Abstract
Purpose:
The majority of broad-panel tumor genomic profiling has used a gene-centric approach, although much of that data is unused in clinical decision-making. We hypothesized that a pathway-centric approach using next-generation sequencing, combined with conventional clinicopathologic features, may better predict disease-free survival (DFS) in early-stage lung adenocarcinoma.
Experimental Design:
Utilizing our prospectively maintained database, we analyzed 492 patients with primary, untreated, completely surgically resected lung adenocarcinoma. Ten canonical pathways were analyzed using broad-panel next-generation sequencing. The correlations of disease-free survival and number (and type) of pathway (NPA) were analyzed using the Kaplan-Meier method and log-rank test. Associations between altered pathways and clinicopathologic variables, as well as identification of actionable therapeutic strategies, were explored.
Results:
Median NPA for the cohort was 2 (range, 0-5). Smoking status, solid morphologic appearance on preoperative CT, maximal standardized uptake value, pathologic tumor size, aggressive histologic subtype, lymphovascular invasion, visceral pleural invasion, and positive lymph nodes were significantly associated with NPA (P<0.05). Of 543 actionable genetic alterations identified, 455 (84%) were within the RTK/RAS pathway. Eighty-six tumors had actionable therapeutic genomic alterations in >1 pathway. On multivariable analysis, higher NPA was significantly associated with worse DFS (HR, 1.31; P=0.014).
Conclusions:
NPA and specific pathway alterations are associated with clinicopathologic features in patients with surgically resected lung adenocarcinoma. Cell cycle, Hippo, TGFβ, and p53 pathway alterations are associated with poor DFS. Finally, NPA is an independent risk factor for poor DFS in our cohort.
Keywords: Next-generation sequencing, pathway analysis, lung adenocarcinoma, recurrence
Introduction
Broad-panel genomic sequencing is increasingly used to select targeted therapies and precision medicine strategies for patients with lung cancer (1). However, a recent study found that <5% of patients who underwent broad-panel genomic sequencing benefited from it, compared with patients who received only EGFR and/or ALK testing (2). This raises the possibility that much of the information included in genomic sequencing panels is currently not well-utilized. Heretofore, the majority of tumor genomic analyses have focused on gene-centric approaches that detect driver alterations and identify functionally irrelevant passenger events, particularly in solid tumors with higher mutational burdens (3). Cancer cell biology and its resulting clinical phenotypes are driven by different pathways, many of which have upstream oncogenic alterations that lead to similar downstream alterations and phenotypes within the same pathway (3,4). Therefore, a pathway-centric approach has been proposed to better identify functional alterations in selected oncogenic pathways, to explore co-occurrence and mutual exclusivity between pathways, and to identify relevant pathway alteration(s) that may be therapeutically exploited. We recently examined The Cancer Genome Atlas (TCGA) and comprehensively identified 10 canonical pathways covering 89% of 9125 different solid tumor types, which now provides a curated and standardized pipeline to perform pathway-centric next-generation sequencing (NGS) studies (5).
Lung adenocarcinoma is a genomically well-annotated malignancy with a high background mutational burden. The majority of lung adenocarcinoma genomic analyses have focused on metastatic disease (6), with the 2014 TCGA analysis the only study to examine genomic changes in earlier-stage lung adenocarcinoma (7). While surgical resection is the preferred treatment for early-stage lung adenocarcinoma, there remains a relatively high recurrence rate, even in pathologic node-negative disease (8–12). Therefore, a significant knowledge gap exists in identifying patients at risk for recurrence following complete surgical resection of lung adenocarcinoma. To address this, we performed a comprehensive analysis to investigate associations between tumor genomic pathway alterations and selected clinicopathologic features and disease-free survival (DFS) in patients with lung adenocarcinoma who have undergone complete resection.
Methods
Patient Cohort
This study was approved by the institutional review board at Memorial Sloan Kettering. Using our prospectively maintained database, we identified patients who underwent complete surgical resection (R0) for lung adenocarcinoma and had targeted NGS (MSK-IMPACT) performed on their primary tumor between February 2008 and January 2018. Exclusion criteria included induction therapy, microscopic or macroscopic residual disease (R1/R2 resection), low-quality NGS, stage IV disease, and mixed tumor type (Supplementary Figure 1).
Clinical characteristics, preoperative computed tomography (CT), positron emission tomography (PET) images, and pathologic reports (8th edition AJCC Cancer Staging Manual) were reviewed. Follow-up was performed in accordance with NCCN guidelines (13). Recurrences were distinguished from metachronous tumors using Martini and Melamed criteria, with confirmation from pathologic and genomic relatedness when available (14).
MSK-IMPACT Sequencing
Sequencing for MSK-IMPACT was performed as previously described (15). Patient clinicopathologic data were matched with genomic data and visualized using the cBioPortal for Cancer Genomics (16,17). Tumor DNA and corresponding patient-matched blood DNA were extracted. All exons and selected introns were sequenced using the MSK-IMPACT panel to identify somatic alterations, copy number alterations, and mutations. Median sequencing coverage was 764X (range, 164-1424). Selected panel sizes have been used over time (341-, 410-, and 468-gene panels for 14, 228, and 250 patients, respectively). Tumor mutational burden (TMB) was defined as the total number of nonsynonymous single-nucleotide or insertion/deletion mutations divided by the number of Mbs in the coding region captured by each panel (0.98, 1.06, and 1.22 Mb in the 341-, 410-, and 468-gene panels, respectively) (18). We have previously shown that TMB calculations using this NGS panel are strongly associated with the TMB assessed by whole-exome sequencing (18). The fraction of genome altered (FGA) was defined as the fraction of log2 copy number variation (gain or loss) >0.2 divided by the size of the genome whose copy number was profiled. FGA was corrected for tumor purity, ploidy and clonal heterogeneity using the FACET method (19).
