Abstract
Background:
The giant sarcomere protein titin is important in both heart health and disease. Mutations in the gene encoding for titin, TTN, are the leading known cause of familial dilated cardiomyopathy. The uneven distribution of these mutations within TTN motivated us to seek a more complete understanding of this gene and the isoforms it encodes in cardiomyocyte sarcomere formation and function.
Methods:
To investigate the function of titin in human cardiomyocytes (CMs), we used CRISPR/Cas9 to generate homozygous truncations in the Z-disk (TTN-Z−/−) and A-band (TTN-A−/−) regions of the TTN gene in human induced pluripotent stem cells. The resulting CMs were characterized using immunostaining, engineered heart tissue mechanical measurements, and single cell force and calcium measurements.
Results:
Following differentiation, we were surprised to find that despite the more upstream mutation, TTN-Z−/−-CMs had sarcomeres and visibly contracted, while TTN-A−/−-CMs did not. We hypothesize that sarcomere formation was due to the expression of a recently discovered isoform of titin, Cronos, which initiates downstream of the truncation in TTN-Z−/−-CMs. Using a custom Cronos antibody we demonstrate that this isoform is expressed and integrated into myofibrils in human cardiomyocytes. TTN-Z−/−-CMs exclusively express Cronos titin, but these cells produce lower contractile force and have perturbed myofibril bundling compared to controls expressing both full-length and Cronos titin. Cronos titin is highly expressed in human fetal cardiac tissue, and when knocked out in hiPSC-CMs these cells exhibit reduced contractile force and myofibrillar disarray, despite the presence of full-length titin.
Conclusions:
We demonstrate that Cronos titin is expressed in developing human cardiomyocytes and is able to support partial sarcomere formation in the absence of full-length titin. Further, Cronos titin is necessary for proper sarcomere function in hiPSC-CMs. Additional investigation is necessary to understand the molecular mechanisms of this novel isoform and how it contributes to human cardiac disease.
Keywords: titin, human induced pluripotent stem cells, cardiomyocytes
Introduction
The giant sarcomere protein titin has a number of important roles in cardiac muscle, where it extends an entire half-sarcomere length from the Z-disc to the M-line and interacts with both the thick and thin filaments1. Titin is a modular protein with four main regions: the N-terminus of titin is anchored to the Z-disk of the sarcomere through interactions with T-cap and α-actinin2,3, as well as with the non-contractile cytoskeleton4,5. The spring-like I-band of titin provides the majority of passive tension in cardiomyocytes6,7, while the rigid A-band region interacts with the thick filament and has been proposed to act as a molecular ruler that establishes the patterning of myosin8–12. The C-terminus of titin is anchored in the M-line by myomesin and contains a kinase domain, which is activated by stretch to promote sarcomere protein turnover13,14. Titin is known to be required for the proper function of sarcomeres, however conflicting evidence exists regarding the necessity of titin in sarcomerogenesis. In cell culture studies, titin is difficult to specifically and completely knockdown due to its genetic complexity and large size15. In vivo studies of early sarcomerogenesis are challenging due to embryonic lethality associated with homozygous truncating mutations of titin16,17. Because of these roadblocks, a major outstanding question is whether titin is crucial for sarcomere formation or only necessary for proper function once sarcomeres are fully formed.
In addition to its important role in healthy cardiomyocytes, heterozygous truncating mutations in the gene encoding for titin (TTN) are the leading known cause of familial dilated cardiomyopathy (DCM), accounting for ~25% of cases18–22. Despite the high prevalence of titin truncating mutations in DCM patients, the pathogenicity of these mutations in the general population is not straightforward. Approximately 0.5–0.7% of healthy, control populations carry truncating mutations in TTN, a rate at least twice that of the total prevalence of non-ischemic DCM17, 22–25. Because of the high prevalence of apparently non-deleterious titin truncating mutations, there has been significant interest in determining if the pathogenicity of variants can be predicted based on their location within TTN. One hypothesis to predict the disease-causing potential of variants is that mutations occurring in constitutively expressed exons are more pathogenic than those in variably spliced exons24,26. Although this hypothesis explains the high odds ratio of 32 for mutations in the A-band region of titin, the majority of which are constitutively spliced in, it does not account for the lower pathogenicity of truncation mutations in other constitutively expressed regions of TTN, such as the Z-disk which has an odds ratio of 19.517. These differences suggest there may be other aspects of the gene architecture of TTN that have not yet been characterized, which contribute to disparate clinical results of truncating mutations.
To elucidate the role of titin during sarcomere development and better understand TTN expression, we have taken the approach of genetically engineering homozygous truncating mutations into human induced pluripotent stem cells (hiPSCs) and studying their function following differentiation into cardiomyocytes (hiPSC-CMs). Genetic engineering in vitro allows for the dissection of titin-specific effects at early developmental stages that would not be possible using animal models. Understanding titin expression and function in hiPSC-CMs is especially important as these cells are often used to study heterozygous titin truncating mutations for disease modeling26–28. Because heterozygous truncating mutations in the A-band region of titin are more pathogenic than those in the Z-disk region, we introduced homozygous truncating mutations in each of these locations to determine if they caused different phenotypes. A previous study of hiPSC-CMs carrying a homozygous A-band titin truncation found the cells lacked sarcomeres26, and due to the embryonic lethality of homozygous titin truncations in both the Z-disk and A-band in animal models16,17, we hypothesized that both mutations would prevent sarcomere formation in hiPSC-CMs. While A-band truncations blocked sarcomere formation, we were surprised to find that cardiomyocytes with Z-disk truncations formed sarcomeres and visibly contracted, albeit much more weakly than wild type (WT) hiPSC-CMs. Sarcomere assembly in Z-disk truncations was associated with the expression of Cronos, a newly described titin isoform with a start site downstream of the truncating mutation in these cells29. In contrast, this isoform is absent (or truncated) in A-band truncations, where sarcomere formation is not observed. We further show that Cronos is highly expressed in developing human hearts and may be involved in sarcomerogenesis. When Cronos is specifically knocked out in hiPSC-CMs, the cells produce lower contractile force and develop sarcomeric disarray, despite the presence of full length titin. We conclude that Cronos titin is expressed in human cardiomyocytes and is necessary for normal sarcomere formation and function.
Methods
The data, analytic methods, and study materials will be made available to other researchers for purposes of reproducing the results or replicating the procedure.
CRISPR/Cas9 targeting of TTN in hiPSCs
Single guide RNAs (sgRNAs) targeting TTN Exons 2 and 326 and the Cronos-specific region were designed using the online CRISPR design tool (crispr.mit.edu) (sgRNA sequences are listed in Table S1) based on the hg19 assembly TTN sequence on the UCSC Genome Browser30 and predicted Cronos start site from ref [29] and used as outlined in the Extended Methods. For all cell lines generated, colonies with homozygous or compound heterozygous mutations causing premature stop codons were also screened for mutations in the top 5 genes predicted to be most susceptible to off-target effects (details in Extended Methods). Mutant cell lines were cryopreserved and karyotyped (Diagnostic Cytogenetics Inc, Seattle, WA).
Cardiac differentiation
Wild type and mutated WTC hiPSCs were differentiated into cardiomyocytes using a previously described monolayer protocol (ref [31], details in Expanded Methods), and maintained in RPMI media (Gibco) supplemented with 2% B-27 with insulin (Life Technologies) and 1% penicillin/streptomycin (Invitrogen). For single cell force, calcium, and morphology measurements and live cell imaging cardiomyocytes were purified using lactate selection by replating cells 14–18 days after the start of differentiation and feeding with DMEM without glucose (Gibco) supplemented with 4 μM sodium lactate (Sigma-Aldrich) on days 18–22 after differentiation32. Cells used for calcium and morphology measurements were cultured in media containing 10 μM cytosine β-D-arabinofuranoside (Sigma-Aldrich) for 2 days following replating as single cardiomyocytes.
Engineered Heart Tissues
Engineered heart tissues (EHTs) were cast 23–32 days after the start of differentiation using a previously described fibrin scaffold system33. Each EHT was seeded with 4×105 hiPSC-CMs and 4×104 HS27a bone marrow stromal cells (ATCC) resuspended in 100μL of fibrin solution and cast between one rigid and one flexible post made from silicone rubber (PDMS, Sylgard 184). EHTs were maintained in RPMI media supplemented with 2% B-27 with insulin, 1% penicillin/streptomycin, and 5mg/mL aminocaproic acid (Sigma-Aldrich), which was changed every 2–3 days. After 3 weeks in culture, the deflection of the flexible posts during 1.5 Hz pacing were tracked by light microscopy. Force was calculated by multiplying the flexible post stiffness (k = 0.95 μN/μm) by the measured post deflection, and the twitch kinetics were obtained from the force profiles.
