SUMMARY
Complement dysregulation is a feature of many retinal diseases, yet mechanistic understanding at the cellular level is limited. Given this knowledge gap about which retinal cells express complement, we performed single-cell RNA sequencing on ~92,000 mouse retinal cells and validated our results in five major purified retinal cell types. We found evidence for a distributed cell-type-specific complement expression across 11 cell types. Notably, Müller cells are the major contributor of complement activators c1s, c3, c4, and cfb. Retinal pigment epithelium (RPE) mainly expresses cfh and the terminal complement components, whereas cfi and cfp transcripts are most abundant in neurons. Aging enhances c1s, cfb, cfp, and cfi expression, while cfh expression decreases. Transient retinal ischemia increases complement expression in microglia, Müller cells, and RPE. In summary, we report a unique complement expression signature for murine retinal cell types suggesting a well-orchestrated regulation of local complement expression in the retinal microenvironment.
In Brief
Overshooting complement activity contributes to retinal degeneration. Pauly et al. demonstrate a distinct complement expression profile of retinal cell types that changes with aging and during retinal degeneration. This prompts the intriguing concept of a local retinal complement activation possibly independent of the systemic components typically produced by the liver.
Graphical Abstract

INTRODUCTION
Single-nucleotide polymorphisms in complement genes are associated with a number of retinal diseases, including glaucoma (Scheetz et al., 2013), age-related macular degeneration (AMD) (Weber et al., 2014), and diabetic retinopathy (Yang et al., 2016; Wang et al., 2013). The immune-privileged retina is among others under regular immune surveillance by proteins of the complement system. Although systemic complement is known to perform homeostatic functions that include opsonization for phagocytosis, formation of membrane attack complexes (MACs), and recruitment of immune cells (Merle et al., 2015), the local regulation of complement within the cellular architecture of the neurosensory retina is poorly understood. Current evidence suggests that complement components are locally expressed in the retinal pigment epithelium (RPE) (Schäfer et al., 2017; Luo et al., 2011; Anderson et al., 2010; Tian et al., 2015; Li et al., 2014; Rutar et al., 2012) as well as microglia (Rutar et al., 2012) and could be independent of the systemic complement, which is produced in hepatocytes and distributed via the bloodstream. A retinal complement system may help facilitate a rapid response to microbial invasion and disposal of damaged cells despite an intact blood-retina barrier.
Upregulation of complement expression, subsequent protein deposition, and MAC formation have been demonstrated in the normal aging (Chen et al., 2010; Ma et al., 2013; Chen et al., 2008) and diseased retina (Crabb, 2014; Sudharsan et al., 2017; Radu et al., 2011; Zhang et al., 2002; Kuehn et al., 2008). In fact, complement components present in extracellular deposits (termed “drusen”) are the hallmark of AMD (Crabb, 2014). Consequently, it is tempting to speculate that a source of complement components during aging could be the retina/RPE itself, as animal studies have shown increased retinal expression of c1q, c3, c4, and cfb in older mice (Ma et al., 2013; Chen et al., 2010). Complement upregulation has also been observed in retinitis pigmentosa (Sudharsan et al., 2017), Stargardt disease (Radu et al., 2011), and conditions associated with transient ischemic tissue damage, viz. diabetic retinopathy (Zhang et al., 2002) and glaucoma (Andreeva et al., 2014; Kuehn et al., 2008; Kim et al., 2013). Despite a clear indication for a fundamental role of the complement system in the retina, it remains unknown which retinal cell populations shape complement homeostasis in the healthy, aging, and diseased retina.
The retina consists of more than 40 different cell types, which cooperate to capture, process, and transmit visual signals to the brain (Macosko et al., 2015; Tian et al., 2015; Rheaume et al., 2018; Shekhar et al., 2016). Our understanding of the healthy and diseased retina and its supporting tissues like the RPE and choriocapillaris has grown recently (Tian et al., 2015; Pinelli et al., 2016). Transcriptomic studies have focused on the whole retina or RPE but miss information about cell-type-specific transcription (Pinelli et al., 2016; Tian et al., 2015). Droplet-based single-cell RNA sequencing (scRNA-seq) has identified the molecular differences among retinal ganglion cells (Rheaume et al., 2018), bipolar cells (Shekhar et al., 2016), and Müller cells (Roesch et al., 2008), but these studies provided little insight into complement expression of the major retinal cell types and changes occurring with aging and degeneration.
Here, we profile complement expression at the single-cell level in the major 11 retinal cell types of the mouse and further validate these results in enriched Müller cells, vascular cells, microglia, neurons, and RPE cells. We observed a characteristic contribution of complement transcripts from distinct retinal cell populations. Our data suggest that the classical and alternative complement pathway could be activated solely by local complement production and thereby could induce C3 cleavage. CFH is the major complement inhibitor in the mouse retina; retinal stress consistently downregulates cfh expression. Moreover, cell-type-specific changes in complement expression differed in aging and acute retinal degeneration induced by transient ischemia, implying a stress-dependent and cell-type-specific modulation of retinal complement homeostasis mediated by the tissue itself.
RESULTS
Single-Cell RNA Sequencing Reveals Complement Component Expression across Multiple Cell Types
Retinal cells, ~92,000 total, were isolated from six male healthy C57BL/6J mice and separated for scRNA-seq (Macosko et al., 2015; Shekhar et al., 2016; Cheng et al., 2013b; Kim et al., 2008). Following sequencing, the data were analyzed using 30 principal components as input to the t-Distributed Stochastic Neighbor Embedding (t-SNE) method for dimension reduction and data visualization (Figures 1A and S1). Cells were classified into eleven major types based on established markers (Tables S1 and S2). We then mapped the expression of complement genes across all 11 cell types and observed a distributed expression of complement components across various resident cells in the retina (Figure 1B; Table S3). We detected cell-type-specific complement expression mainly in the classical pathway via scRNA-seq. Moreover, we mapped the cell-type-dependent expression of both soluble and membrane-bound complement regulators (Figure 1C; Table S3) and found main soluble regulators cfh, vtn, and clu. Cell types expressing complement regulators at the highest levels were Müller cells, pericytes, and endothelial cells. Complement receptors, which detect complement activation (anaphylatoxins or opsonins), were only expressed in microglia cells (Figure 1B; Table S3).