Pathway Alteration and Therapeutic Actionability Identification
We evaluated 10 canonical signaling pathways using the templates provided in the signaling pathways manuscript from the TCGA PanCancer Atlas project (5). The pathways analyzed were (1) cell cycle, (2) Hippo, (3) Myc, (4) Notch, (5) oxidative stress response/Nrf2, (6) PI3K, (7) receptor-tyrosine kinase (RTK)/RAS/MAPK, (8) TGFβ, (9) p53, and (10) β-catenin/Wnt. In total, 109 genes were identified at the intersection of the a priori pathway templates (5) and the MSK-IMPACT panel (Supplementary Table 1). A tumor was considered “altered” in the specific pathway when ≥1 gene relative to control in the corresponding pathway template was altered. The status of specific pathways was determined to be either altered or wild-type for each patient. Number of pathway alterations (NPA) was calculated as the total number of altered pathways out of the 10 identified pathways for each patient.
Therapeutic actionability information was annotated using OncoKB on June 5, 2018 (20). Each potentially actionable genomic alteration was stratified into 1 of 6 levels. Detailed methods used for stratification of therapeutic actionable genetic events are included in the Supplementary Methods. CDK4 and MDM2 genomic data from the TCGA PanCancer Atlas lung adenocarcinoma cohort (6) were accessed using cBioPortal.
Statistical Methods
Associations between clinicopathologic and genomic characteristics, specific pathway alterations, and NPA were analyzed using Fisher’s exact test or Cochran-Armitage test for categorical variables and Wilcoxon rank-sum test or Spearman’s rank correlation test for continuous variables. When comparisons were repeatedly performed across 10 pathways, P values were adjusted using the false discovery rate (FDR) method when needed.
The primary outcome of interest was DFS, defined as the duration between surgery and recurrence or death without recurrence. Patients were censored at the last follow-up. Median follow-up was calculated on the basis of the reverse Kaplan-Meier approach (21). DFS was estimated using the Kaplan-Meier approach and compared between clinicopathologic characteristics and pathway alterations using log-rank tests, stratified by pathologic stage where appropriate. For the DFS analysis, NPA was considered a linear factor, as there was inadequate evidence to reject the linearity assumption (Wald F-Statistic X2=0.09; P=0.8) using restricted cubic splines.
The primary objective was to quantify the prognostic value of NPA. The relationships between NPA and clinically relevant factors were quantified using Cox proportional hazards models, stratified by pathologic stage (except in the cases of stage, tumor size, lymph node status, and visceral pleural invasion [VPI]). A multivariable model was constructed starting with all factors with P<0.1 in the univariable analyses. Multiple imputations were conducted to address missing data (details in online supplement). To avoid loss of information by using the simple summary of NPA, we compared an NPA-only model with a model that included all 10 pathways as individual variables in a multivariable penalized Cox model. Performance of each model was quantified as discrimination (C-index) or calibration (details in online supplement).
Pathway or gene mutual exclusivity or co-occurrence was analyzed using Fisher’s exact test, and P values were adjusted (FDR method). All analyses were two-sided, and P<0.05 was considered statistically significant. Statistical analyses were conducted using Stata 13.1 (Stata, College Station, Texas) and R 3.5.1 (Vienna, Austria).
Results
In total, 492 patients met the inclusion criteria, of whom 356 (72%) had stage I disease. Median follow-up was 19 months (95% CI, 18.4-20.1). Alterations in common lung adenocarcinoma driver genes included 40% KRAS, 26% EGFR, 5% BRAF, 1.6% ALK, 0.8% ROS1, and 0.6% MET (Figure 1A). Alterations in these driver genes were found to be evenly distributed across all NPA groups. The alteration frequencies of the 109 genes utilized for pathway analysis are included in Supplementary Table 2. Median NPA was 2 (range, 0-5) (Figure 1A, Table 1). The most and least frequently altered pathways were RTK/RAS (n=415 patients [84%]) and TGFβ (n=13 patients [3%]). Of the 164 patients with only 1 pathway alteration, 141 (86%) had the RTK/RAS pathway altered. Comparatively, 90% of patients (160/178) with NPA=2, 91% (86/94) with NPA=3, 85% (23/27) with NPA=4, and 83% (5/6) with NPA=5 had the RTK/RAS pathway altered. The RTK/RAS pathway was divided into two pathways as a function of NPA to better visualize the alteration frequencies of individual genes in the RTK and RAS/RAF pathways (Supplementary Figure 2). Median normalized TMB was 4.7 (range, 0-164.2), and median FGA was 4.3% (range, 0%-49.8%).
Figure 1. Association between pathway alterations and genomic features.
A, Oncoprint of all patients. B, Violin plot of tumor mutational burden (TMB) vs. number of pathway alterations (NPA). C, Violin plot of fraction of genome altered (FGA) versus NPA. D, Scatterplot of TMB vs. FGA. Bar plots of (E) TMB and (F) FGA versus pathway alteration status (altered and wild-type [WT]). Standard error displayed as error bar. *shows logarithmic scale.