Live cell imaging
hiPSC-CMs were transduced with ELF1a-mCherry-a-actinin lentivirus ranging from day 23–35 post-differentiation. Viral media was changed 24 hours post-transduction. Cells recovered for 6 days with regular maintenance, and then replated for imaging as described in the Extended Methods.
Statistical analysis
For assays with two groups, student’s t-tests were performed in Excel. For assays with more than two groups, one-way ANOVAs were performed in MATLAB using the “anova1” function, and if the returned p-value was less than 0.01 Tukey’s post-hoc analysis was performed using the “multcompare” function. Adjusted p-values were computed using the Benjamini & Hochberg method.
IRB
The studies for this manuscript did not include any human or animal studies, and thus IRB approval was not required.
Results
Human iPSC line generation, differentiation, & titin expression
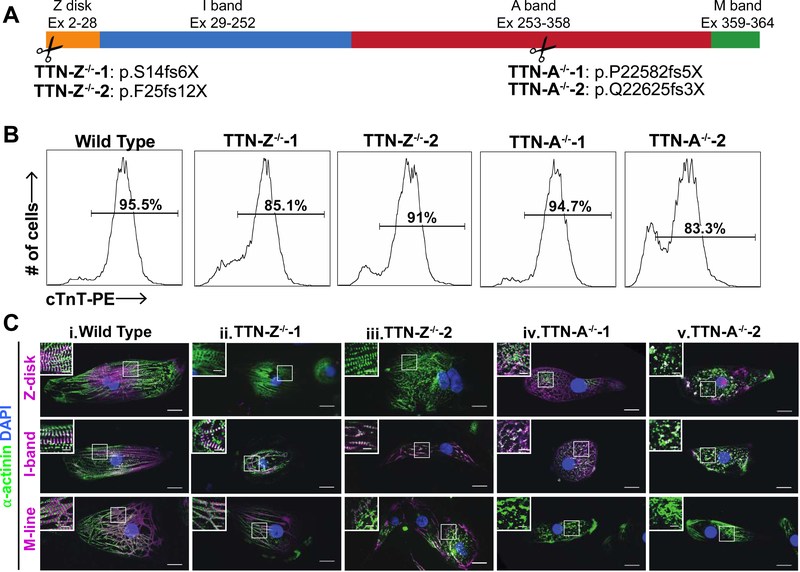
To investigate the role of titin truncation mutations in cardiomyocyte differentiation and sarcomerogenesis, the CRISPR/Cas9 system was used to introduce truncation mutations via non-homologous end joining into the WTC hiPSC line34. Mutations were targeted in Exon 2 (in the Z-disk domain; TTN-Z−/−) and Exon 326 (in the A-band region; TTN-A−/−) of TTN, using 2 different guide RNAs and generating 2 separate clonal lines for each locus (Figure 1A). Both of these exons are constitutively expressed in cardiomyocytes but not in pluripotent cells26. Single-cell colonies were expanded and screened for homozygous mutations that introduce a stop codon and are thus expected to cause truncations. All mutant cell lines had normal karyotypes, and normal colony morphology and growth (Figure S1). After induction with activin A, BMP4 and small molecule Wnt modulators, all lines differentiated into high purity populations of cardiomyocytes as measured by flow cytometry after staining for cardiac troponin T (cTnT) (Figure 1B). Given that we had hypothesized that none of the mutant cell lines would develop sarcomeres, we were surprised to find that the TTN-Z−/− lines, which cause the most proximal truncation, formed monolayers of beating cardiomyocytes. The TTN-A−/− lines did not visibly contract, even in populations that had a high percentage of cTnT+ cells.
Figure 1. Mutant cell line overview.
(A) Exons of the TTN gene that encode for each domain of titin, and the locations along the titin meta-protein of homozygous mutations specifically engineered into hiPSC lines. (B) Representative cTnT flow cytometry results of 30–31 day old hiPSC-CMs show that all cell lines can differentiate into high-purity populations of cardiomyocytes. (C) Immunohistochemistry of hiPSC-CMs from each cell line using titin antibodies that recognize the Z-disk, distal I-band, and M-line regions of the protein. Column i: WT-CMs stain positively for all titin epitopes and show sarcomere formation. Columns ii-iii: TTN-Z−/− CMs have sarcomere formation but only stain positively for titin domains downstream of the Cronos start site. Columns iv-v: TTN-A−/− CMs stain positively for N-terminal antibodies and do not form sarcomeres. In each panel, large image: scale bar = 20μm, inset image: scale bar = 5μm. White boxes indicate regions magnified in inset image.
Because TTN-Z−/−-CMs visibly contracted we hypothesized that these cells were expressing some form of titin that allowed them to form functioning sarcomeres. Levels of full-length titin transcript were not significantly different across any of the cell lines, indicating nonsense-mediated decay of the TTN transcript does not occur (Figure S2B). To investigate titin expression we performed immunocytochemistry on hiPSC-CMs with antibodies to probe for the Z-disk, distal I-band, and M-line domains of titin (Figure 1C). Wild type samples stained positively for all three titin epitopes, and the antibodies localized within the sarcomere in the expected patterns. TTN-Z−/−-CMs exhibited striated staining with α-actinin, and interestingly these sarcomeres only stained positively for distal I-band and M-line titin, both of which are downstream of the mutation introduced into this cell line. Finally, TTN-A−/−-CMs did not form distinct sarcomeres, and diffuse α-actinin, Z-disk, and I-band titin signals were observed. The M-line domain of titin, which is downstream of the mutation in these cells, was not detected. These data indicate that despite inducing a truncating mutation in a constitutively expressed N-terminal exon, the TTN Z−/− cells expressed distal elements of TTN that supported myofibrillogenesis and contractility.
Titin Z-Disk truncations are partially rescued by Cronos titin
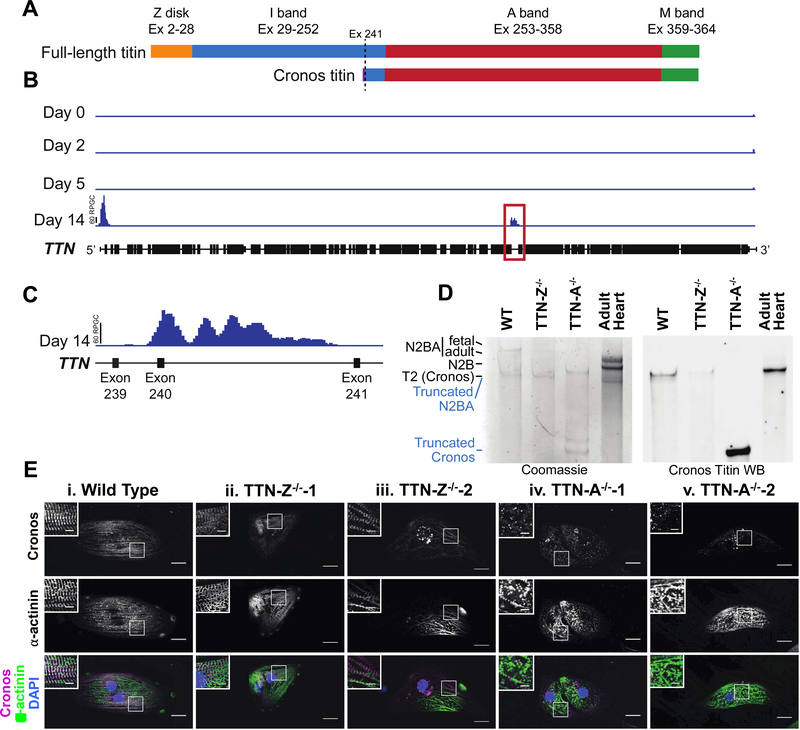
The expression of distal titin epitopes with Z-disk truncation mutations suggested that an alternate transcript was initiated 3’ to our mutations. Zou et al. recently identified an internal promoter in zebrafish ttn, which initiates transcription from a previously unrecognized transcriptional start site (TSS) located in the distal I-band portion29. Named Cronos, this C-terminal isoform of titin is predicted to be approximately two-thirds the size of the full-length isoforms, and to contain the distal I-band, A-band, and M-line regions, consistent with our epitope maps in the Z-disk truncation lines (Figure 2A). Gene targeting approaches disrupting Cronos titin expression in zebrafish cause significant myofibrillar disarray29. Although the transcript has been detected in mammalian cardiac samples29, until now there have not been studies identifying the Cronos titin protein or investigating its role in human cardiomyocytes. We hypothesized that the TTN-Z−/−-CMs express Cronos titin, which allowed for some sarcomere formation in these cells.
