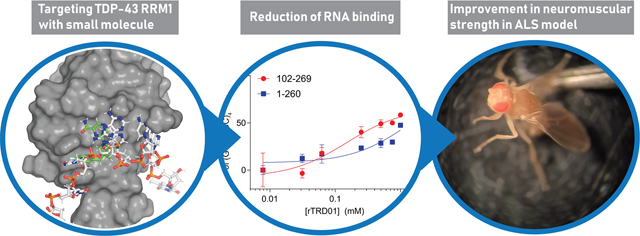
Abstract
RNA dysregulation likely contributes to disease pathogenesis of amyotrophic lateral sclerosis (ALS) and other neurodegenerative diseases. A pathological form of the transactive response (TAR) DNA binding protein (TDP-43) binds to RNA in stress granules and forms membraneless, amyloid-like TDP-43 aggregates in the cytoplasm of ALS motor neurons. In this study, we hypothesized that by targeting the RNA recognition motif (RRM) domains of TDP-43 that confer a pathogenic interaction between TDP-43 and RNA, motor neuron toxicity could be reduced. In silico docking of 50000 compounds to the RRM domains of TDP-43 identified a small molecule (rTRD01) that (i) bound to TDP-43’s RRM1 and RRM2 domains, (ii) partially disrupted TDP-43’s interaction with the hexanucleotide RNA repeat of the disease-linked c9orf 72 gene, but not with (UG)6 canonical binding sequence of TDP-43, and (iii) improved larval turning, an assay measuring neuromuscular coordination and strength, in an ALS fly model based on the overexpression of mutant TDP-43. Our findings provide an instructive example of a chemical biology approach pivoted to discover small molecules targeting RNA–protein interactions in neurodegenerative diseases.
Graphical Abstract
Ahallmark feature of amyotrophic lateral sclerosis (ALS) and related neurodegenerative diseases is the altered subcellular localization of transactive response (TAR) DNA binding protein (TDP-43) and subsequent formation of prion-like TDP-43 aggregates in motor neurons in ∼95% of patients.1 TDP-43 mutations have also been identified in patients with familial and sporadic ALS, underscoring the importance of TDP-43 in the pathophysiology of the neurodegeneration seen in this disease.2 Therefore, investigations into TDP-43 offer an opportunity to study ALS disease progression and enable development of chemical probes with potential for therapeutic intervention.
TDP-43 is a 43 kDa protein consisting of an N-terminal domain (NTD) and two tandem RNA recognition motifs, RRM1 and RRM2, followed by a C-terminal glycine-rich region where most disease-associated mutations converge. TDP-43 binds to nucleic acids via its RRM domains (Figure 1A) and contributes to RNA processing, including but not limited to splicing, translation,3 and cytoplasmic stress granule response, protein complexes that sequester mRNAs to minimize stress-related damage.4
Figure 1.
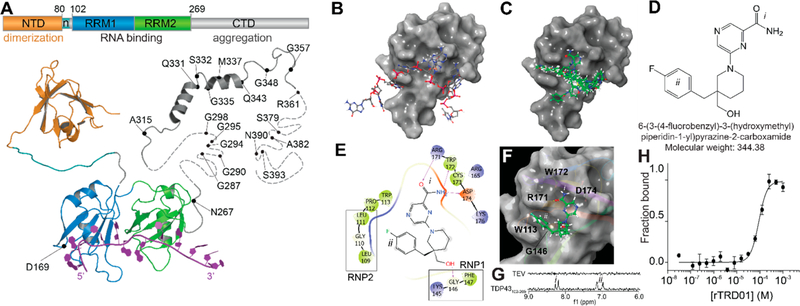
In silico docking to the RNA recognition motif of TDP-43 identifies compound rTRD01. (A) Composite linear and structure representation of TDP-43. The N-terminal domain (NTD) of TDP-43 (orange) is important for dimerization and also contains a nuclear localization signal, n (cyan) (PDB code 2n4p25). The RNA recognition motifs 1 and 2 (RRM1 and 2, blue and green, respectively, PDB code 4bs217) interact with RNA (purple). The C-terminal domain (CTD, gray) is largely unstructured and is considered a “prion-like” domain. Residues M311–Q360 from the CTD were resolved using NMR (PDB 2n3x41). In unstructured regions, each dash represents a residue. ALS-associated mutations are noted. (B) TDP-43 RRM1 domain (surface representation) in complex with DNA (stick and ball representation) (PDB code 4iuf13). (C) Top 10 compounds (green, sticks and balls representation) from in silico docking on TDP-43 RRM1. (D) Structure of rTRD01 (6-(3-(4-fluorobenzyl)-3-(hydroxymethyl)piperidin-1-yl)pyrazine-2-carboxamide), where i is a pyrazinamide group and ii is a fluorobenzyl group shown to interact with TDP-43 by STD NMR (8.2 ppm and 6.9–7.2 ppm, respectively). (E) 2D representation of rTRD01 binding pocket, which includes residues from the ribonucleoprotein motif 1 (RNP1) and ribonucleoprotein motif 2 (RNP2) of TDP-43 RRM1. (F) Docking pose of rTRD01 (green, sticks and balls representation) on RRM1 domain of TDP-43 (surface representation). (G) 1D 1H STD NMR showing on-resonance difference spectrum of 500 μM rTRD01 with 5 μM TDP-43102–269. The aromatic region of the NMR spectrum (6–9 ppm) is shown. (H) MST values from thermographs of NT-647 labeled TDP-43102–269 in the presence of increasing concentrations (0.03 μM to 1 mM) of rTRD01were used to determine dissociation constant for binding of rTRD01 to TDP-43102–269. The data were fitted as described in the Methods section yielding an apparent Kd of 89.4 ± 0.8 μM (r2 = 0.93). Data are given as mean ± SEM (n = 3 independent replicates).