Table 1.
Patient characteristics (N=492)
| Characteristic | No. (%) |
|---|---|
| Age at surgery, years, median (range) | 69 (34-87) |
| Sex | |
| Male | 326 (66) |
| Female | 166 (34) |
| Smoking status | |
| Ever | 380 (77) |
| Never | 112 (23) |
| Pack-yearsa | 20 (0-120) |
| Tumor morphologic appearance, CT criteria | |
| Pure ground glass opacity | 72 (15) |
| Mixed ground glass opacity/subsolid | 90 (18) |
| Solid | 330 (67) |
| Tumor maximal standardized uptake value, median (range) | 3.4 (0-31) |
| Procedure type | |
| Lobectomy | 340 (69) |
| Segmentectomy | 47 (9) |
| Wedge | 102 (21) |
| Pneumonectomy | 3 (1) |
| Pathologic tumor size, cm, median (range) | 1.8 (0.08-19.5) |
| Predominant histologic subtype | |
| Lepidic, AIS, MIA | 73 (15) |
| Acinar | 263 (53) |
| Papillary | 37 (8) |
| Micropapillary | 32 (7) |
| Solid | 60 (12) |
| Other | 27 (5) |
| Lymphovascular invasion | |
| Positive | 302 (62a) |
| Negative | 185 (38a) |
| Visceral pleural invasion | |
| Positive | 77 (16a) |
| Negative | 414 (84a) |
| Lymph node status | |
| Positive | 83 (17a) |
| Negative | 394 (83a) |
| Pathologic stage | |
| I | 356 (72) |
| II | 80 (16) |
| III | 56 (11) |
| Pathway alteration | |
| Cell cycle | 77 (16) |
| Hippo | 14 (3) |
| Myc | 36 (7) |
| Notch | 19 (4) |
| Nrf2 | 27 (5) |
| PI3K | 119 (24) |
| TGFβ | 13 (3) |
| RTK/RAS | 415 (84) |
| p53 | 201 (41) |
| Wnt | 19 (4) |
| Number of pathway alterations | |
| 0 | 23 (5) |
| 1 | 164 (33) |
| 2 | 178 (36) |
| 3 | 94 (19) |
| 4 | 27 (6) |
| 5 | 6 (1) |
| TMB, median (range) (IQR) | 4.7 (0-164.2) (2.5-8.2) |
| FGA, median (range) (IQR) | 0.25 (0-1) (0.1-0.5) |
Abbreviations: AIS, adenocarcinoma in situ; FGA, fraction of genome altered; IQR, interquartile range; MIA, minimally invasive adenocarcinoma; TMB, tumor mutation burden.
Denominator excluding patients with missing data.
Pathway Alterations and Genomic Features
TMB and FGA are essential genomic features reported to be associated with survival and recurrence in patients with cancer (22–25). We investigated the correlation of TMB and FGA with pathway alterations and found, as NPA increased, TMB (ρ=0.50; P<0.001) (Figure 1B) and FGA (ρ=0.31; P<0.001) (Figure 1C) increased correspondingly. However, the correlation between TMB and FGA was significant, but the magnitude of correlation (ρ=0.12; P=0.006) is considered negligible on the basis of the interpretation (26) by Hinkle et al. (Figure 1D).
We next examined the association between TMB, FGA, and individual pathway alterations (Supplementary Table 3). Tumors with cell cycle, Hippo, Myc, Notch, Nrf2, PI3K, p53, or Wnt pathway alterations had significantly higher TMB than tumors without these pathway alterations (Figure 1E). Similarly, tumors with cell cycle and p53 pathway alterations had significantly higher FGA than tumors without these pathway alterations (Figure 1F).
Pathway Alterations and Clinicopathologic Features
We then investigated associations between NPA and selected poor-risk clinicopathologic factors (Figure 2A–H). As NPA increased, the rate of ever-smokers also significantly increased (Cochran-Armitage test, P<0.001). Similar analyses showed increasing proportion of solid tumor morphologic appearance on preoperative CT, PET tumor maximal standardized uptake value (SUVmax) (ρ=0.28; P<0.001), pathologic tumor size (ρ=0.20; P<0.001), aggressive histologic subtype (micropapillary or solid) (Cochran-Armitage test, P<0.001), lymphovascular invasion (LVI; Cochran-Armitage test, P<0.001), VPI (Cochran-Armitage test, P=0.04), and lymph node status (Cochran-Armitage test, P=0.001) were all associated with increasing NPA.
Figure 2. Number of pathway alterations (NPA) and clinicopathologic features and disease-free survival.
Association between NPA and (A) smoking status, (B) morphologic appearance on CT, (C) pathologic tumor size, (D) tumor maximal standardized uptake value (SUVmax), (E) predominate histologic subtype, (F) lymphovascular invasion, (G) visceral pleural invasion, and (H) lymph node status. I, Staging and outcome distribution among different NPA groups. DOD, dead of disease; NED, no evidence of disease. J, Association between disease-free survival and NPA.
Finally, we investigated the association between clinicopathologic features and individual pathway alterations (Supplementary Table 3, Supplementary Figure 3). Nrf2 and PI3K pathway alterations were associated with smoking status. Cell cycle, Hippo, Nrf2, PI3K, p53, and Wnt pathway alterations were associated with solid tumor morphologic appearance on CT scan. Cell cycle, p53, and Wnt pathway alterations were associated with higher tumor SUVmax. Cell cycle, p53, Wnt pathways were associated with pathologic tumor size. P53 pathway alteration was associated with aggressive histologic subtype and LVI.