Figure 1. Single-Cell RNA-Seq of Murine Retina Reveals Complement Expression in Different Cell Types.
Distribution of complement expression in normal mouse retina is delineated by single-cell transcriptomics.
(A) Using unsupervised clustering, we detect all 11 major cell classes in the mouse retina; the distinct cell types that passed quality control (described in STAR Methods) are shown in a t-SNE map (out of the 92,343 cells total, 91,798 cells passed the filter of having mitochondrial gene expression <50%, and 200 < unique gene counts <3,500). Percentages of assigned cell types are summarized in the right panel. BC, bipolar cell.
(B) Cell-type-specific expression of complement genes is shown in a heatmap. Mean expression values of the genes were calculated in each cluster by randomly subsampling a population of 100 cells for each cell type. Each row represents a retinal cell type, and each column corresponds to a gene.
(C) The expression of complement regulators among the various retinal cell types is shown by means of a dot plot. Cell types are arranged roughly by their location in the retina, from the inner layer (top) to the outer layer (bottom). The size of each circle (pct.exp) depicts the percentage of cells in which the gene was detected for a given cell type, and its color depicts the average transcript count in the expressing cells (avg.exp.scale).
See also Figure S1 and Tables S1–S3.
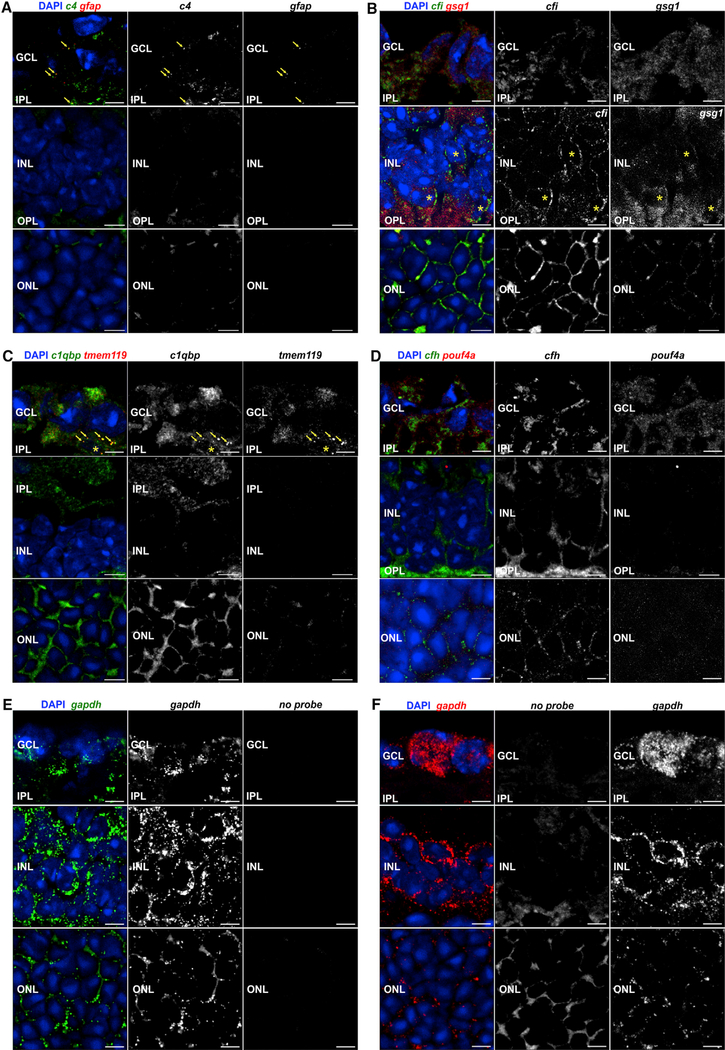
The results from scRNA-seq regarding complement component transcription were exemplary validated using RNA fluorescence in situ hybridization (FISH) (Figure 2). Complement component c4 expression colocalized with gfap-positive astrocytes/Müller cells in the retinal ganglion cell layer (GCL) (Figure 2A), which correlated with scRNA-seq data (Figure 1B). The scRNA-seq data were also confirmed for complement regulator cfi, which was specifically detected in bipolar cells both by transcription analyses (Figure 1C) and via RNA-FISH (Figure 2B). Complement receptor c1qbp was detected in all cell populations in RNA sequencing (RNA-seq) (Figure 1B). In line with that, we found a colocalization of c1qpb transcripts with markers of microglia/macrophages (hereafter termed microglia) (tmem119) in the GCL (Figure 2C), but rather evenly distributed signals were detected in all retinal layers except for the inner nuclear layer (INL).
Figure 2. Localization of Selected Complement Component Transcripts in the Healthy Retina ofPigmented 10-Week-Old Mice via Fluorescence In Situ Hybridization.
(A) Spots indicative of c4 transcripts were detected in the GCL and clearly overlap with the very few particles positive for the astrocyte marker gfap (arrowheads). No transcripts of c4 or, as expected, the astrocyte marker gfap were detected in the INL or outer nuclear layer (ONL).
(B) Cfi expression was very weak in the GCL, with no clear association with cell somata, but was detected at a rather high level in the INL. There, signals partially overlapped with those of the bipolar marker gsg1 (asterisks). In the ONL, probes detecting cfi transcripts produced signals clearly above the autofluorescence background level. A clear distinction regarding whether cfi transcripts are localized in photoreceptors and/or in Müller glia that ensheathe photoreceptor somata in the ONL cannot be made.
