Abstract
Antibody-drug conjugates (ADCs) selectively deliver chemotherapeutic agents to target cells and are important cancer therapeutics. However, the mechanisms by which ADCs are internalized and activated remain unclear. Using CRISPR-Cas9 screens, we uncover many known and novel endolysosomal regulators as modulators of ADC toxicity. We identify and characterize C18ORF8/RMC1 as a regulator of ADC toxicity through its role in endosomal maturation. Through comparative analysis of screens with ADCs bearing different linkers, we show that a subset of late endolysosomal regulators selectively influence toxicity of noncleavable linker ADCs. Surprisingly, we find cleavable valine-citrulline linkers can be processed rapidly after internalization without lysosomal delivery. Lastly, we show that sialic acid depletion enhances ADC lysosomal delivery and killing in diverse cancer cell types, including with FDA (US Food and Drug Administration)-approved trastuzumab emtansine (T-DM1) in Her2-positive breast cancer cells. Together, these results reveal new regulators of endolysosomal trafficking, provide important insights for ADC design and identify candidate combination therapy targets.
Antibody-drug conjugates (ADCs) are an emerging class of targeted cancer therapeutics with immense promise and demonstrated clinical success1,2. ADCs combine monoclonal antibodies with highly toxic small molecules to selectively deliver chemotherapeutic agents to antigen-expressing tumor cells. Four ADCs have been approved by the FDA since 2011, and more than 80 distinct ADCs are currently being tested in clinical trials for a range of cancers2. Despite intense clinical interest, the mechanisms by which ADCs enter and kill the cell—a process comprising many steps, including internalization, intracellular trafficking, catalytic processing and lysosomal escape—remain incompletely understood.
Our current understanding of ADC uptake and trafficking is largely based on studies of endogenous ligands and their receptors. ADCs are thought to bind their target antigen on the cell surface, undergo internalization and traffic to the lysosome. In the lysosome, ADCs are processed by proteolytic and other enzymes that cleave the conjugated drug from the antibody, triggering ‘payload’ release and toxicity. However, antibody binding may alter receptor internalization and trafficking pathways, leading to a decrease in delivery to the lysosome and increased delivery to other cellular compartments3,4, resulting in suboptimal killing. Altered trafficking routes have also been implicated in resistance to ADC treatments4,5, highlighting the need for a comprehensive understanding of how ADC trafficking is controlled.
The chemical linkage between the antibody and the conjugate drug is a critical feature that ensures the stability of ADCs in circulation, while allowing their toxic payload to be released within target cells. Linkers used for ADCs are often classified as either ‘cleavable’ or ‘noncleavable’, and both designs are being used in the clinic1. Cleavable linkers are engineered to be sensitive to enzymes, acidic or reductive conditions, releasing the ADC payload after exposure to these intracellular stimuli1. However, some cleavable linkers have been associated with nonspecific, extracellular release and off-target killing6. In contrast, noncleavable linkers are more stable in circulation, as they require antibody degradation to become cytotoxic1,6. A substantial complication for noncleavable linkers is that their payload remains attached to the linker and the conjugating amino acids, assemblies that tend to be membrane impermeable and require the activity of lysosomal transporters to escape into the cytosol.
The optimal linker type remains an open question, as existing clinical and preclinical data have been decidedly mixed. For example, preclinical studies of trastuzumab emtansine (T-DM1; Kadcyla) showed that the noncleavable thioether linker (SMCC) was more effective than cleavable versions in vivo7. However, noncleavable linkers are ineffective when used in targeting certain cancer antigens8. While further trials and optimization could improve linker efficacy, it is likely that these results depend on intracellular trafficking of the ADC and the genetic landscape of the tumors.
Here, we use a genome-wide CRISPR knockout screen to identify modulators of ADC toxicity in an unbiased fashion, followed by a series of targeted secondary screens to identify genes with differential roles in processing of ADCs with noncleavable linkers versus cleavable valine-citrulline (VC) linkers. We uncovered many known and novel endolysosomal regulators that influence ADC trafficking and processing, revealing potential resistance mechanisms. We identified C18ORF8/RMC1 as a new modulator of ADC toxicity and further characterized its role in general endolysosomal trafficking. Lastly, we found that depletion of sialic acids sensitizes cells to anti-CD22 ADC treatment by boosting the rate of ADC lysosomal delivery, a finding that also applied to the FDA-approved Her2-targeting ADC trastuzumab emtansine (T-DM1). Together, our results elucidate mechanisms controlling ADC trafficking and toxicity, and provide insight for future ADC development.
Results
Genome-wide screen uncovers regulators of ADC toxicity.
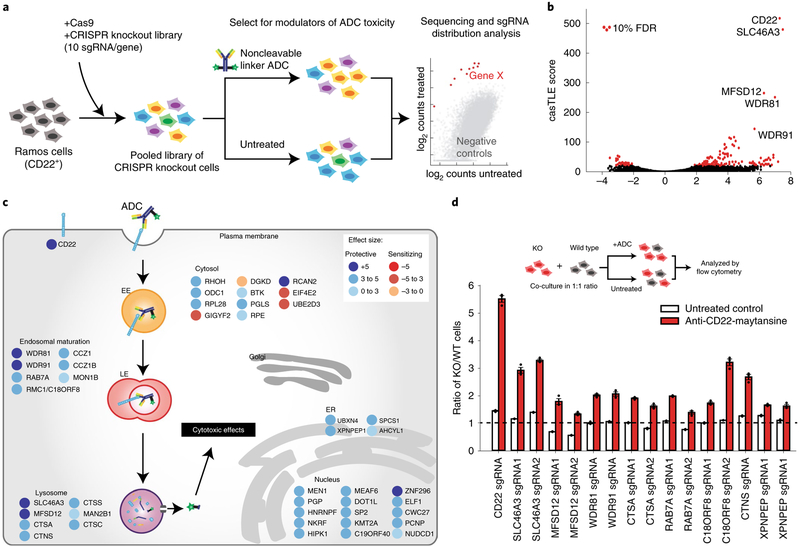
To identify genes regulating ADC toxicity, we conducted a genomewide CRISPR-deletion screen using a CD22-targeting ADC (anti-CD22-asparagine-PEG2-maytansine, hereinafter referred to as anti-CD22-maytansine). The ADC was synthesized using a site-specific conjugation technology based on the aldehyde tag9 and HIPS chemistry10 to attach a microtubule inhibitor payload (maytansine11) coupled through a noncleavable linker to the antibody heavy chain carboxy (C) terminus (Supplementary Fig. 1a). Maytansine is a highly toxic microtubule inhibitor used in many ADCs1. We first tested that the anti-CD22-maytansine ADC inhibits growth of a CD22-positive B lymphocyte Burkitt’s lymphoma cell line (Ramos) in a dose- and antibody-dependent manner (Supplementary Fig. 1b). Then we lentivirally transduced a previously validated single guide RNA (sgRNA) knockout library into Cas9-expressing Ramos cells targeting all protein coding genes with 10 sgRNAs per gene and ~10,000 negative controls12. The cells were split into two conditions in duplicate, one treated with 2 nM anti-CD22-maytansine for three rounds of treatment (each killing ~50% of cells) and the other left untreated. During the 2-week course of the screen, cells with sgRNAs targeting genes required for ADC toxicity are enriched in the ADC treated conditions relative to the untreated conditions, while cells with sgRNAs targeting genes that, when knocked out, sensitize cells to ADC are depleted. The proportion of each sgRNA in the two conditions was measured by deep sequencing and significant regulators of ADC toxicity were identified using casTLE (Fig. 1a)13.
Fig. 1 |. Genome-wide CRISPR screen uncovers diverse endolysosomal regulators of ADC toxicity.
a, Schematic for the genome-wide screen. Ramos cells expressing Cas9 were infected with a lentiviral, genome-wide sgRNA library, the population was split and half the cells were treated with anti-CD22-maytansine ADC for three rounds, killing ~50% cells each round. The resulting populations were subjected to deep sequencing and analysis. The screen was performed in duplicate. b, Volcano plot of all genes indicating effect and confidence scores for genome-wide ADC screen. Effect and confidence scores determined by casTLE. c, Schematic for top 30 and selected endolysosomal trafficking regulator hits in genome-wide screen, color coded by effect size (calculated by casTLE). Subcellular localization is indicated according to gene ontology annotations. EE, early endosomes; LE, late endosomes; ER, endoplasmic reticulum. d, Validation of hits in Ramos cells using competitive-growth assays: cells expressing sgRNAs for knockout (KO) of indicated genes (mCherry+) and control (mCherry−) were cocultured in 1:1 ratio. Cells were either treated with anti-CD22-maytansine ADC or left untreated for 3 d. Resulting ratio of KO:control was determined using flow cytometry. Data are presented as mean ± s.e.m. and are representative of two independent experiments performed in triplicate with consistent results. WT, wild type.