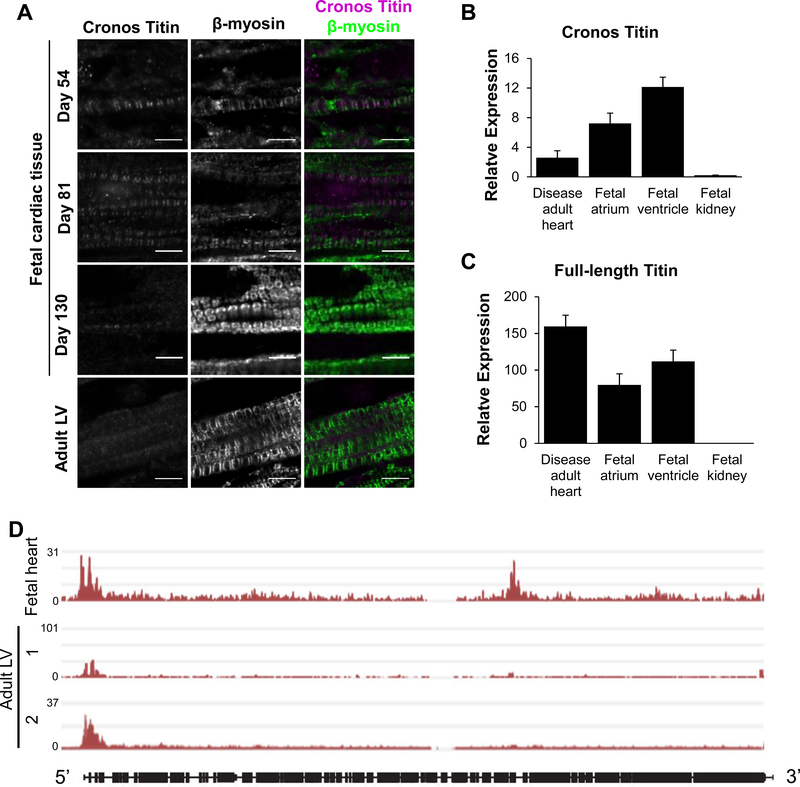
Figure 2. Cronos titin is expressed in hiPSC-CMs.
(A) Schematic of domains expressed in full-length and Cronos titin. Purple indicates a short Cronos-specific region that is intronic to full-length titin. (B) H3K4me3 enrichment in hESC-CMs during differentiation. (C) An enlarged view of the region enclosed in a red box in panel A indicating a prominent peak near the expected Cronos titin transcription start site between exons 239 and 240. (D) Coomassie-stained gel and Western blot of protein samples from wild type and mutant cell lines and adult human heart tissue demonstrate that the Cronos antibody is specific. Blue labels refer to the TTN-A−/− lane, black labels refer to all other samples. (E) Staining for the N-terminal of Cronos titin indicates a doublet pattern around the Z-disks in (i) WT and (ii-iii) TTN-Z−/− CMs and diffuse staining in (iv-v) TTN-A−/− CMs. In each panel, large image: scale bar = 20μm, inset image: scale bar = 5μm. White boxes indicate regions magnified in inset image.
To begin investigating this hypothesis, we revisited ChIP-Seq data from human embryonic stem cells (hESCs) undergoing cardiac differentiation35. We identify two peaks of H3K4me3 (a promoter-specific epigenetic mark) within TTN in definitive cardiomyocytes 14 days after the start of differentiation that are not present during earlier stages of differentiation (Figure 2B). One peak localizes to the 5’ end of TTN corresponding to the transcriptional start site of full-length titin, while the second prominent peak corresponds to the locus previously proposed for the Cronos titin transcriptional start site in the intron between Exon 240 and 241 (Figure 2C). Profiles for the repressive H3K27me3 chromatin mark did not show significant peaks in the TTN locus at any time point studied (Figure S2A). The appearance of H3K4me3 deposition without H3K27me3 is a pattern typically observed in cardiac structural genes31. Together this provides genomic evidence to support the presence of an internal Cronos-specific promoter within the TTN locus in human stem cell-derived cardiomyocytes.
To determine if Cronos titin protein was present in hiPSC-CMs we performed loose gel electrophoresis to distinguish titin isoforms by molecular weight. Analysis of total protein using a Coomassie stain indicates both N2BA and T2 titin are present in WT samples, but only T2 titin is present in TTN-Z−/− cells (Figure 2D). The T2 band has previously been described as a proteolytic degradation product of N2B and N2BA titin36–38, but we and others hypothesized that this band contains the novel isoform Cronos due to its similar expected molecular weight (~2.2MDa)29. To test this idea we raised a custom antibody to recognize the N-terminus of Cronos, the first 13 residues of which are specific to this isoform29 (see Expanded Methods for details). Western blots against hiPSC-CM lysates and human adult left ventricular tissue using this antibody labeled the T2 band but did not detect any of the other titin isoforms present, confirming that this band includes Cronos titin. The Cronos titin band was also recognized by antibodies for the A-band/I-band junction and M-line regions of titin (Figure S2C). Several lower weight titin bands were detected in the TTN-A−/−-CMs on the Coomassie-stained gel and Cronos titin western blot, indicating that only truncated versions of N2BA and Cronos titin are present in these cells. Taken together, these data demonstrate the T2 band includes Cronos titin, indicate that this novel isoform is expressed in hiPSC-CMs as well as human heart tissue, and reveal that TTN-Z−/− CMs are solely expressing this isoform.
To investigate if Cronos titin was being integrated into sarcomeres and establish its localization in the cell lines, we performed immunostaining with our custom antibody (Figure 2E). This revealed doublet patterns surrounding the Z-disks in both wild type and TTN-Z−/− samples, demonstrating that Cronos titin is being expressed and integrated into sarcomeres in these cells. TTN-A−/− samples stained diffusely for Cronos titin, similar to the pattern observed using other titin antibodies that recognize regions upstream of the truncating mutation. This pattern is also consistent with western blot data indicating that the TTN-A−/− cells are expressing a large truncation product of this isoform. Thus, we conclude that the ability of TTN-Z−/− mutants to form sarcomeres likely results from the ability of Cronos to substitute, at least in part, for full-length titin.
Force production in engineered heart tissues and single cells
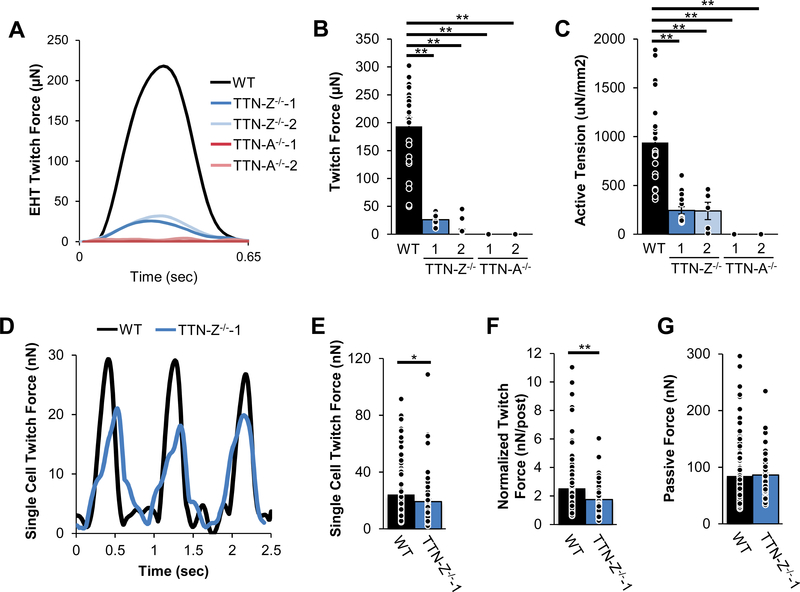
After observing that TTN-Z−/−-CMs can form sarcomeres and contract using Cronos as a substitute for full-length titin, we were interested in establishing whether they had comparable contractility to WT cells. To investigate force production, we characterized the contractile function of wild type and mutant cardiomyocytes in a multicellular context using engineered heart tissues (EHTs)33 and as single cells39. Consistent with observations of cardiomyocytes in monolayers, both wild type and TTN-Z−/− EHTs visibly contracted while TTN-A−/− EHTs did not visibly contract, although they did compact. Measurements of paced twitches 3 weeks after seeding indicate that TTN-Z−/− EHTs produced only ~10% of absolute force produced by wild type EHTs (Figure 3A–B), and approximately 25% of active tension compared to WT (Figure 3C). The time to peak of wild type and TTN-Z−/− EHT twitches was slightly decreased even though the maximum rate of force development was dramatically slower (Figure S3F–G). Neither 50% or 90% relaxation time was significantly different in TTN-Z−/− EHTs, although passive force was significantly decreased (Figure S3B, D–E). Staining for different regions of titin indicated that titin expression was the same in EHTs as in single cells (Figure S4). Taken together, these data indicate that although TTN-Z−/−-CMs are able to form sarcomeres and contract, they produce only a small fraction of the force of wild type cells in engineered tissues.