(C) clqbp transcript was rather evenly distributed across the whole retina. A partial overlap with the microglia marker tmem119 in the GCL could be validated (arrowheads). Note that c1qbp transcripts were detected in the inner plexiform layer (IPL), but not in the INL.
(D) cfh transcripts were detected at low levels in the GCL and INL, and no clear overlap of signals with that of the ganglion cell marker pou4f1 was observed. It seemed to be more enriched in the outer plexiform layer (OPL). Similarly, cfh transcript levels are low in the ONL. The staining pattern could reflect an expression in Müller glia enwrapping photoreceptor somata.
(E and F) As positive control, probes conjugated either with (E) Quasar 670 (green) or (F) Quasar 570 (red) targeting transcripts of gapdh were used. Note the robust detection of the transcript especially in association with cell bodies, which confirms the high expression levels of gapdh detected via scRNA-seq. Autofluorescence background was detected in the recording channels if no appropriate probe was incubated with the tissue in the GCL, while some autofluorescence background was detected in the OPL and ONL if no appropriate probe was incubated with the tissue.
Scale bars, 5 μm.
qPCR-Based mRNA Analyses of Purified Neurons, Müller Cells, Microglia, Vascular Cells, and RPE Decipher the Differentially Transcriptional Relevance of Retinal Cells
Though our single-cell analysis uncovered complement expression in different cell types, some complement components remained undetectable or were found in relatively rare cell types at the single-cell level (Table S3). Therefore, we further validated our results on Müller cells, microglia, vascular cells, and retinal neurons purified by immunomagnetic cell separation using quantitative real-time PCR. RPE was purified through manual scratching of eyecups from male and female albino BALB/c and pigmented C57BL/6 mice (Figure 3A) (Grosche et al., 2016). All five cell populations were characterized by the expression of specific marker genes (Figures 3A–3F).
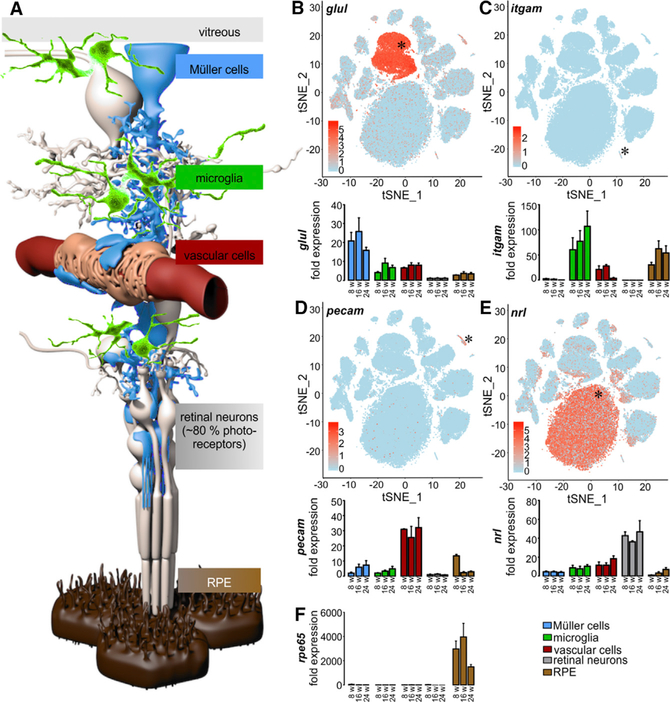
Figure 3. Validation of Enrichment of Different Retinal Cell Types.
(A) Schematic view of distinct retinal cell types. Müller cells (blue), the central glia cells of the retina, are in direct contact with the vitreous and various retinal cell types: microglia (green), vascular cells (red), and neurons (gray). 80% of the retinal neurons are light-responsive photoreceptors that are supported by retinal pigment epithelial cells (RPE, brown).
(B) scRNA-seq data illustrating expression of glutamine synthetase (glul) across retinal cells (top panels). Murine retinal cell populations were enriched by immunomagnetic cell separation and characterized by quantitative real-time PCR using specific markers (bottom panels); Glul is a marker for the Müller cell fraction.
(C) Microglia (and putatively co-enriched macrophages) specifically express cd11b (itgam).
(D) Vascular cell enrichment was proven by strong expression of pecam.
(E) Retinal neurons were characterized by an enhanced detection of the photoreceptor-specific nrl mRNA compared to the other cell populations.
(F) Rpe65 was exclusively expressed in RPE/choroid. Exemplarily shown mean values ± SEM for cell preparations from BALB/c mice at 8, 16, and 24 weeks of age (n = 4–6 for each age).
