Abstract
Many gut functions are attuned to circadian rhythm. Intestinal group 3 innate lymphoid cells (ILC3s) include NKp46+ and NKp46− subsets, which are RORγt dependent and provide mucosal defense through secretion of interleukin-22 (IL-22) and IL-17. Because ILC3s highly express some key circadian clock genes, we investigated whether ILC3s are also attuned to circadian rhythm. We noted circadian oscillations in the expression of clock and cytokine genes, such as REV-ERBα, IL-22, and IL-17, whereas acute disruption of the circadian rhythm affected cytokine secretion by ILC3s. Because of prominent and rhythmic expression of REV-ERBα in ILC3s, we also investigated the impact of constitutive deletion of REV-ERBα, which has been previously shown to inhibit the expression of a RORγt repressor, NFIL3, while also directly antagonizing DNA binding of RORγt. Development of the NKp46+ ILC3 subset was markedly impaired, with reduced cell numbers, RORγt expression, and IL-22 production in REV-ERBα-deficient mice. The NKp46– ILC3 subsets developed normally, potentially due to compensatory expression of other clock genes, but IL-17 secretion paradoxically increased, probably because RORγt was not antagonized by REV-ERBα. We conclude that ILC3s are attuned to circadian rhythm, but clock regulator REV-ERBα also has circadian-independent impacts on ILC3 development and functions due to its roles in the regulation of RORγt.
Introduction
The activity of every cell in the body oscillates within an endogenous period of 24 hours. This circadian rhythm is driven by a molecular clock, which consists of a set of three interlocked transcription-translation feedback loops. The main oscillator is driven by the heterodimer BMAL1:CLOCK that binds E-box motifs, thereby inducing the expression of its inhibitor PER/CRY that completes the feedback loop. BMAL1:CLOCK also induces the expression of REV-ERBs (Nr1d1, Nr1d2), RORs (retinoic acid related orphan receptors: Rora, Rorb, and Rorc), and DBP (D site-binding protein). Through competition at shared ROR response elements (RORE), RORs and REV-ERBs form a second loop of the molecular clock by activating and repressing, respectively, the expression of NFIL3 and BMAL1 (1). Lastly, the activator DBP and the repressor NFIL3 form a similar pair controlling the expression of genes containing a D-box motif such as PER, RORs, and REV-ERBs (2, 3). Each loop of the molecular clock also directs expression of a myriad of clock-controlled genes as the output of circadian rhythm, thus influencing many facets of biology.
The gastrointestinal system is typically tied to a daily rhythm. Feeding behaviors, digestion, absorption, gastric motility, and the microbiota are all attuned to the circadian rhythm (4–6). Recent studies suggest that circadian rhythm and clock genes may may also affect intestinal immune cells. It has been reported that REV-ERBα promotes RORγt expression and T helper 17 (TH17) polarization by repressing the RORγt inhibitor NFIL3 in a diurnal manner (7). Another study found that although REV-ERBα plays an important role in TH17 development, it acts as a negative regulator of the TH17 program by suppressing expression of many core genes (8). Last, a conflicting report indicated that the T cell intrinsic rhythm is dispensable for TH17 polarization (9).
Group 3 innate lymphoid cells (ILC3) are the innate correlates of TH17 based on their shared developmental requirement for the master transcription factor RORγt and secretion of interleukin-17 (IL-17) and IL-22 (10–12). In mice, ILC3s include two lineages based on ontogeny, which can be distinguished using the cell surface markers NKp46 and CCR6. NKp46−CCR6+ (CCR6+) ILC3s are responsible for secondary lymphoid organ formation during fetal development and tertiary lymphoid tissue development in adults (13–16). The second lineage encompasses NKp46−CCR6− (DN) ILC3s and their progeny NKp46+CCR6− (NKp46+) ILC3s (17–19). Whereas NKp46+ ILC3s secrete IL-22 and interferon-γ (IFN-γ), NKp46− ILC3s produce IL-22 and IL-17 (17). Given their abundance in the intestinal mucosa, their resemblance to TH17, and their developmental requirement for a ROR family member, ILC3s are likely to have a strong connection to the biological clock. However, little is known about the role of circadian rhythm and clock genes in ILC3s.
Transcriptional profiling has shown that both ILC3 subsets highly express the circadian nuclear receptor and transcriptional repressor REV-ERBα, encoded by Nr1d1 (20, 21). REV-ERBα, activated by the metabolite heme, forms stable interactions with NCoR (nuclear receptor corepressor) and HDAC3 (histone deacetylase 3) to suppress expression of target genes such as the RORγt repressor NFIL3 (7, 22, 23). REV-ERBα binds to DNA at single RORE elements or a dimer repeat of RORE elements known as RevDR2 through its DNA binding domain (24–26). RORE is also the target of ROR transcriptional activators that include RORγt. The impact of REV-ERBα on RORγt further suggests the link between circadian rhythm and ILC3 biology.
In this study, we addressed two questions: First, are circadian clock-controlled fluctuations in gene expression evident in ILC3s? Second, what is the impact of deleting a highly expressed clock regulator, REV-ERBα, on ILC3 biology, especially given its dual roles in both inhibiting the RORγt repressor NFIL3 and antagonizing the binding of RORγt to DNA? We were able to document circadian oscillations of known clock genes and cytokine expression in ILC3s. Moreover, acute perturbations of the circadian rhythm caused by a shift-work (SW) model curtailed RORγt expression and cytokine production by both NKp46+ and NKp46− ILC3s. Next, using Nr1d1–/– mice, we showed that REV-ERBα deficiency had a marked impact on ILC3 development. Genetic deletion of REV-ERBα impaired NKp46+ ILC3s’ numbers, expression of RORγt, mitochondrial function, and production of IL-22. Mixed bone marrow chimera and conditional Nr1d1–/– mice corroborated that the defect in NKp46+ ILC3s is cell-intrinsic. In contrast, REV-ERBα-deficient NKp46− ILC3s did not exhibit marked defects in numbers, expression of RORγt, and mitochondria possibly due to compensatory expression of other clock regulators. Somewhat paradoxically, NKp46− ILC3s produced excessive IL-17, because RORγt activity was not antagonized by REV-ERBα. Thus, although ILC3s exhibit circadian fluctuations in gene expression and are functionally attuned to circadian rhythms, the key clock protein REV-ERBα is also required for ILC3 development due to its role in regulating RORγt.
Results
Rhythmic expression of cytokines and clock genes in ILC3s
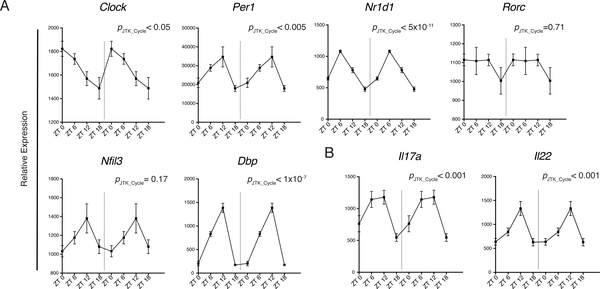
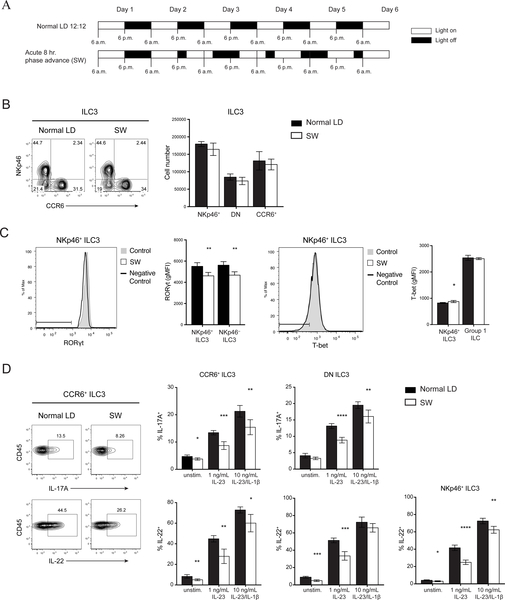
Given that circadian rhythm has been implicated in the polarization of TH17 and their functional similarity with ILC3s, we investigated whether clock genes expression fluctuates in ILC3s over a 24-hour period. We detected cycling transcripts of multiple circadian genes through JTK_CYCLE analysis (p< 0.05) (Fig. 1A) (27, 28). In particular, there were robust rhythmic expressions of Nr1d1 and Dbp. Although Nfil3 transcripts appeared to oscillate, it did not reach statistical significance, and fluctuations of Rorc expression were limited. We also observed cyclic expressions of Il17a and Il22, suggesting that there is also a functional rhythm in ILC3s linked with the molecular clock (Fig. 1B). To confirm that rhythmic gene expression translated to oscillating protein levels, we examined mice at ZT1 (Zeitgeber time 1) and ZT13 close to the nadir and peak, respectively, of mRNA levels for Il17a and Il22. We observed a slight reduction of RORγt levels in ILC3s at ZT13 while T-bet expression was unchanged (Fig. S1A). However, we did not see robust differences in ILC3 cytokine secretion, both in the frequency of cytokine producing cells and in the amount of cytokine production, suggesting that fluctuations at the protein level may be more subtle (Fig. S1B). However, because cells continue their progression through the circadian cycle during the ~4 hours of cytokine stimulation in culture ex vivo, they may reach points in the cycle at which differences in cytokine expression are more attenuated. Thus, to further this investigation using a different strategy, we acutely disrupted the circadian rhythm in mice using a SW model to test whether circadian rhythm regulates ILC3 function. Mice were either maintained in conventional LD 12:12 (12 hours light: 12 hours dark) conditions or subjected to an 8-hour phase advance every 2 days (SW) to simulate circadian disruptions present in SW (Fig. 2A) (29). Altered wheel running activity confirmed disrupted circadian behavior in phase-advanced mice (Fig. S2A). Meanwhile food consumption and body weight suggested that the mice were not under abnormal physiological stress (Fig. S2B, C). Both groups of mice were examined at ZT0 (6:00 a.m.) of day 6. There was no difference in the frequency or number of any ILC3 subsets within the small intestine lamina propria (siLP) (Fig. 2B). However, there was significant reduction in RORγt expression in both NKp46+ and NKp46− ILC3s from the SW cohort (Fig. 2C). Consequently, ILC3s from SW mice produced less IL-17 and IL-22 upon stimulation with varying concentrations of IL-23 or IL-23 plus IL-1β (Fig. 2D). Other relevant transcription factors, such as T-bet, were unaffected. Although we observed limited oscillations in Rorc mRNA (Fig. 1B), the reduction in the protein level of RORγt after acute circadian disruption suggests RORγt expression in ILC3 may also be regulated by posttranscriptional or posttranslational modifications (30, 31). Together, these results suggest that ILC3s have circadian oscillations in cytokine expression and altered circadian rhythm impacts cytokine secretion by all ILC3 subsets.