TDP-43 proteinopathy has been associated with a cytoplasmic accumulation of full-length and fragmented TDP-43 protein in detergent-resistant, ubiquitinated, and hyperphosphorylated aggregates.5 A current working hypothesis is that stress granule formation facilitates or promotes cytoplasmic aggregation of TDP-43 in motor neurons.6–8 Furthermore, localization of TDP-43 to stress granules was shown to be driven by both its RRM1 domain and its C-terminal glycine-rich/prion-like domain.4,9 But, previous efforts to disrupt TDP-43 aggregates using peptides against TDP-43 C-terminus were unsuccessful in decreasing cytotoxicity.10 Interestingly, degenerative ALS phenotypes induced by overexpression of TDP-43 in Drosophila melanogaster were completely reversed by deletion of RRM1 or by mutations (W113A/R151A) that ablated or reduced RNA binding to TDP-43.11 In a cellular model of ALS, TDP35, a pathological cleaved fragment of TDP-43, was shown to sequester TDP-43 through RNA binding, and mutations in RRM1, but not RRM2, dramatically reduced the ability to form inclusions.12
Based on these studies, we hypothesized that targeting TDP-43’s RRM1 domain with small molecules might be a promising strategy to reduce TDP-43 toxicity in ALS models. Computational in silico docking and biochemical experiments identified a compound, 6-(3-(4-fluorobenzyl)-3-(hydroxymethyl)-piperidin-1-yl)pyrazine-2-carboxamide (hereafter designated as rTRD01 for RRM TDP-43 RNA disruptor 01) that bound to TDP-43 in the micromolar range. As it is critical for TDP-43 to maintain normal nucleic acid binding activity in the cell, we tested rTRD01 for its ability to bind normal and disease-linked nucleic acids. rTRD01 was able to reduce TDP-43 interactions with disease-linked nucleic acid while not interfering with a TDP-43 canonical substrate (UG rich binding sequence) in vitro. Finally, rTRD01 reduced ALS-like locomotor phenotypes in a Drosophila model.
RESULTS
In Silico Docking of TDP-43 RNA–Protein Interface Reveals RRM1 Is a Druggable Site.
The crystal structure of RRM1 with single-stranded DNA bound13(Figure 1B), allowed us to identify pockets overlapping with nucleic acid binding sites for targeted in silico docking experiments. Using the Schrödinger modeling program, we created an active site pocket that includes residues Leu109, Gly110, Pro112, Trp113, and Arg171 (Figure 1B,C) in the RRM1 domain. Among several mutations in the RRM1, mutations Trp113Ala and Arg171Ala were shown to be the most deleterious as they increased the estimated dissociation constant of TDP-43 for nucleic acid by 6-fold.13 Moreover, residues Leu109 and Gly110 are known to be part of the ribonucleoprotein domain 2 (RNP2, Leu-Ile-Val-Leu-Gly-Leu),13 that stacks with the bases and the sugar rings of nucleic acids. RNP-1 ([RK]-G-[FY]-[GA]-[FY]-[ILV]-X-[FY]) and RNP-2 ([LI]-[FY]-[VI]-X-[NG]-L) are known consensus sequences in RRM domains,14 usually located in β2 and β3, and stack with the bases and the sugar rings of the nucleic acid.
Virtual screening of a 50 000 compound library from Chembridge resulted in the identification of small molecules that stack with Trp113 on one side and Arg171 on the other side of the active site cleft (Supplementary Figure 1). The docked compounds displayed a significant overlap with nucleic acid binding surface and mimicked several interactions between RRM1 domain and nucleic acid bases. The top 8 compounds from this screen were selected based on Glide score and visual examination15 (Supplementary Table 1) for further studies.
We sourced the top 8 compounds and tested their binding to recombinantly purified TDP-43102–269-His using saturation transfer difference NMR (STD NMR). STD NMR profiling demonstrated that 5 of the 8 compounds bound TDP-43 (Supplementary Figure 2).
Biophysical Characterization of rTRD01.
We selected the top in silico docking hit, rTRD01 (Figure 1D), as it showed the highest XP Glide score16 for TDP-43 RRM1 domain (Supplementary Table 1). The fluorobenzyl group of rTRD01 is predicted to interact with Trp113 and Arg171, while the pyrazinamide group interacts with Asp174, similarly to guanine G6 in the X-ray crystal structure of TDP-43 RRM1 domain with DNA (Figure 1). rTRD01 is also predicted to make hydrogen bonds with Gly146, a residue from the ribonucleoprotein sequence 1 (RNP1) of TDP-43 RRM1,13,17 and with Arg171, Trp172, and Asp174 (Figure 1E,F).
We demonstrated that rTRD01 binds to TDP-43102–269 using STD NMR, while no binding was observed with a control protein (TEV protease) (Supplementary Figure 2 and Figure 1G). The positive peaks for rTRD01 in the presence of TDP-43102–269 were in the region of the fluorobenzyl group (ii, 6.9–7.2 ppm) and the pyrazinamide group (i, 8.2 ppm), confirming the binding implied by virtual screening.
Microscale thermophoresis (MST), an assay that allows sensitive measurement of molecular interactions in solution was used to measure the affinity of rTRD01 binding to TDP-43102–269 (Figure 1H). Thermographs of TDP-43102–269 with a range of concentrations of rTRD01 gave an apparent dissociation constant of 89.4 ± 0.8 μM.