Pathway Alterations and DFS
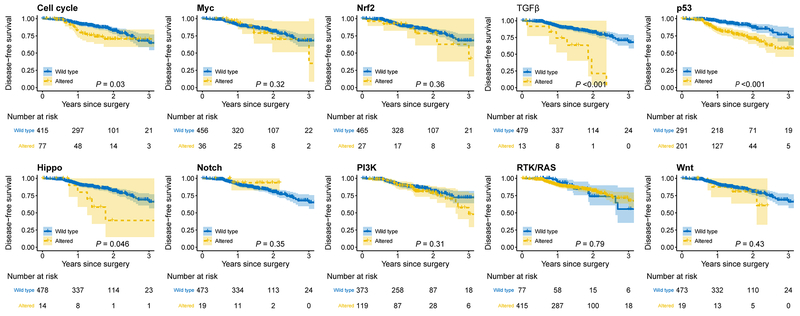
A DFS event occurred in 79 individuals, with 70 (89%) experiencing recurrence of their disease. The recurrence rate was 33% for patients with ≥4 NPA and 9% for those with 0 NPA (Figure 2I). The median DFS for patients with ≥4 NPA was 25.6 months (95% CI, 9.7-41.6); patients with 0 NPA had a median DFS of 89.4 months (95% CI, 10.7-168.1) (Figure 2J). We next examined pathologic stage–specific DFS on the basis of NPA and observed that increasing NPA was associated with worse DFS in stages I, II, and III LUAD (Supplementary Figure 4). Altered pathways were associated with worse 2-year DFS (95% CI) versus wild-type pathways for cell cycle (70.5% [59.0%-84.2%] vs. 82.4% [77.6%-87.5%]; log rank P=0.03), Hippo (38.9% [14.8%-100.0%] vs. 81.6% [77.1%-86.4%]; log rank P=0.046), TGFβ (21.2% [4.0%-100.0%] vs. 82.2% [77.8%-86.8%]; log rank P<0.001), and p53 (70.9% [63.3%-79.4%] vs. 87.0% [81.8%-92.7%]; log rank P<0.001) (Figure 3). On univariable analysis, when stratified by stage, increase in NPA was associated with worse DFS (HR, 1.38 [95% CI, 1.12-1.69]; P=0.002) (Table 2). Other clinicopathologic factors, including solid tumor morphologic appearance, tumor SUVmax, pathologic tumor size, pathologic stage, LVI, predominant histologic subtype, VPI, lymph node status, and FGA, were significantly associated with DFS in univariable models (Table 2). However, subsequent multivariable analysis revealed only NPA (HR, 1.31 [95% CI, 1.06-1.62]; P=0.01), solid tumor morphologic appearance, tumor SUVmax, and pathologic stage were independently associated with DFS (Table 2).
Figure 3.
Associations of individual pathway alterations and disease-free survival
Table 2.
Univariable and multivariable Cox proportional hazards model for disease-free survivala
| Univariable Model | Multivariable Model | |||
|---|---|---|---|---|
| Variable | HR (95% CI) | P value | HR (95% CI) | P value |
| Age at surgery, years | 1.01 (0.99-1.04) | 0.40 | ||
| Female vs. male | 1.28 (0.81-2.04) | 0.29 | ||
| Smoking status: ever | 0.89 (0.53-1.49) | 0.66 | ||
| Solid morphologic appearance on preoperative CT | 5.93 (2.11-16.68) | 0.001 | 4.39 (1.54-12.49) | 0.006 |
| Tumor SUVmax | 1.09 (1.05-1.14) | <0.001 | 1.07 (1.03-1.12) | 0.001 |
| Pathologic stage (8th edition AJCC) | ||||
| II vs. I | 3.33 (1.88-5.89) | <0.001 | 1.59 (0.86-2.95) | 0.14 |
| III vs. I | 7.92 (4.71-13.31) | <0.001 | 3.92 (2.25-6.81) | <0.001 |
| Pathologic tumor size, cm | 1.20 (1.13-1.28) | <0.001 | ||
| Lymphovascular invasion | 2.51 (1.47-4.28) | 0.001 | ||
| Predominate histologic subtype b | 1.41 (0.86-2.33) | 0.18 | ||
| Visceral pleural invasion | 2.76 (1.69-4.50) | <0.001 | ||
| Lymph node status: positive | 4.90 (3.13-7.66) | <0.001 | ||
| Procedure type | ||||
| Sublobar vs. lobectomy | 1.96 (0.93-4.12) | 0.08 | ||
| TMB | 1.01 (0.99-1.03) | 0.26 | ||
| FGA | 2.13 (0.94-4.84) | 0.07 | ||
| Number of pathways altered | 1.38 (1.12-1.69) | 0.002 | 1.31 (1.06-1.62) | 0.01 |
Abbreviations: AJCC, American Joint Committee on Cancer; CI, confidence interval; FGA, fraction of genome altered; HR, hazard ratio; SUVmax, maximal standardized uptake value; TMB, tumor mutation burden.
All models were stratified by pathologic stage except for stage, tumor size, lymph node status and visceral pleural invasion.