Figure 1. Circadian gene expression in ILC3 is associated with rhythmic cytokine expression.
A) Circadian gene and B) Il17a and Il22 expression in sorted ILC3 (all subsets included) over a 24-hour period by RT-qPCR, relative expression to Actb. Double-plotted for better visualization separated by dotted vertical line. Statistical analysis was performed using MetaCycle indicated by PJTK_CYCLEvalue. Points indicate means (±SEM). Data are pooled from two independent experiments (n=4 per time point per experiment).
Figure 2. Acute circadian disruption alters ILC3 cytokine secretion.
A) Diagram of light settings in acute SW model of circadian disruption. B) Representative plots (left) and numbers (right) of siLP ILC3 subsets in control and SW mice. C) Representative histogram and quantification of RORgt (left) and T-bet (right) expressions by intracellular staining in ILC3 subsets and group 1 ILCs (ILC1 and NK cells), horizontal bars in histograms represent negative control gate. gMFI, geometric mean fluorescence intensity. D) Representative plots (left) and quantifications (right) of IL-17A and IL-22 production in ILC3 subsets after in vitro stimulation with indicated concentrations of IL23±IL-1β. Statistical analysis was performed using Student’s t test. Bars indicate means (±SD). * P < 0.05; ** P < 0.01; *** P < 0.001; **** P < 0.0001. Data are representative of two independent experiments (n=5 per group per experiment) (A to D).
REV-ERBα-deficient mice lack intestinal NKp46+ ILC3
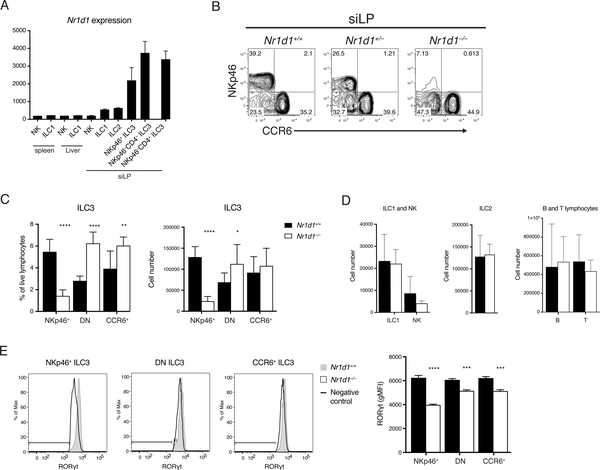
Examination of previously reported ILC3 transcriptome and chromatin profiles revealed that Nr1d1 mRNA (encoding REV-ERBα) is highly expressed in all three ILC3 subsets in comparison to other innate lymphocytes (Fig. 3A) (20). In addition, the robust rhythm of Nr1d1 expression further indicated that REV-ERBα may play a prominent role in ILC3 biology. Moreover, dual roles have been reported for REV-ERBα: it both inhibits the RORγt repressor NFIL3 and directly antagonizes RORγt DNA binding. Thus, we next sought to determine how lack of of REV-ERBα affects ILC3s in the previously described Nr1d1–/– mice (32). Unexpectedly, we observed that Nr1d1–/– mice had reduced frequencies and numbers of NKp46+ ILC3s in the siLP (Fig. 3B, C). A similar but less pronounced reduction of NKp46+ ILC3s was also present in heterozygous Nr1d1+/– mice, suggesting haploinsufficiency. Moreover, in the absence of NKp46+ ILC3, DN ILC3s were increased in both frequency and number. Although there was an increase in the frequency of CCR6+ ILC3s, there was no significant difference in numbers. Other major lymphocyte populations such as Eomes+ natural killer cells (NK), Eomes− ILC1s, ILC2s, B cells, and T cells were not affected (Fig. 3D). Similar results were observed in other tissues enriched for ILC3s including mesenteric lymph nodes (mLNs) and the large intestine (Fig. S3). Because REV-ERBβ, a REV-ERB subtype encoded by the gene Nr1d2, functions similarly and shares many transcription targets with REV-ERBα (33), we asked whether REV-ERBβ might also play a role in ILC3s. However, by examining Vav1iCreNr1d2fl/fl mice, which lack REV-ERBβ in all hematopoietic cells, we found that REV-ERBβ is dispensable in ILC3s in terms of cell numbers, RORγt expression, and cytokine secretion (Fig. S4). This suggests that REV-ERBβ likely plays a redundant role in ILC3s.
Figure 3. REV-ERBa deficiency reduces frequency and number of NKp46+ ILC3.
A) Nr1d1 expression in ILCs of the indicated tissues by microarray analysis, ImmGen Consortium. B) ILC3s from Nr1d1+/+, Nr1d1+/–, and Nr1d1–/– siLP, gated on live CD45+Lin– (CD3/CD5/CD19)RORgt+ cells. C) Frequency of live lymphocytes and cell counts of each ILC3 subsets in the siLP. D) Cell counts of ILC1s, NK cells, ILC2s, B cells, and T cells in siLP. E) Intracellular staining of RORgt in Nr1d1+/+ and Nr1d1–/– ILC3 subsets and quantification of RORgt levels, horizontal bars in histograms represent negative control gate. Statistical analysis was performed using Student’s t test. Bars indicate means (±SD). * P < 0.05; ** P < 0.01; *** P < 0.001; **** P < 0.0001. Data are representative of (B, D, and E) or pooled from at least two independent experiments (C) (n=3 per group per experiment).
Given the crucial role of RORγt in the development and function of ILC3s, we measured RORγt protein levels in Nr1d1–/– and Nr1d1+/+ ILC3s. NKp46+ ILC3s in Nr1d1–/– mice expressed significantly less RORγt than did their Nr1d1+/+ counterparts (Fig. 3E). RORγt expression in the CCR6+ and DN ILC3 subsets was less affected. We conclude that REV-ERBα plays a notable role in regulating RORγt expression in NKp46+ ILC3s, while other ILC3 subsets are less affected. These results, combined with the increase in DN ILC3s, suggest a blockade in developmental pathway leading from DN to NKp46+ ILC3s.