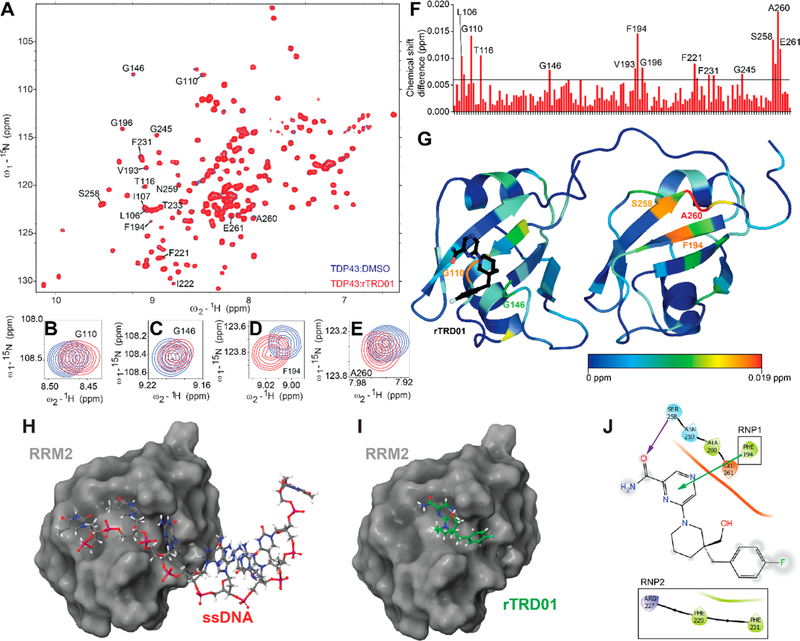
To further confirm the binding site for rTRD01, we utilized 15N–1H heteronuclear single quantum correlation spectroscopy (HSQC) using 15N-labeled TDP-43102–269. 15N–1H HSQC spectra of TDP-43102–269 were obtained free and in complex with rTRD01 (1:4 TDP-43102–269/rTRD01 ratio) (Figure 2A). Using our recently assigned NMR structure of apo-TDP-43102–26918 (Biological Magnetic Resonance Bank ID 27613), we were able to map the residues shifted based on the interaction of rTRD01 in increasing concentrations compared with apo-TDP-43102–269. Specific chemical shift perturbations were observed in the 1H–15N HSQC spectra of TDP-43102–269 upon addition of rTRD01, thus permitting calculation of the geometrical distance covered by each peak as described in the Methods section. In particular, Gly110 and Gly148 (from TDP-43 RRM1) and Phe194, Ala260, and Glu261 (from TDP-43 RRM2) peaks were shifted (Figure 2B–F).
Figure 2.
Mapping the binding of rTRD01 on TDP-43. (A) Superposition of 1H–15N heteronuclear single quantum correlation spectroscopy (HSQC) spectra of 15N-labeled human TDP-43102–269 (150 μM), free (blue) and in complex with rTRD01 in a TDP-43102–269/rTRD01 ratio of 1:4 (red). Close-up of shifts around TDP-43 residues G110 (B), G146 (C), F194 (D), and A260 (E). (F) Average chemical shift changes for assigned residues of the 15N-labeled TDP-43102–269 upon complex formation with rTRD01. The average chemical shift changes of cross-peaks were calculated as described in the Methods section. The horizontal line (0.09 ppm) is the threshold, calculated as described in the Methods section, above which a shift is considered significant. (G) Residues with chemical-shift perturbations upon rTRD01 binding are plotted onto the solution structure of TDP-43 (PDB code 4bs2) using a color gradient indicative of extent of the perturbation. (H) TDP-43-RRM2 domain (surface representation) crystallized with DNA (stick and ball representation) (PDB code 3d2w). (I) TDP-43-RRM2 domain (surface representation) docked with rTRD01 (stick and ball representation). (J) 2D representation of rTRD01 binding pocket that includes residues from the RNP1 and RNP2 of TDP-43 RRM2. The arrows represent predicted bonds (π–cation interaction, green, and halogen bond, purple).
To get a better understanding of rTRD01 binding site(s) to TDP-43, we mapped the chemical-shift perturbations induced by rTRD01 interaction onto the solution crystal structure of TDP-43 (PDB 4bs2) using a color gradient (Figure 2G). rTRD01 is able to bind Gly146, a residue from the ribonucleoprotein motif 1 (RNP-1) and Gly110 from the RNP-213 (Figure 2C,G) of TDP-43 RRM1, thus validating our predicted pocket (Figure 2C). However, additional TDP-43 residues that were shifted upon rTRD01 binding included Phe194 from the RNP-2 of RRM2 and Ala260 and Glu261. We docked rTRD01 on the crystal structure of RRM2 of TDP-43 (Figure 2H–J), and the compound binding site overlaps with the nucleic acid binding site in RRM2. This is not surprising since we targeted RNP sequences, which are highly conserved among RRM domains. Since the RRM domain is an RNA recognition motif shared by greater than 500 proteins with at least one RRM domain (up to six per protein), which have important normal, nonpathological functions in the cell,19 there is a critical need to assess the potential of rTRD01 to bind to other RRM domains.
rTRD01 Reduces TDP-43/RNA Interactions.
Since our target was the TDP-43–nucleic acid interface, we tested the effect of rTRD01 on TDP-43 binding to two different nucleic acid sequences: (i) the canonical substrate of TDP-43, namely, (UG)6 repeats;17,20,21 (ii) the (GGGGCC)4 hexanucleotide repeat expansion (HRE) from the gene c9orf 72, which is a disease-linked RNA known to bind TDP-43.22,23
Two different constructs of TDP-43 were tested: TDP-43102–269, which only contains the RRM domains and TDP-431–260, which also encompass the N-terminal domain, previously shown to participate in nucleic acid binding24 (Figure 1).