Figure 3. Force mechanics of mutant cell lines.
(A) Representative twitches of EHTs seeded with hiPSC-CMs show that WT and TTN-Z−/− EHTs produce measurable force while TTN-A−/− EHTs do not. Measurements of EHTs 3 weeks after seeding while pacing at 1.5Hz show that compared to WT, TTN-Z−/− (B) average absolute twitch forces and (C) active tension normalized to cross-sectional area are significantly reduced. (D) Sample twitch force traces of single cell micropost measurements. TTN-Z−/−-CMs produce lower (E) whole-cell twitch force and (F) twitch force when normalized to cell size. (G) There was no significant difference in passive force. Panels (B)-(C): WT: n=23; TTN-Z-/−−1: n=16; TTN-Z-/−−2: n=5; TTN-A-/−−1: n=16; TTNA-/−−1: n=17. P-values were calculated using ANOVAs and follow-up Tukey’s post-hoc tests. Adjusted p-values were computed using the Benjamini & Hochberg method. Panels (E)-(G): WT: n=107; TTN-Z-/−−1: n=71. P-values were calculated using student’s t-test. Adjusted p-values were computed using the Benjamini & Hochberg method. Error bars indicate standard error. *: adjusted p-value<0.05; **: adjusted p-value<0.01.
To determine if the weak force production of TTN-Z−/− EHTs was caused by attenuated twitches of individual cells, we characterized single cell contractility using a micropost-based assay system39 (Figures 3D–F and S5). TTN-Z−/−-CMs produce significantly lower force on both a whole-cell level and when normalized for cell area (Figure 3D–F). Interestingly, maximum twitch velocity, twitch power, cell size, and passive force were not different between cell types, although both upstroke and relaxation times were decreased in TTN-Z−/−-CMs compared to controls (Figures 3G and S5). Notably, the force difference between single WT-CMs and TTNZ−/−-CMs is less dramatic than that observed in the EHT system. This may be partially due to the younger age and less mature state of cells used in the micropost system, along with the fact that multicellular forces are generated in series in the EHTs, which would be expected to increase the difference between WT and knockout cells (see Discussion).
To determine if the reduced force produced by TTN-Z−/−-CMs was caused by differences in calcium handling, we measured Ca2+ transients during electrical pacing of single cells plated on fibronectin-coated glass slides (Figure S6). Interestingly, we found that the magnitude of calcium transients in TTN-Z−/− and TTN-A−/− CMs was not lower than wild type controls, although some calcium release was slightly slower and reuptake slightly faster in some groups. Based on these data we conclude that differences in calcium transients do not explain attenuated force production observed in individual TTN-Z−/−-CMs.
Cell morphology and sarcomere formation
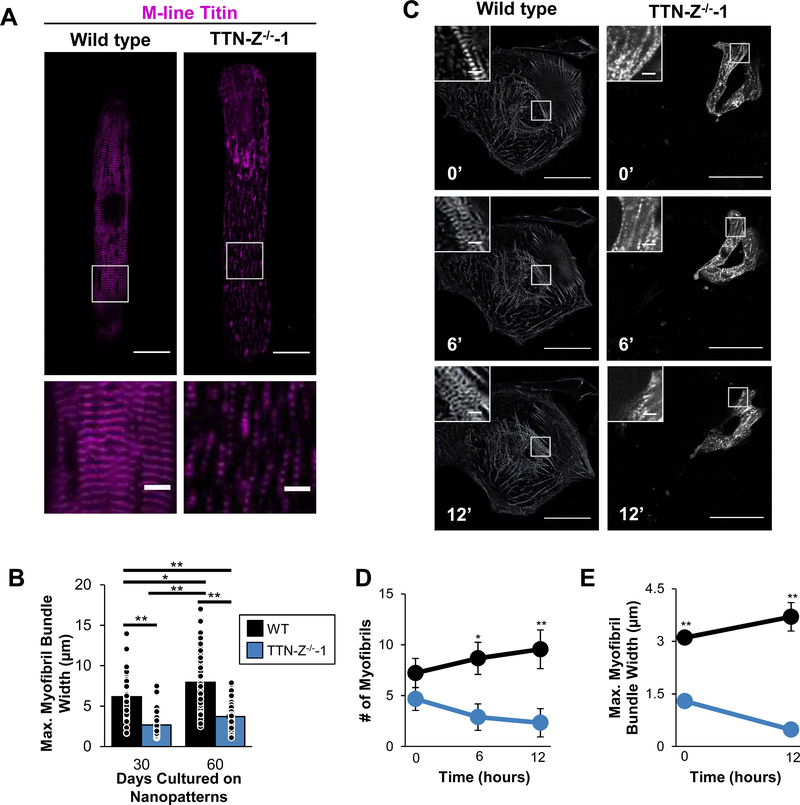
To measure whether sarcomere morphology was distinct between WT and TTN-Z−/−-CMs, we fixed and stained cells after 30 and 60 days of culture on nanopatterned substrates, which provide external cues to improve elongation and sarcomere alignment40,41. Myofibrils in TTN-Z−/− cells were noticeably sparser compared to WT controls, and maximum myofibril bundle width within each cell was less than half of control at both time points (Figure 4A–B). Additionally, while WT myofibril bundle width significantly increased between 30 and 60 days cultured on the nanopatterns, indicative of continued hypertrophy and increasing organization, it did not change in TTN-Z−/−-CMs. This could indicate that TTN-Z−/− myofibril bundling or stability is compromised. Interestingly, sarcomere length, circular variance (a measure of sarcomere disarray), cell area, maximum cell width, and cell length were not significantly different between wild type and TTN-Z−/−-CMs at either time point (Figure S7A–C, F, G). In addition, the TTN-Z−/−CMs exhibited higher multinucleation than wild type controls at the 60 day time point. Although overall rates of multinucleation were non-significantly higher in TTN-Z−/− cells at this time point, there were striking incidences of cells that contained 4 or 5 nuclei (Figure S7D–E). This was never observed in the wild type samples and may be explained by a nuclear role of full-length titin or a cytoplasmic role in inhibiting nuclear division42–44.
Figure 4. Myofibril morphology in WT and TTN-Z−/− CMs.
(A) Representative image of TTN-Z−/−-CM plated on nanopatterns for 60 days show significantly smaller myofibrillar bundles compared to wild type. (B) Morphological analysis of hiPSC-CMs cultured on nanopatterned surfaces for 30 or 60 days indicate that maximal myofibril bundle width was decreased in TTNZ−/−-CMs compared to WT at both time points. (C) Time-lapse images of hiPSC-CMs expressing mCherry-tagged α-actinin show that (D) WT consistently build myofibrils while TTN-Z−/− CMs have unstable myofibrils and (E) WT increase myofibril bundle width while TTN-Z−/− CMs decrease bundle width. Large images: scale bars = 50μm; inset images: scale bars = 5μm. In panel (B): WT: n=43–57; TTN-Z−/−: n=66–70. Nominal P-values were calculated using ANOVAs. Adjusted p-values were calculated using Tukey’s post-hoc test and are reported in this figure. In panel (D-E) WT and TTN-Z−/− n=9. P-values were calculated using student’s t-tests. Adjusted p-values were computed using the Benjamini & Hochberg method. In all panels error bars indicate standard error. *: adjusted p-value<0.05; **: adjusted p-value<0.01.