The developmental defect of REV-ERBα-deficient NKp46+ ILC3s is cell intrinsic
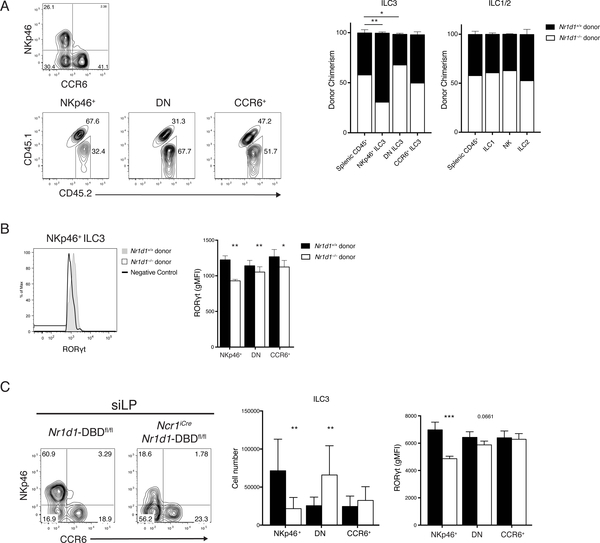
We next investigated whether the defect in ILC3s is cell-intrinsic by generating mixed bone marrow chimeras. A 50:50 mixture of bone marrow hematopoietic progenitors from congenically marked Nr1d1+/+ (CD45.1/2) and Nr1d1–/– (CD45.2) was injected into irradiated Nr1d1+/+ (CD45.1) mice to distinguish donors from radioresistant recipient ILCs. After 8 weeks of engraftment, chimerism of splenic CD45+ cells indicated roughly equivalent engraftment of Nr1d1+/+ and Nr1d1–/– donors with a slight predominance of Nr1d1−/− cells. CCR6+ ILC3 and other innate lymphocytes were reconstituted in ratios consistent with the bone marrow chimerism. In contrast, the majority of NKp46+ ILC3s were derived from Nr1d1+/+ hematopoietic progenitors rather than Nr1d1–/– donors in a clear reversal of the bone marrow chimerism (Fig. 4A). In addition, there was an increase in Nr1d1−/− donor-derived DN ILC3s relative to donor chimerism. Furthermore, in accordance with ex vivo data at steady state (see Fig. 3E), NKp46+ ILC3s derived from Nr1d1–/– progenitors also expressed significantly less RORγt than those derived from Nr1d1+/+ progenitors (Fig. 4B). These results indicated that the decrease in NKp46+ ILC3 numbers and their reduced expression of RORγt in Nr1d1−/− mice are cell intrinsic.
Figure 4. Reduction of NKp46+ ILC3 and RORgt expression in Nr1d1–/– mice is cell intrinsic.
A) Representative plots of ILC3 chimerism, gated on live CD45.2+Lin–(CD3/CD19)RORgt+ cells (left). Quantified donor chimerisms (right) for indicated siLP cell populations compared with chimerism of splenic CD45+ cells. B) Representative histogram of RORgt expression in NKp46+ ILC3s from Nr1d1+/+ and Nr1d1–/– donors (left) and quantification of RORgt expressions in ILC3 subsets (right), horizontal bars in histograms represent negative control gate. C) Representative plot (left), cell count (middle) and quantification of RORgt levels (right) of siLP ILC3 from Nr1d1-DBDfl/fl and Ncr1iCreNr1d1-DBDfl/fl mice. Statistical analysis was performed using one-way ANOVA with multiple comparisons (A) or Student’s t test (B and C). Bars indicate means (±SD). * P < 0.05; ** P < 0.01; *** P < 0.001. Data are representative of three independent experiments (n = 3 to 5 per group per experiment) (A, B, and C, right) or pooled from three independent experiments (n = 3 to 4 per group per experiment) (C, middle).
DN ILC3s have been shown to give rise to NKp46+ ILC3s (17, 18). The increase in Nr1d1–/– DN ILC3s in conjunction with sharply reduced Nr1d1–/– NKp46+ ILC3s in the mixed bone marrow chimeras and Nr1d1–/– mice (see Fig. 3B, C and 4A) suggested a possible blockade in the differentiation of NKp46+ ILC3s. To test this hypothesis, we crossed Ncr1iCre mice with Nr1d1fl/fl mice (also known as Nr1d1-DBDfl/fl due to reported in-frame deletion of the DNA binding domain) to examine the conditional deficiency of REV-ERBα in cells that express NKp46 (33, 34). The Ncr1iCreNr1d1-DBDfl/fl mouse perfectly phenocopied the Nr1d1–/– mouse; we observed a similar reduction in the frequency and numbers of NKp46+ ILC3s and significantly reduced RORγt expression on those remaining (Fig. 4C). Correspondingly, there is a significant accumulation of DN ILC3. This suggests that during the development of NKp46+ ILC3s, the deletion of Nr1d1 driven by the expression of Ncr1iCre resulted in the inability to maintain RORγt expression, thus halting their development.
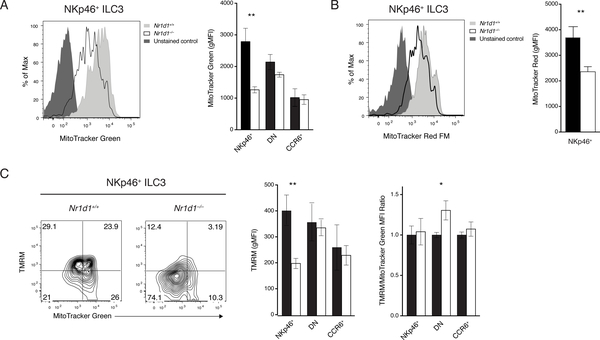
REV-ERBα is required to sustain mitochondria in NKp46+ ILC3s
REV-ERBα has been shown to be intimately involved in cellular metabolism, especially mitochondrial functions. Nr1d1–/– mice have reduced mitochondrial mass and impaired oxidative capacity in skeletal muscle. Overexpression of REV-ERBα in a skeletal muscle cell line and fibroblasts improved mitochondrial respiration and protected cells from oxidative stress (35, 36). Therefore, we hypothesized that Nr1d1–/– NKp46+ ILC3s may also experience mitochondrial dysfunction. Nr1d1–/– NKp46+ ILC3s had significantly reduced mitochondrial mass by MitoTracker Green staining (Fig. 5A). DN and CCR6+ ILC3s were unaffected. The reduction in mitochondrial content of Nr1d1–/– NKp46+ ILC3s may result in insufficient energy production. Mitochondrial membrane potential, assessed by both MitoTracker Red and tetramethylrhodamine methyl ester (TMRM) staining, was significantly reduced in NKp46+ ILC3s, but not in DN and CCR6+ ILC3s (Fig. 5B, C). To determine whether the loss of total mitochondrial membrane potential is a result of reduced mitochondrial mass or defects in mitochondrial respiration, we costained ILC3s with MitoTracker Green and TMRM. By normalizing the TMRM signal to MitoTracker Green signal, we concluded that the loss of mitochondrial membrane potential is due to reduced mitochondrial content (Fig. 5C). Furthermore, this regulation is likely independent of circadian rhythm because there were no marked differences in mitochondrial content and membrane potential after acute circadian disruption (Fig. S5). Therefore, REV-ERBα plays a crucial role in the regulation of mitochondria in NKp46+ ILC3s in a circadian-independent manner. However, the mechanism linking REV-ERBα to mitochondria metabolism specifically in NKp46+ ILC3 will require further investigation.
Figure 5. Requirement of REV-ERBa for mitochondrial content of NKp46+ ILC3s.
A) Mitochondrial content in Nr1d1+/+ and Nr1d1–/– ILC3s by MitoTracker Green FM staining (left, representative histogram; right, gMFI). B) Mitochondrial potential of Nr1d1+/+ and Nr1d1–/– NKp46+ ILC3s by MitoTracker Red FM staining (left, representative histogram; right, gMFI). C) Functional mitochondrial content and per unit mitochondrial potential of Nr1d1+/+ and Nr1d1–/– ILC3s measured by TMRM (mitochondrial potential) and MitoTracker Green FM (mitochondrial mass) co-staining (left, representative plot of NKp46+ ILC3s; middle, TMRM gMFI; right, normalized TMRM/MitoTracker Green ratio). Statistical analysis was performed using Student’s t test. Bars indicate means (±SD). * P < 0.05; ** P < 0.01; *** P < 0.001; **** P < 0.0001. Data are representative of three independent experiments, (n=3 per group per experiment).
REV-ERBα does not influence NKp46+ ILC3 plasticity
NKp46+ ILC3s have the intrinsic ability to undergo cellular transformation into ILC1s through the graded acquisition of T-bet and downregulation of RORgγt (17, 37). Thus, the depletion of NKp46+ ILC3s in Nr1d1–/– mice may also reflect an increased transition into ILC1-like cells. To test this hypothesis, we developed a REV-ERBα-deficient RORγt reporter and fate-mapping mouse (Nr1d1–/– RorceGFP/+Rorc-CretgR26RTdTomato/+) based on the previously published RORγt fate-map mouse design (37). Because the Rorc-Cretg locus appears genetically linked to the Nr1d1 locus, it was only possible to examine Nr1d1+/+ and Nr1d1+/– fate-map mice. Given that Nr1d1+/– mice have a moderate but obvious reduction in NKp46+ ILC3, we hypothesized that increased plasticity would be appreciated in the Nr1d1+/– fate-map mice. Nevertheless, we did not detect more fate-mapped positive siLP ILC1s in Nr1d1+/– than in Nr1d1+/+ fate-map mice (Fig. S6). These results indicate that increased plasticity likely does not contribute to the reduction of NKp46+ ILC3s in the Nr1d1–/– mice.