This initial screen identified 171 known and novel regulators of ADC toxicity at a 10% false discovery rate (FDR) (Fig. 1b,c and Supplementary Fig. 1d; see Supplementary Table 1 for the full list). Top hits were validated with individual sgRNA-expressing knockout cell lines in competitive-growth assays (Fig. 1d). Among the strongest hits was CD22, the target antigen of the ADC, as well as SLC46A3, a previously reported lysosomal transporter required for maytansine conjugates with noncleavable linkers to be able to enter the cytosol after proteolytic processing14. Moreover, we identified a number of endolysosomal genes as modifiers of ADC toxicity, including RAB7A (a key regulator of late endolysosomal trafficking15), MON1 and CCZ1 (both activators of RAB7A activity16). The screen revealed that WDR81 and WDR91, two recently characterized regulators of endosomal maturation17,18, modulate ADC toxicity, suggesting that they also mediate ADC lysosomal delivery. Deletion of lysosomal cathepsins A, C and S was protective against ADC killing; these enzymes are likely to be important for proteolytic degradation of the antibody. The screen also uncovered a number of B-cell signaling genes (CD22, CD79b, BTK, SYK, BLNK and PLCG2 (ref.19)), which may influence CD22 expression or B-cell survival, and thus regulate toxicity to the anti-CD22 ADC. We also identified many nuclear-localized genes, such as chromatin modifiers MEN1 (ref.20) and DOT1L21, which may be required for the anti-mitotic activity of maytansine or could indirectly influence ADC trafficking and processing.
Noncleavable linker ADCs require lysosomal delivery.
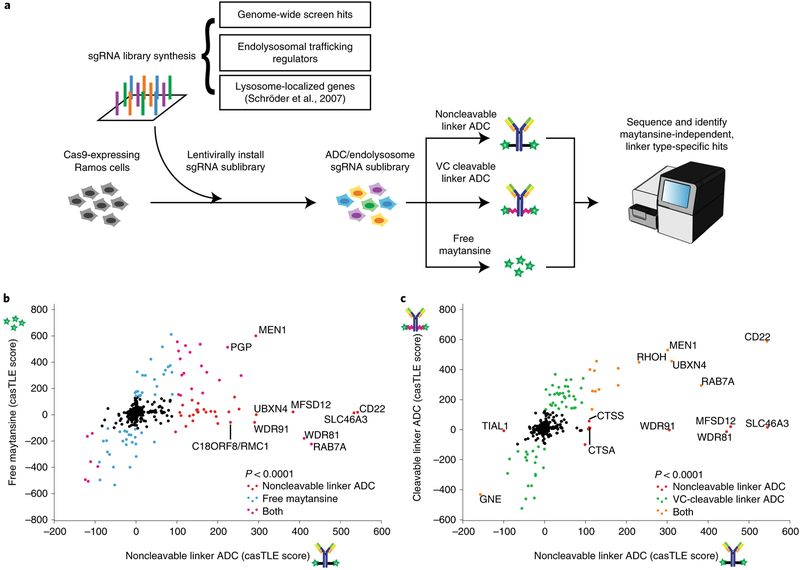
We next aimed to evaluate whether the genetic modifiers identified are important for ADC trafficking, processing or the activity of maytansine. To do this, we performed high cell coverage targeted screens using either an ADC with a noncleavable linker (anti-CD22-maytansine as used above), an ADC with a cleavable linker (anti-CD22-glutamate-PEG2-valine-citrulline-maytansine, hereinafter referred to as anti-CD22-VC-maytansine, see Supplementary Fig. 1b for the structure) or free maytansine. These three treatments were screened in parallel using a focused sublibrary of sgRNAs targeting (1) ~160 of the top hits identified in the genome-wide screen, (2) manually curated endolysosomal regulators and (3) lysosome-localized genes22 (Fig. 2a; see Supplementary Table 5 for list of genes and sgRNAs). In contrast to noncleavable linkers, VC linkers are sensitive to cysteine proteases, such as cathepsin B (ref.23) (Supplementary Fig. 1c), but were recently shown to also be sensitive to other cathepsins24. Proteolytic processing of anti-CD22-VC-maytansine releases a payload that is expected to freely diffuse across endosomal membranes. Similar to the genome-wide screen, screens for each condition were performed in duplicate and either left untreated or treated for three rounds of selection with the ADCs or free maytansine at concentrations that killed ~50% of the cells. Hits were identified using casTLE13 by comparing each treated condition to the untreated control (see Supplementary Table 2 for a complete list of hits and Supplementary Fig. 1e for a comparison with the genome-wide screen).
Fig. 2 |. A subset of endolysosomal trafficking regulators are critical only for toxicity of the noncleavable linker ADC.
a, Schematic for sublibrary targeted screen using anti-CD22-maytansine (noncleavable), anti-CD22-VC-maytansine (cleavable) and free maytansine. The targeted sublibrary was designed, synthesized and lentivirally installed into Cas9-expressing Ramos cells. The Ramos cells were either treated with (1) anti-CD22-maytansine, (2) anti-CD22-VC-maytansine or (3) free maytansine or left untreated. The resulting populations were subjected to deep sequencing and analysis. The screens were performed in duplicate and at 1,000× coverage. b, Comparative analysis of results from anti-CD22-maytansine (noncleavable) and free maytansine sublibrary screens. Signed casTLE scores are reported. c, Comparative analysis of results from anti-CD22-maytansine (noncleavable) and anti-CD22-VC-maytansine (cleavable) sublibrary screens. Signed casTLE scores are reported.
To distinguish genes involved in the activity of maytansine from those influencing ADC trafficking and processing, we compared hits identified in the free maytansine condition with hits obtained using noncleavable anti-CD22-maytansine (Fig. 2b and Supplementary Fig. 2). Genes identified in both conditions, including many nuclear-localized genes, are likely to mediate activity of maytansine. Hits unique to the ADC are likely to influence mechanisms of ADC trafficking, processing or lysosomal escape. These include the target antigen CD22, intracellular trafficking regulators (RAB7A, WDR81 and WDR91), lysosomal cathepsins (CTSS and CTSA) and lysosomal transporters (SLC46A3 and MFSD12), highlighting the importance of lysosomal delivery and processing in ADC toxicity.
To investigate how particular linkers influence ADC toxicity, we compared the hits identified using the noncleavable linker ADC versus the cleavable VC linker ADC (Fig. 2c and Supplementary Fig. 2). As expected, CD22 and hits identified in the free maytansine condition were common hits for both ADCs. We did not identify specific proteases essential for toxicity of anti-CD22-VC-maytansine, likely due to redundancy, as the VC linker can be cleaved by multiple cathepsins24. Strikingly, a subset of endolysosomal trafficking regulators were identified that were only critical for toxicity of the noncleavable linker ADC, but were dispensable for the cleavable linker ADC, suggesting that lysosomal delivery may not be required for processing of VC linkers.
Cleavable linkers are rapidly processed after ADC uptake.
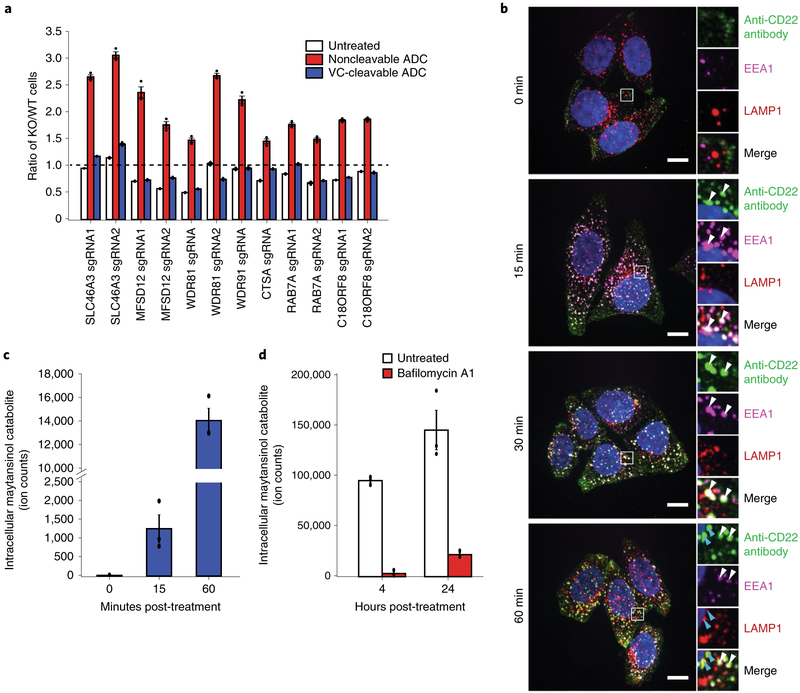
Next, we sought to elucidate the mechanistic differences of proteolytic processing and payload release of noncleavable linker ADC and cleavable linker ADC. We first validated that the subset of endosomal and lysosomal genes identified in our targeted screen are indeed specific to the noncleavable linker ADC (SLC46A3, MFSD12, WDR81, WDR91, CTSA, CTSS and C18ORF8/RMC1) and have little effect on toxicity of the VC cleavable linker ADC in competitive-growth assays (Fig. 3a).