To further investigate sarcomere formation in TTN-Z−/−-CMs, we lentivirally transduced CMs with mCherry-tagged alpha-actinin, replated the cells and performed live-cell imaging every 30 minutes for 12 hours. WT CMs exhibited the expected peripheral flow of alpha-actinin in stress fiber-like structures at the edge of the cell, which then became centripetal as the alpha-actinin condensed and formed z-disks near the center of the cell, as previously reported45. During the 12 hours of imaging a drastic increase in myofibril number and bundle width was observed in WTC-CMs (Figure 4C–E, Video S1). In contrast, TTN-Z−/−-CMs did not show any peripheral flow of alpha-actinin, and myofibril number and bundle width decreased (Video S2). This indicates that TTN-Z−/− are not able to properly bundle myofibrils, and the small myofibrils that are formed are not stable.
Expression of Cronos in human fetal and adult hearts
Having established Cronos titin expression and integration into sarcomeres in hiPSC-CMs, we wanted to examine if this isoform was similarly expressed in human tissue. To study the expression levels and localization of Cronos titin in development we performed immunohistochemistry on human fetal and adult cardiac ventricular samples using our Cronos titin-specific antibody (Figure 5A). Staining for β-myosin heavy chain revealed clear striations in all samples studied, with increasing density and alignment of myofibrils in older fetal and adult samples. Cronos titin staining was clearly visible throughout the tissue of day 54 and 81 fetal heart samples. As in the hiPSC-CMs, Cronos formed doublets within the sarcomere of fetal hearts, corresponding to the space between the thick filament and the Z-disk. Myofibril density and Cronos titin staining were noticeably higher in the day 81 than the day 54 sample. Interestingly, Cronos titin staining was drastically reduced in the day 130 fetal sample, where it was barely visible in isolated portions of the tissue. In sections that Cronos titin was observed, it remained in a striated pattern but had lost the doublet pattern detected in earlier fetal samples. Adult tissue did not show a clear pattern of Cronos titin localization, and the signal was barely above background fluorescence observed in unstained tissue (Figure S8). Cronos and full-length titin transcripts were detected in adult and fetal cardiac samples, and while low sample numbers limit statistical analysis, Cronos appears to be more highly expressed in fetal than adult tissue (Figure 5B–C). Additionally, ChIP-Seq data indicates that a fetal whole heart sample has H3K4me3 peaks at both the full-length and Cronos transcription start sites in TTN, similar to that observed in hPSC-CM samples, whereas adult LV samples only have peaks at the full-length start site (Figure 5D). These data confirm that Cronos titin is highly expressed in human fetal cardiac tissue.
Figure 5. Cronos expression in human cardiac samples.
(A) β-myosin staining shows the presence of myofibrils in all samples. Cronos titin stains strongly in day 54 and 81 fetal samples and indicates localization to the edge of the thick filament. In day 130 samples, Cronos titin is still visible but much sparser and has lost the doublet pattern observed in younger fetal samples. Cronos titin is barely detectable in adult left ventricle (LV) samples. Scale bars = 5μm. (B) Cronos titin transcript is expressed in fetal atrium and ventricle samples and adult LV samples, but not in fetal kidney. (C) Full-length titin transcript is expressed in all cardiac samples studied but not fetal kidney. (D) H3K4me3 marks along TTN in fetal whole heart (GEO accession number GSM772735) and adult left ventricle (LV) tissue samples (GEO accession numbers GSM910580 and GSE101357) expressed as fold change over baseline. Data from ref [46], details in Methods. In panels (B-C) Disease human heart n=3; fetal atrium n=4; fetal ventricle n=2; fetal kidney n=3. Expression calculated relative to HPRT using ΔΔCT method. Error bars represent standard error.
Cronos titin is necessary for organized sarcomere formation
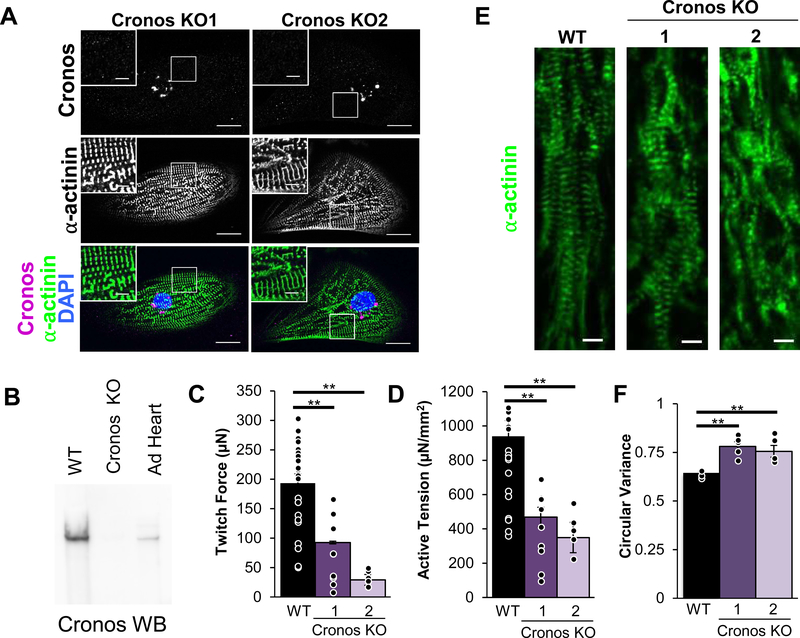
To investigate the role of Cronos titin in cardiomyocyte function and sarcomere formation, we generated two Cronos knock-out (KO) hiPSC lines, both of which expressed full-length titin. Both hiPSC lines differentiated into visibly beating CMs, and immunostaining for Cronos indicated that none was integrated into sarcomeres (Figure 6A), while staining for domains present in full-length titin were still present and correctly localized relative to alpha-actinin (Figure S9). A western blot using the custom Cronos titin antibody indicated this isoform was absent in the Cronos KO-CM (Figures 6B and S10). EHTs prepared with Cronos KO-CM produce less than half the twitch force (92.6μN and 29.1μN in Cronos KO vs 193.4μN in WT) and active tension (468.6 μN/mm2 and 349.2 μN/mm2 in Cronos KO vs. 939.6 μN/mm2 in WT) compared to WT, and have less than half the maximum twitch force velocity (642.7 μm/sec and 260.4 μm/sec in Cronos KO vs. 1330.4 μm/sec in WT; Figures 6B–C; S10C–D). Interestingly, passive force and twitch kinetics were not different in Cronos KO compared to WT (Figure S10A-B). To elucidate the cause of reduced force development, we performed immunohistochemistry on the WT and Cronos EHTs and measured sarcomere length and circular variance, a measure of myofibril alignment. While sarcomere length was no different between the groups, the myofibrils of the Cronos KO EHTs had significantly more disarray compared to WT (Figure 6D–E). Additionally, single cell measurements indicated there was no significant difference in the magnitude of calcium transients, though the maximum rate of calcium release was slower (Figure S11). This indicates that the reduced force production in hiPSC-CMs lacking Cronos titin may result from myofibril disarray.
Figure 6. Characterization of Cronos KO hiPSC-CMs.
(A) Cronos KO CMs do not stain for Cronos titin but do form sarcomeres. In each panel, large image: scale bar = 20μm, inset image: scale bar = 5μm. White boxes indicate regions magnified in inset image. EHTs seeded with Cronos KO CMs produce lower (B) twitch force and (C) active tension normalized to cross-sectional area compared to WT. (D) Representative images of EHT immunohistochemistry show myofibrillar disarray in Cronos KO samples, and when this is (E) quantified by measuring circular variance it is found to be statistically significant. (F) Calcium transient magnitudes of single Cronos KO CMs are not significantly different compared to WT. Panels (B-C): WT: n=23; Cronos KO-1: n=12; Cronos KO-2: n=6. Panel (E): WT: n=6; Cronos KO-1: n=5; Cronos KO-2: n=5. Panel (F): WT: n=35; Cronos KO-1: n=32; Cronos KO-2: n=22. Error bars indicate standard error. Nominal P-values were calculated using ANOVAs. Adjusted p-values were calculated using Tukey’s post-hoc test and are reported in this figure. **: adjusted p-value<0.01.