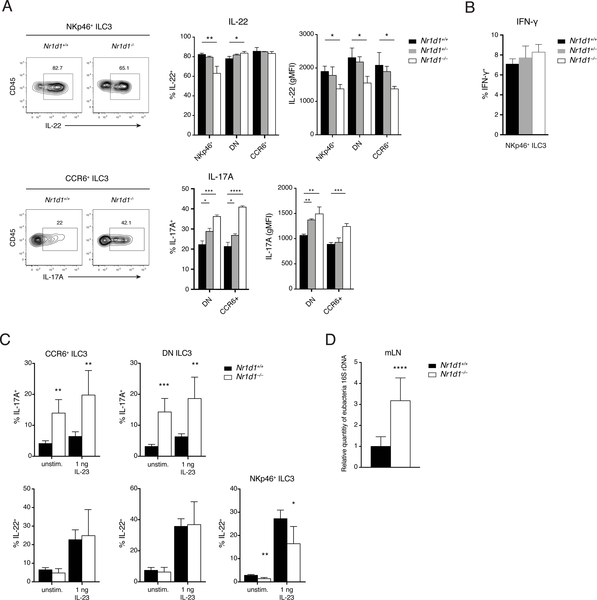
REV-ERBα deficiency differentially affects cytokine production in ILC3 subsets
We next examined the effect of REV-ERBα deficiency on cytokine secretion by ILC3 subsets. After in vitro stimulation with IL-23 and IL-1β, significantly fewer IL-22-producing cells and reduced amount of IL-22 per-cell were present among the residual NKp46+ ILC3s from Nr1d1–/– mice. Nr1d1–/– DN and CCR6+ ILC3s were less affected, exhibiting only a slight reduction in the amount of IL-22 per cell (Fig. 6A). In contrast, more IL-17-producing cells and higher IL-17 production on a per-cell level were present among CCR6+ and DN ILC3s from Nr1d1–/– mice than among their Nr1d1+/+ counterparts. Last, lack of REV-ERBα had no effect on the production of IFN-γ by NKp46+ ILC3s after stimulation with a combination of IL-12, IL-18, IL-1β, and IL-23 (Fig. 6B). Given the role of IL-17A and IL-22 in mucosal immunity, we next tested the impact of altered ILC3 function in a model of Clostridium difficile infection (CDI). Similar effects of REV-ERBα deficiency on ILC3 function were observed. On day 2 after infection, Nr1d1–/– DN and CCR6+ ILC3s produced significantly more IL-17A than did Nr1d1+/+ ILC3s both ex vivo and after in vitro stimulation. Whereas we did not observe differences in IL-22 secretion by DN and CCR6+ ILC3s, Nr1d1–/– NKp46+ ILC3s secreted less IL-22 (Fig. 6C). Furthermore, we detected more bacterial translocation to the mLNs in Nr1d1–/– mice than in Nr1d1+/+ mice (Fig. 6D). Previous reports have suggested that IL-17A drives inflammatory responses in CDI and implicated increased IL-17A in poorer disease outcomes (38–40). Therefore, hyperresponsive IL-17 secretion by REV-ERBα-deficient ILC3s may contribute to more severe inflammation and bacterial burden.
Figure 6. REV-ERBα deficiency differentially affects cytokine production in ILC3 subsets.
A) IL-17A and IL-22 production in ILC3 subsets in response to in vitro stimulation with IL1β and IL-23 (10 ng/ml). Left: Representative intracellular staining of IL-22 and IL-17A. Right: Frequency and expression levels (gMFI) of IL-22+ and IL-17A+ ILC3s (right). B) IFN-g production in NKp46+ ILC3s in response to in vitro stimulation with a mixture of IL1β, IL-23, IL-12, and IL-18. C) IL-17 and IL-22 secretion by ILC3 subsets on day 2 of CDI. D) qPCR analysis of relative bacterial load translocated to mLNs on day 2 of CDI. rDNA, ribosomal DNA. Statistical analysis was performed using one-way ANOVA with multiple comparisons test (A and B) or Student’s t-test (C and D). Bars indicate means (±SD). * P < 0.05; ** P < 0.01; *** P < 0.001; **** P < 0.0001. Data are representative of two independent experiments (n = 3 to 5 per group per experiment) (A to C) or pooled from two independent experiments (n = 4 or 5 per group per experiment) (D).
Lack of REV-ERBα in CCR6+ and DN ILC3s elicits compensatory mechanisms
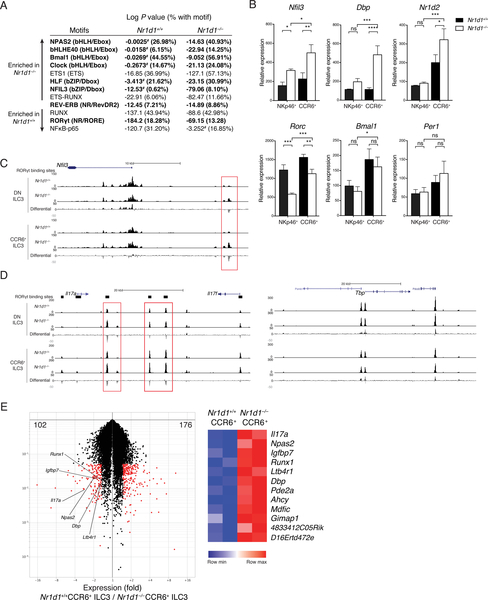
We next asked why ILC3 subsets are differentially affected by REV-ERBα deficiency despite sharing similar levels of expression. Given REV-ERBα’s role in regulating chromatin conformation, we tested whether lack of REV-ERBα affected chromatin accessibility by ATAC-seq (assay for transposase-accessible chromatin using sequencing) of DN and CCR6+ ILC3s from Nr1d1+/+ and Nr1d1–/– mice. After identifying genomic regions that are differentially accessible in Nr1d1+/+ and Nr1d1–/– cells, we analyzed the enrichment of transcription factor motifs in these regions using HOMER to determine the transcriptional programs affected by REV-ERBα deficiency (41). In Nr1d1–/– DN and CCR6+ ILC3s, significantly enriched motifs included bHLH/E-box and bZIP/D-box motifs that are central to circadian transcriptional programs (Fig. 7A, Table S3, S4) (42). In contrast, preferential enrichment of nuclear factor κB (NFκB) and RUNX motifs, as well as an increase in RORE motifs, was evident in Nr1d1+/+ DN and CCR6+ ILC3s (Table S1, S2). The RevDR2 motif (REV-ERB) did not differ dramatically between Nr1d1+/+ and Nr1d1–/– ILC3s. This indicates that in Nr1d1–/– DN and CCR6+ ILC3s, the epigenetic landscape may be more open to the regulation by the other transcription factors of the molecular clock. ATAC-seq showed increased accessibility at the Nfil3 locus and reverse transcription quantitative polymerase chain reaction (RT-qPCR) showed increased expression of Nfil3, Dbp, and Nr1d2 in Nr1d1–/– CCR6+ ILC3 compared with Nr1d1+/+ CCR6+ ILC3 (Fig. 7B, C, Fig. S7A). In contrast, Nr1d1–/– NKp46+ ILC3 failed to significantly upregulate Dbp and Nr1d2 compared to Nr1d1+/+ NKp46+ ILC3. These results suggested that Nr1d1–/– CCR6+ ILC3 have the increased clock gene expression and receptive epigenetic landscape to provide compensation for the loss of REV-ERBα.
Figure 7: REV-ERBa deficiency alters the epigenetics landscape and differentially affects clock gene expression in ILC3 subsets.
A) Transcription factor motifs enriched in regions that are differentially accessible in Nr1d1–/–NKp46– (DN and CCR6+) ILC3s vs. Nr1d1+/+NKp46– ILC3s, log P values for enrichment and frequency of target sequences with motif. Circadian related motifs shown in bold; #, not significant or possible false-positive result. B) Gene expressions of sorted NKp46+ and CCR6+ ILC3s from Nr1d1+/+ and Nr1d1–/– siLP by RT-qPCR, relative expression to Actb. C and D) UCSC Genome Browser view of ATAC-seq tracks of mouse C) Nfil3, D) Il17, and Tbp loci in the indicated ILC3 subsets. Black bars on top represent locations of RORgt binding sites based on published RORgt hromatin immunoprecipitation sequencing in TH17 cells (GEO: GSE56020). The differential tracks are subtraction plots showing differences in peak accessibility between Nr1d1+/+ and Nr1d1–/– ILC3. E) Volcano plot of genes with >1.5-fold differential expression (red) in Nr1d1+/+ vs. Nr1d1–/– CCR6+ ILC3s (left); Heatmap of known RORgt target genes differentially expressed in Nr1d1+/+ and Nr1d1–/– CCR6+ ILC3s. Statistical analysis was performed using one-way ANOVA with multiple comparisons test (B). Bars indicate means (±SD). ns, not significant; * P < 0.05; ** P < 0.01; *** P < 0.001; **** P < 0.0001. Data are representative of at least two independent experiments (n=3 per group per experiment) (B).
