Abstract
Purpose
The burden of esophageal cancer (EC) continues to rise, and non-invasive screening tools are needed. Methylated DNA markers (MDMs) assayed from plasma show promise in detection of other cancers. For EC detection, we aimed to discover and validate MDMs in tissue, and determine their feasibility when assayed from plasma.
Experimental Design
Whole-methylome sequencing was performed on DNA extracted from 37 tissues (28 EC; 9 normal esophagus) and 8 buffy coat samples. Top MDMs were validated by methylation specific PCR on tissue from 76 EC (41 adeno, 35 squamous cell) and 17 normal esophagus. Quantitative allele-specific real-time target and signal amplification was used to assay MDMs in plasma from 183 patients (85 EC, 98 controls). Recursive partitioning (rPART) identified MDM combinations predictive of EC. Validation was performed in silico by bootstrapping.
Results
From discovery, 23 candidate MDMs were selected for independent tissue validation; median area under the receiver operating curve (AUC) for individual MDMs was 0.93. Among 12 MDMs advanced to plasma testing, rPART modeling selected a 5 MDM panel (FER1L4, ZNF671, ST8SIA1, TBX15, ARHGEF4) which achieved an AUC of 0.93 (95% CI: 0.89–0.96) on best-fit and 0.81 (95% CI: 0.75–0.88) on cross-validation. At 91% specificity, the panel detected 74% of EC overall, and 43, 64, 77, and 92% of stages I, II, III, and IV, respectively. Discrimination was not affected by age, sex, smoking, or body mass index.
Conclusion
Novel MDMs assayed from plasma detect EC with moderate accuracy. Further optimization and clinical testing are warranted.
Keywords: DNA methylation, esophageal Neoplasms/prevention and control, biomarker, epigenomics, sequence analysis, DNA
INTRODUCTION
Esophageal cancer (EC) ranks as the 6th most common cause of cancer death worldwide, with 456,000 new cases and 400,000 deaths each year(1). Two main subtypes of EC, esophageal adenocarcinoma (EAC) and esophageal squamous cell carcinoma (ESCC), are both increasing in incidence in different parts of the world (2).
Unfortunately, patients often present when symptomatic with advanced stage disease (3). Consequently, 5-year survival rates are less than 20% in developed countries and less than 5% in many developing countries (1,3–5). Early stage disease, however, is associated with substantially improved survival of 80–90% at 5 years (6,7). Therefore, pre-symptomatic early detection of EC could change outcomes with this deadly disease (8).
Screening for EC has yet to be implemented for the general population. Early detection by endoscopy may have a role in geographic regions with high prevalence, but is difficult to administer in most countries due to its invasiveness and high cost (9). Blood testing offers the appeal of noninvasiveness and ready distribution, but sufficiently accurate markers have yet to be developed.
Blood-based biomarkers for EC have been investigated in small studies; these include circulating tumor cells, autoantibodies, microRNA, and aberrantly methylated DNA (10–14). Acquired aberrant covalent DNA methylation is an epigenetic modification, which promotes oncogenesis by altered gene expression. Hypermethylation of CpG islands in promoter regions can silence tumor suppressor genes, while hypomethylation of repetitive gene elements may lead to genomic instability and activation of oncogenes (15). DNA methylation events appear to be less heterogeneous than mutations; for example, studies in primary tumor tissues show that as few as 4 methylated DNA markers (MDMs) achieve nearly perfect discrimination between GI neoplasms and controls (16–22). Furthermore, technological advances in assay chemistry have dramatically improved the analytical sensitivity for MDMs in plasma to allow detection of low copy numbers associated with early stage cancer, notably of the liver and stomach (19,23).
Our group and others have systematically applied next-generation sequencing techniques to expand the list of candidate MDMs of gastro-intestinal cancers and demonstrated feasibility for plasma testing in several phase I and II clinical studies (19,23). Additionally, novel assay chemistry, specifically quantitative allele-specific real-time target and signal amplification assay (QuARTS), achieves very high analytical sensitivity for circulating cell-free DNA, but has not yet been applied to EC (20,23).
We hypothesized that: 1) next-generation DNA sequencing would identify novel and highly discriminant MDMs of EC (EAC and ESCC); 2) MDMs would be confirmed in independent tissues; and, 3) MDMs would show clinical feasibility for detection of EC when assayed from plasma-extracted DNA.
METHODS
Overview
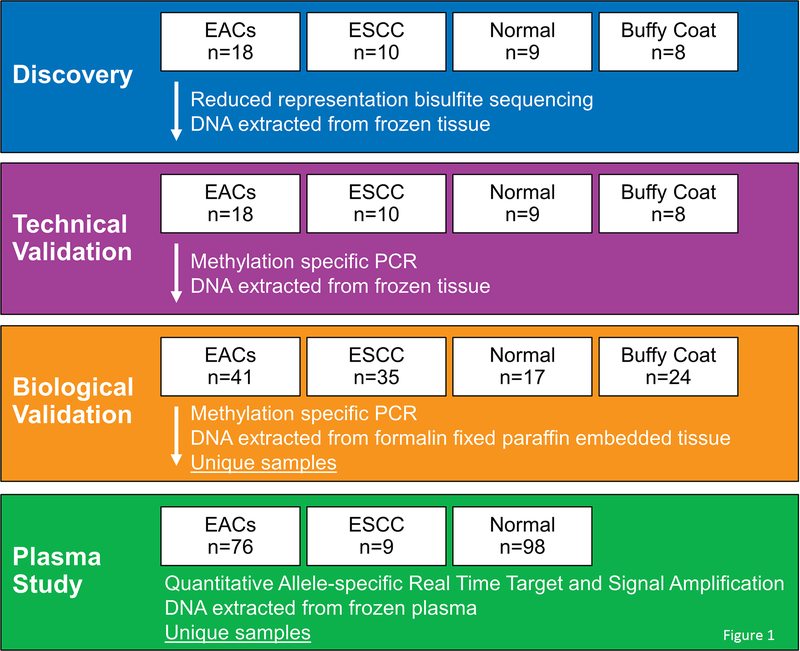
The investigative design comprised four sequential case-control studies (Figure 1). First, we performed unbiased whole methylome sequencing by reduced representation bisulfite sequencing (RRBS) on DNA extracted from frozen primary tumor and control tissues to identify novel differentially methylated regions (DMRs) of the genome, followed by technical validation using quantitative methylation specific PCR (qMSP) on the same frozen samples. Technically validated DMRs were then biologically validated on independent formalin-fixed paraffin-embedded (FFPE) tissue samples. DMRs showing highest discrimination by a priori criteria were carried forward as candidate MDMs for plasma testing. Finally, QuARTS was used to assay DNA extracted from archival plasma samples from EC case and control patients. For each sample, QuARTS products were normalized by plasma volume and β-actin (ACTB).
Figure 1.
Study flow diagram.