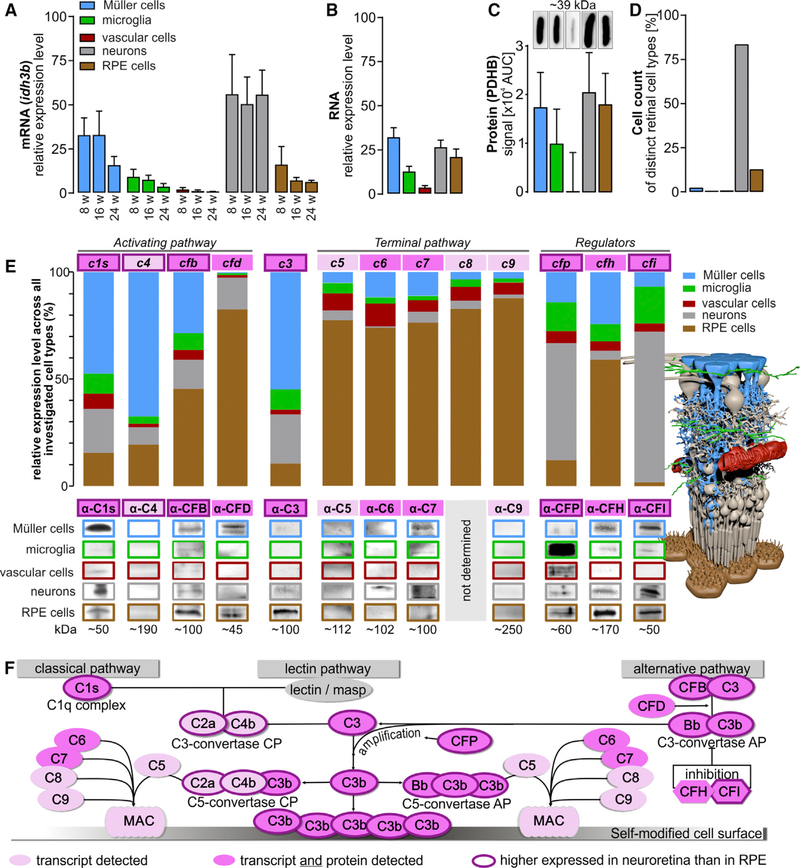
Commonly used housekeeping genes showed high transcriptional and translational variability across different cell types (Figures S2A–S2D) except pyruvate dehydrogenase E1 component subunit beta (pdhb) (Figures S2E and S2F) and isocitrate dehydrogenase 3 (NAD+) beta (idh3b) (Figures S2G and S2H), which had relatively homogeneous expression levels. We decided to use idh3b to determine how distinct cell populations proportionally contributed to the total retinal complement transcriptome, because it showed expression levels similar to those of the complement genes, while gapdh, for example, was expressed at much higher levels and thus appeared to be less appropriate (Figure S2C). Based on idh3b, we estimate that the neuronal fraction contributes 60% of the total retina transcriptome and Müller cells contribute 25% (Figure 4A). Vascular cells and microglia expressed lower levels of idh3b, indicating low cell numbers and/or low transcription activity of these cell types in the mouse retina. Quantifying total RNA by RNA picochip analysis, we found similar RNA quantities in Müller cells (33%), neurons (26%), and the RPE/choroid fraction (22%) per mouse (Figure 4B). In accordance with these results, PDHB protein levels were relatively similar across Müller cells, neurons, and RPE cells (Figure 4C), whereas microglia and vascular cells showed weaker PDHB signals. In agreement with previous reports of cell counts in the mouse, we found neurons make up 85%, RPE cells 13%, Müller cells 2%, and microglia and vascular cells are less than 1% of all retinal cell types (Figure 4D; Table S2) (Jeon et al., 1998). While Müller cells are a comparatively infrequent cell type (2%), they contribute up to one third of the total mRNA content in the retina (Figures 1A, 4A, 4B, and 4D; Table S2). Accordingly, our study provides important insight into the proportional transcriptional activities of these five major cell populations in the retina.
Figure 4. Contribution of Retinal Cell Types to the Retinal Architecture, Expressome, and Complement Homeostasis.
(A) mRNA expression of the housekeeping gene idh3b as determined in samples from retinal cell populations enriched from retinae of 8−, 16, and 24-week-old albino mice without adjusting the RNA input amount per cell type. This enables an estimate of the contribution of each cell type to the retinal transcriptome. Bars represent mean values ± SEM (n = 4–6).
(B) The total RNA amount isolated from retinal cell populations enriched from albino mice (8 to 16 weeks old) was investigated using picochip analysis. Bars represent mean values ± SEM (n = 5–8).
(C) Quantification of PDHB protein expression via western blots performed on five retinal cell types purified from 4–6 albino mice.
(D) Previously published (Jeon et al., 1998) and our own retinal cell counts in the healthy mouse retina.
(E) Expression levels of indicated complement components were determined from cells of albino mice at mRNA (bars, 8-week-old mice) and at protein level (western blot, 8− to 24-week -old mice). The overall contribution of each cell population to the local complement homeostasis was determined by analyzing the total yield of mRNA or protein derived from the respective cell population so that both are reflected by the data (expression level per cell type and the number of cells per cell type present in the retina).
(F) Scheme of the complement system that can be activated via three different mechanisms and is enhanced by an amplification loop. Note that complement components only detected at the transcript level are delineated in pink, and those that were also confirmed at the protein level are shown in dark pink. Complement components with higher expression in retinal cell types compared to the RPE/choroid fraction are pinpointed by a thick outline. CP, classical pathway; AP, alternative pathway.
See also Figures S2 and S3 and Table S7.
Retinal Cell Populations Express Unique Complement Signatures
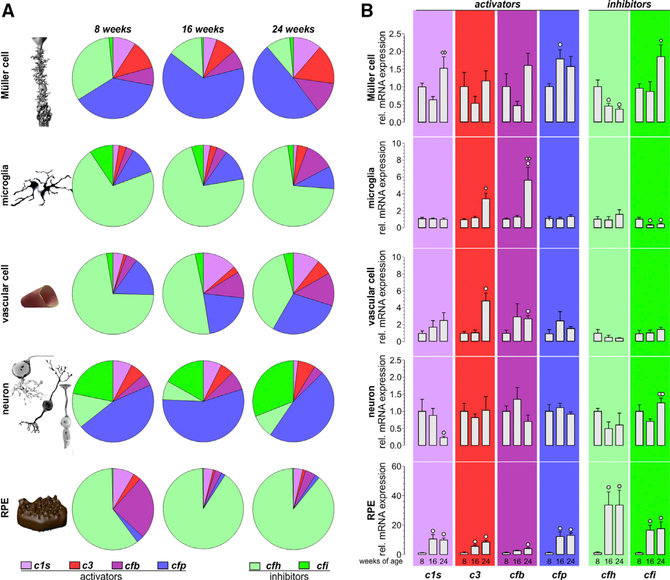
Encouraged by the distributed complement expression pattern across 11 cell types (Figures 1B and 1C; Table S3), we hypothesized that specific retinal cell types shape the intraretinal complement homeostasis through expression of specific complement components. We selected six disease-associated genes (Schäfer et al., 2017; Weber et al., 2014) (c1s, c3, cfb, cfp, cfh, and cfi) and six supporting complement genes (c4, cfd, c5, c6, c7, c8, and c9) for further validation via quantitative real-time PCR and western blot (Figures 4E and 4F) and found that Müller cells contributed the most complement activator transcripts, expressing 47% of c1s, 67% of c4, and 54% of c3 retinal transcripts in 8-week-old mice (Figure 4E). Retinal neurons dominated the expression of the complement regulators cfi and cfp, while 59% of the cfh, 45% of the cfb, and 82% of the cfd mRNA and c5-c9 transcripts responsible for the terminal MAC were detected in RPE. Despite the relatively low number of microglia, the resident immune cells of the retina contributed proportionally more cfh mRNA and a similar amount of cfb transcripts to the retinal complement population compared with retinal neurons.