Dbp and Nr1d2 compensatory up-regulations are noteworthy. Given that DBP is an antagonistic partner of NFIL3 that recognizes the same D-box motif and induces the transcription of target genes (43) and that NFIL3 inhibits RORγt in Th17 cells (7), increased DBP in Nr1d1–/– CCR6+ ILC3s can counteract the inhibitory effect of NFIL3 to maintain the expression of RORγt. In contrast, in Nr1d1–/– NKp46+ ILC3s, increased NFIL3 without increased DBP likely contributed to the repression of RORγt expression. Moreover, Nr1d1–/– CCR6+ ILC3s also up-regulated the expression of Nr1d2 much more markedly than did Nr1d1–/– NKp46+ ILC3s. Thus, up-regulation of REV-ERBβ may be another compensatory mechanism enabled in CCR6+ but not NKp46+ Nr1d1–/– ILC3s that leads to the dichotomous effects of REV-ERBα deficiency on RORγt expression in the two ILC3 subsets.
REV-ERBα deficiency results in derepression and unimpeded RORγt activity at targets
The compensation by DBP or REV-ERBβ could explain the limited reduction of RORγt expression but not the increased production of IL-17 in Nr1d1–/– DN and CCR6+ ILC3s. Because REV-ERBα regulates chromatin structure and competes against RORγt for RORE binding sites to inhibit gene expression, we hypothesized that loss of REV-ERBα may lead to increased accessibility at the Il17 locus and RORγt may bind regulatory regions of Il17 gene unimpeded in Nr1d1–/– DN and CCR6+ ILC3s. The Il17 locus was consistently more accessible in Nr1d1–/– ILC3s at putative RORγt binding sites across multiple biological replicates (Fig. 7D, Fig. S7B). Meanwhile, the control Tbp locus was not preferentially open (Fig. 7D, Fig S7C). Furthermore, we isolated and compared the transcriptomes of Nr1d1+/+ with Nr1d1–/– CCR6+ ILC3s. Among the genes up-regulated in Nr1d1–/– CCR6+ ILC3s were some known RORγt targets (44), including Il17a, consistent with an amplified RORγt activity at this locus (Fig. 7E). We conclude that the paradoxical increased production of IL-17 by Nr1d1–/– DN and CCR6+ ILC3s depends on the loss of inhibition by REV-ERBα and an augmented RORγt activity.
Discussion
In this study, we report two major findings. We first found limited but detectable circadian fluctuation in homeostatic cytokine secretion by ILC3s. The degree to which ILC3 cytokine fluctuations are controlled by an ILC3 intrinsic molecular clock or driven by extrinsic signals regulated by circadian rhythms such as nutrition-derived signals remains to be established. Regardless, intestinal ILC3 function attuned to feeding behavior, digestion, and intestinal motility may facilitate nutrient absorption, given the impact of ILC3-secreted IL-22 in regulating transport of lipids and lipophilic vitamins in intestinal epithelial cells (45, 46). Acute perturbations of the normal circadian rhythm hindered expression of RORγt and optimal secretion of IL-17 and IL-22 by ILC3s. Thus, disruption of circadian fluctuations may impair the capacity of ILC3s to preserve homeostatic interactions with nutrients and commensal bacteria (47, 48), thereby facilitating malabsorption and dysbiosis. Future studies will specify which intestinal functions are impacted by ILC3 daily fluctuations.
Beyond the role of circadian rhythm in the regulation of ILC3 functions, our second finding is that REV-ERBα is a core ILC3 transcription factor that directly impacts the development of ILC3s independent of circadian regulation. The impact of REV-ERBα on ILC3 development is complex and varies in distinct ILC3 subsets. In NKp46+ ILC3s, lack of REV-ERBα led to increased NFIL3 that reduced RORγt expression. A similar mechanistic sequence was reported in TH17 (7). Conditional Nr1d1 knockout analyses further demonstrated that reduced NKp46+ ILC3s numbers are likely related to a block of DN differentiation into NKp46+ ILC3s. Although trafficking of circulating immune cells has been recently linked to circadian biology (49), the reduction of NKp46+ ILC3 in Nr1d1–/– mice is unlikely to result from a homing defect as they develop from DN ILC3s present in tissue
In contrast to what was observed in NKp46+ ILC3, the increase in NFIL3 expression in REV-ERBα-deficient CCR6+ ILC3s was not paralleled by a marked reduction in RORγt. These seemingly opposing impacts of REV-ERBα deficiency in distinct ILC3 subsets depended on activation of feedback loops involving DBP and REV-ERBβ in CCR6+ ILC3s that compensated, at least in part, for the perturbations due to the REV-ERBα defect. It will be important to validate this concept in the REV-ERBα/β double knockout mice. However, because REV-ERBα is also a competitive antagonist of RORγt, REV-ERBα deficiency in CCR6+ ILC3s did enhance the ability of RORγt to freely bind (unopposed by REV-ERBα) to RORE elements. This mechanism, corroborated by our chromatin analysis of the Il17 locus, resulted in upregulation of IL-17A, similar to what has been described in TH17 cells (8). Overall, our results highlight that since clock genes are involved in complex feedback loops, constitutive lack of any one of them is likely to have distinct developmental defects in each cell type or subset, depending on the expression of other clock genes. In line with this concept, we do not exclude the possibility that REV-ERBα deficiency may also affect the amplitude or phase of circadian gene oscillations in ILC3s as previously observed in other cell types (50, 51).
REV-ERBα has been shown to be involved in many aspects of physiology in multiple tissues, including regulation of lipid metabolism in the liver, energy expenditure in skeletal muscle, and behavior through activities in the central nervous system (35, 52, 53). Our study shows that REV-ERBα impacts the metabolism of an immune cell type by controlling mitochondrial content of NKp46+ ILC3s. Nr1d1–/– NKp46+ ILC3s had significantly reduced mitochondrial content and consequently inadequate mitochondrial energy production. These observations are consistent with studies of REV-ERBα-deficient fibroblasts and skeletal muscle cells in which REV-ERBα was shown to promote mitochondrial biogenesis and suppress mitophagy (35, 36). Previous studies have demonstrated the importance of mitochondrial dynamics in regulating memory T cell fate and function (54). BNIP3- and BNIP3L-mediated mitophagy was also reported to promote NK cell survival and memory formation following viral infection (55). Because ILC3s are long-lived cells like memory T cells, they may similarly depend on mitochondrial oxidative phosphorylation. Loss of mitochondrial content and energy production through oxidative phosphorylation may contribute to reduced development of NKp46+ ILC3s or deviate their metabolism towards glycolysis. Given the role of REV-ERBα as developmental regulator of ILC3s that we report here, and the availability of REV-ERBα agonists and antagonists, REV-ERBα could prove an impactful target for therapeutic interventions aimed at modulating ILC3s in pathology.
Materials and Methods
Study Design
The objective of this study was 1) to characterize the circadian rhythm in ILC3s and 2) to investigate the role of REV-ERBα in the transcriptional programs that regulate ILC3 development and function. We used previously reported mouse models to perform experiments in cellular immunology, metabolism, and bioinformatics. The number of independent experiments and sample sizes are outlined in the figure legends.
Animals
Nr1d1–/– (Nr1d1tm1Ven/J), RorceGFP (B6.129P2(Cg)-Rorctm2Litt/J), B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J, and C57BL/6J mice were obtained from Jackson Laboratory. Nr1d1fl/fl (or Nr1d1-DBDfl/fl) mice (33) were provided by E.S.M. Nr1d2fl/fl mice (56) were provided by L.A.S. Rorc-Cretg mice (16) were provided by A. Tumanov. Ncr1iCre mice (57) were provided by E. Vivier. All mice were backcrossed to the C57BL/6 background. Sex- and age-matched littermates or cohoused males and females were used within each experiment and were randomly assigned to experimental groups. Mice were bred and maintained in specific pathogen-free facilities at Washington University in Saint Louis. For circadian studies, mice were housed in light-controlled cabinets. All studies were conducted in accordance with the Washington University Animal Studies Committee.
Tissue dissociation
Small intestines (either whole length or the proximal 50%) and large intestines were flushed to remove luminal contents, and Peyer’s patches were removed. Intestines were opened lengthwise, gently agitated for 20 min in Hanks’ balanced salt solution (HBSS) containing HEPES, bovine calf serum, and EDTA, and then vortexed. Small intestines were subjected to a second round of gentle agitation and vortexing in EDTA. The tissue was then rinsed with HBSS prior to digestion with collagenase IV (Sigma-Aldrich) in complete RPMI-1640 for 40 min (small intestine) or 60 min (large intestine) at 37°C under agitation. Digests were filtered through 100-μm mesh and subjected to density gradient centrifugation using 40% and 70% Percoll solutions. To generate single cell suspensions of mLNs, tissues were mashed against a 70-μm filter.