EAC, esophageal adenocarcinoma; ESCC, esophageal squamous cell carcinoma; PCR, polymerase chain reaction; QuARTS, quantitative allele-specific real-time target and signal amplification.
To distinguish differentially methylated regions of genomic DNA extracted from tissues (DMRs) from circulating fragments of methylated DNA used as tumor markers, we refer to the latter as methylated DNA markers (MDMs). All laboratory testing was performed by personnel blinded to clinical data. For the phase I clinical study, all clinical data were reviewed by a single clinician (YQ) prior to un-blinding of MDM assay results by the lead statistician (DWM).
Patients and samples
All study procedures were conducted after approval from the Mayo Clinic Institutional Review Board. Studies were conducted in accordance with ethical principles from the Declaration of Helsinki. Tissues used in RRBS & technical validation steps were obtained from the Esophageal Adenocarcinoma and Barrett’s Esophagus registry (EABE), which enrolled patients with EAC, ESCC, and healthy controls, under informed consent between December 2010 and September 2015. Esophageal tumor tissue was sampled in treatment-naive patients prior to endoscopic therapy, surgery, radiation, or chemotherapy. Healthy control tissues were from individuals free from any prior esophageal neoplasm or Barrett’s esophagus. De-identified buffy coat samples were also sequenced to control for and then exclude regions of leukocyte DNA with elevated methylation, due to their potential to cause high background in plasma (24). FFPE tissues used for biological validation were obtained from the Mayo Clinic Tissue Registry, an archive of clinical tissue specimens maintained by the Mayo Clinic Department of Anatomic Pathology. All frozen and FFPE tissues underwent research histopathology review by an expert gastrointestinal pathologist (TCS) prior to macro-dissection and DNA extraction.
For the plasma study, frozen archives of the EABE Registry provided EDTA-buffered plasma samples from EC patients. A second archive of patients free from cancer was enrolled from a 7-county regional population between December 2015 and July 2017. From these, we selected plasma samples from age- and sex-matched control patients.
All plasma samples were collected in EDTA buffer and processed by the Mayo Clinic Biospecimens Accession and Processing laboratory. Referent to the time of plasma collection, cases were required to be free from exposure to local/regional therapy or systemic chemotherapy, and free from any primary cancer outside of the esophagus, excluding non-melanoma skin cancers; controls were required to be free from any cancer within 5-years of sample collection. Staging referent to the time of plasma sample collection was determined using the 8th edition of the American Joint Committee on Cancer (AJCC) classifications (25).
Laboratory methods and assays
Discovery & technical validation
DNA was isolated from macro-dissected frozen tissue sections using the DNeasy 96 blood and tissue kit (Qiagen, Hilden Germany). DNA from buffy coat samples was purified using the QIAamp DNA Mini kit (Qiagen). DNA yield was measured fluorometrically on a plate reader with the Quant-iT Pico-Green dsDNA assay reagents (Invitrogen, Carlsbad CA). Samples were bisulfite-converted using the Zymo EZ-96 DNA Methylation kit (Zymo Research, Irvine, CA) and amplified with SYBR Green I detection using the LightCycler 480 (Roche Diagnostics, Indianapolis IN). RRBS was performed as previously described (19).
Sequencing results were processed by Illumina (San Diego CA) pipeline modules for image analysis and base calling. Secondary analysis was performed using SAAP-RRBS (26). Briefly, reads were cleaned-up using Trim-Galore (Babraham Institute, Cambridge UK) and aligned to the GRCh37/hg19 reference genome build with whole genome bisulfite sequence Mapping program (BSMAP) (27). Methylation ratios were determined by calculating C/(C+T) or conversely, G/(G+A) for reads mapping to the reverse strand, for CpGs with coverage ≥ 10X and base quality score ≥ 20.
For technical validation, qMSP primers for candidate DMRs were designed with Methprimer software (University of California, San Francisco, CA) as previously described (26). Oligonucleotides were synthesized by IDT (Coralville, IA). Prior to use, qMSP assays were quality-control tested on bisulfite converted and unconverted methylation (+/−) controls to verify amplification. Assay standards were dilutions of bisulfite converted and SssI treated genomic DNA, which ensured absolute quantification. Results were expressed as fractional methylation using the bisulfite-treated PCR product of ACTB as a marker for total DNA in each sample, and analyzed logistically.
Biological validation in tissue
The same qMSP assays then were run on DNA extracted from independent FFPE case and control tissues. DNA was purified using the QIAamp FFPE Tissue Kit (Qiagen), bisulfite treated with the Zymo kit, and amplified with SYBR Green I detection using the LightCycler 480 as above.
Plasma study
DNA from independent EC patients and cancer-free controls was purified and bisulfite converted from 3–4mL of plasma using a proprietary semi-automated silica bead DNA extraction method developed to maximize analyte recovery and sample throughput (20). The eluted DNA was subsequently treated with ammonium bisulfite and re-purified to remove polymerase inhibitors. The DMR sequences for 12 MDM candidates were used to construct modified QuARTS triplex assays, as previously described (21). This technology platform was selected for its high analytical sensitivity, which is at least 10-fold higher than that of conventional qMSP. Triplexes were assayed on the LightCycler 480 (Roche) and all results were normalized by plasma volume and by ACTB amplified from each sample.
Statistical Analysis
Discovery
To identify candidate DMRs from the RRBS data set, the percentage of methylation was compared between cases, tissue controls, and buffy coat controls at each mapped CpG. For this comparison, a tiled reading frame within 100 base pairs of each CpG was used to identify DMRs of interest, specifically areas where methylation in controls was <5%. Regions were analyzed only if the variance of methylation percentage across subgroups was >0 and the total depth of coverage per sample group was ≥200 reads (10 reads per subject on average). The sample size requirements were estimated as previously reported (19). In brief, the highest background for methylation in controls was assumed to be 5%, and a >3x increase in the odds ratio was considered biologically relevant. At a coverage depth of 10 reads, ≥18 samples per group were required to achieve 80% power assuming binomial variance inflation factor of 1, with a two-sided test at a significance level of 5%. To account for loss of power in case of greater variance inflation, we further restricted sites with a minimum read depth of 20. Following variance-inflated logistic regression, candidate DMRs were ranked according to p-value, area under the receiver operating characteristics curve (AUC), methylation fold-change ratio (FC), and absolute methylation difference between cases and controls (ΔM).
Biological validation
MDM candidates were first selected from those with AUCs and FC values within the top 40% of those that advanced through technical validation. In addition, MDMs that detected additional tumor samples not represented by the top 40%, or showing high discrimination for other GI cancers, were also included (16–20,23).
For the confirmation of each MDM in independent FFPE samples, those with a lower 95% CI bound for the estimated AUC exceeding 0.7 were deemed to have acceptable accuracy. Assuming an estimated AUC of 0.8, to achieve this bound of acceptance at a one-sided significance level of 0.05, a minimum of 35 cases per cancer subtype were required with a case: control mix of 2:1.