We confirmed expression of complement activator C1s, CFB, and CFP proteins in all enriched murine cell populations (Figure 4E). Complement C3 protein was detected in RPE, Müller cells, and neurons, while the complement inhibitor CFH and CFI proteins were present in all cell types except the vascular cell population. Alternative pathway protease CFD was identified in the Müller cell and RPE fraction, while C6 was only found in neurons and C7 was detected in Müller cells and neurons. We could not detect C4, C5, and C9 proteins in any of the retinal cell populations (Figure 4E). For C8, no specific antibody was commercially available. There was an overlap in the complement transcript levels determined by quantitative real-time PCR and that of the complement proteins detected in various cell populations. For example, the strong c1s mRNA expression found in Müller cells matched the robust C1s protein levels, and cfd mRNA expression corresponded with CFD protein detection in RPE. Interestingly, neurons expressed ~40% of the total cfp mRNA, whereas CFP protein levels were highest in microglia (Figure 4E). This may imply a spatial separation of complement component transcription and complement component accumulation at the protein level within the retina.
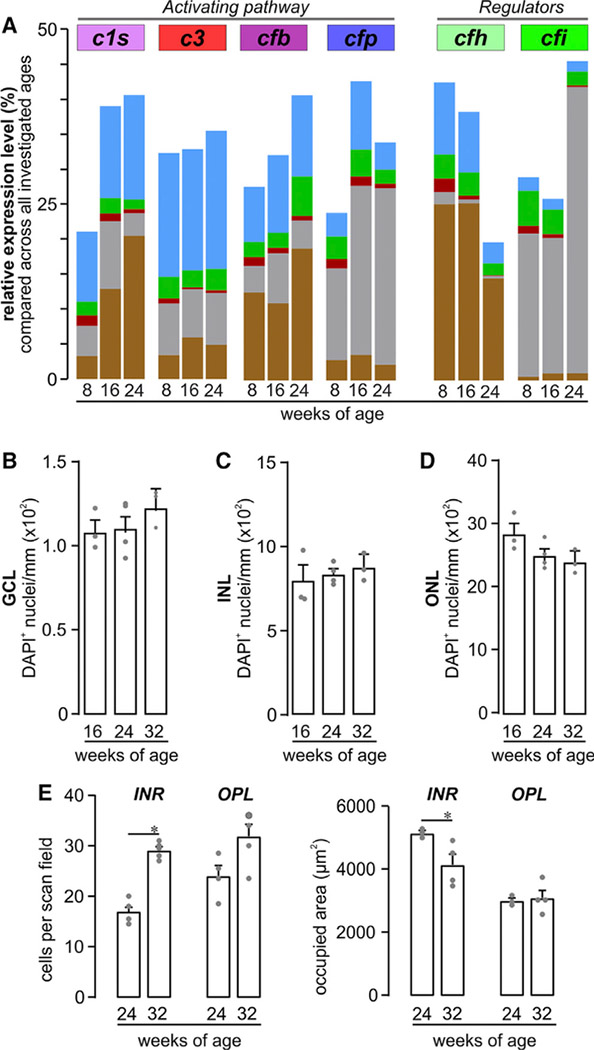
Age-Dependent Changes in the Complement Expression of Different Retinal Cell Populations
We further investigated age-dependent changes in expression levels of known disease associated complement transcripts via quantitative real-time PCR among the different retinal cell populations in mice from 8 to 24 weeks of age. C1s, cfb, cfp, and cfi transcripts increased with age among all cell populations (Figure 5A). Upregulated c1s expression in RPE cells lead to doubling of the total retinal c1s mRNA between 8 and 24 weeks (Figure 5A). Transcripts of the alternative pathway activator cfb, primarily produced by Müller cells and RPE, increased at 16 and 24 weeks of age. The largest increase in cfb transcript levels (2.6-fold) was found in the microglial population between 8 and 24 weeks (Figure 5A). The highest proportional contribution of cfp retinal transcripts came from neurons (55%) at 8 weeks of age, and this contribution further increased to 73% at 16/24 weeks of age (Figure 5A). In contrast, c3 expression remained relatively stable (Figure 4E), except for the vascular cell population, where the c3 levels dropped by 50% between 8- and 24- week-old mice (Figure 5A).
Figure 5. Retinal Phenotype and Complement Homeostasis in Aging Albino Mice.
(A) Expression levels of c1s, c3, cfb, cfp, cfh, and cfi were determined from cells of albino mice at the mRNA level (bars) at the indicated ages. The overall putative contribution of each cell population to the local complement homeostasis was determined by analyzing the total yield of mRNA derived from the respective cell population.
(B-D) The quantities of DAPI+ cell nuclei in the (B) GCL, (C) INL, and (D) ONL were comparable in 16− to 32-week-old albino retinae.