Antibodies and flow cytometry
Fluorochrome-conjugated antibodies to mouse CD45, CD3ε, CD4, CD5, CD19, NK1.1, RORγt, Eomes, IL-22, CD45.1, and CD45.2 were purchased from eBioscience. Fluorochrome-conjugated antibodies to mouse CD90.2, GATA3, CD196, IL-17A, T-bet, CD4, CD19, and T cell receptor β (TCRβ) were purchased from BD Biosciences. Fluorochrome- and biotinconjugated antibodies to mouse CD3ε, CD19, CD45.1, KLRG1, NK1.1, CD196, and NKp46 were purchased from BioLegend. Biotin-conjugated antibody to mouse NKp46 was also generated inhouse. For staining, single cell suspensions were incubated with Fc block for 10 minutes and then stained with antibodies and Fc block for 20 min at 4°C. Dead cells were excluded using either the LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (ThermoFisher Scientific) or 7-aminoactinomycin D. Intracellular proteins were stained using either the BD Biosciences Fixation/Permeabilization Solution Kit or the eBioscience Transcription Factor Staining Kit. Cells were run on a FACSCanto II or LSRFortessa (BD Biosciences) and analyzed using FlowJo (FlowJo LLC). Cells counts were conducted with counting beads (eBioscience). ILCs were identified as live CD3ε−CD5−CD19− lymphocyte-sized cells that were either GATA3hiKLRG1+ (ILC2), RORγt+ (ILC3), RORγt−NKp46+NK1.1+ (ILC1), or RORγt− NKp46+NK1.1+Eomes+ (NK). In experiments with intracellular cytokine or mitochondrial staining, ILC3 were identified as CD3ε−CD5−CD19−CD90.2hiCD45int live lymphocytes and then were further subdivided using NKp46 and CCR6. ILC3s were purified by sorting live, lymphocyte-sized cells that were negative for CD3ε, CD5, CD19, TCRβ, KLRG1; positive for CD90.2; and intermediate for CD45.
Mixed bone marrow chimeras
Recipient Nr1d1+/+ CD45.1 mice were irradiated once with 11 Gy. The next day, 3×106 Nr1d1+/+ CD45.1/2 and 3×106 Nr1d1–/– CD45.2 donor bone marrow cells were injected intravenously. Mice were analyzed eight weeks after irradiation.
Mitochondrial staining
Cells were stained with each or combinations of 100 to 200nM MitoTracker Green FM, 200nM MitoTracker Red FM, and 100nM TMRM (Invitrogen) in HBSS buffer for 30 min at 37°C.
Cell stimulations and cytokine analysis
Cells were stimulated ex vivo with no cytokine (unstimulated), IL-23 (1 ng/ml), IL-1β and IL-23 (10 ng/ml), or a mixture [IL-1β (10 ng/ml), IL-23 (10 ng/ml), IL-18 (12.5 ng/ml), and IL-12 (3.2 ng/ml)] for 3.5 hours as indicated. For intracellular cytokine staining, brefeldin A was present for the last 3 hours of stimulation.
Acute SW model
Mice were singly housed, synchronized to standard lighting condition of LD 12:12, and allowed to adjust to its new enclosure. Then, mice were randomly assigned to either remain in this control condition or undergo circadian disruption. In the SW model, mice were sub- jected to 8-hour phase advance every 2 days for 5 days (see Fig. 2A). All mice were studied at ZT0/6:00 a.m. of day 6. Wheel running activity was recorded throughout the experiment. Each mouse was weighed at the beginning and at the end of the experiment. Food consumption was measured after the first phase advance and at the end of the experiment.
Circadian expression study
Because of time requirement of small intestine tissue dissociation, the following procedures were designed to allow for optimal sam- ple collection and measurements within a 24-hour period. For the 24-hour gene expression study, mice were randomly divided into two groups. One group was housed in standard LD 12:12 condition (light on from 6:00 a.m. to 6:00 p.m.). The other group was housed in LD 12:12 condition with a 6-hour delay (light on from 12:00 p.m. to 12:00 a.m.). Both groups were allowed to adjust and synchronize to lighting condition for a minimum of 3 weeks. Then, mice from both groups were euthanized at 6:00 a.m. and 6:00 p.m. on the same day to allow collection of samples at ZT0, ZT6, ZT12, and ZT18 within the same day. ZT0 denotes time of lights on, and ZT12 denotes time of lights off. For study of ILC3 protein expression at ZT1 and ZT13, mice were divided into two groups. One group was housed in standard LD 12:12 condition (light on from 6:00 a.m. to 6:00 p.m.), and the other group was housed in inverted DL 12:12 condition (light on from 6:00 p.m. to 6:00 a.m.). Both groups were allowed to adapt to lighting condition for a minimum of 3 weeks and then euthanized at 7:00 a.m. for sample collection at ZT1 and ZT13.
C. difficile Infection
The C. difficile VPI 10463 strain was cultured in brain heart infusion agar with blood at 37°C in anaerobic conditions (AnaeroGen, Oxoid; ThermoFisher Scientific; Waltham, MA, USA). Infection in mice was performed as previously described (58). Briefly, mice were pre-treated with antibiotics [kanamycin (0.4 mg/ml), gentamicin (0.035 mg/ml), colistin (0.035 mg/ml), metronidazole (0.215 mg/ml), and vancomycin (0.045 mg/ml) from Sigma-Aldrich] in drinking water for 4 days. Twenty-four hours later, mice received a single intraperitoneal dose of clindamycin (10 mg/kg, intraperitoneally) (Sigma-Aldrich). Then after 1 day, mice were orally gavaged with 1 × 108 colony forming units (CFUs) of C. difficile. For bacterial translocation, DNA from mLN was extracted using QIAamp Fast DNA Stool Mini Kit (Qiagen). Microbial load was quantified by measuring Eubacteria 16S ribosomal DNA using qPCR. Results were calculated using a standard curve of serial diluted E. coli genomic DNA and normalized to tissue mass.
Microarray analysis
RNA was extracted from sorted siLP ILC3s with an RNeasy Plus Micro Kit (QIAGEN). RNA amplification and hybridization to Affymetrix Mouse Gene 2.0 ST array was conducted by the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine in Saint Louis. Data were analyzed using Multiplot by GenePattern.
Assay for transposase-accessible chromatin using sequencing
Transposase reactions and library preparation for ATAC-seq was preformed as previously described (59), according to manufacturer’s instructions (Illumina), and sequenced using Illuminia HiSeq 3000 2×150. Libraries were processed using NovaAlign (alignment), DeepTool’s BamCoverage [reads per kilobase of transcripts per million (RPKM) normalization] and the UCSC (University of California, Santa Cruz) Genome Browser (visualization) (60). The UCSC Genome Browser was used to generate differential accessibility track collections (61). TH17 RORγt peaks were generated from previously published data [Gene Expression Omnibus (GEO): GSE56020] (44).
Transcription factor motif analysis
Differentially accessible genomic regions between Nr1d1+/+ and Nr1d1–/– ILC3s from ATAC-seq were defined using DESeq2 binomial enrichment as peaks with >1.5 fold change in peak intensity and p value <0.05 and then extracted for analysis by HOMER motif pipeline with default settings and size parameter set to peak size (41, 62).
Reverse transcription quantitative polymerase chain reaction
ILC3s were purified by fluorescence-activated cell sorting, lysed, and frozen in RLT buffer (Qiagen). RNA was extracted using the RNeasy Plus Micro Kit (Qiagen). Complementary DNA (cDNA) was synthesized with qScript cDNA Synthesis Kit (Quantabio). cDNA expression was analyzed by qPCR using iTaq Universal SYBR Green Supermix (Bio-Rad Laboratories) on the StepOnePlus system (Applied Biosystems). The expression of target genes was calculated and normalized to the expression of the control gene Actb.
Statistical analysis
Data were analyzed with either Sudent’s t tests or one-way analysis of variance (ANOVA) as indicated in figure legends using GraphPad Prism 7. Circadian periodicity was evaluated in R using MetaCycle as previously described (28). Bars indicate mean (±SD). ns, not significant; * P < 0.05; ** P < 0.01; *** P < 0.001; **** P < 0.0001.
Supplementary Material
Fig. S1. Protein levels of cytokine and transcription factors in ILC3s at different points of the circadian cycle.
Fig. S2. Acute SW model disrupts circadian rhythm without altering food intake or body weight.
Fig. S3. REV-ERBα deficiency results in reduced NKp46+ ILC3 in multiple tissues
Fig. S4. REV-ERBβ deficiency alone does not affect ILC3s.
Fig. S5. Acute circadian disruption does not affect ILC3 mitochondria.
Fig. S6. Reduction of NKp46+ ILC3 in Nr1d1–/– mice is not due to increase in plasticity.
Fig. S7. Accessibility of Nfil3, Il17, and Tbp loci in Nr1d1+/+ and Nr1d1–/– ILC3s.
Table S1. Motif enrichment of known motifs by HOMER in Nr1d1+/+ NKp46− ILC3s.
Table S2. De novo motif enrichment by HOMER in Nr1d1+/+ NKp46− ILC3s.
Table S3. Motif enrichment of known motifs by HOMER in Nr1d1–/– NKp46− ILC3s.