Discussion
To assess the role of titin in the formation of sarcomeres, we generated two hiPSC lines with homozygous mutations in constitutively expressed exons of TTN. We hypothesized that proximal truncations (TTN-Z−/−) would produce more severe phenotypes than distal truncations (TTN-A−/−). Instead, we found that instead the proximal truncation mutations were compatible with myofibril assembly and contraction, whereas the distal truncations had neither myofibrils nor contractility. Using a custom-generated antibody, we determined that TTN-Z−/−-CMs express the Cronos titin isoform, which appears to initiate transcription from an internal promoter distal to the truncation and is sufficient to support some sarcomere formation. In contrast, TTN-A−/−-CMs express what are likely N-terminal truncations of both full-length and Cronos titin and do not form myofibrils, as has been previously reported26. This study also reported that hiPSC-CMs carrying a homozygous truncation in the early I-band region of titin can also form sarcomeres. However, this mutation was in an exon that is only sometimes spliced in, and the sarcomere formation was associated with splicing out the mutated exon26. In the current study, the engineered Z-disk mutations are in a constitutively expressed exon, ruling out variable splicing as a mechanism of escape. Thus, we conclude that Cronos titin is sufficient for some sarcomere formation and the absence of both full-length and Cronos titin inhibits sarcomerogenesis. While a dominant-negative effect of the large N-terminal fragments in TTN-A−/−-CMs cannot be ruled out from our data, experiments in zebrafish using a combined CRISPR/Cas9 knockout and morpholino knockdown implicate a deficiency of titin rather than a dominant-negative effect29. Additionally, alternative splicing or other internal start sites in titin could not have been detected by the methods in this study, and will require follow-up experiments to assess in this system.
Our data are consistent with a structural role for titin, and in particular the C-terminal regions, in sarcomere formation. Signaling roles cannot be ruled out, however, given that titin has multiple functions. For example, it is possible that the absence of the M-line region of titin disrupts signaling pathways dependent upon the titin kinase domain, which is involved in protein turnover and hypertrophy14. However, there are several pieces of evidence to suggest that this is not the case. Previously, a mouse embryonic stem cell line with a homozygous truncating mutation in the M-line region of titin preventing expression of both the kinase domain and C-terminal of the protein was found to lack sarcomeres when differentiated into cardiomyocytes47. However, a mouse model harboring a mutation that deletes the kinase domain but not the C-terminal of the protein was observed to form sarcomeres early in embryonic development48. This indicates that it may be the anchoring function of the carboxy terminus of titin, rather than the kinase function, that is required for sarcomere formation. This notion is consistent with our finding that cTnT and α-actinin protein are both diffusely present in TTN-A−/−-CMs and indicates that sarcomeric proteins may still be expressed but cannot assemble into functional units. This suggests that signaling pathways relying on M-line titin may not be crucial for sarcomeric protein expression, while other functions of this domain, such as anchoring to myomesin, may be important for this stage of development.
TTN-Z−/−-CMs produced lower force as both multicellular EHTs and single cells compared to wild type CMs, although calcium transients had similar magnitudes. Interestingly, the force deficit of individual cells was not as dramatic as in EHTs. One potential explanation for this could be the greater maturity of the cardiomyocytes in EHTs compared to monolayers49, such that differences in the rate and extent of myofibril formation and bundling in cells become more pronounced. Another possibility is the summation of cellular forces in series in tissue compared to single cell assays are compounding differences in force production, or that there is more compliance between coupled cells in TTN-Z−/− EHTs compared to WT.
Staining of human tissue indicated that Cronos titin is most highly expressed in early fetal cardiac tissue, suggesting that it is predominantly a developmental isoform. The T2 band, which we demonstrate includes Cronos titin, has been shown to consistently decrease in intensity in cardiac samples as animals mature36 and as hiPSC-CMs are aged in culture26, supporting the notion that Cronos titin is predominantly a developmental isoform. Because hiPSC-CMs mature more rapidly in three-dimensional culture compared to two-dimensional50, cells in the EHTs may be more mature and thus expressing more full-length titin, while single cells on microposts are younger and thus may be expressing mostly Cronos titin.
The passive tension measured in individual TTN-Z−/−-CMs was not significantly different compared to wild type cells (Fig 5D). This could be explained by the immaturity of the contractile lattice in these cells, as fetal cardiac tissue has very low passive tension, especially from titin contributions36,51,52. Our recent studies have indicated that hiPSC-CMs matured for 80100 days on patterned substrates have similar contractile properties to 74 day fetal tissue41, and because the cells used in the micropost assay were significantly younger than this (35–40 days post differentiation) it is likely they were less mature. Thus, it is possible that the passive tension being measured from single-cell hiPSC-CMs on the microposts is predominantly due to other sources, such as non-muscle actin filaments or intermediate filaments within the cells6.
Morphological analysis of the myofibrils indicated that TTN-Z−/− had smaller myofibril bundle widths at the Z-disk compared to wild type (Figure 4B), suggesting either smaller diameters of individual myofibrils or fewer myofibrils in parallel. Previously, conflicting data have been reported regarding the necessity of N-terminal titin for the formation of Z-disks2,53, and the current study indicates that Z-disks are able to form in the absence of the N-terminal of titin. However, the live cell images of cells expressing tagged alpha-actinin indicate the stability or bundling ability of these Z-disks is compromised, and they quickly degrade after they are formed. We found that sarcomere length was not significantly different between wild type and TTN-Z−/−-CMs, which could indicate that full-length titin is not a major contributor to determining sarcomere length in cells at this stage of maturation. Supporting this notion, the length of I-band titin, which is missing in TTN-Z−/−-CMs, does not influence thin filament length, even in adult mice54.
For the first time in human cells we have knocked out Cronos titin. The resulting cardiomyocytes exhibit reduced force production and sarcomeric disarray, similar to what was observed in zebrafish29. Cronos is highly expressed in fetal cardiac tissue samples and appears to have a role in sarcomere formation or stability, particularly in the thick filament, given its likely interaction through the included A-band domains. Although further experiments are necessary to understand the role of this isoform in normal human development, these findings indicate that Cronos is an important player in sarcomerogenesis.
It is striking that cardiomyocytes are able to form sarcomeres in the absence of full-length titin in TTN-Z−/−-CMs, given that these mutations are not compatible with survival in rodent models16,17. We conclude that sarcomeres form in TTN-Z−/−-CMs and not TTN-A−/−-CMs due to the presence of Cronos titin, an isoform of titin that we demonstrate for the first time is expressed and integrated into myofibrils of human cardiomyocytes. Based on the findings of this study, it is likely that Cronos is a developmental isoform of titin. We conclude that Cronos titin is necessary for normal sarcomere development and function as full Cronos KO CMs produce lower contractile force and disarrayed sarcomeres. The discovery of this isoform in human samples motivates a closer look at how DCM is caused by truncating mutations in TTN and the potential role of Cronos in DCM pathogenesis. In particular, the presence of Cronos titin should be taken into account when evaluating hiPSC models of DCM, as this isoform clearly plays a significant role in hiPSC-CM function. Elucidating the functions of Cronos titin in human development and DCM will be crucial for fully understanding heart development and disease.
Supplementary Material
Clinical Perspective.
What is new?
Our studies using genetically engineered human induced pluripotent stem cell-derived cardiomyocytes experimentally confirm that the gene encoding the giant sarcomere protein titin, TTN, includes an internal promoter and start site.
This internal start site encodes for the isoform Cronos, which we demonstrate is able to support some sarcomere formation in hiPSC-CMs.
To our knowledge, this is the first study to experimentally demonstrate Cronos is expressed in fetal and adult cardiac tissue and is necessary for proper human cardiomyocyte function.
What are the clinical implications?
Truncating mutations in TTN are the leading known cause of familial dilated cardiomyopathy (DCM).
Cronos titin is a previously unstudied form of titin that we show is necessary for normal human cardiomyocyte function and could be contributing to these titinopathies.
Truncating mutations in TTN are most often found in the regions of the gene included in Cronos titin, warranting further investigation into its role in DCM pathogenesis.