Plasma study
Marker selection for the plasma study was limited to 12 candidates which were representative of both EAC and ESCC, had the lowest amplification of leukocyte DNA, and could be assayed from the shortest (<100bp) amplicon size. Recursive partitioning (rPart) was used to develop a prediction model for cancer based on the QuARTS assay products of individual MDM and combinations of multiple MDMs. AUCs for individual MDMs and the predicted probability of cancer from the fitted rPart tree were used to assess accuracy. It was estimated that a minimum of 97 patients in the case group and 97 in the control group would provide 80% power to distinguish an estimated AUC of ≥0.8 from a null value of 0.7 with a two-sided test of significance set at the 5% level.
In silico cross-validation was performed by generating 500 boot-strapped subsets (with a training: testing ratio of 2:1) of the original data. The reproducibility of the entire panel of MDMs was estimated by averaging sensitivity, specificity, and AUC across the 500 simulated datasets. The potential influence of clinical subtypes on prediction accuracy was assessed by comparing stratified AUCs. All tests were performed at the 5% level of significance.
To measure the biological relevance of MDMs brought into the plasma study, expectation maximization values of RNA sequencing data were obtained from publically available data sets of EAC primary tumor samples (The Cancer Genome Atlas, TCGA) and normal esophagus tissues (Genotype Tissue Expression Project) using the XENA browser and the DESeq2 analysis (28,29).
In order to evaluate the specificity of the MDMs for esophageal cancer in comparison to other common cancers, we downloaded DNA methylation data from TCGA from Genomic Data Commons (GDC) portal (https://portal.gdc.cancer.gov/), June 2018 (46–50), and extracted data for cancers from the esophagus, colon/rectum, liver, and lung. Methylation data in TCGA was derived from tissue based studies using Infinium Human Methylation 450K BeadChip containing 450 CpG sites. EAC and ESCC were evaluated both separately and combined. Because of the platform differences, we mapped the CpG sites from the 450k array to the genomic coordinates of our selected MDMs, and those with at least one CpG site mapped were kept for evaluation (average methylation was obtained if more than one CpG site were mapped in a region). Tumor tissue was used as site-specific cases, and tumor-adjacent normal tissue was used as site-specific controls. Methylation was measured as an average beta value, which is equivalent to methylation ratio from bisulfite sequencing. Box plots of case versus control were generated for each cancer type. One way ANOVA along with TukeyHSD was used to determine overall and pair-wise methylation differences among different cancer types for each MDM; mean methylation difference and adjusted p-values for multiple testing were obtained accordingly.
RESULTS
Discovery & technical validation
DNA from frozen esophageal tissue was available from 28 EC cases (18 EAC, 10 ESCC) and 17 normal controls (9 normal esophagus without synchronous Barrett’s esophagus or EC and 8 normal buffy coat samples from patients without history of cancer)(Figure 1). Sequencing yielded on average 25 million reads that mapped to the GRCh37/hg19 genome assembly with 3 million CpGs having ≥10X coverage. Of these, approximately one million occurred in regions designated as CpG islands. Pre-specified filtering criteria for candidate DMRs included AUC ≥ 0.90, ,FC ≥ 50, ΔM ≥ 0.15, p-value ≤ 0.01, CpG density of 0.05 – 0.3, and a minimum range of 40 base pairs. Our software identified ~185,000 EAC and ~99,000 ESCC DMRs with a variance >0, DMRs were reduced to 94 (EAC) and 88 (ESCC) by these criteria.
Thirty-four EAC and 34 ESCC assays were selected for technical validation; qMSP testing on the sequenced cases and controls resulted in AUCs which ranged from 0.60 to 0.99 (median 0.88), FCs of 4 to >1000 (median 26), and ΔM of 0.0082 to 0.54 (median 0.19). We also calculated intra-class correlation to assess the reliability of the MDM results. Coefficients ranged from 0.00016 – 0.98 (median 0.95) indicating high reproducibility for the majority of markers.
Thirteen MDMs hypermethylated in both EAC and ESCC (ARHGEF4, Chr10.102, Chr19.402, DLX4, DMRTA2, GRIN2D, KCNA3, TBX15, TSPYL5, ZNF132, ZNF610, ZNF671, ZNF781), 5 EAC specific MDMs (Chr9.999, ELMO1, FGF14, ST8SIA1, and ZNF568), and 5 MDMs that were recurrently methylated in other cancers (BMP3, FER1L4, IKZF1, NDRG4, OPLAH) were chosen for biological validation (16–20,23).
Biological validation in tissue
The independent tissue cohort included 41 patients with EAC, 35 patients with ESCC, and 17 normal controls without EC or BE (Figure 1). Groups were age- and sex-balanced. Buffy coat samples from 24 patients free from cancer were also run as controls.
Discrimination by candidate MDMs on independent tissue validation is summarized (Table 1). AUCs for the independent EAC tissue comparison against normal esophageal mucosa were 0.44 – 0.99 (median 0.93); and 0.33 – 1.00 (median 0.87) for ESCC tissue. AUCs for EAC compared to buffy coat were 0.85 – 1.00 (median 0.97); AUCs for ESCC compared to buffy coat were 0.80 – 0.99 (median 0.91). Median FCs were 54 for EAC compared to normal esophagus, 10 for ESCC compared to normal, 811 for EAC compared to buffy, and 454 for ESCC compared to buffy.
Table 1.
Discrimination of methylated DNA markers for esophageal cancer during biological validation in DNA extracted from tissues.
| MDM | AUC (EAC) | p-value | AUC (ESCC) | p-value |
|---|---|---|---|---|
| ELMO1* | 0.99 | <.0001 | 0.74 | 0.0003 |
| FGF14 | 0.93 | <.0001 | 0.45 | 0.0223 |
| Chr9.999 | 0.83 | <.0001 | 0.51 | 0.0102 |
| ST8SIA1* | 0.98 | <.0001 | 0.59 | 0.0003 |
| ZNF568 | 0.91 | 0.1588 | 0.75 | 0.8952 |
| ARHGEF4* | 0.79 | 0.0001 | 0.81 | <.0001 |
| DLX4 | 0.75 | 0.0389 | 0.70 | 0.5563 |
| DMRTA2* | 1.00 | <.0001 | 1.00 | <.0001 |
| GRIN2D | 0.99 | <.0001 | 0.88 | <.0001 |
| KCNA3 | 0.92 | <.0001 | 0.60 | 0.0288 |
| Chr10.102 | 0.44 | 0.8974 | 0.33 | 0.7419 |
| Chr19.403 | 0.66 | 0.0124 | 0.61 | 0.0154 |
| TBX15* | 0.93 | <.0001 | 0.91 | <.0001 |
| TSPYL5* | 0.95 | <.0001 | 0.90 | <.0001 |
| ZNF132 | 0.88 | <.0001 | 0.75 | <.0001 |
| ZNF610 | 0.97 | <.0001 | 0.87 | <.0001 |
| ZNF671* | 0.89 | <.0001 | 0.89 | <.0001 |
| ZNF781 | 0.99 | <.0001 | 0.94 | <.0001 |
| OPLAH* | 0.94 | <.0001 | 0.77 | 0.0004 |
| IKZF1* | 0.92 | <.0001 | 0.37 | 0.0086 |
| FER1L4* | 0.92 | <.0001 | 0.69 | 0.0002 |
| NDRG4* | 0.96 | <.0001 | 0.54 | 0.0537 |
| BMP3* | 0.96 | <.0001 | 0.50 | 0.0181 |
Markers advanced to further testing in plasma study.