Table S4. De novo motif enrichment by HOMER in Nr1d1–/– NKp46− ILC3s.
Table S5. Sequences of primers used for qPCR.
Acknowledgments
We thank Eric Vivier for the Ncr1iCre mice; P. Murray for Nfil3 plasmids; M. Cella, W. Song, and V. Cortez for helpful discussion; P. Sheehan for help with MetaCycle analysis; and T. Ulland and H. Miller for help with bone marrow chimera experiments. REV-ERBβ floxed mice (Nr1d2fl/fl) are available from L.A.S. under a material transfer agreement with The Scripps Research Institute. The Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine assisted with microarray and ATAC-seq experiments. All flow cytometry work was conducted in the Flow Cytometry and Fluorescence Activated Cell Sorting Core in the Department of Pathology and Immunology at Washington University School of Medicine.
Funding
This study was supported by NIH grants AI095542, DE025884, and AI134236 (to M.C.), AI134035 (to M.C. and E.M.O.), MH092769 (to T.P.B.), K99 DK118110 (to J.K.B.), and T32 GM007200 (to Q.W.). J.L.F. was supported by FAPESP (2018/10165–0). M.C. receives research support from Pfizer, Crohn’s & Colitis Foundation, and anonymous donors, NY.
Footnotes
References
- 1.Guillaumond F, Dardente H, Giguere V, Cermakian N, Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J. Biol. Rhythms. 20, 391–403 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Curtis AM, Bellet MM, Sassone-Corsi P, O’Neill LAJ, Circadian Clock Proteins and Immunity. Immunity. 40, 178–186 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Takahashi JS, Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 18, 164–179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bechtold DA, Loudon ASI, Hypothalamic clocks and rhythms in feeding behaviour. Trends Neurosci. 36, 74–82 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Tahara Y, Shibata S, Chronobiology and nutrition. Neuroscience. 253, 78–88 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Asher G, Sassone-Corsi P, Time for food: The intimate interplay between nutrition, metabolism, and the circadian clock. Cell. 161, 84–92 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Yu X, Rollins D, Ruhn KA, Stubblefield JJ, Green CB, Kashiwada M, Rothman PB, Takahashi JS, Hooper LV, TH17 cell differentiation is regulated by the circadian clock. Science (80-. ). 342, 727–730 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amir M, Chaudhari S, Wang R, Campbell S, Mosure SA, Chopp LB, Lu Q, Shang J, Pelletier OB, He Y, Doebelin C, Cameron MD, Kojetin DJ, Kamenecka TM, Solt LA, REV-ERBα Regulates T H 17 Cell Development and Autoimmunity. Cell Rep. 25, 3733–3749.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hemmers S, Rudensky AY, The Cell-Intrinsic Circadian Clock Is Dispensable for Lymphocyte Differentiation and Function. Cell Rep. 11, 1339–1349 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vivier E, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie ANJ, Mebius RE, Powrie F, Spits H, Innate Lymphoid Cells: 10 Years On. Cell. 174, 1054–1066 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Robinette ML, Colonna M, Immune modules shared by innate lymphoid cells and T cells. J. Allergy Clin. Immunol. 138, 1243–1251 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sciumè G, Shih HY, Mikami Y, O’Shea JJ, Epigenomic views of innate lymphoid cells. Front. Immunol. 8 (2017), doi: 10.3389/fimmu.2017.01579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsuji M, Suzuki K, Kitamura H, Maruya M, Kinoshita K, Ivanov II, Itoh K, Littman DR, Fagarasan S, Requirement for Lymphoid Tissue-Inducer Cells in Isolated Follicle Formation and T Cell-Independent Immunoglobulin A Generation in the Gut. Immunity. 29, 261–271 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Mebius RE, Rennert P, Weissman IL, Developing Lymph Nodes Collect CD4+CD3− LTβ+ Cells That Can Differentiate to APC, NK Cells, and Follicular Cells but Not T or B Cells. Immunity. 7, 493–504 (1997). [DOI] [PubMed] [Google Scholar]
- 15.Sun Z, Unutmaz D, Zou Y-R, Sunshine MJ, Pierani A, Brenner-Morton S, Mebius RE, Littman DR, Science (80-. )., in press, doi: 10.1126/science.288.5475.2369. [DOI] [PubMed] [Google Scholar]
- 16.Eberl G, Thymic Origin of Intestinal T Cells Revealed by Fate Mapping of RORγt+ Cells. Science (80-. ). 305, 248–251 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Klose CSN, a Kiss E, Schwierzeck V, Ebert K, Hoyler T, D’Hargues Y, Göppert N, Croxford AL, Waisman A, Tanriver Y, Diefenbach A, A T-bet gradient controls the fate and function of CCR6-RORγt+ innate lymphoid cells. Nature. 494, 261–265 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Rankin LC, Groom JR, Chopin M, Herold MJ, a Walker J, a Mielke L, McKenzie ANJ, Carotta S, Nutt SL, Belz GT, The transcription factor T-bet is essential for the development of NKp46+ innate lymphocytes via the Notch pathway. Nat. Immunol. 14, 389–395 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Viant C, Rankin LC, Girard-Madoux MJH, Seillet C, Shi W, Smyth MJ, Bartholin L, Walzer T, Huntington ND, Vivier E, Belz GT, Transforming growth factor-β and Notch ligands act as opposing environmental cues in regulating the plasticity of type 3 innate lymphoid cells. Sci. Signal. 9 (2016), doi: 10.1126/scisignal.aaf2176. [DOI] [PubMed] [Google Scholar]
- 20.Robinette ML, Fuchs A, Cortez VS, Lee JS, Wang Y, Durum SK, Gilfillan S, Colonna M, Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat. Immunol. 16, 306–317 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koues OI, Collins PL, Cella M, Robinette ML, Porter SI, Pyfrom SC, Payton JE, Colonna M, Oltz EM, Distinct Gene Regulatory Pathways for Human Innate versus Adaptive Lymphoid Cells. Cell. 165, 1134–1146 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB, Burris LL, Khorasanizadeh S, Burris TP, Rastinejad F, Identification of heme as the ligand for the orphan nuclear receptors REV-ERBα and REV-ERBβ. Nat. Struct. Mol. Biol. 14, 1207–1213 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin L, Wu N, Lazar MA, Nuclear receptor Rev-erbalpha: a heme receptor that coordinates circadian rhythm and metabolism. Nucl. Recept. Signal. 8, e001 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harding HP, Lazar MA, The orphan receptor Rev-ErbA alpha activates transcription via a novel response element. Mol. Cell. Biol. 13, 3113–21 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forman BM, Chen J, Blumberg B, Kliewer SA, Henshaw R, Ong ES, Evans RM, Cross-talk among ROR alpha 1 and the Rev-erb family of orphan nuclear receptors. Mol. Endocrinol. 8, 1253–1261 (1994). [DOI] [PubMed] [Google Scholar]
- 26.Harding HP, Lazar MA, The monomer-binding orphan receptor Rev-Erb represses transcription as a dimer on a novel direct repeat. Mol. Cell. Biol. 15, 4791–4802 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hughes ME, Hogenesch JB, Kornacker K, JTK-CYCLE: An efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J. Biol. Rhythms. 25, 372–380 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu G, Anafi RC, Hughes ME, Kornacker K, Hogenesch JB, MetaCycle: An integrated R package to evaluate periodicity in large scale data. Bioinformatics. 32, 3351–3353 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hadden H, Soldin SJ, Massaro D, Circadian disruption alters mouse lung clock gene expression and lung mechanics. J. Appl. Physiol. 113, 385–392 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim C, Allada R, Emerging roles for post-transcriptional regulation in circadian clocks. Nat. Neurosci. 16, 1544–1550 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Gallego M, Virshup DM, Post-translational modifications regulate the ticking of the circadian clock. Nat. Rev. Mol. Cell Biol. 8, 139–148 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Chomez P, Neveu I, Mansén A, Kiesler E, Larsson L, Vennström B, Arenas E, Increased cell death and delayed development in the cerebellum of mice lacking the rev-erbA(alpha) orphan receptor. Development. 127, 1489–98 (2000). [DOI] [PubMed] [Google Scholar]