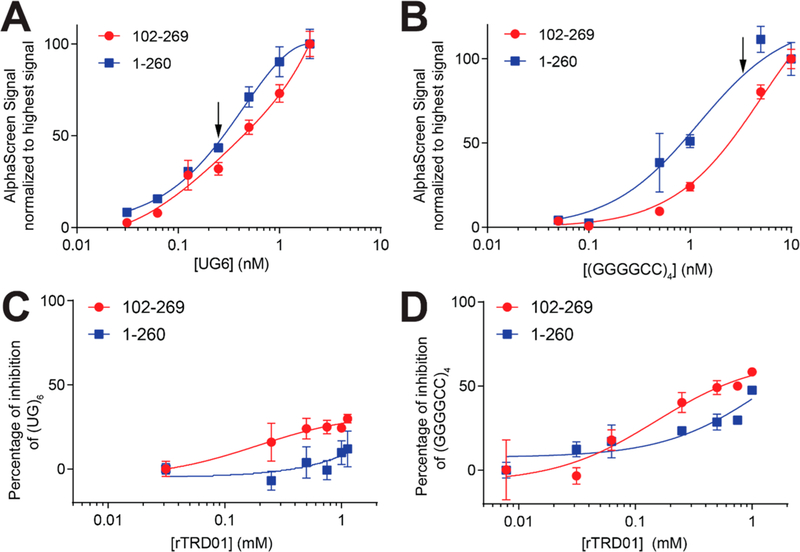
First, we used an amplified luminescent proximity homogeneous alpha assay (ALPHA) to measure binding between biotinylated (UG)6 RNA and TDP-43. Similar apparent Kd values were observed for both TDP-43 constructs binding to (UG)6: 0.73 ± 0.1 nM for TDP-43102–269 and 0.4 ± 0.04 nM for TDP-431–260 (Figure 3A). Binding was also observed between TDP-43 and (GGGGCC)4, albeit with a lower affinity than (UG)6, at 5.1 ± 0.6 nM for TDP-43102–269 and 1.21 ± 0.24 nM for TDP-431–260 (Figure 3B). Interestingly, TDP-431–260 bound (GGGGCC)4 with a higher affinity than the RRM-only construct, indicating the involvement of the NTD in RNA binding. The ability of the NTD to bind nucleic acids, albeit weakly, has been described previously, and two regions within this domain have been suggested as binding nucleic acids.24,25 A recent study showed the ability of TDP-43 NTD to undergo liquid–liquid phase separation induced by ssDNA in a length-dependent manner26 and could explain why (GGGGCC)4 binds TDP-431–260 better than TDP-43102–269, while (UG)6 had similar affinity for both constructs.
Figure 3.
RNA/TDP-43 disruption by rTRD01. Proteins TDP-43102–269-His or TDP-431–260-His (0.75 nM) were incubated with increasing concentrations of biotinylated UG6 (A) or (GGGGCCC)4 (B). The Y-axis displays the ALPHA signal. A nonlinear regression was fit to the data, using a one-site specific binding equation. UG6 RNA bound to TDP-43102–269 with an apparent Kd of ∼0.73 ± 0.1 nM. Some error bars are smaller than the symbols. (C, D) Concentration-dependent curves were obtained for rTRD01’s disruption of nucleic acid–TDP-43 interaction at a single RNA concentration (arrow) indicated on the panels above. Data is represented as mean ± SEM (n = 3).
Next, we measured inhibition of TDP-43/RNA binding at a single concentration of RNA with increasing concentration of rTRD01; we used both protein constructs (102–260 and 1–260), as well as (UG)6 and (GGGCC)4 RNA. rTRD01 had limited effect on TDP-43 association with (UG)6 sequence for both TDP-43 constructs (Figure 3C). In contrast, rTRD01 inhibited (GGGGCC)4 interaction for TDP-43102–269 to a maximum of 50% inhibition with an IC50 of ∼150 μM. For the TDP-431–260 construct, rTRD01 was able to inhibit 50% of (GGGGCC) 4 interaction at 1 mM (Figure 3D).
rTRD01 Reduces ALS-like Locomotor Defects in a Model of ALS in Fruit Fly.
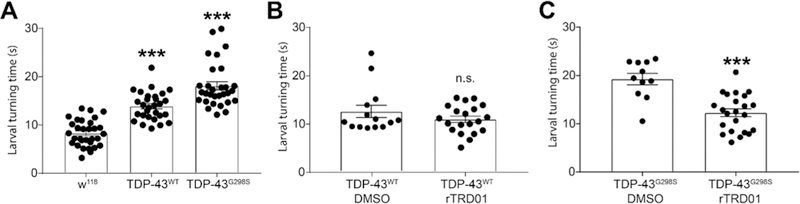
To assay possible in vivo neuroprotective effects of rTRD01, we used a Drosophila model of ALS, based on TDP-43 overexpression, which has been shown to recapitulate several aspects of the disease.27–29 Larval turning time, an assay that measures neuromuscular coordination and strength, was used as described.30 Larvae expressing TDP-43WT or TDP-43G298S in motor neurons using the GAL–UAS bipartite expression system31 exhibited locomotor defects in turning time compared to w1118 (genetic background control) larvae (Figure 4A). While rTRD01 was not able to rescue locomotion in TDP-43WT expressing larvae (Figure 4B), it significantly improved larval turning time in TDP-43G298S expressing larvae, from 19.3s to 12.3s (P value = 0.0001) (Figure 4C).