AUC (EAC) Tissue: Median area under the receiver operating curve for discrimination of esophageal adenocarcinoma from normal esophagus.
AUC (ESCC) Tissue: Median area under the receiver operating curve for discrimination between esophageal squamous cell carcinoma from normal esophagus.
We selected 12 MDMs (ARHGEF4, ELMO1, ST8SIA1, OPLAH, FER1L4, TBX15, ZNF671, IKZF1, TSPYL5, NDRG4, BMP3, and DMRTA2) for the plasma study. Gene expression analysis of publically available RNA sequencing data in EAC and normal esophagus show significant up- or down-regulation for 9 of 11 MDMs which annotate to coding regions (Supplemental Table 1). Oligonucleotides were redesigned in a modified QuARTS format to hybridize to the exact regions addressed with the qMSP primers.
Plasma study
Plasma samples from 85 independent and unique EC cases and 98 normal controls without cancer or BE were tested. Patient characteristics are summarized in Table 2. Clinical TNM staging algorithm was used according to AJCC 8th edition classification.
Table 2.
Clinical characteristics of patients in the plasma study.
| Case (N=85) | Control (N=98) | p-value | |
|---|---|---|---|
| Age, median (IQR) | 67 (59, 74) | 68 (58, 73) | 0.888 |
| Male sex, n (%) | 77 (90.6%) | 84 (85.7%) | 0.433 |
| Caucasian race, n (%) | 81 (95.3%) | 82 (83.7%) | 0.155 |
| History of smoking, n (%) | 54 (63.5%) | 36 (36.7%) | <0.01 |
| Current alcohol use, n (%) | 35 (41.2%) | 37 (37.8%) | 0.192 |
| BMI, median (IQR) | 29.8 (25.4–33.1) | 27.7 (25.2–30.8) | 0.221 |
| EAC, n (%) | 76 (89%) | ||
| ESCC, n (%) | 9 (11%) | ||
| Stage | |||
| I, n (%) | 14 (16%) | ||
| II, n (%) | 14(16%) | ||
| III, n (%) | 31 (37%) | ||
| IV, n (%) | 26 (31%) | ||
IQR, interquartile range; BMI, body mass index; CI, confidence interval; EAC, esophageal adenocarcinoma; ESCC, esophageal squamous cell carcinoma
Cases included 76 EAC, 9 ESCC, and represented all stages: I (n=14), II (n=14), III (n=31) and IV (n=26). Cases and controls were well-balanced for age, sex, and BMI, but there was a significantly higher number of former smokers among case patients (p<0.001).
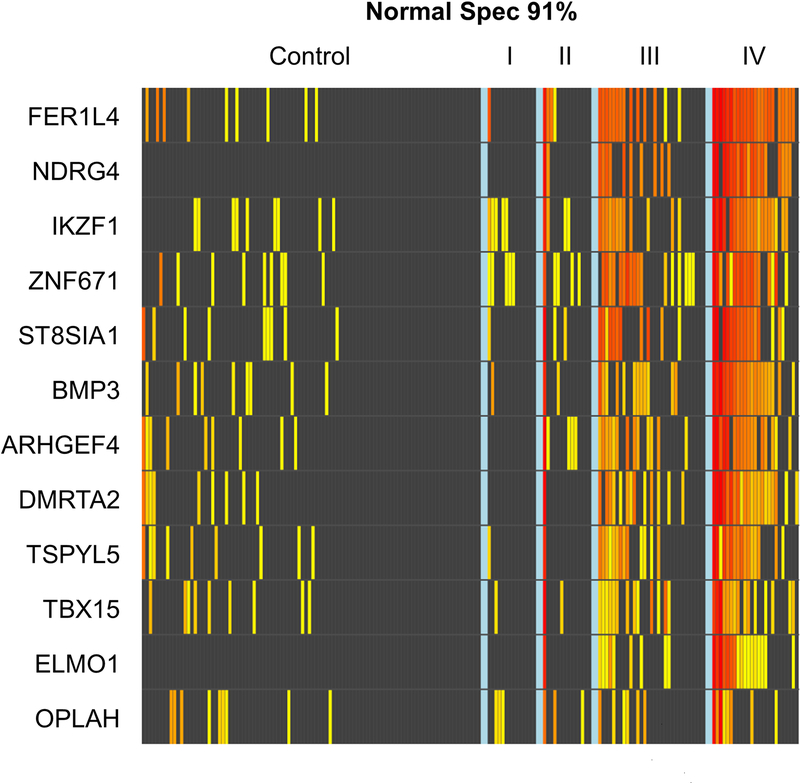
After normalization to the ACTB internal control, individual MDMs had an AUC of 0.64– 0.78. ELMO1 and NDRG4 were 100% specific (Figure 2). Plots of MDM level distributions show substantial separation between cases and controls for most individual candidates (Supplemental Figure 1).
Figure 2.
Beta-actin corrected methylation intensity in DNA markers assayed from plasma.
Each column on the x-axis represents an individual patient’s sample; each row on the y-axis represents the methylation specific PCR product by QuARTS assay for each marker on a logarithmic scale. A 91% specificity threshold was selected by the rPart model. Increasing yellow-red color spectrum reflects increasing methylation intensity above the 91st percentile value of the control patient samples.