Fig. 3 |. Lysosomal delivery is not required for processing of VC cleavable linkers.
a, Validation of hits in Ramos cells using competitive-growth assays: cells expressing sgRNAs for KO of indicated genes (mCherry+) and control (mCherry−) were cocultured in 1:1 ratio. Cells were either treated with anti-CD22-maytansine, anti-CD22-VC-maytansine (blue) or left untreated (white) for 3 d. Resulting ratio of KO:control was determined using flow cytometry. Data are presented as mean ± s.e.m. and are representative of two independent experiments performed in triplicate with consistent results. b, Internalization of anti-CD22 antibody conjugated to Alexa Fluor 488 (anti-CD22-AF488) was monitored by immunofluorescence microscopy in HeLa cells. Cells were incubated on ice with anti-CD22-AF488 for 20 min. Cells were then washed once with cold PBS and internalization was initiated by prewarmed media. Cells were washed and fixed at indicated time points after initiation of internalization. The bottom row is a magnified view of the white boxes, which shows the colocalization of the anti-CD22 antibodies with early endosomal marker, EEA1, and lysosomal marker, LAMP1. White arrows indicate colocalization of anti-CD22 antibody and EEA1; blue arrows indicate colocalization of anti-CD22 antibody and LAMP1. Scale bars, 10 μm. Images are representative of two independent experiments performed in triplicate. Quantification of colocalization is shown in Supplementary Fig. 3b. c, Maytansinol release from anti-CD22-VC-maytansine in CD22-expressing HeLa cells. Anti-CD22-VC-maytansine (10 nM) was added to cells and reactions were quenched at the indicated time points. The level of intracellular maytansinol catabolite was determined by LC-MS/MS; see Methods for detailed extraction and detection protocol. Maytansinol signal is normalized by cell number and internal standard MMAE. Data are presented as mean ± s.e.m. and are representative of three independent experiments performed in triplicate with consistent results. d, Maytansinol release from anti-CD22-VC-maytansine in CD22-expressing HeLa cells treated with bafilomycin A1. Cells were treated with 100 nM bafilomycin A1 or left untreated for 20 h. Cells were then incubated with anti-CD22-VC-maytansine for the indicated times. The level of intracellular maytansinol catabolite was determined by LC-MS/MS; see Methods for detailed extraction and detection protocol. Maytansinol signal is normalized by cell number and internal standard MMAE. Data are presented as mean ± s.e.m. and are representative of three independent experiments performed in triplicate with consistent results.
Because toxicity of the cleavable VC linker ADCs does not depend on genes regulating endosomal maturation or specific lysosomal proteases, we hypothesize that the VC cleavable linker can be processed in earlier endosomal compartments, and may be able to bypass defects in lysosomal delivery. To test this, we stably expressed CD22 in HeLa cells (Supplementary Fig. 3a) and monitored trafficking of a fluorescently labeled CD22 antibody. We found that the antibody colocalizes with early endosomal marker EEA1 at 15 min after treatment and began to colocalize with lysosomal marker LAMP1 in approximately 1 h (Fig. 3b; see Supplementary Fig. 3b for quantification). We then used liquid chromatography-tandem mass spectrometry (LC-MS/MS) to detect intracellular levels of the maytansinol cleavage product25 in cells treated with the cleavable anti-CD22 ADC at the corresponding time points (Supplementary Fig. 3c,d). The cleaved maytansinol product began to appear as early as 15 min posttreatment (Fig. 3c and Supplementary Fig. 3e), coinciding with the accumulation of anti-CD22 antibodies in early endosomes, as assayed by microscopy, but before accumulation in the lysosome (Fig. 3b and Supplementary Fig. 3b). The maytansinol catabolite continued to accumulate over the course of the experiment (Supplementary Fig. 3e). Release of maytansinol catabolite in Ramos cells followed similar kinetics (Supplementary Fig. 3f). Notably, inhibition of V-ATPase using bafilomycin A1 (ref.26) prevented maytansinol payload release, indicating that cleavage of the VC linker is dependent on proteases that require proper acidification of endosomes and lysosomes (Fig. 3d). Together, our genetic screening and LC-MS/MS results suggest that cleavable versus noncleavable linker ADCs have distinct trafficking requirements, and that lysosomal delivery is less critical for the toxicity of ADCs with cleavable VC linkers.
C18ORF8/RMC1 modulates endolysosomal trafficking of ADCs.
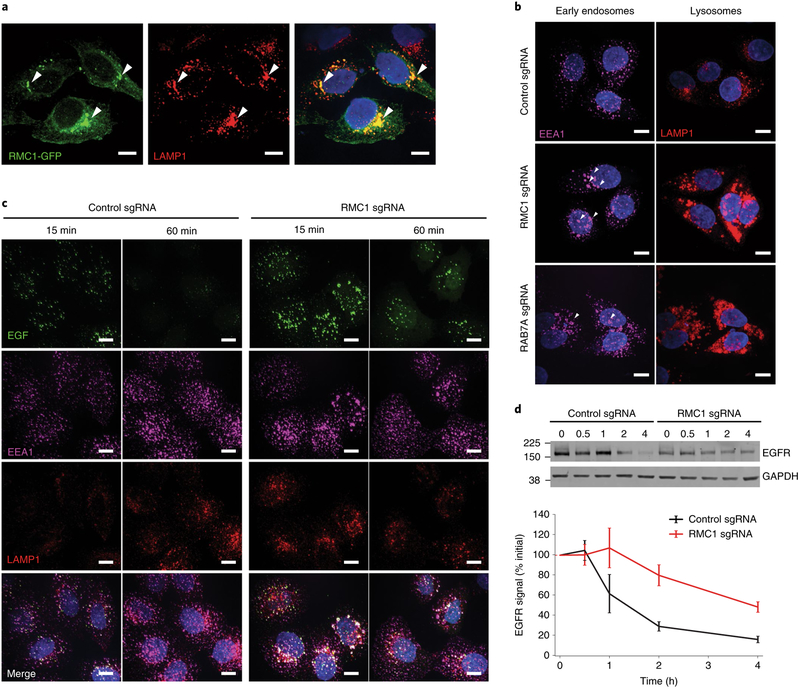
A top maytansine-independent hit in our screen was C18ORF8/RMC1 (hereinafter referred to as RMC1), a gene recently identified as an autophagy regulator acting in concert with MON1 and CCZ1 to activate RAB7 (ref.27). As knockout of either RAB7A or RMC1 protects cells from toxicity of noncleavable linker ADC (Fig. 3a), we hypothesized that RMC1 plays a role in endosomal maturation through regulation of RAB7A activity. We first examined the localization of a C-terminal GFP-fused RMC1 in HeLa cells and found that it colocalizes with the lysosomal marker LAMP1 (Fig. 4a). Next, we knocked down RMC1 or RAB7A using CRISPR interference28 (CRISPRi) in HeLa cells; both perturbations led to enlarged early endosomes and lysosomes (Fig. 4b), accompanied by increased EEA1 and LAMP1 staining (Supplementary Fig. 4a). To investigate the role of RMC1 in general endolysosomal trafficking, we monitored trafficking of epidermal growth factor (EGF), a canonical endocytic cargo that traffics to the lysosome. Depletion of RMC1 resulted in delayed EGF lysosomal delivery and EGF accumulation in early endosomes (Fig. 4c and Supplementary Fig. 4b). Consistently, lysosomal degradation of EGF receptor (EGFR) after EGF treatment was also impaired (Fig. 4d). Finally, to determine how RAB7A activity influences the role of RMC1 in ADC toxicity, we tested whether overexpression of constitutively active RAB7-Q67L re-sensitizes RMC1-deleted cells to noncleavable linker ADCs. Indeed, RMC1-deleted cells overexpressing RAB7-Q67L were more sensitive to ADC treatment than cells expressing GFP control (Supplementary Fig. 4c,d). Together, these results support the hypothesis that RMC1 modulates ADC delivery and toxicity in a RAB7-dependent manner, and, more generally, demonstrate a role for RMC1 in delivery of endocytic substrates to the lysosome.