- 33.Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong L, DiTacchio L, Atkins AR, Glass CK, Liddle C, Auwerx J, Downes M, Panda S, Evans RM, Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature. 485, 123–127 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Fang B, Emmett MJ, Damle M, Sun Z, Feng D, Armour SM, Remsberg JR, Jager J, Soccio RE, Steger DJ, Lazar MA, Discrete functions of nuclear receptor Rev-erb couple metabolism to the clock. Science (80-. ). 348, 1488–1492 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woldt E, Sebti Y, Solt LA, Duhem C, Lancel S, Eeckhoute J, Hesselink MKC, Paquet C, Delhaye S, Shin Y, Kamenecka TM, Schaart G, Lefebvre P, Nevière R, Burris TP, Schrauwen P, Staels B, Duez H, Rev-erb-α modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nat. Med. 19, 1039–1046 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sengupta S, Yang G, O’Donnell JC, Hinson MD, McCormack SE, Falk MJ, La P, Robinson MB, Williams ML, Yohannes MT, Polyak E, Nakamaru-Ogiso E, Dennery PA, The circadian gene Rev-erbα improves cellular bioenergetics and provides preconditioning for protection against oxidative stress. Free Radic. Biol. Med. 93, 177–189 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vonarbourg C, Mortha A, Bui VL, Hernandez PP, Kiss EA, Hoyler T, Flach M, Bengsch B, Thimme R, Hölscher C, Hönig M, Pannicke U, Schwarz K, Ware CF, Finke D, Diefenbach A, Regulated expression of nuclear receptor RORγt confers distinct functional fates to NK cell receptor-expressing RORγt+ innate lymphocytes. Immunity. 33, 736–751 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu H, Chen K, Sun Y, Carter M, Garey KW, Savidge TC, Devaraj S, Tessier ME, von Rosenvinge EC, Kelly CP, Pasetti MF, Feng H, Cytokines Are Markers of the Clostridium difficile-Induced Inflammatory Response and Predict Disease Severity. Clin. Vaccine Immunol. 24, 1–11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saleh MM, Frisbee AL, Leslie JL, Buonomo EL, Cowardin CA, Ma JZ, Simpson ME, Scully KW, Abhyankar MM, Petri WA, Colitis-Induced Th17 Cells Increase the Risk for Severe Subsequent Clostridium difficile Infection. Cell Host Microbe. 25, 756–765.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakagawa T, Mori N, Kajiwara C, Kimura S, Akasaka Y, Ishii Y, Saji T, Tateda K, Endogenous il-17 as a factor determining the severity of Clostridium difficile infection in mice. J. Med. Microbiol. 65, 821–827 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK, Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol. Cell. 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fang B, Everett LJ, Jager J, Briggs E, Armour SM, Feng D, Roy A, Gerhart-Hines Z, Sun Z, Lazar MA, Circadian enhancers coordinate multiple phases of rhythmic gene transcription in vivo. Cell. 159, 1140–1152 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mitsui S, Yamaguchi S, Matsuo T, Ishida Y, Okamura H, Antagonistic role of E4BP4 and PAR proteins in the circadian oscillatory mechanism. Genes Dev. 15, 995–1006 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiao S, Yosef N, Yang J, Wang Y, Zhou L, Zhu C, Wu C, Baloglu E, Schmidt D, Ramesh R, Lobera M, Sundrud MS, Tsai PY, Xiang Z, Wang J, Xu Y, Lin X, Kretschmer K, Rahl PB, Young RA, Zhong Z, Hafler D, Regev A, Ghosh S, Marson A, Kuchroo VK, Small-molecule RORγt antagonists inhibit T helper 17 cell transcriptional network by divergent mechanisms. Immunity. 40, 477–489 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mao K, Baptista AP, Tamoutounour S, Zhuang L, Bouladoux N, Martins AJ, Huang Y, Gerner MY, Belkaid Y, Germain RN, Innate and adaptive lymphocytes sequentially shape the gut microbiota and lipid metabolism. Nature. 554, 255–259 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Spencer SP, Belkaid Y, Dietary and commensal derived nutrients: Shaping mucosal and systemic immunity. Curr. Opin. Immunol. 24, 379–384 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pickard JM, Maurice CF, Kinnebrew MA, Abt MC, Schenten D, V Golovkina T, Bogatyrev SR, Ismagilov RF, Pamer EG, Turnbaugh PJ, V Chervonsky A, Rapid fucosylation of intestinal epithelium sustains host–commensal symbiosis in sickness. Nature. 514, 638 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Belkaid Y, Harrison OJ, Homeostatic Immunity and the Microbiota. Immunity. 46, 562–576 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He W, Holtkamp S, Hergenhan SM, Kraus K, de Juan A, Weber J, Bradfield P, Grenier JMP, Pelletier J, Druzd D, Chen C-S, Ince LM, Bierschenk S, Pick R, Sperandio M, Aurrand-Lions M, Scheiermann C, Circadian Expression of Migratory Factors Establishes Lineage-Specific Signatures that Guide the Homing of Leukocyte Subsets to Tissues. Immunity, 1–16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Preitner N, Damiola F, Luis-Lopez-Molina J Zakany, D. Duboule, U. Albrecht, U. Schibler, The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 110, 251–260 (2002). [DOI] [PubMed] [Google Scholar]
- 51.Duez H, van der Veen JN, Duhem C, Pourcet B, Touvier T, Fontaine C, Derudas B, Baugé E, Havinga R, Bloks VW, Wolters H, van der Sluijs FH, Vennström B, Kuipers F, Staels B, Regulation of Bile Acid Synthesis by the Nuclear Receptor Rev-erbα. Gastroenterology. 135, 689–698 (2008). [DOI] [PubMed] [Google Scholar]
- 52.Kim K, Lee EJ, Son GH, Hwang O, Dluzen DE, Park S-B, Yun S, Son HJ, Lee I, Chung S, Kim K-S, Choe HK, Impact of Circadian Nuclear Receptor REV-ERBα on Midbrain Dopamine Production and Mood Regulation. Cell. 157, 858–868 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, Liu XS, A Lazar M, A Circadian Rhythm Orchestrated by Histone Deacetylase 3 Controls Hepatic Lipid Metabolism. Science (80-. ). 331, 1315–1319 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Buck MDD, O’Sullivan D, Klein Geltink RII, Curtis JDD, Chang CH, Sanin DEE, Qiu J, Kretz O, Braas D, van der Windt GJJW, Chen Q, Huang SCC, O’Neill CMM, Edelson BTT, Pearce EJJ, Sesaki H, Huber TBB, Rambold ASS, Pearce ELL, Mitochondrial Dynamics Controls T Cell Fate through Metabolic Programming. Cell. 166, 63–76 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O’Sullivan TE, Johnson LR, Kang HH, Sun JC, BNIP3- and BNIP3L-Mediated Mitophagy Promotes the Generation of Natural Killer Cell Memory. Immunity. 43, 331–342 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Banerjee S, Wang Y, Solt LA, Griffett K, Kazantzis M, Amador A, El-Gendy BM, Huitron-Resendiz S, Roberts AJ, Shin Y, Kamenecka TM, Burris TP, Pharmacological targeting of the mammalian clock regulates sleep architecture and emotional behaviour. Nat. Commun. 5, 1–13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Narni-Mancinelli E, Chaix J, Fenis A, Kerdiles YM, Yessaad N, Reynders A, Gregoire C, Luche H, Ugolini S, Tomasello E, Walzer T, Vivier E, Fate mapping analysis of lymphoid cells expressing the NKp46 cell surface receptor. Proc. Natl. Acad. Sci. 108, 18324–18329 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen X, Katchar K, Goldsmith JD, Nanthakumar N, Cheknis A, Gerding DN, Kelly CP, A Mouse Model of Clostridium difficile-Associated Disease. Gastroenterology. 135, 1984–1992 (2008). [DOI] [PubMed] [Google Scholar]
- 59.Collins PL, Cella M, Porter SI, Li S, Gurewitz GL, Hong HS, Johnson RP, Oltz EM, Colonna M, Gene Regulatory Programs Conferring Phenotypic Identities to Human NK Cells. Cell. 176, 348–360.e12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T, deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haeussler M, Zweig AS, Tyner C, Speir ML, Rosenbloom KR, Raney BJ, Lee CM, Lee BT, Hinrichs AS, Gonzalez JN, Gibson D, Diekhans M, Clawson H, Casper J, Barber GP, Haussler D, Kuhn RM, Kent WJ, The UCSC Genome Browser database: 2019 update. Nucleic Acids Res. 47, D853–D858 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Protein levels of cytokine and transcription factors in ILC3s at different points of the circadian cycle.
Fig. S2. Acute SW model disrupts circadian rhythm without altering food intake or body weight.
Fig. S3. REV-ERBα deficiency results in reduced NKp46+ ILC3 in multiple tissues
Fig. S4. REV-ERBβ deficiency alone does not affect ILC3s.
Fig. S5. Acute circadian disruption does not affect ILC3 mitochondria.
Fig. S6. Reduction of NKp46+ ILC3 in Nr1d1–/– mice is not due to increase in plasticity.
Fig. S7. Accessibility of Nfil3, Il17, and Tbp loci in Nr1d1+/+ and Nr1d1–/– ILC3s.
Table S1. Motif enrichment of known motifs by HOMER in Nr1d1+/+ NKp46− ILC3s.
Table S2. De novo motif enrichment by HOMER in Nr1d1+/+ NKp46− ILC3s.
Table S3. Motif enrichment of known motifs by HOMER in Nr1d1–/– NKp46− ILC3s.
Table S4. De novo motif enrichment by HOMER in Nr1d1–/– NKp46− ILC3s.
Table S5. Sequences of primers used for qPCR.