Abstract
Background
The tumor suppressor role of lncRNA PTCSC3 has been reported in papillary thyroid carcinoma, our study aimed to investigate its involvement in gastric cancer.
Methods
Tumor tissues and adjacent healthy tissues were collected from gastric cancer patients. Expression of PTCSC3 and lncRNA Linc-pint in these tissues was analyzed by RT-qPCR. The interaction between PTCSC3 and Linc-pint was analyzed by overexpression experiments. Cell proliferation and stemness were analyzed by CCK-8 assay and cell stemness assay, respectively.
Results
PTCSC3 and lncRNA Linc-pint were both downregulated in tumor tissues than in adjacent healthy tissues of gastric cancer patients. Low levels of PTCSC3 and Linc-pint were closely correlated with poor survival. PTCSC3 and Linc-pint overexpression inhibited tumor growth and cancer cell stemness, while Linc-pint knockdown played an opposite role an attenuated the effects of PTCSC3 overexpression. Expression levels of PTCSC3 and Linc-pint were significantly correlated in tumor tissues but not in adjacent healthy tissues. Overexpression of PTCSC3 and Linc-pint upregulated the expression of each other.
Conclusion
PTCSC3 inhibits tumor growth and cancer cell stemness in gastric cancer by interacting with lncRNA Linc-pint.
Keywords: gastric cancer, lncRNA PTCSC3, lncRNA Linc-pint, growth, stemness
Introduction
Non-coding RNAs (ncRNAs) are emerging classes of non-protein coding RNA transcripts with pivotal roles in gene regulation.1 It has been reported that more than 98% of all human genome transcriptional output is ncRNAs and most ncRNAs are expressed in specific types of tissues and cells or under certain stress conditions or during specific developmental stages, indicating their extensive involvement in human body growth and development.2 Long non-coding RNAs, or lncRNAs, are a subgroup of ncRNAs longer than 200 nt.3 Functional characterization has revealed that lncRNAs not only participate in almost all important physiological processes, but also have critical functions in the development of diseases,4 such as different types of cancer.5,6 However, function of most lncRNAs is still unknown.
Gastric cancer is one of the most frequently diagnosed malignancies and is also the third leading cause of cancer-related deaths worldwide.7 Most gastric cancer patients at early stages can be cured by surgical operations. However, more than half of patients with advanced gastric cancer will die of carcinoma recurrence even after curative gastrectomy.8 Therefore, more studies are needed to further characterize the molecular alterations involved in the pathogenesis of gastric cancer.9,10 Our genome-wide transcriptome identified a large number of differentially expressed lncRNAs in gastric cancer patients (data not shown). Among those differential lncRNAs, lncRNA PTCSC3 which has been reported to be downregulated in papillary thyroid carcinoma,11 is also downregulated in gastric cancer and is positively correlated with lncRNA Linc-pint, a characterized tumor suppressor in lymphoblastic leukemia.12 Our study was therefore carried out to investigate the involvement of PTCSC3 and Linc-pint in gastric cancer and explored the possible interactions between them.
Materials and Methods
Research Subjects
Our study included 78 patients with gastric cancer were diagnosed and treated in Fourth Hospital of Hebei Medical University from March 2010 to March 2015. Inclusion criteria: 1) gastric cancer patients who were diagnosed by pathological biopsy; 2) patients who were diagnosed for the first time and no treatment was performed; 3) patients willing to participate. Exclusion criteria: 1) patients who received treatment before admission or who were transferred from other hospitals; 2) patients who were complicated with other severe diseases, such as other types of cancer and systemic infection. The 78 patients included 43 females and 35 males, and age ranged from 29 to 66 years, with a mean age of 48.6±6.1 years. According to AJCC staging, there were 16 cases at stage I, 22 cases at stage II, 24 cases at stage III and 16 cases at stage IV. This study had been approved by Ethic Committee of Fourth Hospital of Hebei Medical University before patient admission and all participants signed informed consent.