Acknowledgments
We thank Dr. Bruce Conklin (Gladstone Institute) for providing the WTC11 hiPSC line and Dr. Siegfried Labeit (University of Heidelberg) for providing titin antibodies. This research was supported by the Cell Analysis Facility Flow Cytometry and Imaging Core in the Department of Immunology at the University of Washington. We would like to acknowledge the Mike and Lynn Garvey Cell Imaging Lab at the Institute for Stem Cells and Regenerative Medicine (UW) and its director Dr. Dale Hailey for assistance with sample imaging and analysis. We thank the Birth Defects Research Laboratory at the University of Washington and its director Dr. Ian Glass for providing the fetal tissue used in this study.
Sources of Funding
This work was supported by National Institute of Health grants T32 EB1650 (R.J.Z., K.B.), F32 HL126332 (A.L.), T32 HL007312 (P.F.), R01 HL135143 and R01HL146436 (D.-H.K.), R01 HL111197 and R01 HD048895 (M.R.), and R01 HL128362, P01 HL094374, R01 HL084642, U54 DK107979, and P01 GM081619 (C.E.M.); National Science Foundation CBET-1509106 (N.S.); German Research Foundation grant SFB1002/TPA08 (W.A.L.); Foundation Leducq Transatlantic Network of Excellence Award (C.E.M.), and a gift from the Robert B. McMillen Foundation (C.E.M.). The Birth Defects Research Laboratory is supported by NIH grant R24HD000836 to Dr. Ian A. Glass.
Non-standard Abbreviations and Acronyms
- CMs
cardiomyocytes
- cTnT
cardiac troponin T
- DCM
dilated cardiomyopathy
- EHT
engineered heart tissue
- hESC
human embryonic stem cells
- hiPSC
human induced pluripotent stem cells
- hiPSC-CMs
human induced pluripotent-derived cardiomyocytes
- KO
knock out
- sgRNA
single guide RNA
- TSS
transcriptional start site
- WT
wild type
Footnotes
Disclosures
Charles Murry is a scientific founder and equity holder in Cytocardia. Deok-Ho Kim is a scientific founder and equity holder in NanoSurface Biomedical Inc.
References
- 1.Fürst DO, Osborn M, Nave R, Weber K. The organization of titin filaments in the half-sarcomere revealed by monoclonal antibodies in immunoelectron microscopy: a map of ten nonrepetitive epitopes starting at the Z line extends close to the M line. J Cell Biol. 1988;106:1563–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gregorio CC, Trombitás K, Centner T, Kolmerer B, Stier G, Kunke K, Suzuki K, Obermayr F, Herrmann B, Granzier H, et al. The NH2 terminus of titin spans the Z-disc: Its interaction with a novel 19-kD ligand (T-cap) is required for sarcomeric integrity. J Cell Biol. 1998;143:1013–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Young P, Ferguson C, Bañuelos S, Gautel M. Molecular structure of the sarcomeric Zdisk: Two types of titin interactions lead to an asymmetrical sorting of α-actinin. EMBO J. 1998;17:1614–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Witt CC, Burkart C, Labeit D, Mcnabb M, Wu Y, Granzier H, Labeit S. Nebulin regulates thin filament length, contractility, and Z-disk structure in vivo. EMBO J. 2006;25:3843–3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Labeit S, Lahmers S, Burkart C, Fong C, McNabb M, Witt S, Witt C, Labeit D, Granzier H. Expression of Distinct Classes of Titin Isoforms in Striated and Smooth Muscles by Alternative Splicing, and Their Conserved Interaction with Filamins. J Mol Biol. 2006;362:664–681. [DOI] [PubMed] [Google Scholar]
- 6.Granzier HL, Irving TC. Passive tension in cardiac muscle: contribution of collagen, titin, microtubules, and intermediate filaments. Biophys J. 1995;68:1027–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Linke WA, Popov VI, Pollack GH. Passive and active tension in single cardiac myofibrils. Biophys J. 1994;67:782–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whiting A, Wardale J, Trinick J. Does titin regulate the length of muscle thick filaments? J Mol Biol. 1989;205:263–268. [DOI] [PubMed] [Google Scholar]
- 9.Müller S, Lange S, Gautel M, Wilmanns M. Rigid Conformation of an Immunoglobulin Domain Tandem Repeat in the A-band of the Elastic Muscle Protein Titin. J of Mol Biol. 2007;371:469–480. [DOI] [PubMed] [Google Scholar]
- 10.Bennett PM, Gautel M. Titin domain patterns correlate with the axial disposition of myosin at the end of the thick filament. J Mol Biol. 1996;259:896–903. [DOI] [PubMed] [Google Scholar]
- 11.Tskhovrebova L, Bennett P, Gautel M, Trinick J. Titin ruler hypothesis not refuted. Proc Natl Acad Sci. 2015;112:E1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Granzier HL, Hutchinson KR, Tonino P, Methawasin M, Li FW, Slater RE, Bull MM, Saripalli C, Pappas CT, Gregorio CC, et al. Deleting titin’s I-band/A-band junction reveals critical roles for titin in biomechanical sensing and cardiac function. Proc Natl Acad Sci. 2014;111:14589–14594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Obermann WM, Gautel M, Weber K, Fürst DO. Molecular structure of the sarcomeric M band: mapping of titin and myosin binding domains in myomesin and the identification of a potential regulatory phosphorylation site in myomesin. EMBO J. 1997;16:211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lange S, Xiang F, Yakovenko A, Vihola A, Hackman P, Rostkova E, Kristensen J, Brandmeier B, Franzen G, Hedberg B, et al. The kinase domain of titin controls muscle gene expression and protein turnover. Science. 2005;308:1599–1603. [DOI] [PubMed] [Google Scholar]
- 15.Person V, Kostin S, Suzuki K, Labeit S, Schaper J. Antisense oligonucleotide experiments elucidate the essential role of titin in sarcomerogenesis in adult rat cardiomyocytes in long-term culture. J Cell Sci. 2000;113:3851–3859. [DOI] [PubMed] [Google Scholar]
- 16.Gramlich M, Michely B, Krohne C, Heuser A, Erdmann B, Klaassen S, Hudson B, Magarin M, Kirchner F, Todiras M, et al. Stress-induced dilated cardiomyopathy in a knock-in mouse model mimicking human titin-based disease. J Mol Cell Cardiol. 2009;47:352–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schafer S, de Marvao A, Adami E, Fiedler LR, Ng B, Khin E, Rackham OJL, van Heesch S, Pua CJ, Kui M, et al. Titin-truncating variants affect heart function in disease cohorts and the general population. Nat Genet. 2017;49:46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akinrinade O, Alastalo T- P, Koskenvuo JW. Relevance of truncating titin mutations in dilated cardiomyopathy. Clin Genet. 2016;90:49–54. [DOI] [PubMed] [Google Scholar]
- 19.Norton N, Li D, Rampersaud E, Morales A, Martin ER, Zuchner S, Guo S, Gonzalez M, Hedges DJ, Robertson PD, et al. Exome sequencing and genome-wide linkage analysis in 17 families illustrate the complex contribution of TTN truncating variants to dilated cardiomyopathy. Circ Cardiovasc Genet. 2013;6:144–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pugh TJ, Kelly M, Gowrisankar S, Hynes E, Seidman M, Baxter SM, Bowser M, Harrison B, Aaron D, Mahanta LM, et al. The landscape of genetic variation in dilated cardiomyopathy as surveyed by clinical DNA sequencing. Genet Med. 2014;16:601–608. [DOI] [PubMed] [Google Scholar]
- 21.van Spaendonck-Zwarts KY, Posafalvi A, van den Berg MP, Hilfiker-Kleiner D, Bollen IAE, Sliwa K, Alders M, Almomani R, van Langen IM, van der Meer P, et al. Titin gene mutations are common in families with both peripartum cardiomyopathy and dilated cardiomyopathy. Eur Heart J. 2014;35:2165–2173. [DOI] [PubMed] [Google Scholar]
- 22.Haas J, Frese KS, Peil B, Kloos W, Keller A, Nietsch R, Feng Z, Muller S, Kayvanpour E, Vogel B, et al. Atlas of the clinical genetics of human dilated cardiomyopathy. Eur Heart J. 2015;36:1123–1135. [DOI] [PubMed] [Google Scholar]