(E) Left: microglia were quantified in the inner retinal layers (INR) such as ganglion cell and inner plexiform layer and additionally in the outer plexiform layer (OPL) on basis of Iba1 labeling in mice of the indicated age. Right: the area occupied by processes of a single microglia was measured as exemplarily depicted by the dashed circles of different color for OPL microglia in (D). Bars represent mean values ± SEM from 2–4 animals. *p < 0.05, **p < 0.01, Mann-Whitney U test. See also Figure S2 and Table S7.
CFH is the main negative regulator of the complement system. We found that cfh expression decreased by 50% in all cell populations in 24-week-old mice compared to 8-week-old mice. In 24-week-old mice, RPE cells produced the majority of the retinal cfh (Figure 5A), although the majority of the cfi transcripts (which act together with CFH) were produced by neurons, specifically rod bipolar cells (Figures 1C and 5A). Together with its functional counterpart, cfp, the expression of cfi also increased in neurons of 24-week-old mice compared to the 8- and 16-week-old mice (Figure 5A).
These divergent changes in the local complement expression in the retina of aging mice were not accompanied by any detectable retinal cell loss (Figures 5B–5D) but were accompanied by increased microglia numbers (Figure 5E) and enhanced microglial activation, as suggested by a decreased occupied area of microglial processes (Figure 5E).
A Characteristic Proportion of Activating and Inhibiting Complement Transcripts in Distinct Retinal Cell Types
Having noted a cell-type-specific complement expression in our data, we sought to understand the balance of complement activator and inhibitor expression in the different cell types by normalizing complement expression levels to the housekeeping gene to allow a comparison between cell populations independent of cell counts (Figure 6). Strikingly, we discovered that certain cell types like RPE and microglia mainly express inhibitory complement components (cfh and cfi), whereas other cell populations, such as neurons and Müller cells, mostly express complement activators (c1s, c3, cfb, and cfp) (Figure 6A). Interestingly, neurons expressed more cfi than cfh than the remaining cell types (Figure 6A).
Figure 6. Comparison of Complement Component Expression between Retinal Cell Types of Aging Mice.
(A) Expression of complement components was determined by quantitative real-time PCR. Diagrams represent the relative amount of transcripts per cell (normalized to the idh3b housekeeper expression) of the different complement components in the respective cell type enriched from mice at the indicated age. Note the high expression level of inhibitory complement factors in RPE/choroid samples as well as in microglial and vascular cells, while complement-activating genes appear to dominate in Müller cells and neurons. Data were collected from 4–6 wild-type albino mice (numbers are given in Table S4).
(B) Complement expression analysis by quantitative real-time PCR was performed on enriched retinal cell types from 8-, 16-, and 24-week-old mice. Bars represent mean values ± SEM of cells purified from 4–6 animals. Mann-Whitney U testing was performed on all data. A circle indicates a significant difference compared to the expression level at 8 weeks of age; whereas a diamond indicates a significant difference compared to the expression level at 16 weeks of age. °/◇ p < 0.05; ° °/◇◇p < 0.01.
Finally, we checked forage-dependent changes in the expression of the respective complement components—now, in contrast to results presented in Figure 5A, independent of putative changes in cell numbers. We could confirm a significant upregulation of complement activators such as c1s, c3, cfb, and cfp with increasing age (Figure 6B). Most of these changes were detected in RPE cells but at later stages (e.g., 24-week-old mice) also in microglia, vascular, and Müller cells. Expression changes of complement inhibitors were not consistent across cell types. While cfh was significantly downregulated in Müller glia in mice at 16 weeks of age, it was upregulated in RPE. Moreover, cfi was significantly downregulated in microglia but upregulated in Müller glia, retinal neurons, and RPE.
The spatial distribution of complement activators and regulators signifies a unique complement signature for each retinal cell type that was dynamically changing, even though relatively short intervals of aging were investigated.
Acute Ischemic Retinal Injury Triggers Robust Cell-Type-Specific Complement Expression
Retinal tissue injury is a common manifestation of retinal disease. To evaluate how tissue injury might change cell-type-specific complement expression, we used a retinal ischemia/reperfusion (I/R) injury model to induce acute retinal degeneration (Wagner et al., 2017). We found a significant increase in the expression of complement activators 24 h post-ischemia in the different isolated cell populations (Figure 7). Consistent with our previous results, cfi appeared to be the main complement inhibitor in neurons, whereas cfh was the major complement inhibitor expressed in the remaining cell populations. Compared to aging retina (Figures 5 and 6), the upregulation of c1s, c3, cfb, and cfi transcript expression of was more pronounced in post-ischemic retina at the mRNA level (Figures 7A–7C). Interestingly, this response in c1s, c3, and cfb expression was provoked by changes in the RPE. Moreover, cfh and cfi showed again a mutually opposite pattern of expression changes whereby cfh mRNA decreased and cfi increased in I/R retinae (Figure 7C). Detection of C3 (Figure 7D) and C1s (Figure 7E) at protein level via immunolabeling 3 days after the ischemic tissue injury was performed to enable detection of newly formed protein. C1s puncta were evenly distributed over all retinal layers, with a slight enrichment in the GCL and RPE (Figure 7D). Similarly, C3 labeling was observed in all retinal layers, and a moderate rise in labeling intensity could be observed 3 days after the ischemic tissue injury, especially in the outer retina affecting photoreceptors and RPE (Figure 7E). Complement components are typically secreted by the producing cells. In line with this fact, we did not find a perfect cell-type-specific match of protein distribution compared to the transcript signatures described above. It has to be considered that ischemic damage may lead to transient breakdown of the blood retinal barrier and that the complement components detected via this approach could also be derived from the systemic complement system, at least in post-ischemic tissue.
Figure 7. Transient Ischemic Stress Results in Cell-Type-Specific Upregulation of Transcripts from Activating Complement Components and Downregulation of cfh.