Figure 4.
rTRD01 improves larval turning in a Drosophila model of ALS. (A) Larvae expressing D42 GAL4-driven human wild-type (TDP-43WT) or G298S mutant TDP-43 (TDP-43G298S) take significantly longer to turn over following a ventral-up inversion. D42 GAL4 crossed with w1118 larvae were used as control. Asterisks indicate statistical difference assessed by Kruskal-Wallis (p < 0.0001, n = 20–30). (B) rTRD01 has no significant effect on the turning time of larvae expressing TDP-43WT. (C) rTRD01 reduced the turning time of larvae expressing TDP-43G298S from ∼19s to less than ∼13s, on average from n ≥ 20 larvae tested. rTRD01 was fed at a concentration of 20 μM in the food. Data is presented as mean ± SEM. Asterisks indicate statistically significant difference as assessed by a Mann-Whitney test (p < 0.0001, n ≥ 20).
Reducing pathological interactions without impacting normal RNA function of TDP-43 is challenging. Nevertheless, rTRD01 is able to attenuate ALS locomotor defects in a Drosophila model of ALS (Figure 4) while not affecting TDP-43’s interaction with (UG)6 substrate in vitro (Figure 3). rTRD01 had limited toxicity at 50 μM in NSC-34 motoneuron-like cells (Supplementary Figure 3). It is important to note that while Drosophila does not seem to express (GGGGCC)4, rTRD01 could reduce other non-canonical RNA–TDP-43 interactions relevant in ALS. Moreover, the mutation G298S, used in this study, was shown to be associated with enlarged TDP-43 enriched neuronal RNA granules,32 but another study did not find an altered RNA binding for TDP-43G298S.11 It is possible that rTRD01 helps restore some normal level of TDP-43 function. We cannot rule out the possibility that binding of the c9orf 72 RNA to TDP-43 is the only contributing factor important for c9orf 72-linked ALS as c9orf 72 expansions have many other effects (e.g., loss of C9orf72 protein, RNA foci formation, etc.), which are likely to at least contribute to ALS.
A limitation of our study is that the reduction in motor defects caused by rTRD01 is unlikely to occur only through disruption of specific TDP-43/RNA interactions as initially hypothesized. Other mechanisms may also be at play since the RRM domain is the site of several pathological modifications. Acetylation of TDP-43, seen in spinal cord of ALS patients,33 and oxidation of TDP-43, found in frontal cortex extracts of FTLD-TDP brains,34 are both consequences of oxidative stress, and both enhance TDP-43 aggregation. Furthermore, both mechanisms have been suggested for therapeutic targeting. Importantly, both modifications, K14 for acetylation and C173 for oxidation, are within the rTRD01 binding pocket (Figure 2C). Quantifying acetylation and oxidation of TDP-43 in the presence of rTRD01 in a context of oxidative stress is likely to provide additional insights into the compound’s mode of action.
Future efforts will also focus on determining if a pan effect of rTRD01, that is, hitting several RRM domains, in ALS is part of the mechanism by which the compound reduces ALS symptoms or if off-targeting is to be expected.
Although the full mechanism of action of rTRD01 is currently under investigation, our work provides evidence that (i) RNA–protein interfaces are druggable in ALS and (ii) TDP-43 is a valid target for development of small molecules. An advantage of rTRD01 is its ability to selectively inhibit interactions with RNA derived from c9orf72 while sparing physiologically relevant canonical interactions (UG)6 RNA. This approach is quite exciting as it will be of therapeutic interest for ALS in addition to several other TDP-43 related neurodegenerative diseases including the very recently described limbic-predominant age-related TDP-43 encephalopathy (LATE), a type of dementia mimicking Alzheimer’s dementia.35 Our study provides an instructive example of a chemical biology approach pivoted to discover small molecules targeting RNA–protein interactions in a broad spectrum of neurodegenerative diseases.
METHODS
Materials.
All reagents were purchased from Sigma (St. Louis, MO, USA) and Fisher Scientific (Hampton, NH) unless otherwise indicated. 6-(3-(4-Fluorobenzyl)-3-(hydroxymethyl)piperidin-1-yl)-pyrazine-2-carboxamide (rTRD01) was purchased from Chembridge Inc. (www.hit2lead.com).
rTRD01 (log P = 1.3) has not been previously reported as an aggregator, or to be similar to an aggregator, as predicted by Aggregator Advisor.36 A similar query of rTRD01 in the Zinc15 database revealed no hits to molecules containing PAINS chemotypes.
Cloning of TDP-43 Subdomains.
The coding sequence of human TDP-431–260 was amplified by PCR using the primers Fw 5′-AATGGGTCGCGGATCCATGTCTGAATATATTCGGGTAACC-3′ and Rv 5′-GTGCGGCCGCAAGCTTCTAGGCATTGGATA-TATGAACGCTGA-3′) (Eurofins Genomics, Louisville, KY) and subcloned using the CloneTech in-Fusion kit into the pET28a (+) vector between BamHI and HindIII restriction sites, resulting in a construct with an N-terminal 6×His tag. Amplified sequences were verified by DNA sequencing.
Purification of Recombinant TDP-43 Subdomains.
Human TDP-431–260 and TDP-43102–269 were expressed in E. coli BL21(DE3) cells (Novagen) in LB rich or M9 minimal media supplemented with 15NH4Cl. TDP-43102–269 and TDP-431–260 were purified exactly as described in ref 18.
In Silico Docking Using Schrodinger.