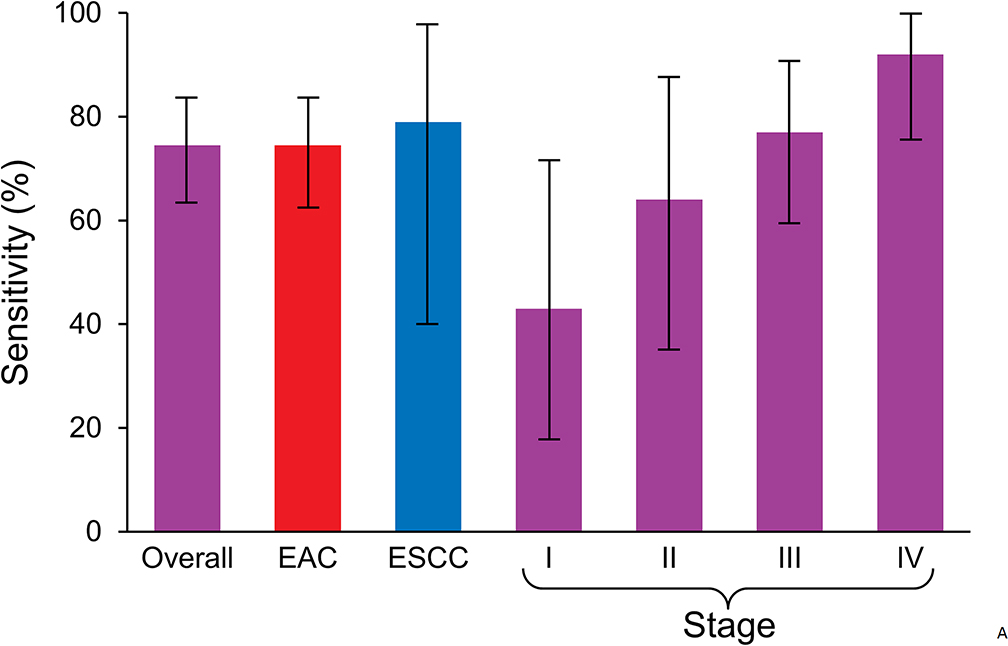
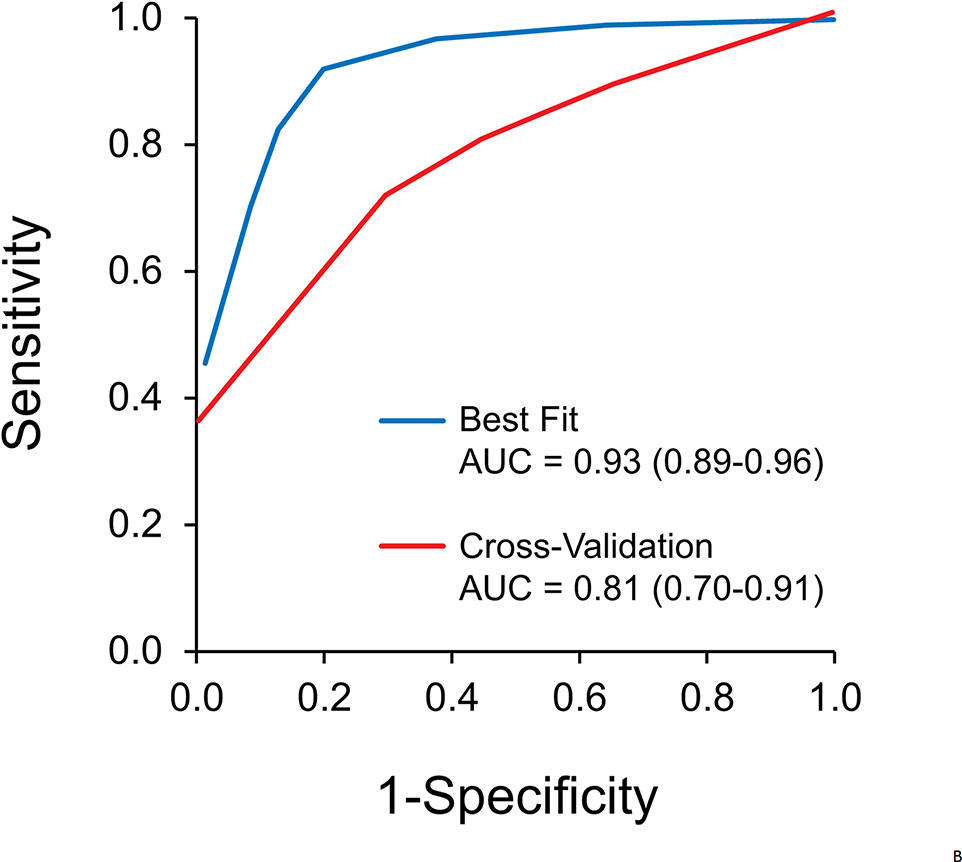
The rPART modeling process selected a 5-marker combination (FER1L4, ZNF671, ST8SIA1, TBX15, ARHGEF4) normalized by ACTB as the most discriminant panel. At a specificity cut-off of 91% by the model, the MDM panel detected 74% of EC overall, 74% of EAC, and 78% of ESCC (Figure 3A). By stage, the panel was able to detect 43% stage I, 64% stage II, 77% stage III, and 92% stage IV disease (Figure 3A). The rPART model yielded an area under the receiver operating characteristics curve of 0.93 (95% CI: 0.89–0.96) on best-fit and 0.81 (95% CI: 0.75–0.88) on cross-validation (Figure 3B).
Figure 3.
A. At a specificity cut-off of 91% by the model, the 5-marker panel was able to detect 43% stage I, 64% stage II, 77% stage III, and 92% stage IV disease. The marker panel detected 74% of esophageal cancer overall, 74% of esophageal adenocarcinoma, and 78% of esophageal squamous cell carcinoma. Confidence intervals are shown. B. Overall discrimination of esophageal cancer by methylated DNA marker panel assayed from plasma: areas under the receiver operating characteristic curves for best-fit and cross-validation analyses (and 95% confidence intervals) are shown.
AUC, area under the receiver operating characteristics curve.
Clinical covariates (age, sex, smoking, and weight) had no significant effect on the performance of our panel of biomarkers (Table 3).
Table 3.
Effect of clinical covariates in plasma study on area under the receiver operating curve (AUC) of the 5-marker panel, AUCs and 95% confidence intervals are shown.
| No | Yes | p-value | |
|---|---|---|---|
| Age >65 | 0.95 (0.9–1) | 0.91 (0.86–0.96) | 0.30 |
| Male | 0.97 (0.92–1) | 0.92 (0.88–0.96) | 0.12 |
| Tobacco Use | 0.92 (0.87–0.97) | 0.92 (0.86–0.98) | 0.97 |
| Obese | 0.93 (0.87–0.98) | 0.92 (0.87–0.98) | 0.92 |
Site-specificity of MDMs for esophageal cancers
Using microarray data from TCGA, 7 of our 12 markers were successfully mapped. Using this information, we were able to compare the methylation levels of these MDMs in tissue from EAC and ESCC, versus other common cancer types (Supplemental Figure 2). Supplemental Table 2 shows pair-wise comparisons of methylation between EC with each of the other cancer types. TBX15 was the only marker from the 5-MDM panel that mapped to the TCGA data. TBX15 methylation ratios were significantly higher for both EAC and ESCC compared to all other cancers in the comparison, except colorectal cancer. For DMRTA2, methylation ratios were significantly higher for EAC and ESCC combined than for all other cancers.
DISCUSSION
Current strategies for early detection and prevention of EC rely on demographic stratification followed by endoscopy in high-risk individuals. However, this approach is cost-prohibitive and insensitive in the general population, as over 80% of EAC is diagnosed de novo (30). An inexpensive, non-invasive, and standardized screening tool such as a blood test that could be applied to a wider population at risk for EC would be transformative.