Fig. 4 |. C18ORF8/RMC1 is required for endosomal trafficking and lysosomal delivery of ADCs.
a, C-terminal GFP-tagged RMC1 localizes to LAMP1-positive, late endosome/lysosomes, visualized by immunofluorescence and confocal microscopy. White arrows indicate sites of colocalization. The experiment was independently repeated twice, with similar results. Scale bars, 10 μm. b, CRISPRi-mediated knockdown of RMC1 or RAB7A in HeLa cells results in enlarged endosomes (EEA1) and lysosomes (LAMP1). The experiment was independently repeated twice, with similar results. Scale bars, 10 μm. c, Trafficking of EGF in RMC1-knockdown HeLa cells. Cells were serum-starved for 16 h, then were placed on ice and treated with EGF for 10 min. Cells were then washed once with cold PBS and internalization was initiated by prewarmed media. Cells were fixed at the indicated time and visualized by immunofluorescence and confocal microscopy. The experiment was independently repeated twice, with similar results. Scale bars, 10 μm. d, EGFR degradation in RMC1-knockdown HeLa cells. Cells were treated with EGF for 10 min, and then EGFR levels were examined at the indicated time points by western blot. EGFR levels were quantified with ImageJ software. Data are presented as mean ± s.e.m. and are representative of two independent experiments performed in triplicate with consistent results. See Supplementary Fig. 8 for uncropped western blot.
Sialic acid depletion sensitizes cells to ADC toxicity.
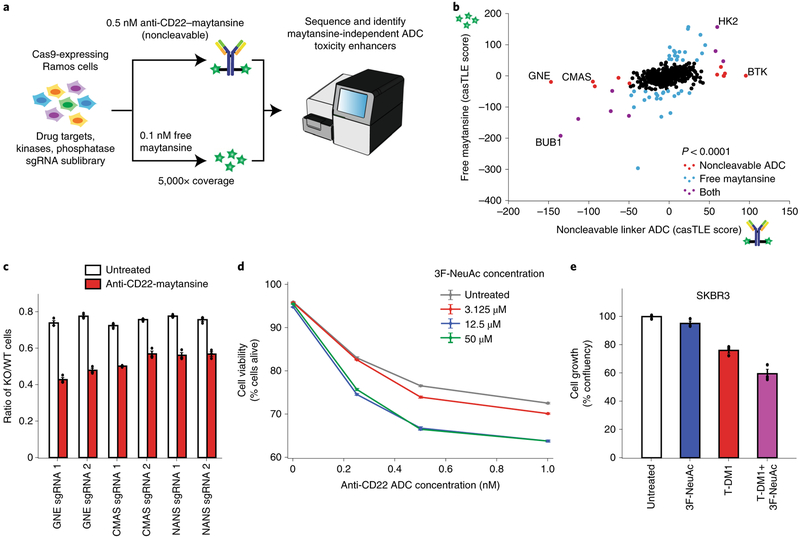
Genes whose deletion sensitizes cancer cells to ADCs could potentially serve as targets for combination therapies. Although the genome-wide and follow-up screens identified many genes and pathways required for ADC toxicity, we identified relatively few genes whose deletion robustly sensitized cells to ADC treatment. We hypothesized that this was due to either insufficient cell coverage (~1,000 cells per sgRNA) or the high dose of ADC used in the screens, both of which could limit the dynamic range of sensitization. We thus repeated the screen at a higher cell coverage (~5,000 cells per sgRNA) and with lower dose of the noncleavable linker ADC (0.5 nM) to search specifically for sensitizing targets (Fig. 5a). To prioritize the identification of candidate combination therapy targets, we screened a targeted sublibrary enriched in known drug targets, kinases and phosphatases. As we were most interested in finding genes that influence ADC trafficking and processing, a free maytansine treatment was also used as a counter-screen to filter out maytansine-dependent genes. As expected, this design identified many strong sensitizing hits that were missed in the genome-wide screen (Supplementary Fig. 5a–c; see Supplementary Table 3 for a complete list of hits).
Fig. 5 |. Inhibition of sialic acid synthesis sensitizes cells to CD22 ADC toxicity.
a, Schematic for targeted screen for identifying sensitizing hits. Ramos cells expressing Cas9 were infected with a lentiviral, sgRNA sublibrary targeting drug targets, kinases and phosphatases. Cells were then treated with either 0.5 nM anti-CD22-maytansine, 0.1 nM free maytansine or left untreated. The resulting populations were subjected to deep sequencing and analysis. The screen was performed in duplicate and at 5,000× coverage. b, Comparative analysis of results from anti-CD22 noncleavable linker ADC (0.5 nM) and free maytansine (0.1 nM) sublibrary screens performed at 5,000× coverage. c, Validation of hits in Ramos cells using competitive-growth assays. Cells expressing sgRNAs for KO of indicated genes (mCherry+) and control (mCherry−) were cocultured in 1:1 ratio. Cells were either treated with anti-CD22 noncleavable ADC or left untreated for 3 d. Resulting ratio of KO:control was determined using flow cytometry. Data are presented as mean ± s.e.m. and are representative of two independent experiments performed in triplicate with consistent results. d, Cell viability of Ramos cells treated with noncleavable ADC and 3F-NeuAc at different concentrations. Cells were first treated with 3F-NeuAc at indicated concentrations for 48 h, then ADCs at indicated concentrations were added. Cell viability, determined by forward and side scatter gating for live cells, was assayed 72 h post-ADC addition by flow cytometry. Data are presented as mean ± s.e.m. and are representative of three independent experiments performed in triplicate with consistent results. e, Cell growth (normalized confluency percentage) of SKBR3 treated with 100 μM 3F-NeuAc, 2 nM T-DM1, or combination of both. Cells were pretreated with 100 μM 3F-NeuAc for 48 h, followed by 72-h incubation with 2 nM T-DM1. Confluency was determined by IncuCyte S3 Live cells analysis system and normalized to maximum confluency of the untreated condition at the end of the 5-d experiment. Data are presented as mean ± s.e.m. and are representative of two independent experiments performed in triplicate with consistent results.
Interestingly, the top two maytansine-independent sensitizing hits, GNE and CMAS, are both members of the sialic acid biosynthesis pathway (Fig. 5b and Supplementary Fig. 5d). Using competitive-growth assays, we validated that disruption of sialylation, by knocking out GNE, CMAS or NANS (an additional pathway member not represented in the sublibrary), sensitizes Ramos cells to ADC treatment (Fig. 5c). In addition, chemical inhibition of sialylation with a fluorinated sialic acid analog, peracetylated 3Fax-NeuAc29 (3F-NeuAc), or enzymatic removal of cell-surface sialic acids by Vibrio cholerae sialidase30, also sensitized Ramos cells to ADC killing (Fig. 5d and Supplementary Fig. 5e). Reduction of cell-surface sialic acids in these cells was confirmed by labeling with Sambucus nigra agglutinin and Erythrina crista-galli lectin, which preferentially bind to α2,6-linked sialic acids29 and ‘uncapped’ glycans31, respectively (Supplementary Fig. 5f–h). Notably, deletion of GNE, CMAS or NANS also sensitized cells to the cleavable linker ADC anti-CD22-VC-maytansine (Supplementary Fig. 5i), suggesting that sensitization by inhibiting sialylation may be broadly useful for ADCs with both noncleavable linkers and cleavable VC linkers. Importantly, pretreatment with 3F-NeuAc does not lead to increased anti-CD22 ADC killing in the CD22-negative chronic myeloid leukemia cell line K562 (Supplementary Fig. 6c), indicating that the increased toxicity observed with sialic acid inhibition is dependent on the specificity of ADC binding.
To investigate whether the sensitizing effects of sialic acid depletion can be generalized to ADCs with different targets and in different cell types, we tested whether pretreatment with 3F-NeuAc also sensitizes Her2-positive cancer cell lines to Her2-targeted ADC trastuzumab emtansine (T-DM1). Indeed, 3F-NeuAc treatment sensitized breast cancer lines SKBR3 and ZR-75-1, and the gastric carcinoma line NCI-C87 to T-DM1 (Fig. 5e and Supplementary Fig. 6a,b), indicating that the strategy of combining ADC and depletion of sialic acids may be useful for multiple cancer antigen targets.
Depletion of sialic acid enhances ADC lysosomal delivery.
To identify which steps of ADC internalization and processing are modulated by sialylation, we considered several models. Inhibition of sialic acid synthesis may: (1) increase ADC target antigen cellsurface expression and/or ADC binding, (2) enhance complement-mediated killing activity or (3) increase the rate of lysosomal delivery. First, we measured CD22 expression and antibody accessibility in Ramos cells, but 3F-NeuAc treatment did not alter cell-surface binding of anti-CD22 ADCs or whole-cell CD22 protein levels (Supplementary Fig. 6d,e). Second, we tested whether the increased killing is dependent on complement activity, but depletion of active complement factors in cell culture media did not abolish the sensitizing effect of 3F-NeuAc on Ramos cells or the breast cancer cell lines SKBR3 and ZR-75-1 (Supplementary Fig. 6f–h).