(A) The relative amount of complement transcripts per retinal cell type indicated that complement-activating transcripts are more abundant in pigmented mice than in albino mice in which transcripts of complement inhibitors dominate at the same age. Note the strong relative upregulation and the resulting shift toward transcripts from complement activators 24 h after transient ischemic retinal stress in all retinal cell types of C57BL/6 mice (numbers are given in Table S5).
(B) Major changes of local complement expression (normalized to the housekeeper) were detected by quantitative real-time PCR 24 h after transient ischemia. The most pronounced upregulation of complement activators was found in Müller cells (c3 and cfb), microglia (c3, cfb, and cfp) and RPE (c1s and c3). Cfh was downregulated in Müller cells, while cfi was upregulated in all investigated cell types. Significantly different expression as compared to that cells from healthy control eyes is indicated (*p < 0.05, **p < 0.01, Mann-Whitney U test).
(C) Complement transcript contribution of the different retinal cell populations (no normalization to the housekeeper and no adjustment of RNA input) indicated a pro-inflammatory milieu in the post-ischemic retina. Müller cells, microglia, and RPE cells mainly contributed to the changed complement homeostasis in post-ischemic retinae.
(D) Representative immunostaining of complement component C1S and C3 in the healthy and post-ischemic pigmented retina 3 days after injury. Sections were co-labeled for the Müller cell marker glutamine synthetase (GLUL), and nuclei were visualized by DAPI co-labeling.
(E) Representative micrographs of retinal sections incubated with the combination of secondary antibodies and DAPI only.
GCL, ganglion cell layer; IPL, inner plexiform layer; INL, inner nuclear layer; OPL, outer plexiform layer; ONL, outer nuclear layer; RPE, retinal pigment epithelium.
Scale bars, 20 μm. In (A) and (B), data were collected from 3 – 5 animals. See also Figure S2 and Table S7.
DISCUSSION
Since the retina is an immune-privileged tissue, understanding its local complement system is critical to our understanding of retinal inflammation. To identify the retinal cell types expressing complement, we isolated and sequenced ~92,000 cells from healthy mouse retinae. scRNA-seq was validated by RNA-FISH analysis for selected genes and 12 complement components by assays of RNA and protein in enriched cell populations of Müller cells, microglia, vascular cells, retinal neurons, and RPE. Collectively, our results show that complement components are locally expressed by different retinal cell populations, challenging the conventional belief that the complement source in the retina is confined to resident immune cells. We detected complement transcripts and proteins that were produced at higher levels in the neuroretina than in RPE that would be sufficient to activate the classical and/or alternative complement pathway. In consequence, cleavage of C3 into its anaphylatoxin C3a and opsonin C3b could be performed independently of blood-derived complement components (Figure 4F). Complement transcripts of the components that are key to MAC assembly were primarily detected in RPE and rarely found as proteins in the neuroretina. The RPE forms the outer retina-blood barrier, which is able to secrete substrates not only toward the subretinal space (apical) but also toward the choroidal side (basal). Considering its complement expression profile and the largely understudied idea of a context-specific apical or basal secretion pattern of complement by RPE sitting at the blood-retinal interface, it is tempting to speculate that by doing so, the RPE could actively shape the complement homeostasis of the retina, which needs to be addressed in future studies.
Given the cell-specific ratios of activating and inhibiting complement component expression levels, each cell type appears to have a specific role in maintaining retinal complement homeostasis. Moreover, cell populations present in the retina at relatively low numbers, such as microglia and Müller cells (Jeon et al., 1998), seem to have a major impact on retinal complement expression levels. We found that these cells contribute substantially to the total retinal transcriptome. This suggests that quantification of cell numbers alone is insufficient to draw conclusions about the contribution of cellular activity to the global expression profile of the retina. Nonetheless, it has been shown that transcription is tightly regulated according to cell size, and cells with larger cell bodies, such as Müller cells, can provide more mRNA than the more abundant cells with a smaller volume (e.g., neurons and RPE) (Marguerat and Bähler, 2012; Kempe et al., 2015).
The complement system helps maintain normal ocular functions (Sohn et al., 2003, 2000), and its dysregulation significantly influences retinal disease (Sudharsan et al., 2017; Radu et al., 2011; Weber et al., 2014; Yang et al., 2016; Scheetz et al., 2013). Although earlier studies have found low background expression of complement in retinal microglia (Luo et al., 2011; Schäfer et al., 2017; Anderson et al., 2010; Rutar et al., 2012) and RPE/choroid (Schäfer et al., 2017; Luo et al., 2011, 2013), our results unequivocally show that other cell types are capable of local complement production. We did observe expression differences between the scRNA-seq and the MACS-enriched cell populations that can be explained by the lower sensitivity to detect gene expression compared to quantitative real-time PCR. For instance, compared to the single-cell analysis, quantitative real-time PCR allowed us to more readily detect the expression of c1s and c3 in Müller cells, cfp in microglia and vascular cells, cfb in all major retinal cell types, and cfd, c5-c9 mainly in RPE cells. Still, both approaches converged in their assessment of cell-type-specific expression for most complement components. Our results also add to previous work on retinal complement in humans (Tian et al., 2015; Li et al., 2014; Anderson et al., 2010), mice (Schäfer et al., 2017; Luo et al., 2011), and rats (Rutar et al., 2012) by also detecting protein expression for nine complement components that reflect their RNA expression.