Molecular docking studies were performed using Schrodinger suite of programs including Glide docking. X-ray structure of the human TDP-43 RRM1 (PDB code 4iuf13) was used for virtual screening of small molecule libraries. Docking grids were generated using TDP-43 active sites. We focused our docking on a 10 Å3 pocket on Leu109, Gly110, Pro112, Trp113, and Arg171 in the RRM1 domain. Virtual screening was performed using stepwise virtual screening protocol in Glide program. DIVERSet-CL library, a small molecule library of 50 000 compounds from Chembridge Inc., was used as input for virtual screening. Resulting docking poses were analyzed using docking score, and the top 8 compounds were selected for further screening. The resulting complexes were ranked using Glide score and other energy related terms.
Saturation Transfer Difference Nuclear Magnetic Resonance (STD NMR) Spectroscopy for Small Molecule Binding.
One-dimensional 1H saturation transfer difference nuclear magnetic resonance (STD NMR) spectra with a spectral width of 12 ppm were collected as previously done for small molecule binding.15
Heteronuclear Single Quantum Correlation Nuclear Magnetic Resonance (HSQC-NMR).
All NMR data were collected on Bruker Avance NEO 800 MHz spectrometer with TCI-H&F/C/N probe at 25 °C. A transverse relaxation optimized spectroscopy (TROSY) with a solvent suppression pulse sequence was used to acquire all HSQC data. 15N labeled TDP-43102–269-His (100 μM) was incubated with rTRD01. NMR data processing and analysis was performed using programs NMRPipe37 and Sparky (Goddard and Kneller, Sparky 3, University of California, San Francisco). Chemical shift differences were calculated using the following equation from ref 38:
The population standard deviation from the average change38 for the entire protein was calculated using
Chemical shifts that were greater than 1 standard deviation (σ) were considered significant.38
Microscale Thermophoresis.
Purified TDP-43102–269-His was labeled using the Monolith Protein Labeling Kit RED-NTA (Nanotemper, Germany) according to the manufacturer’s instructions. Microscale thermophoresis experiments were performed as reported previously.39 Briefly, 50 nM of labeled protein was mixed with a range of concentrations of rTRD01 in PBST buffer. The thermographs were recorded using MST premium capillaries at 20% LED and medium MST power. Data analysis was performed with the MO Affinity Analysis software (Nanotemper), and the data was fit using the specific binding with Hill model in GraphPad.
Amplified Luminescent Proximity Homogeneous Alpha Assay (ALPHA).
ALPHA experiments were conducted as reported previously.40 TDP-43 proteins (0.75 nM) were mixed with 10 μg/mL of anti-His acceptor beads (PerkinElmer) and RNA sequences (biotinylated-UG6 or biotinylated-(GGGGCC)4) at various concentrations were incubated with streptavidin donor beads (PerkinElmer). The protein and the RNA were then mixed in a final volume of 20 μL of 25 mM Tris, pH 7.4, 0.1% BSA, 0.1% Triton X-100, 0.2 mM DTT in white, opaque low volume 384-well plates. After 90 min incubation at RT, the AlphaScreen signal was measured using an EnSpire Alpha instrument (PerkinElmer).
For measuring the effect of rTRD01 on TDP-43 interaction with RNA sequences, the proteins were preincubated 30 min with ranging concentrations of rTRD01, in the presence of anti-His beads. The RNA with the streptavidin beads was then added and incubated for 90 min at RT.
Larval Turning Assay.
Assays were performed as previously described.27 Briefly, wandering third instar larvae were placed on a grape juice plate, containing rTRD01 or vehicle, at RT. After becoming acclimated, crawling larvae were gently turned ventral side up and monitored until they were able to turn back (dorsal side up) and continue their forward movement. The amount of time that it took each larva to complete this task was recorded.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from 2014 Arizona Health Sciences Career Development Award and Arizona Disease Control Research Commission ADHS16–162407 Arizona grant (to M.K.) and R01 NS091299 (to D.C.Z.). We thank Katie Freeman, Amy Banks, Rick Scott, and Allen Reitz of Fox Chase Chemical Diversity Center for guidance on the alpha-screen assay protocol.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschembio.9b00481.