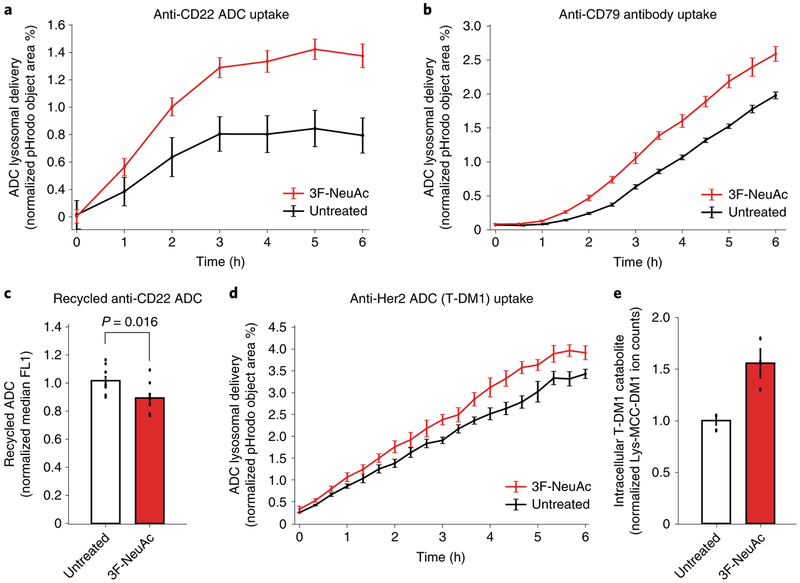
Lastly, we examined whether inhibiting sialylation alters ADC lysosomal delivery. We assayed ADC lysosomal delivery using pHrodo-labeled anti-CD22 ADC, which only fluoresces in acidic environments, such as the lysosome. Cells depleted of sialic acids, either by pretreatment with 3F-NeuAc (Fig. 6a) or by genetic deletion of GNE (Supplementary Fig. 7a), demonstrated increased rates of ADC lysosomal delivery. Treatment of Ramos cells with 3F-NeuAc also increased the rate of maytansinol payload release from the cleavable linker ADC anti-CD22-VC-maytansine, as measured by LC-MS/MS (Supplementary Fig. 7b). To determine whether the increased lysosomal delivery is specific to CD22, we measured the lysosomal trafficking rate of an antibody targeting CD79b, another commonly investigated ADC target32. As a control, we measured uptake of pHrodo-labeled dextran as a marker of general fluid-phase endocytosis33. Interestingly, 3F-NeuAc treatment increased the lysosomal delivery rate of the anti-CD79 antibodies (Fig. 6b) but not that of dextran in Ramos cells (Supplementary Fig. 7c), suggesting that depletion of sialic acids alters the lysosomal delivery of selective substrates.
Fig. 6 |. Depletion of sialic acid enhances rate of ADC lysosomal delivery.
a, Internalization of pHrodo-labeled anti-CD22 ADC in wild-type Ramos cells pretreated with 12.5 μM 3F-NeuAc for 48 h (red) or untreated (black). pHrodo signal was measured using the IncuCyte S3 Live cells analysis system and normalized to cell area. Data are presented as mean ± s.e.m. and are representative of three independent experiments performed in triplicate with consistent results. b, Internalization of pHrodo-labeled anti-CD79 antibody in wild-type Ramos cells pretreated with 12.5 μM 3F-NeuAc for 48 h (red) or untreated (black). pHrodo signal was measured using the IncuCyte S3 Live cells analysis system and normalized to cell area. Data are presented as mean ± s.e.m. and are representative of two independent experiments performed in triplicate with consistent results. c, Recycling of anti-CD22 ADCs in Ramos cells pretreated with 12.5 μM 3F-NeuAc for 48 h (red) or untreated (white). Cells were treated with unlabeled anti-CD22 ADC for 1 h, acid washed to remove surface-bound ADCs and returned to 37 °C for 30 min to allow for ADC recycling. ADCs recycled back to the cell surface are detected by Alexa Fluor 488-labeled anti-human IgG antibodies and analyzed using flow cytometry. Data are presented as mean ± s.e.m. and are representative of two independent experiments with six replicates. d, Internalization of pHrodo-labeled T-DM1 in wild-type SKBR3 cells pretreated with 100 μM 3F-NeuAc for 48 h (red) or untreated (black). pHrodo signal was measured using the IncuCyte S3 Live cells analysis system and normalized to cell area. Data are presented as mean ± s.e.m. and are representative of two independent experiments performed in triplicate with consistent results. e, Accumulation of T-DM1 catabolite Lys-MCC-DM1 in wild-type SKBR3 cells treated with 100 μM 3F-NeuAc. SKBR3 cells were either pretreated with 100 μM 3F-NeuAc or left untreated for 48 h, followed by addition of 10 nM T-DM1. After 24 h, the level of intracellular T-DM1 catabolite, Lys-MCC-DM1, was determined by LC-MS/MS; see Methods for detailed extraction and detection protocol. Lyc-MCC-DM1 signal is normalized by cell number and the internal standard MMAE. Data are presented as mean ± s.e.m. and are representative of three independent experiments performed in triplicate with consistent results.
Sialic acid depletion reduces anti-CD22 ADC recycling.
After internalization, ADCs are either trafficked to the lysosome or recycled back to the plasma membrane4. Therefore, an increase in ADC lysosomal delivery rate could be caused by an increased rate of ADC internalization or a decreased rate of ADC recycling. We first measured uptake of fluorescently labeled anti-CD22 antibodies in Ramos cells, but found that 3F-NeuAc treatment did not alter the rate of antibody internalization (Supplementary Fig. 7d).
Next, we tested whether depletion of sialic acids reduces the rate of recycling of anti-CD22 ADC in Ramos cells. Cells were allowed to take up unlabeled anti-CD22 ADC for 1 h, acid washed to remove surface-bound ADCs and returned to 37 °C to allow for ADC recycling. ADCs that recycled back to the plasma membrane were then detected by a fluorescently labeled anti-human IgG. Indeed, depletion of sialic acids, either by 3F-NeuAc or Vibrio cholerae sialidase treatments, lowered the rate of ADC recycling (Fig. 6c and Supplementary Fig. 7e,f). Together, these results suggest that the increased rate of anti-CD22 ADC lysosomal trafficking after sialic acid depletion is, at least in part, due to a reduction in ADC recycling.
Inhibiting sialylation increases T-DM1 lysosome delivery.
Finally, to determine whether the effects of sialic acid depletion on ADC lysosomal trafficking can be generalized to other ADCs and cancer cell types, we measured the rate of T-DM1 lysosomal delivery and payload release using LC-MS/MS34 in SKBR3 cells. Indeed, enhanced T-DM1 lysosomal delivery and increased T-DM1 catabolite (Lys-MCC-DM1) accumulation were observed in 3F-NeuActreated SKBR3 cells (Fig. 6d,e). Together, these results show that inhibition of sialylation is a generalizable strategy for increasing ADC lysosomal delivery, and suggests that combining ADCs and sialic acid depletion may enhance the efficacy of ADC treatment.
Discussion
Here, we used CRISPR screens to evaluate the mechanism of toxicity for antibody-drug conjugates bearing noncleavable and cleavable valine-citrulline linkers, which are known to have critically different efficacies toward different tumors6–8,35. We identified many known and novel genetic modifiers of ADC toxicity, and uncovered critical roles for endolysosomal regulators in mediating ADC toxicity. We identified C18ORF8/RMC1 as a gene required for endosomal maturation and lysosomal delivery of ADC. Surprisingly, only a subset of the identified endolysosomal regulators are required for toxicity of ADCs with cleavable VC linkers. We showed that this is because payload release from cleavable VC linkers is not dependent on lysosomal delivery, which contrasts with the prevailing wisdom in the field. Finally, we show that depletion of sialic acids sensitizes cells to ADC toxicity by boosting ADC lysosomal delivery. This may enable a novel combination therapy strategy that combines ADCs with modulation of sialic acid levels to increase ADC efficacy. In proof-of-principle studies, this strategy was effective in increasing in vitro killing with both CD22- and Her2-targeting ADCs in multiple cancer cell lines, including lymphoma, breast and gastric cancer.
Although ADCs with noncleavable linkers are more stable and thus elicit less off-target toxicity when used in vivo1,35, some tumors show marked resistance to these agents8,35,36. Our work shows that noncleavable linker ADCs show greater reliance on late endosomal trafficking regulators (RAB7, RMC1, WDR81 and WDR91) and lysosomal cathepsins (cathepsins A, C and S) for toxicity, whereas, surprisingly, ADCs with VC cleavable linkers (designed to be cleaved by cathepsin B) do not. Indeed, payload release from VC-linked ADCs occurs within tens of minutes after ADC internalization, suggesting that resistance to noncleavable ADCs due to impaired lysosomal delivery may be circumvented by ADCs bearing cleavable VC linkers. While similar payload release kinetics have been previously reported37, our genetic screen results provide additional evidence that lysosomal delivery is not essential for toxicity of ADCs with cleavable VC linkers. These results may help explain why ADCs with cleavable linkers are efficacious in a broader range of target cancers6,8 and remain toxic to cell lines resistant to noncleavable linker ADCs38,39. Importantly, these genes regulating late endosomal trafficking and processing of ADCs may serve as potential predictive biomarkers for resistance against noncleavable linker ADCs.