One means by which this could be achieved is through identification of highly discriminant markers, such as MDMs, for EC that could be applied to liquid biopsies. Ours was the first comprehensive whole-methylome discovery effort on differential methylation across both ESCC and EAC. In the first phase of our study, thousands of novel DMRs for EC were discovered through whole-methylome sequencing on DNA from frozen tissue. The top performing DMRs were then validated technically using a more clinically applicable platform, MSP. Carrying forward, the DMRs were biologically validated in independent FFPE tissue, which confirmed high discrimination. Through this rigorous filtering and independent validation process, DMRs were carefully selected as candidate MDMs with minimal risk of false discovery.
The 12 MDMs with highest discrimination in tissue were subsequently tested in a large, independent plasma study of EC case patients and normal controls. While the purpose of this investigation was to identify MDMs based on their discrimination for EC and not to evaluate their function, it is noteworthy that each of the top candidate MDMs has biological associations with tumorigenesis (cell cycle, signaling, and transcriptional regulation) which lends confidence that our discovery process led to bona fide epigenetic biomarker candidates (Supplemental Table 1). Similar to reports in other cancers, we have observed that these MDMs alter gene expression through both up and down-regulation in esophageal cancer. Although DNA methylation canonically has been thought to silence gene expression through hypermethylation in promoter regions, more recent studies have found that methylation can occur anywhere in the genome; regulation of transcription can even be influenced by remote enhancers or repressors that only contact a gene via chromatin folding (36–39).
In plasma, a panel of 5 MDMs accurately detected EC, demonstrating feasibility for a non-invasive MDM plasma assay for detection of EC. Unlike earlier studies on biomarkers for esophageal cancer, updated clinical staging was determined for all cases according to AJCC 8th edition, and all stages were included in our study (25). Although our study was not designed to study the effect of MDMs on prognosis, our markers are in fact highly correlated with stage of disease, which predicts survival. MDMs may also be potentially useful as markers of residual disease or cancer recurrence following surgery for curative intent. Furthermore, MDMs are broadly informative and can be site specific (32).
Not only could an MDM panel be used to target those at risk for EC on the basis of BE or endemic ESCC, it may potentially be used for screening in the general population as part of a universal cancer screening tool. By aggregating the prevalence of multiple cancers, such a universal tool could revolutionize our approach to screening less-prevalent cancers, and prediction of tumor site can efficiently direct the evaluation of test-positive patients (31, 32).
Optimal site prediction for EAC and ESCC was not the primary goal of this study. It was anticipated from prior experiments that markers selected for EC detection in the present study would be methylated in other GI cancers (23,40). It is also known that there are additional MDMs that readily distinguish between foregut and hindgut neoplasms in tissue studies, plasma assays, and in stool assays (22, 41). In TCGA data examined in the present study, the MDMs we interrogated in plasma are methylated in other GI cancers, most commonly colorectal cancer. However, DMRTA2 showed significantly greater hypermethylation in esophageal cancers relative to all other aerodigestive cancers studied. It is therefore anticipated that a panel of MDMs can be further optimized for site-specific cancer detection, as is the focus of our ongoing research.
Although the MDM panel was less sensitive for stage I disease (43%, 95% CI 18–71%), where detection would be the most impactful, it is worth noting that the sensitivity for other blood-based tests, and even endoscopy has not been specifically reported for stage I disease. In addition, our samples were collected and processed on generic platforms that were developed before recent advances in nucleic acid preservation technology, and therefore not optimized for recovery of cell-free tumor DNA. This could have led to DNA degradation, decreased recovery of markers, and, accordingly, an artefactual reduction in sensitivity. Newer nucleic acid preservation media are now known to prevent degradation and improve recovery of target DNA while also reducing plasma contamination from leukocyte lysis during processing, transport and storage (33). Sample collection and processing optimization will therefore be a critical goal of future prospective studies.
Although our study suggests the feasibility of a plasma-based test for detecting EC, there are several important limitations to consider. We were constrained by our relatively small regional population for ESCC cases. To maximize statistical power, ESCC and EAC were incorporated in the same cohort for the training of a 5-marker model in plasma. This was supported by the significant overlap of MDMs observed between the two cancers. All MDMs discovered in ESCC performed well in EAC (Table 1). Because our plasma cohort consisted mostly of EAC, MDMs specific for EAC were chosen for biological validation. Mapping 7 of our MDMs to methylation data available in TCGA (Supplemental Figure 2) showed that similar to our results, all MDMs except those chosen specifically for EAC were similarly methylated in both cancers. Therefore, although we agree that EAC and ESCC are biologically distinct cancers, there is significant overlap in their methylation signatures which may reflect that MDMs are more representative of the host organ epithelium, rather than the underlying tumor biology. Similar patterns have also been observed in liver tumors arising in different underlying chronic disease states (42). These epigenetic similarities allowed us to select representative candidates for both cancer types in combination.
Our plasma cohort did not include patients with Barrett’s esophagus as a disease-control group. This is because detection of MDMs in plasma appears to be dependent on invasive characteristics of tumors such as angiogenesis, tumor necrosis and metastatic potential; thus, we do not anticipate false positive MDM results in plasma assay due to underlying non-dysplastic BE (35). Support of this hypothesis comes from studies of methylated SEPTIN9 in plasma for detection of colorectal neoplasms; the adenoma detection rate by this plasma marker fell at/below the false positive threshold, indicating that invasive cancer is most likely required to introduce cancer-specific methylated DNA into plasma (43). The markers we have piloted in plasma for EAC do include recently described markers of BE (34).
While the population of the present study is 80% white, we have tested the ESCC markers in larger independent cohorts of patient tissues from geographically and ethnically distinct regions (China, Iran and India)(44). DNA samples obtained from minimally invasive balloon cytology scrapings have also confirmed that MDMs for ESCC discovered in this study are represented in high grade dysplasia and ESCC in patients from central China (45).
Finally, the cross-validation AUC, while strong (0.81) was lower than that of the best fit (0.93). A larger clinical study is needed to measure MDM performance with greater precision. Prospective collection will also permit optimization and standardization of storage and processing conditions, referent to older EDTA collected archives, anticipated to improve recovery of MDM targets in early stage cancers.
In conclusion, our study demonstrates early feasibility for the noninvasive detection of EC by assay of informative MDMs in plasma. This approach has potential value in EC screening, prognosis, and surveillance applications. Further investigation is indicated to optimize sample collection and processing, assay configuration, and MDM panel selection, as well as to validate findings in larger prospective clinical studies.
Supplementary Material
Translational Relevance.
Esophageal cancer is a lethal disease for which no effective screening tool exists at a population level. An inexpensive, automatable, and non-invasive screening tool could transform the future of this disease. Ours was the first whole-methylome sequencing of both esophageal adenocarcinoma and squamous cell carcinoma. Through our discovery in tissue, differentially methylated regions highly discriminant for both types of cancer were identified. These subsequently underwent robust rounds of validation in independent tissue samples. Ultimately, plasma assays for candidate methylated DNA markers were developed; a 5-marker panel achieved an AUC of 0.81 on cross-validation, and 0.93 on best-fit for discrimination of esophageal cancer from normal controls, when assayed from plasma. In addition, marker sensitivity in plasma was directly related to cancer stage, suggesting potential for prognosis and surveillance. These data suggest feasibility of a non-invasive test for detection of esophageal cancer and open the door for further optimization and clinical investigation.