Specimen Collections and Cell Lines
Biopsy was performed to collect cancer and paracancerous tissues from each patient. Tissues were stored in liquid nitrogen before use.
SNU-1 and Hs 746T human gastric cancer cell lines were used in this study to perform in vitro experiments. Cells of these two cell lines were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). ATCC-formulated RPMI-1640 Medium (ATCC) supplemented with 10% fetal bovine serum (FBS, ATCC) was used to cultivate the cells of both cell lines at 37°C in a 5% CO2 incubator.
Follow-Up
All patients were followed up for 5 years after admission to record their overall conditions. Patients failed to complete follow-up and patients who died of other causes during follow-up were not included in this study.
Cell Transfections
PcDNA3.1 vector (Invitrogen) expressing PTCSC3 or Linc-pint was designed and constructed by Sangon (Shanghai, China). Linc-pint siRNA and negative control siRNA were also designed and synthesized by Sangon. Cells of SNU-1 and Hs 746T cell lines were cultivated overnight to each 70–80% confluence. All cell transfections were performed using Lipofectamine 2000 reagent (cat no. 11668-019; Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) with all operations performed in strict accordance with manufacturer’s instructions. Doses of vectors and siRNAs were 10 and 45 nM, respectively. Un-transfected cells were control cells, and cell transfected with empty pcDNA3.1 vectors or negative control siRNAs were negative control cells. Cells were harvested at 24 h after transfection for subsequent experiments.
Total RNA Extraction and Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR)
Tissues were ground in liquid nitrogen, followed by the addition of RNAzol reagent (Sigma-Aldrich, St. Louis, MO, USA) to extract total RNA. RNAzol reagent was also directly mixed with in vitro cultivated cells to extract total RNA. Total RNAs were reversely transcribed using SuperScript IV Reverse Transcriptase (Thermo Fisher Scientific, lnc.). To detect the expression of PTCSC3 and Linc-pint, PCR reaction systems were prepared using Applied Biosystems™ PowerUp™ SYBR™ Green Master Mix (Thermo Fisher Scientific, lnc.) and all PCR reactions were performed on CFX96 Touch Deep Well™ Real-Time PCR Detection System (Bio-Rad) with 18S RNA as the endogenous control. Primers of PTCSC3 and Linc-pint as well as 18S RNA were designed and synthesized by Sangon (Shanghai, China). Expression of TCSC3 and Linc-pint was normalized to 18S RNA using 2−ΔΔCq method.
In vitro Cell Proliferation Assay
Cells were harvested at 24 h after transfection and in vitro cell proliferation assay was performed using Cell Counting Kit-8 (CCK-8) kit (Sigma-Aldrich) to explore the effects of TCSC3 and Linc-pint on gastric cancer cell proliferation. Briefly, cells were mixed with ATCC-formulated RPMI-1640 Medium containing 10% FBS to make single-cell suspensions. Cell density was adjusted to 4×104 cell/mL and cell suspensions were transferred to a 96-well plate with 100 μL cell suspension in each well. Cells were cultivated (37°C and 5% CO2) and 10 μL CCK was added into each well 24, 48, 72 and 96 hrs later. After that, cells were cultivated from additional 4 hrs and 10 μL DMSO was added. OD values at 450 nm were measured to reflect cell proliferation rates.
Flow Cytometry
Cells were harvested at 24 hrs after transfection and flow cytometry was performed to explore the effects of PTCSC3 and Linc-pint on gastric cancer cell stemness. Cells were harvested by trypsinization. IgG1-PE or CD133-PE antibody (130-093-193, Meltenyi Biotec, Germany) was used to incubate with cells (5×105 cells per mL) at 4°C for 20 mins. After that, cells were harvested and dissolved in PBS. FACS Aria system (BD Immunocytometry Systems, San Jose, CA, USA) was then used to detect signals and data were processed using Cell Quest software (Becton Dickinson Ltd).