Tumor histologic subtypes collapsed as micropapillary + solid vs. other subtypes.
We next compared the simple pathway analysis model (simple-PA model; includes only NPA) with the complex pathway analysis model (complex-PA model; includes the 10 pathways) (Supplementary Table 4) in terms of performance by quantifying their discrimination and calibration. The cross-validated C-indexes were 0.658 (95% CI, 0.576-0.732) and 0.657 (95% CI, 0.530-0.750) for the simple-PA and complex-PA models. Bias-corrected calibration curves confirmed the simple-PA model was well-calibrated, whereas the complex-PA model tended to underestimate DFS (Supplementary Figure 4).
Mutual Exclusivity, Co-occurrence, and Therapeutic Actionabilities
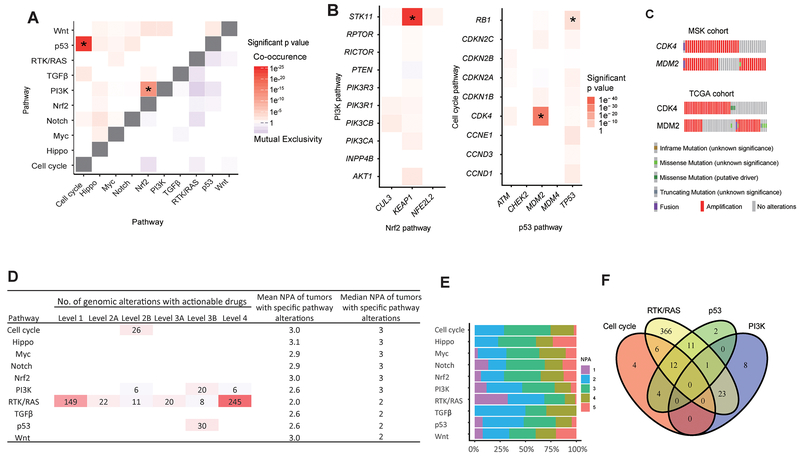
The mutual exclusivity of a pathway or gene may reflect functional redundancy or synthetic lethality (27,28), whereas co-occurrence(s) may reflect functional synergies important in resistance to targeted therapies (29). Of the 10 pathways investigated, 2 pairs, p53–cell cycle (OR, 5.78; P<0.001) and PI3K-Nrf2 (OR, 7.17; P<0.001), co-occurred with statistical significance (Figure 4A). We found no mutual exclusivity among pathways in our cohort. Within the p53–cell cycle and PI3K-Nrf2 pathways, alterations in three pairs of genes—CDK4-MDM2 (Log2 OR>3; P<0.001), RB1-TP53 (Log2 OR=2.81; P=0.03), and STK11-KEAP1 (Log2 OR>3; P<0.001)—co-occurred with statistical significance (Figure 4B).
Figure 4. Mutual exclusivity, co-occurrence, and therapeutic actionability.
A, Co-occurrence and mutual exclusivity between pathways. Asterisk indicates the P value is significant (P<0.001, adjusted using the false discovery rate method). Color scale shows the significance of co-occurrence or mutual exclusivity (-log P value). B, Co-occurrence and mutual exclusivity of single genes within PI3K-Nrf2 pathways and p53–cell cycle pathways. Asterisk indicates the P value is significant (CDK4-MDM2, P<0.001; RB1-TP53, P=0.03; STK11-KEAP1, P<0.001). C, Oncoprint of the CDK4-MDM2 altered patients in this study and the TCGA pan-cancer atlas cohort. D, Actionable genetic alterations within 10 pathways annotated using OncoKB and mean/median number of pathway alterations (NPA) for each pathway. E, Stacked bar plot of percentage of NPA (x-axis) in patients with specific pathway (y-axis) altered. F, Venn diagram showing patients who have actionable alterations in the 4 indicated pathways. Intersections of the circles show the number of patients who have >1 actionable pathway.
Co-occurrences of CDK4 (cell cycle pathway) and MDM2 (p53 pathway) amplification were significant (n=26 cell cycle, n=30 p53, n=16 both) (Figure 4C). This pair of genes also has corresponding level 2B (palbociclib and abemaciclib for CDK amplification) and level 4 (DS-3032b and RG7112 for MDM2 amplification) actionable drugs. This observation suggests patients with tumors with co-occurent CDK4 and MDM2 amplifications may benefit from combined therapies that target both pathways. Supporting our observation, analysis of the TCGA lung adenocarcinoma cohort (5) reveals these genes have a significant co-occurrence (Log2 OR>3; P<0.001) (Figure 4C).
Finally, using OncoKB, we investigated the therapeutic actionability of selected genomic alterations. We identified 543 actionable genomic alteration events across 437 tumors (Figure 4D). A list of tumors with targetable alterations at the pathway and gene level with associated therapies and levels of evidence as curated using OncoKB can be found in Supplementary Table 5. Of these 543 actionable alterations, 455 (84%) were within the RTK/RAS pathway—including all level 1 and level 2A actionable alterations. Interestingly, tumors with RTK/RAS pathway alterations had the lowest mean NPA (n=2) and the highest proportion of tumors with 1 NPA (Figures 4D and 4E). Of the 437 tumors with actionable alterations, 351 had actionable therapeutic alterations in only 1 pathway; the other 86 had actionable therapeutic alterations in >1 pathway (Figure 4F).