Acknowledgments
Grant Support: This work was supported by the Maxine and Jack Zarrow Family Foundation of Tulsa Oklahoma (to JBK), R37CA214679 (to JBK), a gift from Eddie Gong and Dana Clay (to DAA), the Carol M. Gatton endowment for Digestive Diseases Research (to DAA). This publication was also made possible by UL1 TR002377 from the National Center for Advancing Translational Sciences (NCATS). RRBS sequencing costs and QuARTS assays were provided by Exact Sciences (Madison WI).
Abbreviations
- AUC
area under the receiver operating characteristics curve
- BE
Barrett’s esophagus
- BMI
body mass index
- BSMAP
whole genome bisulfite sequence mapping program
- CI
confidence interval
- CpG
5’—Cytosine—phosphate—Guanine—3’
- DMR
differentially methylated region
- DNA
Deoxyribonucleic acid
- EC
esophageal cancer
- EAC
esophageal adenocarcinoma
- EABE
Esophageal Adenocarcinoma and Barrett’s Esophagus Registry, Mayo Clinic, Rochester, MN
- EDTA
Ethylenediaminetetraacetic acid
- ESCC
esophageal squamous cell carcinoma
- FFPE
formalin fixed paraffin embedded
- FC
fold change
- GDC
Genomics Data Commons
- MDMs
methylated DNA markersqMSP: quantitative methylation specific polymerase chain reaction
- qMSP
quantitative methylation specific PCR
- QuARTS
quantitative allele-specific real time target and signal amplification
- PCR
polymerase chain reaction
- RRBS
Reduced Representation Bisulfite Sequencing
- TCGA
The Cancer Genome Atlas
Footnotes
Disclosures:
Drs. Wu, Kisiel & Ahlquist, Messrs. Taylor & Mahoney, and Ms. Yab are employees of Mayo Clinic who are listed as inventors in an intellectual property agreement between Mayo Clinic and Exact Sciences, under which they could receive royalties, in accordance with Mayo Clinic policy. Dr. Iyer is a research collaborator with Exact Sciences. Drs. Allawi and Lidgard are employees of Exact Sciences Development Company (Madison WI). Drs. Qin, Sawas, Buttar, Katzka and Mrs. Burger have nothing to disclose.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA: a cancer journal for clinicians 2011;61(2):69–90. [DOI] [PubMed] [Google Scholar]
- 2.Thrift AP. The epidemic of oesophageal carcinoma: where are we now? Cancer epidemiology 2016;41:88–95. [DOI] [PubMed] [Google Scholar]
- 3.Arnal MJD, Arenas ÁF, Arbeloa ÁL. Esophageal cancer: Risk factors, screening and endoscopic treatment in Western and Eastern countries. World journal of gastroenterology: WJG 2015;21(26):7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Codipilly DC, Qin Y, Dawsey SM, Kisiel J, Topazian M, Ahlquist D, et al. Screening for esophageal squamous cell carcinoma: recent advances. Gastrointestinal endoscopy 2018;88(3):413–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hiripi E, Jansen L, Gondos A, Emrich K, Holleczek B, Katalinic A, et al. Survival of stomach and esophagus cancer patients in Germany in the early 21st century. Acta oncologica 2012;51(7):906–14. [DOI] [PubMed] [Google Scholar]
- 6.Buttar NS, Wang KK, Lutzke LS, Krishnadath KK, Anderson MA. Combined endoscopic mucosal resection and photodynamic therapy for esophageal neoplasia within Barrett’s esophagus. Gastrointestinal endoscopy 2001;54(6):682–8. [DOI] [PubMed] [Google Scholar]
- 7.Wang G-Q, Jiao G-G, Chang F-B, Fang W-H, Song J-X, Lu N, et al. Long-term results of operation for 420 patients with early squamous cell esophageal carcinoma discovered by screening. The Annals of thoracic surgery 2004;77(5):1740–4. [DOI] [PubMed] [Google Scholar]
- 8.Wei W-Q, Chen Z-F, He Y-T, Feng H, Hou J, Lin D-M, et al. Long-term follow-up of a community assignment, one-time endoscopic screening study of esophageal cancer in China. Journal of Clinical Oncology 2015;33(17):1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang J, Wei W-Q, Niu J, Liu Z-C, Yang C-X, Qiao Y-L. Cost-benefit analysis of esophageal cancer endoscopic screening in high-risk areas of China. World Journal of Gastroenterology: WJG 2012;18(20):2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu YW, Peng YH, Chen B, Wu ZY, Wu JY, Shen JH, et al. Autoantibodies as potential biomarkers for the early detection of esophageal squamous cell carcinoma. The American journal of gastroenterology 2014;109(1):36–45 doi 10.1038/ajg.2013.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang S, Du H, Li G. Significant prognostic value of circulating tumor cells in esophageal cancer patients: A meta-analysis. Oncotarget 2017;8(9):15815–26 doi 10.18632/oncotarget.15012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu F, Tian T, Xia LL, Ding Y, Cormier RT, He Y. Circulating miRNAs as novel potential biomarkers for esophageal squamous cell carcinoma diagnosis: a meta-analysis update. Diseases of the esophagus : official journal of the International Society for Diseases of the Esophagus 2017;30(2):1–9 doi 10.1111/dote.12489. [DOI] [PubMed] [Google Scholar]
- 13.Moinova HR, LaFramboise T, Lutterbaugh JD, Chandar AK, Dumot J, Faulx A, et al. Identifying DNA methylation biomarkers for non-endoscopic detection of Barrett’s esophagus. Science translational medicine 2018;10(424) doi 10.1126/scitranslmed.aao5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chettouh H, Mowforth O, Galeano-Dalmau N, Bezawada N, Ross-Innes C, MacRae S, et al. Methylation panel is a diagnostic biomarker for Barrett’s oesophagus in endoscopic biopsies and non-endoscopic cytology specimens. Gut 2017. doi 10.1136/gutjnl-2017-314026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dickinson BT, Kisiel J, Ahlquist DA, Grady WM. Molecular markers for colorectal cancer screening. Gut 2015;64(9):1485–94 doi 10.1136/gutjnl-2014-308075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imperiale TF, Ransohoff DF, Itzkowitz SH, Levin TR, Lavin P, Lidgard GP, et al. Multitarget stool DNA testing for colorectal-cancer screening. New England Journal of Medicine 2014;370(14):1287–97. [DOI] [PubMed] [Google Scholar]
- 17.Kisiel JB, Klepp P, Allawi HT, Taylor WR, Giakoumopoulos M, Sander T, et al. Analysis of DNA Methylation at Specific Loci in Stool Samples Detects Colorectal Cancer and High-grade Dysplasia in Patients with Inflammatory Bowel Disease. Clinical Gastroenterology and Hepatology 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kisiel JB, Raimondo M, Taylor W, Yab TC, Mahoney DW, Sun Z, et al. New DNA methylation markers for pancreatic cancer: discovery, tissue validation, and pilot testing in pancreatic juice. Clinical cancer research 2015:clincanres. 2469.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kisiel JB, Dukek BA, Kanipakam RV, Ghoz HM, Yab TC, Berger CK, et al. Hepatocellular Carcinoma Detection by Plasma Methylated DNA: Discovery, Phase I Pilot, and Phase II Clinical Validation. Hepatology 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allawi HT, Giakoumopoulos M, Flietner E, Oliphant A, Volkmann C, Aizenstein B, et al. Detection of lung cancer by assay of novel methylated DNA markers in plasma. AACR; 2017. [Google Scholar]