Additional methods, structures, chemical names, docking scores, and docking poses of top 8 hits from in silico docking, 1H STD NMR of binding of small molecules to TDP-43 RRM, phenotypic screening of rTRD compounds, and cytotoxicity in NSC34 cells of rTRD01 (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Kwong LK, Uryu K, Trojanowski JQ, and Lee VMY. (2007) TDP-43 Proteinopathies: Neurodegenerative Protein Misfolding Diseases without Amyloidosis. Neurosignals 16, 41–51. [DOI] [PubMed] [Google Scholar]
- (2).Buratti E. (2015) Functional Significance of TDP-43 Mutations in Disease. Adv. Genet 91, 1–53. [DOI] [PubMed] [Google Scholar]
- (3).Buratti E, and Baralle FE. (2008) Multiple Roles of TDP-43 in Gene Expression, Splicing Regulation, and Human Disease. Front. Biosci., Landmark Ed 13, 867–878. [DOI] [PubMed] [Google Scholar]
- (4).Colombrita C, Zennaro E, Fallini C, Weber M, Sommacal A, Buratti E, Silani V, and Ratti A. (2009) TDP-43 Is Recruited to Stress Granules in Conditions of Oxidative Insult. J. Neurochem 111, 1051–1061. [DOI] [PubMed] [Google Scholar]
- (5).Scotter EL, Chen HJ, and Shaw CE. (2015) TDP-43 Proteinopathy and ALS: Insights into Disease Mechanisms and Therapeutic Targets. Neurotherapeutics 12, 352–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Khalfallah Y, Kuta R, Grasmuck C, Prat A, Durham HD, and Vande Velde C. (2018) TDP-43 Regulation of Stress Granule Dynamics in Neurodegenerative Disease-Relevant Cell Types. Sci. Rep 8, 7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Aulas A, and Vande Velde C. (2015) Alterations in Stress Granule Dynamics Driven by TDP-43 and FUS: A Link to Pathological Inclusions in ALS? Front. Cell. Neurosci 9, 423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Fernandes N, Eshleman N, and Buchan JR. (2018) Stress Granules and ALS: A Case of Causation or Correlation? Adv. Neurobiol 20, 173–212. [DOI] [PubMed] [Google Scholar]
- (9).Dewey CM, Cenik B, Sephton CF, Dries DR, Mayer P, Good SK, Johnson BA, Herz J, and Yu G. (2011) TDP-43 Is Directed to Stress Granules by Sorbitol, a Novel Physiological Osmotic and Oxidative Stressor. Mol. Cell. Biol 31, 1098–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Liu R, Yang G, Nonaka T, Arai T, Jia W, and Cynader MS. (2013) Reducing TDP-43 Aggregation Does Not Prevent Its Cytotoxicity. Acta Neuropathol. Commun 1, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Ihara R, Matsukawa K, Nagata Y, Kunugi H, Tsuji S, Chihara T, Kuranaga E, Miura M, Wakabayashi T, Hashimoto T, and Iwatsubo T. (2013) RNA Binding Mediates Neurotoxicity in the Transgenic Drosophila Model of TDP-43 Proteinopathy. Hum. Mol. Genet 22, 4474–4484. [DOI] [PubMed] [Google Scholar]
- (12).Che MX, Jiang LL, Li HY, Jiang YJ, and Hu HY. (2015) TDP-35 Sequesters TDP-43 into Cytoplasmic Inclusions through Binding with RNA. FEBS Lett. 589, 1920–1928. [DOI] [PubMed] [Google Scholar]
- (13).Kuo PH, Chiang CH, Wang YT, Doudeva LG, and Yuan HS. (2014) The Crystal Structure of TDP-43 RRM1-DNA Complex Reveals the Specific Recognition for UG- and TG-Rich Nucleic Acids. Nucleic Acids Res. 42, 4712–4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Burd CG, and Dreyfuss G. (1994) Conserved Structures and Diversity of Functions of RNA-Binding Proteins. Science (Washington, DC, U. S.) 265, 615–621. [DOI] [PubMed] [Google Scholar]
- (15).François-Moutal L, Jahanbakhsh S, Nelson ADL, Ray D, Scott DD, Hennefarth MR, Moutal A, Perez-Miller S, Ambrose AJ, Al-Shamari A, Coursodon P, Meechoovet B, Reiman R, Lyons E, Beilstein M, Chapman E, Morris QD, Van Keuren-Jensen K, Hughes TR, Khanna R, Koehler C, Jen J, Gokhale V, and Khanna M. (2018) A Chemical Biology Approach to Model Pontocerebellar Hypoplasia Type 1B (PCH1B). ACS Chem. Biol 13, 3000–3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, and Mainz DT. (2006) Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein-Ligand Complexes. J. Med. Chem 49, 6177–6196. [DOI] [PubMed] [Google Scholar]
- (17).Lukavsky PJ, Daujotyte D, Tollervey JR, Ule J, Stuani C, Buratti E, Baralle FE, Damberger FF, and Allain FHT. (2013) Molecular Basis of UG-Rich RNA Recognition by the Human Splicing Factor TDP-43. Nat. Struct. Mol. Biol 20, 1443–1449. [DOI] [PubMed] [Google Scholar]
- (18).Scott DD, Francois-Moutal L, Kumirov VK, and Khanna M. (2019) 1H, 15 N and 13 C Backbone Assignment of Apo TDP-43 RNA Recognition Motifs. Biomol. NMR Assignments 13, 163–167. [DOI] [PubMed] [Google Scholar]
- (19).Maris C, Dominguez C, and Allain FHT. (2005) The RNA Recognition Motif, a Plastic RNA-Binding Platform to Regulate Post-Transcriptional Gene Expression. FEBS J. 272, 2118–2131. [DOI] [PubMed] [Google Scholar]
- (20).Colombrita C, Onesto E, Megiorni F, Pizzuti A, Baralle FE, Buratti E, Silani V, and Ratti A. (2012) TDP-43 and FUS RNA-Binding Proteins Bind Distinct Sets of Cytoplasmic Messenger RNAs and Differently Regulate Their Post-Transcriptional Fate in Motoneuron-like Cells. J. Biol. Chem 287, 15635–15647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Bhardwaj A, Myers MP, Buratti E, and Baralle FE. (2013) Characterizing TDP-43 Interaction with Its RNA Targets. Nucleic Acids Res. 41, 5062–5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Kanai Y, Dohmae N, and Hirokawa N. (2004) Kinesin Transports RNA: Isolation and Characterization of an RNA-Transporting Granule. Neuron 43, 513–525. [DOI] [PubMed] [Google Scholar]