Statistical Analysis
All experiments were performed 3 times and data were recorded as mean ± standard deviation. Comparisons of expression levels of TCSC3 and Linc-pint between cancer and paracancerous tissues were performed by paired t test. Correlations between expression levels of PTCSC3 and Linc-pint were analyzed by Pearson’s correlation coefficient. Comparisons among multiple groups were performed by one-way ANOVA followed by Tukey’s test. Based on the expression levels of PTCSC3 and Linc-pint in tumor tissues, patients were divided into high (n=36) and low (n=42) PTCSC3 level groups, as well as high (n=38) and low (n=40) Linc-pint level groups according to Youden’s index. Survival curves were plotted based on Kaplan–Meier method and compared by log-rank t test. Differences with p<0.05 were statistically significant.
Results
PTCSC3 and Linc-pint Were Both Downregulated in Cancer Tissues of Gastric Cancer Patients
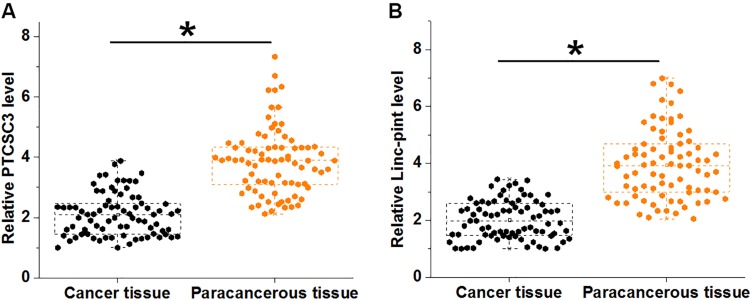
RT-qPCR was performed to detect the expression of PTCSC3 and Linc-pint in both cancer and paracancerous tissues of 78 gastric cancer patients. Compared with cancer tissues, expression levels of PTCSC3 were significantly lower in paracancerous tissues (Figure 1A, p<0.05). In addition, Linc-pint was also downregulated in cancer tissues than in paracancerous tissues (Figure 1B, p<0.05).
Figure 1.
PTCSC3 and Linc-pint were both downregulated in cancer tissues of gastric cancer patients. RT-qPCR results showed that PTCSC3 (A) and Linc-pint (B) were both downregulated in cancer tissues of gastric cancer patients compared with paracancerous tissues (*p<0.05).
Low Levels of PTCSC3 and Linc-pint in Cancer Tissue Indicate Poor Survival
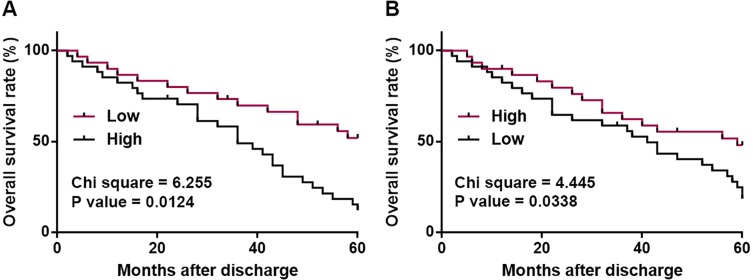
Based on the expression levels of PTCSC3 and Linc-pint in tumor tissues, patients were divided into high (n=36) and low (n=42) PTCSC3 level groups, as well as high (n=38) and low (n=40) Linc-pint level groups according to Youden’s index. Survival curves were plotted based on Kaplan–Meier method and compared by log-rank t test. As shown in Figure 2A, patients in low-level PTCSC3 group showed significantly lower overall survival rate compared with patients in high PTCSC3 group (Figure 2A). In addition, the overall survival rate of patients in low Linc-pint level group was also significantly lower than that of patients in high Linc-pint level group (Figure 2B).
Figure 2.
Low levels of PTCSC3 and Linc-pint in cancer tissue indicate poor survival. Survival curve analysis on 5-year follow-up data showed that low levels of PTCSC3 (A) and Linc-pint (B) in cancer tissue indicate poor survival.