- 21.Ghoz HM, Aboelsoud MM, Yab TC, Berger CK, Taylor WR, Cao X, et al. dna Methylation Markers for Detection of Extrahepatic Cholangiocarcinoma: Discovery, Tissue validation, and Pilot Testing in Biliary Brush Samples: 169. Hepatology 2015;62:295A. [Google Scholar]
- 22.Kisiel JB, Taylor WR, Yab TC, Ghoz HM, Foote PH, Devens ME, et al. Accurate site prediction of gastrointestinal cancer by novel methylated DNA markers: Discovery & Validation. AACR; 2015. [Google Scholar]
- 23.Anderson BW, Suh Y-S, Choi B, Lee H-J, Yab TC, Taylor W, et al. Detection of Gastric Cancer with Novel Methylated DNA Markers: Discovery, Tissue Validation, and Pilot Testing in Plasma. Clinical Cancer Research 2018:clincanres. 3364.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li L, Choi J-Y, Lee K-M, Sung H, Park SK, Oze I, et al. DNA methylation in peripheral blood: a potential biomarker for cancer molecular epidemiology. Journal of epidemiology 2012;22(5):384–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rice TW, Ishwaran H, Ferguson MK, Blackstone EH, Goldstraw P. Cancer of the esophagus and esophagogastric junction: an eighth edition staging primer. Journal of Thoracic Oncology 2017;12(1):36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kisiel JB, Yab TC, Taylor WR, Chari ST, Petersen GM, Mahoney DW, et al. Stool DNA testing for the detection of pancreatic cancer: assessment of methylation marker candidates. Cancer 2012;118(10):2623–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xi Y, Li W. BSMAP: whole genome bisulfite sequence MAPping program. BMC bioinformatics 2009;10(1):232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goldman M, Craft B, Brooks A, Zhu J, Haussler D. The UCSC Xena Platform for cancer genomics data visualization and interpretation. BioRxiv. 2018. [Google Scholar]
- 29.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Visrodia K, Singh S, Krishnamoorthi R, Ahlquist D, Wang K, Iyer PG, et al. Systematic review with meta‐analysis: prevalent vs. incident oesophageal adenocarcinoma and high‐grade dysplasia in Barrett’s oesophagus. Alimentary pharmacology & therapeutics 2016;44(8):775–84. [DOI] [PubMed] [Google Scholar]
- 31.Ahlquist DA. Universal cancer screening: revolutionary, rational, and realizable. NPJ precision oncology 2018;2(1):23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018:eaar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pottekat A, Allawi HT, Boragine GT, Kaiser MW, Sander T, Krueger C, et al. A comprehensive assessment of the impact of preanalytical variables on cell free DNA and circulating tumor cells in blood. AACR; 2018. [Google Scholar]
- 34.Iyer PG, Taylor WR, Johnson ML, Lansing RL, Maixner KA, Yab TC, et al. Highly Discriminant Methylated DNA Markers for the Non-endoscopic Detection of Barrett’s Esophagus. The American journal of gastroenterology 2018:1. [DOI] [PubMed] [Google Scholar]
- 35.Ahlquist DA, Zou H, Domanico M, Mahoney DW, Yab TC, Taylor WR, et al. Next-generation stool DNA test accurately detects colorectal cancer and large adenomas. Gastroenterology 2012;142(2):248–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lokk K, Modhukur V, Rajashekar B, Martens K, Magi R, Kolde R, et al. DNA methylome profiling of human tissues identifies global and tissue-specific methylation patterns. Genome biology. 2014;15(4):r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 2010;466(7303):253–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, et al. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nature biotechnology. 2009;27(4):361–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lorincz MC, Dickerson DR, Schmitt M, Groudine M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nature structural & molecular biology. 2004;11(11):1068–75. [DOI] [PubMed] [Google Scholar]
- 40.Raimondo M, Yab TC, Mahoney DW, Taylor WR, Anderson KS, Ahlquist DA. 487 Methylated DNA Markers in Pancreatic Juice Discriminate Pancreatic Cancer From Chronic Pancreatitis and Normal Controls. Gastroenterology. 2013;144(5):S-90. [Google Scholar]
- 41.Kisiel JB, Wu CW, Taylor WR, Allawi HT, Yab TC, Foote PH, et al. 393 - Multi-Site Gastrointestinal Cancer Detection by Stool DNA. Gastroenterology. 2018;154(6):S-95. [Google Scholar]
- 42.Hlady RA, Tiedemann RL, Puszyk W, Zendejas I, Roberts LR, Choi JH, et al. Epigenetic signatures of alcohol abuse and hepatitis infection during human hepatocarcinogenesis. Oncotarget. 2014;5(19):9425–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahlquist DA, Taylor WR, Mahoney DW, Zou H, Domanico M, Thibodeau SN, et al. The stool DNA test is more accurate than the plasma septin 9 test in detecting colorectal neoplasia. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association. 2012;10(3):272–7.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iyer PG, Buglioni A, Cao X, Foote PH, Yab TC, Taylor WR, et al. Mo1748 - Validation of Novel Methylated DNA Markers for the Detection of Esophageal Squamous Cell Carcinoma and Dysplasia: Multi-National Tissue Study. Gastroenterology. 2018;154(6):S-794–S-5. [Google Scholar]
- 45.Qin Y, Taylor WR, Foote PH, McGlinch M, Cao X, Wu CW, et al. Methylated Dna Markers Detect Esophageal Squamous Dysplasia in Mucosal Tissue and When Sampled by Nonendoscopic Esophageal Balloons: An Exploratory Study. Gastroenterology. 2019;156(6):S-227–S-8. [Google Scholar]
- 46.TCGA Network. Integrated genomic characterization of oesophageal carcinoma. Nature. 2017; 541:169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.TCGA Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012; 487:330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.TCGA Network. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell. 2017; 169(7):1327–1341.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.TCGA Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014; 511:540–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.TCGA Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012; 489:519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.