Discussion
To our knowledge, this is the first study to comprehensively investigate the association between selected pathway alterations and detailed clinicopathologic features and DFS in patients with completely resected lung adenocarcinoma using broad-panel NGS. Ninety-five percent of our cohort had at least 1 pathway alteration, and our analysis used 109 genes, of which 57 were altered in <2% of patients. This “long tail” distribution (30–32) of altered genes may complicate the identification of the functionally relevant driver genes from the passenger genes. However, as we show, aggregating these genes into biologically relevant molecular pathways permits the identification of associations with relevant clinicopathologic features and DFS.
It is unknown which pathway alteration(s) initiate the progressive evolution of normal tissue along the continuum toward a neoplastic state. Our results revealed that, among patients with 1 NPA, 86% had an alteration in the RTK/RAS pathway. Of the 10 oncogenic pathways studied, the RTK/RAS pathway comprised the greatest number of clinically relevant genes (n=38), including three common driver genes in lung adenocarcinoma (KRAS, EGFR, BRAF). Activation of the RTK/RAS pathway leads to cell proliferation and growth (33), which are fundamental for tumor cell development (34,35). This may explain the high frequencyof RTK/RAS pathway alteration in our study cohort. Similar findings were reported in the lung adenocarcinoma TCGA cohort (5). Furthermore, the proportion of patients with alterations in the RTK/RAS pathway remained stable across all NPA groups. This suggests RTK/RAS pathway activation may be an early event in lung adenocarcinoma. However, this finding warrants further validation from dynamic tumor genomic profiling and multisite tumor sequencing.
Increases in NPA were significantly associated with increases in TMB (ρ=0.50; P<0.001) and FGA (ρ=0.31; P<0.001). This suggests that, as more mitogenic signaling pathway alterations occur, there may also be more gene replication errors (such as gene mutations or copy number alterations). However, although significant, the magnitude of correlation (Spearman correlation coefficient=0.12) between TMB and FGA is considered negligible on the basis of the interpretation (26) by Hinkle et al. Neither was shown to be an independent predictor of DFS on multivariable analysis. In a pathway-centric analysis, some pathways, such as cell cycle, are more copy number–driven, whereas other pathways, such as TGFβ, are more mutation-driven (5). This suggests TMB and FGA can each play a unique role in assessing therapeutic response and clinical outcomes in patients with lung adenocarcinoma. For example, higher TMB is associated with potentially better efficacy of immunotherapy (18), whereas higher FGA may be associated with worse outcomes following immunotherapy (36). Finally, high TMB has been associated with conflicting prognoses in lung cancer (23,25), whereas increasing FGA has been associated with worse outcomes in prostate, breast, and colorectal cancers (24,37,38).
Increases in NPA were significantly associated with tumor SUVmax, solid morphologic appearance on preoperative CT, pathologic tumor size, aggressive histologic subtype, LVI, VPI, and pathologic lymph node status. All these clinicopathologic features have been previously associated with poor outcomes following complete resection of lung adenocarcinoma (11,39,40). Finally, we found that increasing NPA was independently associated with worse DFS.
Analysis of mutual exclusivity and co-occurrence revealed interplay within and across pathways. When these findings were combined with therapeutic actionability from a pathway-centric perspective, we found that 84% of actionable alterations were located within the RTK/RAS pathway, and tumors with RTK/RAS pathway alterations had the lowest mean NPA. Since activation of alternative pathways is an important mechanism of acquired resistance to targeted therapy (41), our results suggest the success of drugs in targeting tumors with an isolated RTK/RAS pathway alteration may be secondary to the intrinsically low number of alternative pathways. As MDM2 and CDK4 are found in a similar location on chromosome 12q13-15, the amplification of these two genes often occurs together. Using OncoKB to investigate therapeutic actionability, we found that both genes are actionable (level 2B and level 4). This implies the combination of targeted therapies may be effective in patients with alterations in both pathways, which is supported by preclinical studies (42).
Limitations of our study include using DNA sequencing data alone, whereas previous pathway analyses were curated using RNA-seq data (5). Therefore, if a gene is epigenetically silenced through promoter hypermethylation, it would be missed in our analysis. Moreover, the lack of RNA-seq or reverse phase protein array data precludes validation of somatic alterations that can affect the transcriptomic and protein level(s). We also did not evaluate tumor clonality. When two pathways are co-altered, we are unable to distinguish between clonal and subclonal driver mutations. Our ability to assess spatial heterogeneity of pathway alterations over time is limited, as we obtained a single sample per patient. More longitudinal data (i.e., multisampling over time) would allow investigation of the temporal ordering of pathway alterations. We analyzed primarily mitogenic signaling pathways; pathways of other oncogenic processes, such as DNA repair, ubiquitination, and metabolic pathways, were not examined. Most of our cohort are early-stage; thus, our results may not be applicable to patients with more-advanced-stage disease.
Oncogenic pathway analysis using a 10-pathway template characterized by 109 genes from broad-panel NGS revealed important associations with clinicopathologic features. NPA is an independent risk factor for DFS following complete resection of lung adenocarcinoma and is associated with potential therapeutic actionability. Integration of pathway alteration annotation into broad-panel NGS may help stratify patients’ risk of recurrence and aid in selection of optimized induction or adjuvant therapeutic strategies.
Supplementary Material
Translational Relevance.