Expression Levels of PTCSC3 and Linc-pint Were Significantly Correlated in Cancer Tissues
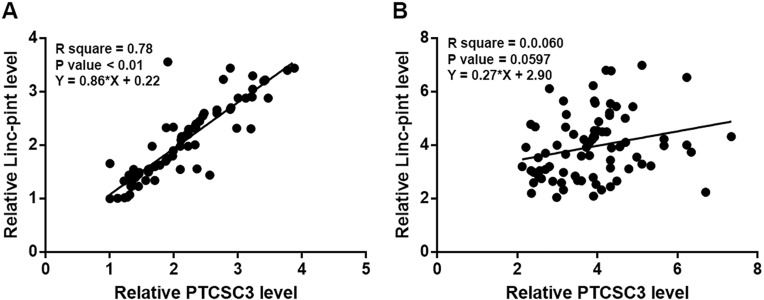
Correlations between expression levels of PTCSC3 and Linc-pint were analyzed by Pearson’s correlation coefficient. It was observed that expression levels of PTCSC3 and Linc-pint were significantly and positively correlated in cancer tissues (Figure 3A). In contrast, the correlation between expression levels of PTCSC3 and Linc-pint was not significant in paracancerous tissues (Figure 3B).
Figure 3.
Expression levels of PTCSC3 and Linc-pint were significantly correlated in cancer tissues. Pearson’s correlation coefficient analysis showed that expression levels of PTCSC3 and Linc-pint were significantly correlated in cancer tissues (A) but not in paracancerous tissues (B).
PTCSC3 and Linc-pint Upregulated Each Other in Gastric Cancer Cells
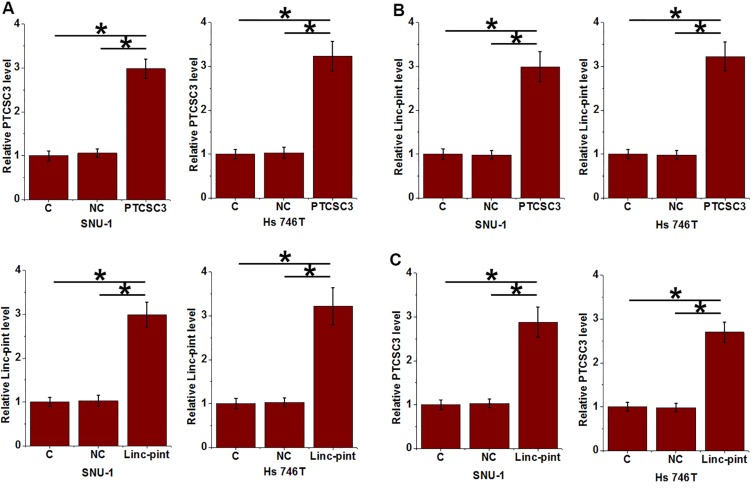
PTCSC3 and Linc-pint were overexpressed in cells of both SNU-1 and Hs 746T cell line to further investigate the interactions between PTCSC3 and Linc-pint (Figure 4A, p<0.05). Compared with control (C) and negative control (NC), overexpression of PTCSC3 led to significantly upregulated Linc-pint expression in cells of both cell lines (Figure 4B, p<0.05). In addition, overexpression of Linc-pint also mediated the upregulation of PTCSC3 in those cells (Figure 4C, p<0.05).
Figure 4.
PTCSC3 and Linc-pint upregulated each other in gastric cancer cells. Overexpression of PTCSC3 and Linc-pint was reached in cells of both SNU-1 and Hs 746T cell line at 24 h after transfection (A). Overexpression of PTCSC3 led to significantly upregulated Linc-pint expression in cells of both cell lines (B). In addition, overexpression of Linc-pint also mediated the upregulation of PTCSC3 in those cells (C) (*p<0.05).
