanians by intradermal injection of aseptic, purified, cryopreserved Plasmodium falciparum sporozoites. Am J Trop Med Hyg. 2014;91:471–480. doi: 10.4269/ajtmh.14-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Flowchart summarizing identification of GEO datasets and publications.
Data Availability Statement
Data sharing is not applicable to this article as no datasets were generated or analysed during the current study