Broad-panel tumor genomic profiling typically uses a gene-centric approach for analysis and to guide clinical decision-making, although most of the genomic data remain unused. We hypothesized that a pathway-centric approach using next-generation sequencing may offer additional information to identify patients at high risk of recurrence following resection of lung adenocarcinoma. We interrogated 492 lung adenocarcinoma specimens and identified number of pathway alterations (NPA) to be independently associated with disease-free survival. Moreover, NPA and specific pathway perturbations were associated with clinicopathologic features of tumors at high risk of recurrence. Finally, we identified specific pathway alteration co-occurrences that implicate possible functional synergies for targeting both pathways in patients at high risk of recurrence. Collectively, these observations highlight the prognostic importance and potential discovery of therapeutic vulnerabilities when a tumor genomic pathway-centric analysis is utilized using broad-panel next-generation sequencing.
Acknowledgments
We gratefully acknowledge the members of the Molecular Diagnostics Service in the Department of Pathology. David B. Sewell, of the MSK Department of Surgery, provided superb editorial assistance.
Financial support: This work was supported by the National Cancer Institute (grant numbers R01CA217169 to D.R.J., R01CA240472 to D.R.J., and R01CA236615 to P.S.A.); the Department of Defense (grant number LC160212 to P.S.A); and the National Institutes of Health (grant number T32CA009501 to W.S.B.). In addition, this work was funded in part by the Marie-Josée and Henry R. Kravis Center for Molecular Oncology and by the National Cancer Institute/National Institutes of Health (Cancer Center Support Grant P30CA008748).
Disclosure statement: P.S.A. has received funding from ATARA Biotherapeutics and OSE Immunotherapies and has patents on a mesothelin chimeric antigen receptor and PD-1 dominant negative receptor. M.J.B. is a consultant for AstraZeneca Pharmaceuticals. J.M.I. has stock ownership in LumaCyte. V.W.R. receives grant support from Stand Up To Cancer and Genelux. M.L. has served on ad hoc advisory boards at Astra-Zeneca, Bristol-Myers Squibb, Takeda, Bayer, and Merck and has received research support from Loxo Oncology and Helsinn Healthcare. D.B.S. serves as a consultant for Pfizer, Loxo Oncology, Vivideon Therapeutics, Lilly Oncology, and Illumina. M.F.B. has consulted for Roche and received research support from Illumina. D.R.J. serves as a senior medical advisor for Diffusion Pharmaceuticals and a consultant for Merck and AstraZeneca. All other authors have nothing to disclose. The funding sources had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
The order of this author has changed.
References
- 1.Korf BR, Rehm HL. New approaches to molecular diagnosisnew approaches to molecular diagnosis. JAMA 2013;309(14):1511–21. [DOI] [PubMed] [Google Scholar]
- 2.Presley CJ, Tang D, Soulos PR, Chiang A, Longtine J, Adelson K, et al. Association of broad-based genomic sequencing with survival among patients with advanced non–small cell lung cancer in the community oncology setting. JAMA 2018;320(5):469–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castellano E, Santos E. Functional specificity of ras isoforms: so similar but so different. Genes Cancer 2011;2(3):216–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yuen ST, Davies H, Chan TL, Ho JW, Bignell GR, Cox C, et al. Similarity of the phenotypic patterns associated with BRAF and KRAS mutations in colorectal neoplasia. Cancer Res 2002;62(22):6451–5. [PubMed] [Google Scholar]
- 5.Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, et al. Oncogenic signaling pathways in The Cancer Genome Atlas. Cell 2018;173(2):321–37. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jordan EJ, Kim HR, Arcila ME, Barron DA, Chakravarty D, Gao J, et al. Prospective comprehensive molecular characterization of lung adenocarcinomas for efficient patient matching to approved and emerging therapies. Cancer Discov 2017:CD-16-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511(7511):543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hung JJ, Hsu WH, Hsieh CC, Huang BS, Huang MH, Liu JS, et al. Post-recurrence survival in completely resected stage I non-small cell lung cancer with local recurrence. Thorax 2009;64(3):192–6. [DOI] [PubMed] [Google Scholar]
- 9.Kelsey CR, Marks LB, Hollis D, Hubbs JL, Ready NE, D’Amico TA, et al. Local recurrence after surgery for early stage lung cancer: an 11-year experience with 975 patients. Cancer 2009;115(22):5218–27. [DOI] [PubMed] [Google Scholar]
- 10.Saynak M, Veeramachaneni NK, Hubbs JL, Nam J, Qaqish BF, Bailey JE, et al. Local failure after complete resection of N0-1 non-small cell lung cancer. Lung Cancer 2011;71(2):156–65. [DOI] [PubMed] [Google Scholar]
- 11.Taylor MD, Nagji AS, Bhamidipati CM, Theodosakis N, Kozower BD, Lau CL, et al. Tumor recurrence after complete resection for non-small cell lung cancer. Ann Thorac Surg 2012;93(6):1813–20; discussion 20-1. [DOI] [PubMed] [Google Scholar]
- 12.Brandt WS, Bouabdallah I, Tan KS, Park BJ, Adusumilli PS, Molena D, et al. Factors associated with distant recurrence following R0 lobectomy for pN0 lung adenocarcinoma. J Thorac Cardiovasc Surg 2018;155(3):1212–24. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding L, Bailey MH, Porta-Pardo E, Thorsson V, Colaprico A, Bertrand D, et al. Perspective on oncogenic processes at the end of the beginning of cancer genomics. Cell 2018;173(2):305–20 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martini N, Melamed MR. Multiple primary lung cancers. J Thorac Cardiovasc Surg 1975;70(4):606–12. [PubMed] [Google Scholar]