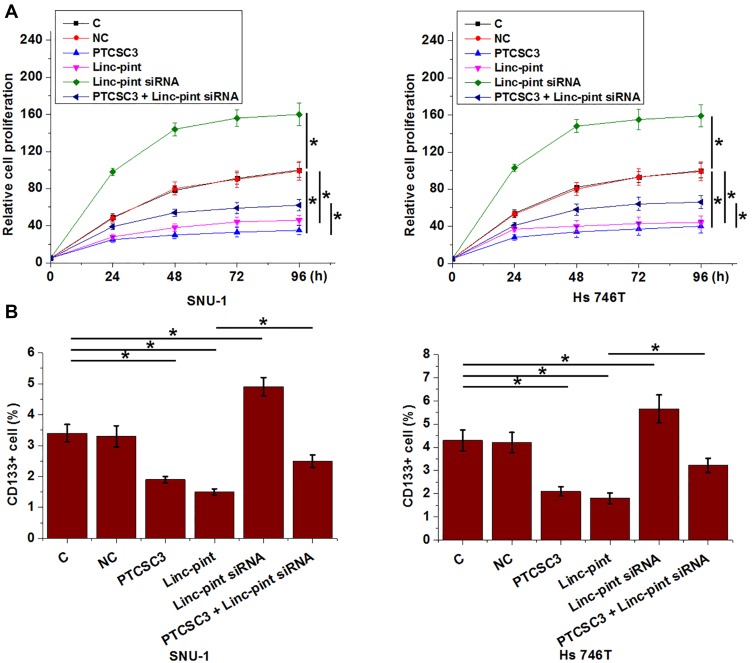
PTCSC3 and Linc-pint Regulated Gastric Cancer Cell Proliferation and Stemness
Compared with control (C) and negative control (NC), overexpression of PTCSC3 and Linc-pint led to significantly inhibited proliferation (Figure 5A) and decreased percentage of CD133+ cells (Figure 5B) (p<0.05). Linc-pint siRNA played an opposite role and attenuated the effects of PTCSC3 overexpression on cancer cell proliferation and stemness (p<0.05).
Figure 5.
PTCSC3 and Linc-pint regulated gastric cancer cell proliferation and stemness. Overexpression of PTCSC3 and Linc-pint led to significantly inhibited proliferation (A) and decreased percentage of CD133+ cells (B). Linc-pint siRNA played an opposite role and attenuated the effects of PTCSC3 overexpression on cancer cell proliferation and stemness (*p<0.05).
Discussion
LncRNAs are critical determinants in cancer biology, while function of most lncRNAs remains unknown. The key finding of the present study is that PTCSC3 and Linc-pint two lncRNAs play a role in tumor suppressor in gastric cancer by forming a positive feedback regulation circle.
Survival of advanced gastric cancer patients is still poor.13 Therefore, development of novel prognostic markers is always needed to design individualized post-surgery treatment and care strategies. Some prognostic markers, such as ImmunoScore signature,14 have shown potentials of clinical application. Our studies identified PTCSC3 and Linc-pint as two promising prognostic biomarkers for gastric cancer. RNA expression detection as an easy disease prediction approach may be superior to most other approaches in terms of the input of time and cost. However, more clinical studies are needed to further verify the clinical values of these two lncRNAs.
LncRNAs regulate gene expression at different levels, such as methylation and posttranscriptional and translational regulation.15,16 However, interactions between different lncRNAs are largely unknown. In the present study, we proved that PTCSC3 and Linc-pint may form a positive feedback regulation circle in gastric cancer, and this positive feedback regulation is involved in the regulation of gastric cancer cell proliferation and stemness. Interestingly, no significant correlation between expression levels of PTCSC3 and Linc-pint was observed in paracancerous tissue. Therefore, certain pathological factors may be involved to mediate the feedback regulation between PTCSC3 and Linc-pint.
It is also worth noting that Linc-pint silencing only partially attenuated the inhibitory effects of PTCSC3 overexpression on cancer cell proliferation and stemness. Therefore, PTCSC3 may interact with multiple factors to participate in the pathogenesis of gastric cancer. Instead of the interaction between two lncRNAs, PTCSC3 is more likely a component of regulation network. More studies are needed to further characterize this network. In this study, we only used CD133+ marker to analyze cancer cell stemness. Our future studies will try to include more stemness markers to further verify our conclusions.