- (23).Ishiguro A, Kimura N, Watanabe Y, Watanabe S, and Ishihama A. (2016) TDP-43 Binds and Transports G-Quadruplex-Containing MRNAs into Neurites for Local Translation. Genes to Cells 21, 466–481. [DOI] [PubMed] [Google Scholar]
- (24).Qin H, Lim L-Z, Wei Y, and Song J. (2014) TDP-43 N Terminus Encodes a Novel Ubiquitin-like Fold and Its Unfolded Form in Equilibrium That Can Be Shifted by Binding to SsDNA. Proc. Natl. Acad. Sci. U. S. A 111, 18619–18624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Mompean M, Romano V, Pantoja-Uceda D, Stuani C, Baralle FE, Buratti E, and Laurents DV. (2016) The TDP-43 N-Terminal Domain Structure at High Resolution. FEBS J. 283, 1242. [DOI] [PubMed] [Google Scholar]
- (26).Wang L, Kang J, Lim L, Wei Y, and Song J. (2018) TDP-43 NTD Can Be Induced While CTD Is Significantly Enhanced by SsDNA to Undergo Liquid-Liquid Phase Separation. Biochem. Biophys. Res. Commun 499, 189–195. [DOI] [PubMed] [Google Scholar]
- (27).Estes PS, Boehringer A, Zwick R, Tang JE, Grigsby B, and Zarnescu DC. (2011) Wild-Type and A315T Mutant TDP-43 Exert Differential Neurotoxicity in a Drosophila Model of ALS. Hum. Mol. Genet 20, 2308–2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Joardar A, Menzl J, Podolsky TC, Manzo E, Estes PS, Ashford S, and Zarnescu DC. (2015) PPAR Gamma Activation Is Neuroprotective in a Drosophila Model of ALS Based on TDP-43. Hum. Mol. Genet 24, 1741–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Estes PS, Daniel SG, Mccallum AP, Boehringer AV, Sukhina AS, Zwick RA, and Zarnescu DC. (2013) Motor Neurons and Glia Exhibit Specific Individualized Responses to TDP-43 Expression in a Drosophila Model of Amyotrophic Lateral Sclerosis. Dis. Models & Mech 6, 721–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Joardar A, Menzl J, Podolsky TC, Manzo E, Estes PS, Ashford S, and Zarnescu DC. (2015) PPAR Gamma Activation Is Neuroprotective in a Drosophila Model of ALS Based on TDP-43. Hum. Mol. Genet 24, 1741–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Pan Y, Zhou Y, Guo C, Gong H, Gong Z, and Liu L. (2009) Differential Roles of the Fan-Shaped Body and the Ellipsoid Body in Drosophila Visual Pattern Memory. Learn. Mem 16, 289–295. [DOI] [PubMed] [Google Scholar]
- (32).Liu-Yesucevitz L, Lin AY, Ebata A, Boon JY, Reid W, Xu Y-F, Kobrin K, Murphy GJ, Petrucelli L, and Wolozin B. (2014) ALS-Linked Mutations Enlarge TDP-43-Enriched Neuronal RNA Granules in the Dendritic Arbor. J. Neurosci 34, 4167–4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Cohen TJ, Hwang AW, Restrepo CR, Yuan CX, Trojanowski JQ, and Lee VMY. (2015) An Acetylation Switch Controls TDP-43 Function and Aggregation Propensity. Nat. Commun 6, 5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Cohen TJ, Hwang AW, Unger T, Trojanowski JQ, and Lee VMY. (2012) Redox Signalling Directly Regulates TDP-43 via Cysteine Oxidation and Disulphide Cross-Linking. EMBO J. 31, 1241–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Nelson PT, Dickson DW, Trojanowski JQ, Jack CR, Boyle PA, Arfanakis K, Rademakers R, Alafuzoff I, Attems J, Brayne C, Coyle-Gilchrist ITS, Chui HC, Fardo DW, Flanagan ME, Halliday G, Hokkanen SRK, Hunter S, Jicha GA, Katsumata Y, Kawas CH, Keene CD, Kovacs GG, Kukull WA, Levey AI, Makkinejad N, Montine TJ, Murayama S, Murray ME, Nag S, Rissman RA, Seeley WW, Sperling RA, White CL III, Yu L, and Schneider JA. (2019) Limbic-Predominant Age-Related TDP-43 Encephalopathy (LATE): Consensus Working Group Report. Brain 142, 1503–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Irwin JJ, Duan D, Torosyan H, Doak AK, Ziebart KT, Sterling T, Tumanian G, and Shoichet BK. (2015) An Aggregation Advisor for Ligand Discovery. J. Med. Chem 58, 7076–7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, and Bax A. (1995) NMRPipe: A Multidimensional Spectral Processing System Based on UNIX Pipes. J. Biomol. NMR 6, 277–293. [DOI] [PubMed] [Google Scholar]
- (38).Williamson MP. (2013) Using Chemical Shift Perturbation to Characterise Ligand Binding. Prog. Nucl. Magn. Reson. Spectrosc 73, 1–16. [DOI] [PubMed] [Google Scholar]
- (39).Francois-Moutal L, Dustrude ET, Wang Y, Brustovetsky T, Dorame A, Ju W, Moutal A, Perez-Miller S, Brustovetsky N, Gokhale V, Khanna M, and Khanna R. (2018) Inhibition of the Ubc9 E2 SUMO-Conjugating Enzyme-CRMP2 Interaction Decreases NaV1.7 Currents and Reverses Experimental Neuropathic Pain. Pain 159, 2115–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Cassel JA, McDonnell ME, Velvadapu V, Andrianov V, and Reitz AB. (2012) Characterization of a Series of 4-Aminoquinolines That Stimulate Caspase-7 Mediated Cleavage of TDP-43 and Inhibit Its Function. Biochimie 94, 1974–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Jiang LL, Zhao J, Yin XF, He WT, Yang H, Che MX, and Hu HY. (2016) Two Mutations G335D and Q343R within the Amyloidogenic Core Region of TDP-43 Influence Its Aggregation and Inclusion Formation. Sci. Rep 6, 23928. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.