- 15.Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn 2015;17(3):251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio Cancer Genomics Portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2(5):401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6(269):pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rizvi H, Sanchez-Vega F, La K, Chatila W, Jonsson P, Halpenny D, et al. Molecular determinants of response to anti-programmed cell death (PD)-1 and anti-programmed death-ligand 1 (PD-L1) blockade in patients with non-small-cell lung cancer profiled with targeted next-generation sequencing. J Clin Oncol 2018;36(7):633–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res 2016;44(16):e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol 2017;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schemper M, Smith TL. A note on quantifying follow-up in studies of failure time. Control Clin Trials 1996;17(4):343–6. [DOI] [PubMed] [Google Scholar]
- 22.Ciriello G, Gatza Michael L, Beck Andrew H, Wilkerson Matthew D, Rhie Suhn K, Pastore A, et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell 2015;163(2):506–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Devarakonda S, Rotolo F, Tsao M-S, Lanc I, Brambilla E, Masood A, et al. Tumor mutation burden as a biomarker in resected non–small-cell lung cancer. J Clin Oncol 2018;36(30):2995–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hieronymus H, Schultz N, Gopalan A, Carver BS, Chang MT, Xiao Y, et al. Copy number alteration burden predicts prostate cancer relapse. Proc Natl Acad Sci U S A 2014;111(30):11139–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Owada-Ozaki Y, Muto S, Takagi H, Inoue T, Watanabe Y, Fukuhara M, et al. Prognostic impact of tumor mutation burden in patients with completely resected non-small cell lung cancer: brief report. J Thorac Oncol 2018;13(8):1217–21. [DOI] [PubMed] [Google Scholar]
- 26.Hinkle DE, Wiersma W, Jurs SG. Applied statistics for the behavioral sciences. http://catalog.hathitrust.org/api/volumes/oclc/50716608.html 2003.
- 27.Etemadmoghadam D, Weir BA, Au-Yeung G, Alsop K, Mitchell G, George J, et al. Synthetic lethality between CCNE1 amplification and loss of BRCA1. Proc Natl Acad Sci U S A 2013;110(48):19489–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mina M, Raynaud F, Tavernari D, Battistello E, Sungalee S, Saghafinia S, et al. Conditional selection of genomic alterations dictates cancer evolution and oncogenic dependencies. Cancer Cell 2017;32(2):155–68 e6. [DOI] [PubMed] [Google Scholar]
- 29.Nissan MH, Pratilas CA, Jones AM, Ramirez R, Won H, Liu C, et al. Loss of NF1 in cutaneous melanoma is associated with RAS activation and MEK dependence. Cancer Res 2014;74(8):2340–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Armenia J, Wankowicz SAM, Liu D, Gao J, Kundra R, Reznik E, et al. The long tail of oncogenic drivers in prostate cancer. Nature Genet 2018;50(5):645–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leiserson MDM, Vandin F, Wu H-T, Dobson JR, Eldridge JV, Thomas JL, et al. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nature Genet 2015;47(2):106–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garraway LA, Lander ES. Lessons from the cancer genome. Cell 2013;153(1):17–37. [DOI] [PubMed] [Google Scholar]
- 33.Imperial R, Toor OM, Hussain A, Subramanian J, Masood A. Comprehensive pancancer genomic analysis reveals (RTK)-RAS-RAF-MEK as a key dysregulated pathway in cancer: Its clinical implications. Semin Cancer Biol 2019;54:14–28. [DOI] [PubMed] [Google Scholar]
- 34.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144(5):646–74. [DOI] [PubMed] [Google Scholar]
- 35.Feitelson MA, Arzumanyan A, Kulathinal RJ, Blain SW, Holcombe RF, Mahajna J, et al. Sustained proliferation in cancer: mechanisms and novel therapeutic targets. Semin Cancer Biol 2015;35 Suppl:S25–S54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davoli T, Uno H, Wooten EC, Elledge SJ. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 2017;355(6322). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bao L, Qian Z, Lyng MB, Wang L, Yu Y, Wang T, et al. Coexisting genomic aberrations associated with lymph node metastasis in breast cancer. J Clin Invest 2018;128(6):2310–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang L, Feizi N, Chi C, Hu P. Association analysis of somatic copy number alteration burden with breast cancer survival. Front Genet 2018;9(421). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Isaka M, Kojima H, Takahashi S, Omae K, Ohde Y. Risk factors for local recurrence after lobectomy and lymph node dissection in patients with non-small cell lung cancer: Implications for adjuvant therapy. Lung Cancer 2018;115:28–33. [DOI] [PubMed] [Google Scholar]
- 40.Uramoto H, Tanaka F. Prediction of recurrence after complete resection in patients with NSCLC. Anticancer Res 2012;32(9):3953–60. [PubMed] [Google Scholar]
- 41.Nagano T, Tachihara M, Nishimura Y. Mechanism of resistance to epidermal growth factor receptor-tyrosine kinase inhibitors and a potential treatment strategy. Cells 2018;7(11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laroche-Clary A, Chaire V, Algeo MP, Derieppe MA, Loarer FL, Italiano A. Combined targeting of MDM2 and CDK4 is synergistic in dedifferentiated liposarcomas. J Hematol Oncol 2017;10(1):123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.