Our work also identified regulators of endolysosomal trafficking, as well as roles for previously characterized genes. For example, we find that WDR81 and WDR91, genes previously shown to be involved in endosomal maturation and autophagy17,18,40, are also selectively required for trafficking of ADCs to the lysosome. In addition, we demonstrate a role for C18ORF8/RMC1 in intracellular trafficking. This gene was recently identified to play a role in autophagy, and was found in a complex with CCZ1 and MON1 (ref.27), which we also identify in our screen. Here, we show that RMC1 is required for proper endolysosomal trafficking and degradation of EGFR. Additionally, RMC1 mediates ADC delivery and toxicity in a RAB7A-dependent manner, which is consistent with the reported role of RMC1 in RAB7A activation during autophagic flux27.
Many of the hits identified in our study are required for ADC toxicity; therefore loss-of-function of these genes may be potential resistance mechanisms for ADCs used in the clinic. For example, decreased expression of SLC46A3, a lysosomal transporter that we identified as critical for cytotoxicity of the noncleavable linker ADC, has been shown to be a mechanism of innate and acquired resistance to other noncleavable ADCs in patient-derived xenografts and primary multiple myeloma bone marrow samples14,39,41. Moreover, a major mechanism of acquired resistance to ADCs, which has been shown in in vitro models, is altered ADC trafficking and lysosomal function39,42,43, a signature that is also identified in our screens.
We found that depletion of sialic acids renders cells more sensitive to ADCs due to an increased rate of ADC lysosomal delivery in a wide range of cancer cell types. We show that this may, at least in part, be due to a reduction in ADC recycling rates. Previous studies have shown that altering cell-surface sialic acid levels can influence receptor endocytosis and intracellular trafficking through disrupting Siglec receptor clustering44, membrane fluidity45 or binding of cell-surface glycans to the extracellular galectin lattice46. Further research will be required to determine whether these mechanisms play a role in influencing the rate of ADC recycling and lysosomal delivery.
Because hypersialylation in malignant cells has been shown to be a mechanism of immune evasion47, preclinical studies focusing on depleting tumor sialylation have developed several promising therapeutic strategies48. One such example is the fluorinated sialic acid analog used in our study, 3F-NeuAc. Our data show that combining 3F-NeuAc with either anti-CD22 ADCs or the anti-Her2 ADC T-DM1 resulted in increased killing. Notably, 3F-NeuAc is effective in reducing global glycan sialylation in mice but has deleterious ‘on-target’ effects, such as impairing liver and kidney function49. This systemic toxicity can be circumvented by intratumoral injections of 3F-NeuAc, resulting in decreased tumor sialylation levels50. Alternatively, targeted-glycan modification using antibody-sialidase conjugates can be used for removal of target cell-surface sialic acids30. Our work suggests that combining ADCs with these and other methods to deplete sialylation in cancer cells may be a promising therapeutic strategy that warrants further exploration. Together, our studies reveal mechanisms underlying ADC trafficking and toxicity, provide insight for ADC design and a generalizable method to investigate the biology of diverse ADCs and identify candidate combination strategies that may improve therapeutic efficacy.
Methods
Cell culture.
Ramos cells, SKBR3, ZR-75-1 and NCI-N87 cells were obtained from American Type Culture Collection (ATCC) and grown in RPMI 1640 Medium (Life Technologies, catalog no. 11875093) supplemented with 10% fetal bovine serum (FBS, Fisher, catalog no. SH30910), 2 mM l-glutamine (Fisher, catalog no. SH3003401) and 1% penicillin-streptomycin (Fisher, catalog no. SV30010). HeLa cells (ATCC) were grown in DMEM (Life Technologies, catalog no. 11995073) supplemented with 10% FBS, 2 mM l-glutamine and 1% penicillin-streptomycin. All cells were cultured at 37 °C with 5% CO2.
Genome-wide CRISPR-Cas9 screens in Ramos cells.
Our previously established genome-wide, 10 sgRNA per gene CRISPR deletion library1 was synthesized, cloned and infected into Cas9-expressing Ramos cells, as previously described1. Briefly, ~300 million Ramos cells stably expressing SFFV-Cas9-BFP were infected with the CRISPR knockout library at a multiplicity of infection (MOI) of 0.3–0.4. Cells expressing sgRNAs were selected for using puromycin (1 μg ml−1) for 3–4 d such that >90% of cells were mCherry-positive as measured by flow cytometry. Selected cells were then allowed to recover and expand in puromycin-free media for up to 7 d. Deep sequencing was used to confirm sufficient sgRNA representation in the library.
For the screen, cells were split into two conditions, each in duplicate: an untreated control group and ADC treated group (2 nM anti-CD22-Asp-PEG2-maytansine). Cells were incubated with 2 nM ADC for 48 h for each round of treatment, after which the ADC was removed from media by pelleting cells and re-suspending in fresh ADC-free media. This treatment occurred four times over 3 weeks, with cells recovering to ~90% viability after each round of selection. Over the course of the screen, cells were maintained at 500,000 cells ml−1. At the end of the screen, 200 million cells were recovered from each condition and pelleted by centrifugation. Genomic DNA of each condition was extracted using Qiagen DNA Blood Maxi kit (catalog no. 51194). The sgRNA sequences were amplified and prepared for sequencing as previously described2. These libraries were then sequenced using an Illumina NextSeq with ~40 million reads per condition; ~200× coverage per library element). Analysis and comparison of guide composition of ADC treated versus untreated conditions were performed using casTLE as previously described1,3. See Supplementary Table 1 for the complete genome-wide screen result.
Targeted screens in Ramos cells.
The secondary screen library targets a total of 811 genes (10 sgRNA per gene): 159 of 171 genes that passed 10% FDR from the Ramos genome-wide screen, genes previously implicated in endolysosomal trafficking and genes found to localize to lysosomes in a proteomics study4, along with 1,500 negative-control sgRNAs (see Supplementary Table 5 for a complete list of genes and sgRNAs). The library oligonucleotides were synthesized by Agilent Technologies and cloned into pMCB320 using BstXI/BlpI overhangs after PCR amplification. The Cas9-expressing Ramos cells were lentivirally infected with the secondary screen library at an MOI of 0.3–0.4, as previously described. After puromycin selection (1 μg ml−1 for 3 d) and expansion, cells were treated with either 2 nM anti-CD22-maytansine, 0.5 nM anti-CD22-VC-maytansine, 0.2 nM free maytansine or left untreated as control, with each in duplicate at ~3,000× coverage per library element. Each treatment lasted 48 h and was repeated four times over 3 weeks, with cells recovering to ~90% viability after each round of selection. As with the genome-wide screen, cells were maintained at 500,000 cells per ml−1 throughout the screen. At the end of the screen, 30 million cells were used for genomic extraction for each condition, sequenced (5–10 million reads per condition) and analyzed using casTLE. See Supplementary Table 2 for the complete sublibrary screen results.
Targeted screen for uncovering sensitizing hits.
To uncover additional sensitizing hits, we used a library of ~2,000 genes targeting known drug targets, kinases and phosphatase in the human genome1. The library was lentivirally infected into Cas9-expressing Ramos cells at an MOI of 0.3–0.4, as previously described. After puromycin selection (1 μg ml−1 for 3 d) and expansion, cells were treated with either 0.5 nM anti-CD22 noncleavable linker ADC, 0.1 nM free maytansine or left untreated, each in duplicate at ~5,000× coverage per library element. Each treatment lasted 24 h, killing 20–30% of cells. Cells were allowed to recover to 90% survival after each treatment. The treatment was repeated three times over 2 weeks. Cells were maintained at 500,000 cells per ml throughout the screen. At the end of the screen, 50 million cells were used for genomic extraction for each condition and sequenced with 10 million reads per condition. Hits were identified using casTLE. See Supplementary Table 3 for the complete screen results.
Competitive-growth assays in Ramos cells.
To validate results of genome-wide and secondary screens, we infected Cas9-expressing Ramos cells with plasmids expressing both a single specific sgRNA and mCherry, as previously described12 (see Supplementary Table 1 for sgRNA sequences). The infected cells were puromycin-selected for 3 d and allowed to recover for 2 d. Equal numbers of sgRNA/mCherry-positive cells were cocultured with wild-type/mCherry-negative cells, and were subsequently treated with different ADCs at the same concentration used in the screens. The percentage of mCherry-positive cells was quantified using a BD Accuri C6 flow cytometer after 72 h. All results are normalized to ratio of control sgRNA/ mCherry-positive:wild-type/mCherry-negative cells51.