- 23.Herman DS, Lam L, Taylor MRG, Wang L, Teekakirikul P, Christodoulou D, Conner L, DePalma SR, McDonough B, Sparks E, et al. Truncations of titin causing dilated cardiomyopathy. New Engl J Med. 2012;366:619–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roberts AM, Ware JS, Herman DS, Schafer S, Baksi J, Bick AG, Buchan RJ, Walsh R, John S, Wilkinson S, et al. Integrated allelic, transcriptional, and phenomic dissection of the cardiac effects of titin truncations in health and disease. Sci Transl Med. 2015;7:270ra6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ware JS, Li J, Mazaika E, Yasso CM, DeSouza T, Cappola TP, Tsai EJ, Hilfiker-Kleiner D, Kamiya CA, Mazzarotto F, et al. Shared Genetic Predisposition in Peripartum and Dilated Cardiomyopathies. New Engl J Med. 2016;374:233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hinson JT, Chopra A, Nafissi N, Polacheck WJ, Benson CC, Swist S, Gorham J, Yang L, Schafer S, Sheng CC, et al. Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science. 2015;349:982–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schick R, Mekies LN, Shemer Y, Eisen B, Hallas T, Ben Jehuda R, Ben-Ari M, Szantai A, Willi L, Shulman R, et al. Functional abnormalities in induced Pluripotent Stem Cellderived cardiomyocytes generated from titin-mutated patients with dilated cardiomyopathy. alto-Setala K, ed. PLoS One. 2018;13:e0205719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gramlich M, Pane LS, Zhou Q, Chen Z, Murgia M, Schötterl S, Goedel A, Metzger K, Brade T, Parrotta E, et al. Antisense mediated exon skipping: a therapeutic strategy for titin-based dilated cardiomyopathy. EMBO Mol Med. 2015;7:562–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zou J, Tran D, Baalbaki M, Tang LF, Poon A, Pelonero A, Titus EW, Yuan C, Shi C, Patchava S, et al. An internal promoter underlies the difference in disease severity between N- and C-terminal truncation mutations of Titin in zebrafish. eLife. 2015;4:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palpant NJ, Pabon L, Friedman CE, Roberts M, Hadland B, Zaunbrecher RJ, Bernstein I, Zheng Y, Murry CE. Generating high-purity cardiac and endothelial derivatives from patterned mesoderm using human pluripotent stem cells. Nat Protoc. 2016;12:15–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tohyama S, Hattori F, Sano M, Hishiki T, Nagahata Y, Matsuura T, Hashimoto H, Suzuki T, Yamashita H, Satoh Y, et al. Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell. 2013;12:127–137. [DOI] [PubMed] [Google Scholar]
- 33.Bielawski KS, Leonard A, Bhandari S, Murry CE, Sniadecki NJ. Real-Time Force and Frequency Analysis of Engineered Human Heart Tissue Derived from Induced Pluripotent Stem Cells Using Magnetic Sensing. Tissue Eng Part C Methods. 2016;22:932–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kreitzer FR, Salomonis N, Sheehan A, Huang M, Park JS, Spindler MJ, Lizarraga P, Weiss WA, So P, Conklin BR. A robust method to derive functional neural crest cells from human pluripotent stem cells. Am J Stem Cells. 2013;2:119–131. [PMC free article] [PubMed] [Google Scholar]
- 35.Paige SL, Thomas S, Stoick-Cooper CL, Wang H, Maves L, Sandstrom R, Pabon L, Reinecke H, Pratt G, Keller G, et al. A temporal chromatin signature in human embryonic stem cells identifies regulators of cardiac development. Cell. 2012;151:221–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Opitz CA, Leake MC, Makarenko I, Benes V, Linke WA. Developmentally regulated switching of titin size alters myofibrillar stiffness in the perinatal heart. Circ Res. 2004;94:967–975. [DOI] [PubMed] [Google Scholar]
- 37.Opitz CA, Linke WA. Plasticity of cardiac titin/connectin in heart development. J Muscle Res Cell Motil. 2005;26:333–342. [DOI] [PubMed] [Google Scholar]
- 38.Makarenko I, Opitz CA, Leake MC, Neagoe C, Kulke M, Gwathmey JK, Del Monte F, Hajjar RJ, Linke WA. Passive stiffness changes caused by upregulation of compliant titin isoforms in human dilated cardiomyopathy hearts. Circ Res. 2004;95:708–716. [DOI] [PubMed] [Google Scholar]
- 39.Rodriguez ML, Graham BT, Pabon LM, Han SJ, Murry CE, Sniadecki NJ. Measuring the Contractile Forces of Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes With Arrays of Microposts. J Biomech Eng. 2014;136:051005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carson D, Hnilova M, Yang X, Nemeth CL, Tsui JH, Smith AST, Jiao A, Regnier M, Murry CE, Tamerler C, et al. Nanotopography-Induced Structural Anisotropy and Sarcomere Development in Human Cardiomyocytes Derived from Induced Pluripotent Stem Cells. ACS Appl Mater Interfaces. 2016;8:21923–21932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pioner JM, Racca AW, Klaiman JM, Yang KC, Guan X, Pabon L, Muskheli V, Zaunbrecher R, Macadangdang J, Jeong MY, et al. Isolation and Mechanical Measurements of Myofibrils from Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Stem Cell Reports. 2016;6:885–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zastrow MS, Flaherty DB, Benian GM, Wilson KL. Nuclear titin interacts with A- and B-type lamins in vitro and in vivo. J Cell Sci. 2006;119:239–249. [DOI] [PubMed] [Google Scholar]
- 43.Qi J, Chi L, Labeit S, Banes AJ. Nuclear localization of the titin Z1Z2Zr domain and role in regulating cell proliferation. Am J Physiol Cell Physiol. 2008;295:C975–C985. [DOI] [PubMed] [Google Scholar]
- 44.Machado C, Sunkel CE, Andrew DJ. Human autoantibodies reveal titin as a chromosomal protein. J Cell Biol. 1998;141:321–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chopra A, Kutys ML, Zhang K, Polacheck WJ, Sheng CC, Luu RJ, Eyckmans J, Hinson JT, Seidman JG, Seidman CE. Force Generation via β-Cardiac Myosin, Titin, and α-Actinin Drives Cardiac Sarcomere Assembly from Cell-Matrix Adhesions. Dev Cell. 2018;44:87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, Kaul R, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Musa H, Meek S, Gautel M, Peddie D, Smith AJH, Peckham M. Targeted homozygous deletion of M-band titin in cardiomyocytes prevents sarcomere formation. J Cell Sci. 2006;119:4322–4331. [DOI] [PubMed] [Google Scholar]
- 48.Weinert S, Bergmann N, Luo X, Erdmann B, Gotthardt M. M line-deficient titin causes cardiac lethality through impaired maturation of the sarcomere. J Cell Biol. 2006;173:559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tulloch NL, Muskheli V, Razumova MV., Korte FS, Regnier M, Hauch KD, Pabon L, Reinecke H, Murry CE. Growth of engineered human myocardium with mechanical loading and vascular coculture. Circ Res. 2011;109:47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwan J, Campbell SG. Prospects for In Vitro Myofilament Maturation in Stem CellDerived Cardiac Myocytes. Biomarker insights. 2015;10:91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lahmers S, Wu Y, Call DR, Labeit S, Granzier H. Developmental Control of Titin Isoform Expression and Passive Stiffness in Fetal and Neonatal Myocardium. Circ Res. 2004;94:505–513. [DOI] [PubMed] [Google Scholar]
- 52.Warren CM, Krzesinski PR, Campbell KS, Moss RL, Greaser ML. Titin isoform changes in rat myocardium during development. Mech Dev. 2004;121:1301–1312. [DOI] [PubMed] [Google Scholar]
- 53.Bowman AL, Catino DH, Strong JC, Randall WR, Kontrogianni-Konstantopoulos A, Bloch RJ. The rho-guanine nucleotide exchange factor domain of obscurin regulates assembly of titin at the Z-disk through interactions with Ran binding protein 9. Mol Biol Cell. 2008;19:3782–3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kolb J, Li F, Methawasin M, Adler M, Escobar YN, Nedrud J, Pappas CT, Harris SP, Granzier H. Thin filament length in the cardiac sarcomere varies with sarcomere length but is independent of titin and nebulin. J Mol Cell Cardiol. 2016;97:286–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.