Conclusion
In conclusion, PTCSC3 and Linc-pint are downregulated in gastric cancer. PTCSC3 and Linc-pint may inhibit gastric cancer by forming a positive feedback regulation circle to inhibit cancer cell proliferation and reduce cell stemness.
Data Sharing Statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
The present study was approved by the Ethics Committee of Fourth Hospital of Hebei Medical University. The research has been carried out in accordance with the World Medical Association Declaration of Helsinki. All patients provided written informed consent prior to their inclusion within the study.
Author Contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18(1):5–18. doi: 10.1038/nrc.2017.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palazzo AF, Lee ES. Non-coding RNA: what is functional and what is junk? Front Genet. 2015;6:2. doi: 10.3389/fgene.2015.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7–21. doi: 10.1038/nrg3606 [DOI] [PubMed] [Google Scholar]
- 4.Chen X, Yan CC, Zhang X, et al. Long non-coding RNAs and complex diseases: from experimental results to computational models. Brief Bioinform. 2016;18(4):558–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. doi: 10.1016/j.ccell.2016.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ling H, Vincent K, Pichler M, et al. Junk DNA and the long non-coding RNA twist in cancer genetics. Oncogene. 2015;34(39):5003–5011. doi: 10.1038/onc.2014.456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rugge M, Fassan M, Graham DY. Epidemiology of Gastric Cancer. Cham: Springer; 2015:23–34. [Google Scholar]
- 8.Catalano V, Labianca R, Beretta GD, Gatta G, de Braud F, Van Cutsem E. Gastric cancer. Crit Rev Oncol Hematol. 2009;71(2):127–164. doi: 10.1016/j.critrevonc.2009.01.004 [DOI] [PubMed] [Google Scholar]
- 9.Vogiatzi P, Vindigni C, Roviello F, Renieri A, Giordano A. Deciphering the underlying genetic and epigenetic events leading to gastric carcinogenesis. J Cell Physiol. 2007;211(2):287–295. doi: 10.1002/(ISSN)1097-4652 [DOI] [PubMed] [Google Scholar]
- 10.Venerito M, Link A, Rokkas T, et al. Gastric cancer – clinical and epidemiological aspects. Helicobacter. 2016;21:39–44. doi: 10.1111/hel.12339 [DOI] [PubMed] [Google Scholar]
- 11.Xia S, Ji R, Zhan W. Long noncoding RNA papillary thyroid carcinoma susceptibility candidate 3 (PTCSC3) inhibits proliferation and invasion of glioma cells by suppressing the Wnt/β-catenin signaling pathway. BMC Neurol. 2017;17(1):30. doi: 10.1186/s12883-017-0813-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garitano-Trojaola A, San José-Enériz E, Ezponda T, et al. Deregulation of linc-PINT in acute lymphoblastic leukemia is implicated in abnormal proliferation of leukemic cells. Oncotarget. 2018;9(16):12842–12852. doi: 10.18632/oncotarget.24401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen S, Rao H, Liu J, et al. Lymph nodes ratio based nomogram predicts survival of resectable gastric cancer regardless of the number of examined lymph nodes. Oncotarget. 2017;8(28):45585–45596. doi: 10.18632/oncotarget.17276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang Y, Zhang Q, Hu Y, et al. ImmunoScore signature: a prognostic and predictive tool in gastric cancer. Ann Surg. 2018;267(3):504–513. doi: 10.1097/SLA.0000000000002116 [DOI] [PubMed] [Google Scholar]
- 15.Dykes IM, Emanueli C. Transcriptional and post-transcriptional gene regulation by long non-coding RNA. Genomics Proteomics Bioinformatics. 2017;15(3):177–186. doi: 10.1016/j.gpb.2016.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu X, Tan X, Hulshoff M, et al. 5042The role of long non coding RNA in gene-specific promoter methylation during cardiac fibrogenesis. Eur Heart J. 2017;38(suppl_1). doi: 10.1093/eurheartj/ehx493.5042. [DOI] [Google Scholar]