Western blotting.
Cells were lysed for 30 min at 4 °C using protein extraction buffer (300 mM NaCl, 100 ml Tris pH 8, 0.2 mM EDTA, 0.1% Triton X-100, 10% glycerol) supplemented with 1× cOmplete protease inhibitor (Roche, catalog no. 11697498001) and centrifuged to collect the supernatant lysate. Protein concentration was measured by DC Protein Assay Kit (Bio-Rad, catalog no. 5000111EDU), separated on SDS-PAGE gels (Life Technologies, catalog no. NP0322BOX) and transferred to nitrocellulose membranes (Bio-Rad, catalog no. 1620146). The following primary antibodies were used: rabbit monoclonal anti-EGFR (1:2,000, Abcam ab52894) and mouse monoclonal anti-GAPDH (1:10,000, Fisher AM4300). Secondary antibodies used were: goat polyclonal anti-mouse IRDye 680RD (1:10,000, Li-COR 925–68070) and goat polyclonal anti-rabbit IRDye 800CW (1:10,000, Li-COR 925–32211). Images were acquired using the Li-COR Odyssey CLx imaging system.
Confocal microscopy.
Cells were grown on glass cover slips or glass well plates and were stained using standard immunocytochemistry techniques. Briefly, cells were fixed with 4% formaldehyde, permeabilized with 0.1% Triton X-100, blocked with 3% BSA and stained with the following antibodies: mouse monoclonal anti-EEA1 (1:1,000, BD 610457) and rabbit monoclonal anti-LAMP1 (1:1,000, CST 9091S). Cover slips were mounted using VectaShield. Images were acquired using a Nikon Eclipse microscope with a ×100 oil immersion objective.
Sample preparation and measurement of ADC catabolite by LC-MS/MS.
For each condition, 10 million Ramos cells, or 5 million HeLa cells, were plated in 10-cm dishes and allowed to grow for 24 h before addition of 10 nM anti-CD22-VC-maytansine or T-DM1. At appropriate time points, cells were collected (trypsinized in the case of HeLa cells) and pelleted. The cell pellets were washed twice with PBS and extracted with 50% acetonitrile in water containing 10 nM monomethyl auristatin E (MMAE) as internal standard. The cell/acetonitrile mixture was vortexed for 30 s, and allowed to rest on ice for 1 min, and this vortex/rest step was repeated two more times. The mixture was then left to shake at 4 °C for 15 min, incubated at −20 °C for 2 h and centrifuged at 10,000g. The clarified supernatant was transferred to a new tube and dried under N2 gas flow. The dried extract was reconstituted in 100 μl 50% acetonitrile in water before analysis by LC-MS/MS, which was performed using an Agilent 6470 triple quadrupole mass spectrometer with an Agilent Jet Stream electrospray ionization source. Analytes were separated on a ZORBAX RRHD Extend C18 column (1.8 μm particle size × 2.1 mm diameter × 50 mm length) with gradient elution at a flow of 0.6 ml min−1. The mobile phases were A, 0.1% formic acid in water, and B, 0.1% formic acid in acetonitrile. Gradient conditions were: isocratic at 95% A for 0.2 min, followed by a linear gradient from 5% to 95% B from 0.2 to 4 min, followed by an isocratic hold at 95% B for 1 min (4.2–5.2 min) followed by immediate return to 5% A (5.2–5.3 min). The total run time was 6 min. The ion source conditions were: gas temperature 250 °C, gas flow 12 l min−1, nebulizer 25 psi and sheath gas temperature and flow rate 300 °C and 12 l min−1, respectively.
Multiple-reaction monitoring (MRM) was used to monitor the analytes and internal standard. The mass transitions monitored for quantification were m/z 749.4→547.2 for maytansinol payload released from VC linker, m/z 1,103.6→547.2 for Lys-MCC-DM1 and m/z 718.5→686.5 for MMAE. For Lys-MCC-DM1 and MMAE, we confirmed that observed chromatographic peaks eluted at the same time as authentic standards. For the maytansinol payload released from the VC linker, no authentic standard was available. During methods development, we used additional MRMs m/z 749.4→485.2, 1103.6→485.2 and 718.5→152.1 to confirm the identity of the released maytansinol payload, Lys-MCC-DM1 and MMAE, respectively. Peak detection and integration were performed with Agilent MassHunter software. For Figs. 3c and 6d, and Supplementary Figs. 3d,e and 6d, the obtained ion counts in biological samples were normalized to ion counts for the MMAE internal standard, and this ratio was normalized to cell number.
ADC lysosomal delivery assays in IncuCyte.
For lysosomal delivery assays, cells were seeded at 10,000 cells per well in a poly-l-lysine (0.1% w/v)-coated 96-well plate and allowed to adhere overnight. Anti-CD22 ADC, anti-Her2 ADC T-DM1 or anti-CD79b antibodies were labeled with IncuCyte Human FabFluor-pH Red antibody labeling reagent (IncuCyte, catalog no. 4722) by mixing ADC and FabFluor reagent at a molar ratio of 1:3 in RPMI media and incubating for 15 min. The antibody-FabFluor mixture was added to cells to a final concentration of 4 μg ml−1. The cell plate was then transferred into an IncuCyte S3 and images were obtained with a ×20 objective every hour for 24 h. Analysis was performed with IncuCyte S3 integrated software.
Antibody uptake assay using flow cytometry.
For each replicate, 0.5 million Ramos cells were collected, washed once with cold dPBS and incubated on ice with 10 μg ml−1 anti-CD22 labeled with Alexa Fluor 488 for 20 min. Ten times volume of prewarmed media was then added to cells to initiate internalization. Cells were then placed in a water bath at 37 °C. At 0, 2, 5, 10, 15, 60 and 120 min, cells were removed from the water bath and placed on ice. After washing twice with cold dPBS, cells were acid washed (0.2 M acetic acid, 0.5 M NaCl, pH 2.8) with shaking at 4 °C for 10 min to remove surface-bound antibodies. Cells were then washed once more with cold dPBS and antibody internalization was analyzed on a BD Accuri C6 flow cytometer. Median FL1 of live Ramos cells (gated by forward and side scatter) was reported.
ADC recycling assay using flow cytometry.
For each replicate, 0.5 million Ramos cells were collected, washed once with cold dPBS and re-suspended with warm media with 10 μg ml−1 anti-CD22-maytansine ADC. After an hour of ADC internalization, cells were washed twice with cold dPBS and acid washed (0.2 M acetic acid, 0.5 M NaCl, pH 2.8) with shaking at 4 °C for 10 min to remove surfacebound ADCs. Cells were then washed twice with cold dPBS and re-suspended in warm media and placed in a water bath at 37 °C for 30 min to allow for ADC recycling. Cells were then placed on ice and washed twice with cold dPBS, and incubated on ice with Alexa Fluor 488-labeled anti-human IgG for 45 min. Cells were then washed twice with cold dPBS and analyzed on a BD Accuri C6 flow cytometer. Median FL1 of live Ramos cells (gated by forward and side scatter) was reported.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The complete results of genome-wide screens and secondary screens are in Supplementary Datasets 1–4. All data are available from the corresponding author upon reasonable request.
Code availability
casTLE v.1.0 is available at https://bitbucket.org/dmorgens/castle.
Supplementary Material
Acknowledgements
We thank G. Hess, P. Drake, A. Lu and S. Pfeffer for assistance and discussions, A. Gupta for help with LC-MS/MS and M. Dubreuil and E. Jeng for comments on the manuscript. We also thank M. Pegram and W. Liang for generously providing the T-DM1 used in our experiments. This work was funded by grants from NIH (grant nos. F31 GM126688-01A1 to C.K.T., NIH 1DP2HD084069-01 to M.C.B. and NIH R01 CA227942 to C.R.B.).
Footnotes
Competing interests
R.M.B. and D.R. are employees of Catalent Biologics. C.R.B. is a cofounder and Scientific Advisory Board member of Redwood Bioscience, which generated antibody-drug conjugates used in this work. Stanford University has filed a patent application based on the findings in this article.
Supplementary information is available for this paper at https://doi.org/10.1038/s41589-019-0342-2.
Reprints and permissions information is available at www.nature.com/reprints.
Correspondence and requests for materials should be addressed to M.C.B.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, statements of code and data availability and associated accession codes are available at https://doi.org/10.1038/s41589-019-0342-2.
References
- 1.Beck A, Goetsch L, Dumontet C & Corvaïa N Strategies and challenges for the next generation of antibody-drug conjugates. Nat. Rev. Drug Discov 16, 315–337 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Lyon R Drawing lessons from the clinical development of antibody-drug conjugates. Drug Discov. Today Technol 30, 105–109 (2018). [DOI] [PubMed] [Google Scholar]
- 3.St Pierre CA, Leonard D, Corvera S, Kurt-Jones EA & Finberg RW Antibodies to cell surface proteins redirect intracellular trafficking pathways. Exp. Mol. Pathol 91, 723–732 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ritchie M, Tchistiakova L & Scott N Implications of receptor-mediated endocytosis and intracellular trafficking dynamics in the development of antibody drug conjugates. MAbs 5, 13–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loganzo F et al. Tumor cells chronically treated with a trastuzumabmaytansinoid antibody-drug conjugate develop varied resistance mechanisms but respond to alternate treatments. Mol. Cancer Ther 14, 952–963 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Donaghy H Effects of antibody, drug and linker on the preclinical and clinical toxicities of antibody-drug conjugates. MAbs 8, 659–671 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis Phillips GD et al. Targeting HER2-positive breast cancer with trastuzumab-DM1, an antibody-cytotoxic drug conjugate. Cancer Res 68, 9280–9290 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Polson AG et al. Antibody-drug conjugates for the treatment of non-Hodgkin’s lymphoma: target and linker-drug selection. Cancer Res 69, 2358–2364 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Rabuka D, Rush JS, deHart GW, Wu P & Bertozzi CR Site-specific chemical protein conjugation using genetically encoded aldehyde tags. Nat. Protoc 7, 1052–1067 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agarwal P et al. Hydrazino-Pictet-Spengler ligation as a biocompatible method for the generation of stable protein conjugates. Bioconjug. Chem 24, 846–851 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Lopus M et al. Maytansine and cellular metabolites of antibodymaytansinoid conjugates strongly suppress microtubule dynamics by binding to microtubules. Mol. Cancer Ther 9, 2689–2699 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morgens DW et al. Genome-scale measurement of off-target activity using Cas9 toxicity in high-throughput screens. Nat. Commun 8, 15178 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morgens DW, Deans RM, Li A & Bassik MC Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat. Biotechnol 34, 634–636 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamblett KJ et al. SLC46A3 is required to transport catabolites of noncleavable antibody maytansine conjugates from the lysosome to the cytoplasm. Cancer Res 75, 5329–5340 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Hyttinen JMT, Niittykoski M, Salminen A & Kaarniranta K Maturation of autophagosomes and endosomes: a key role for Rab7. Biochim. Biophys. Acta 1833, 503–510 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Poteryaev D, Datta S, Ackema K, Zerial M & Spang A Identification of the switch in early-to-late endosome transition. Cell 141, 497–508 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Liu K et al. Negative regulation of phosphatidylinositol 3-phosphate levels in early-to-late endosome conversion. J. Cell Biol 212, 181–198 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rapiteanu R et al. A genetic screen identifies a critical role for the WDR81-WDR91 complex in the trafficking and degradation of tetherin. Traffic 17, 940–958 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burger JA & Wiestner A Targeting B cell receptor signalling in cancer: preclinical and clinical advances. Nat. Rev. Cancer 18, 148–167 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Mould AW et al. Global expression profiling of murine MEN1-associated tumors reveals a regulatory role for menin in transcription, cell cycle and chromatin remodelling. Int. J. Cancer 121, 776–783 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Wood K, Tellier M & Murphy S DOT1L and H3K79 methylation in transcription and genomic stability. Biomolecules 8, 11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schröder B et al. Integral and associated lysosomal membrane proteins. Traffic 8, 1676–1686 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Dubowchik GM et al. Cathepsin B-labile dipeptide linkers for lysosomal release of doxorubicin from internalizing immunoconjugates: model studies of enzymatic drug release and antigen-specific in vitro anticancer activity. Bioconjug. Chem 13, 855–869 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Caculitan NG et al. Caculitan, N. G. et al. Cathepsin B is dispensable for cellular processing of cathepsin B-cleavable antibody-drug conjugates. Cancer Res 77, 7027–7037 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Zmolek W, Bañas S, Barfield RM, Rabuka D & Drake PM A simple LC/MRM-MS-based method to quantify free linker-payload in antibody-drug conjugate preparations. J. Chromatogr. B 1032, 144–148 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Yoshimori T, Yamamoto A, Moriyama Y, Futai M & Tashiro Y Bafilomycin A1, a specific inhibitor of vacuolar-type H+-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J. Biol. Chem 266, 17707–17712 (1991). [PubMed] [Google Scholar]
- 27.Pontano Vaites L, Paulo JA, Huttlin EL & Harper JW Systematic analysis of human cells lacking ATG8 proteins uncovers roles for GABARAPs and the CCZ1/MON1 regulator C18orf8/RMC1 in macro and selective autophagic flux. Mol. Cell. Biol 38, e00392–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qi LS et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rillahan CD et al. Global metabolic inhibitors of sialyl- and fucosyltransferases remodel the glycome. Nat. Chem. Biol 8, 661–668 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao H, Woods EC, Vukojicic P & Bertozzi CR Precision glycocalyx editing as a strategy for cancer immunotherapy. Proc. Natl Acad. Sci. USA 113, 10304–10309 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu AM et al. Differential affinities of erythrina cristagalli lectin (ECL) toward monosaccharides and polyvalent mammalian structural units. Glycoconj. J 24, 591–604 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Polson AG et al. Antibody-drug conjugates targeted to CD79 for the treatment of non-Hodgkin lymphoma. Blood 110, 616–623 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Li L et al. The effect of the size of fluorescent dextran on its endocytic pathway. Cell Biol. Int 39, 531–539 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Liu Y et al. LC-MS/MS method for the simultaneous determination of Lys-MCC-DM1, MCC-DM1 and DM1 as potential intracellular catabolites of the antibody-drug conjugate trastuzumab emtansine (T-DM1). J. Pharm. Biomed. Anal 137, 170–177 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Lu J, Jiang F, Lu A & Zhang G Linkers having a crucial role in antibody-drug conjugates. Int. J. Mol. Sci 17, 561 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abdollahpour-Alitappeh M et al. Antibody-drug conjugates (ADCs) for cancer therapy: strategies, challenges, and successes. J. Cell Physiol 234, 5628–5642 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Lee B-C et al. FRET reagent reveals the intracellular processing of peptide-linked antibody-drug conjugates. Bioconjug. Chem 29, 2468–2477 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Wang H et al. Aberrant intracellular metabolism of T-DM1 confers T-DM1 resistance in human epidermal growth factor receptor 2-positive gastric cancer cells. Cancer Sci 108, 1458–1468 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li G et al. Mechanisms of acquired resistance to trastuzumab emtansine in breast cancer cells. Mol. Cancer Ther 17, 1441–1453 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Liu K et al. WDR91 is a Rab7 effector required for neuronal development. J. Cell Biol 216, 3307–3321 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kinneer K et al. SLC46A3 as a potential predictive biomarker for antibodydrug conjugates bearing non-cleavable linked maytansinoid and pyrrolobenzodiazepine warheads. Clin. Cancer Res 24, 6570–6582 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Ríos-Luci C et al. Resistance to the antibody-drug conjugate T-DM1 Is based in a reduction in lysosomal proteolytic activity. Cancer Res 77, 4639–4651 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Sung M et al. Caveolae-mediated endocytosis as a novel mechanism of resistance to trastuzumab emtansine (T-DM1). Mol. Cancer Ther 17, 243–253 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Zhang M & Varki A Cell surface sialic acids do not affect primary CD22 interactions with CD45 and surface IgM nor the rate of constitutive CD22 endocytosis. Glycobiology 14, 939–949 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Mathew MP et al. Metabolic flux-driven sialylation alters internalization, recycling, and drug sensitivity of the epidermal growth factor receptor (EGFR) in SW1990 pancreatic cancer cells. Oncotarget 7, 66491–66511 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nabi IR, Shankar J & Dennis JW The galectin lattice at a glance. J. Cell Sci 128, 2213–2219 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Pearce OMT & Läubli H Sialic acids in cancer biology and immunity. Glycobiology 26, 111–128 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Wang L, Liu Y, Wu L & Sun X-L Sialyltransferase inhibition and recent advances. Biochim. Biophys. Acta 1864, 143–153 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Macauley MS et al. Systemic blockade of sialylation in mice with a global inhibitor of sialyltransferases. J. Biol. Chem 289, 35149–35158 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Büll C et al. Sialic acid blockade suppresses tumor growth by enhancing T-cell-mediated tumor immunity. Cancer Res 78, 3574–3588 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Deans RM et al. Parallel shRNA and CRISPR-Cas9 screens enable antiviral drug target identification. Nat. Chem. Biol 12, 361–366 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete results of genome-wide screens and secondary screens are in Supplementary Datasets 1–4. All data are available from the corresponding author upon reasonable request.