The regulation of complement expression in whole-cell populations from the aging retina largely matched the changes we calculated for the normalized cellular expression rates in the distinct cell types. This implies that expression changes were driven by changes at the transcriptional level, not by changes in cell numbers, in accordance with the data from our morphometric analyses performed on aging BALB/c mice. Interestingly, the relative expression of all tested complement components increased in the whole-RPE-cell population with increasing age, except for cfh, whose expression decreased. We speculate that increased single-cell cfh expression alone is unable to counterbalance the overall age-associated RPE dysfunction and/or degeneration (as also indicated by a reduced rpe65 expression). Accordingly, RPE-dependent cfh transcripts decrease at 24 weeks, the putative contribution of cfh in regulating the alternative complement pathway in the retinal microenvironment could be diminished, and the intraretinal milieu could be misbalanced.
We also discovered that healthy neurons produce the complement regulators cfp and cfi. The relevance of these complement components has already been shown for AMD (Fritsche et al., 2016; Weber et al., 2014; Micklisch et al., 2017). In murine models of retinal degeneration, cfi expression increases after polyethylene-glycol-induced insult, whereas cfp expression decreases in the light-damaged retina (Schäfer et al., 2017; Lyzogubov et al., 2014). Our single cell analysis demonstrated that cfi mRNA localizes specifically to rod bipolar cells and protein detection to neurons and Müller cells in mice. CFI inactivates the complement system, and the resulting cleavage products modulate the activity of phagocytes. Rods primarily govern scotopic vision, and mice have a rod-dominated retina similar to humans (except for the fovea centralis). This rod-bipolar-cell-specific cfi transcription in the mouse retina further suggests that the retinal complement system is influenced by functional and anatomical characteristics of the retina.
Age-related anatomical alterations in the retina have been demonstrated in histological analyses (Grossniklaus et al., 2013). Consistent with previous results (Damani et al., 2011; Friedman and Ts’o, 1968), we found increased microglial and decreased RPE marker gene expression with aging. In our study, the expression of complement transcripts c1s, cfb, cfp, and cfi increased while that of cfh decreased in retinal cells between 8 and 24 weeks of age. This indicates a role for complement in retinal adaptation during maturation and with processes of aging (Mukai et al., 2018). Although age-dependent upregulation of complement transcripts, including that of c1q, c3, c4, and cfb, in the retina has been described (Chen et al., 2010), our findings add a role for microglia in the expression of c3 and cfb. Our results also show that Müller cells and neurons provide a substantial proportion of retinal complement transcripts and, thus, their impact on retinal complement homeostasis has likely been underestimated by past studies. Cell culture studies have suggested that Müller glia can produce C1q (Astafurov et al., 2014) and that complement activation products can regulate Müller cell activity via C5a-receptor and influence retinal disease (Cheng et al., 2013a). Our results show a direct involvement of Müller cells in the transcript expression of the retinal complement components, and it is likely that in the retina, similar to the brain, neurons and glia cells orchestrate complement-mediated maturation of nervous tissue via synaptic pruning, progenitor proliferation, and neuronal migration (Tenner et al., 2018). Aging and Alzheimer disease brains increase expression of c1q, c3, and c4 (Walker and McGeer, 1992; Cribbs et al., 2012), which might point to a general mechanism of local complement function in the overall aging of the CNS.
Ischemia and subsequent reperfusion (e.g., upon treatment with anti-VEGF therapy) are associated with diabetic retinopathy (Silva et al., 2015; Traveset et al., 2016; Levin et al., 2017), and polymorphisms in CFB and CFH (Wang et al., 2013) have been implicated in modifying disease progression. We identified Müller cells as a major cell type involved in this process (by downregulating cfh transcripts and upregulating cfb) in the analysis of post-ischemic mouse retina. Gene profiling studies of whole mouse retinas also suggest an important role of the complement pathway in I/R-associated damage (Andreeva et al., 2014). Retinal c1q, c1s, c1r, c2, c3, c4a, and cfh expression has been reported after transient ischemia in mice (Kuehn et al., 2008; Kim et al., 2013; Andreeva et al., 2014). However, it remains undetermined how individual retinal cell types modulate complement activity after retinal I/R injury. Here, we show that mainly Müller cells, microglia, and RPE increase c3 and cfb expression following ischemia, while cfp is upregulated primarily in microglia. The complement inhibitor cfi also showed cell-type-specific enhanced mRNA levels in microglia, RPE, and retinal neurons, whereas expression of the complement inhibitor cfh was significantly reduced in Müller cells following ischemia. These expression changes suggest augmentation of local complement components following ischemia.
Genetic variation in several complement genes, such as CFH and CFI, are associated with AMD, glaucoma, and central serous retinopathy (Fritsche et al., 2016; Weber et al., 2014; Grassmann et al., 2016). CFI, along with cofactor CFH, regulates complement activity by degrading complement components C3b and C4b (Davis et al., 1984), thereby facilitating the cleavage of C3b into inactive fragments (Sim et al., 1993). We discovered a spatially distinct transcription pattern of cfi and its cofactor, cfh. cfi was mainly detected in retinal neurons (specifically rod bipolar cells), while cfh was detected primarily in vascular and RPE cells. Further, we found opposing transcriptional regulation of cfi and cfh during aging and ischemia. These findings hint at a CFH-independent function of CFI in the retina, perhaps in conjunction with other cofactors such as CR1 or CD46 (Sim et al., 1993). To date, there are no known AMD-associated polymorphisms in the cd46 or cr1 genes, but cd46 knockout induces retinal degeneration (Lyzogubov et al., 2016), highlighting its relevant role in retinal physiology.
Given the cell-type-specific expression profile of complement genes in the retina, we propose that a balanced local complement expression is linked to normal retinal integrity. Moreover, our data show that changes in local, cell-type-specific complement expression during aging and acute stress can be induced by cell stress and retinal degeneration and, thus, could in the end also contribute to disease progression. For example, a reaction common to I/R damage and aging appears to be the decrease of intraretinal expression of the complement inhibitor cfh. This working hypothesis of course needs further validation at the functional level by future studies.
Taken together, our cell-type-specific analyses provide an alternative perspective on how expression of complement genes, such as those identified by a genome-wide association study (GWAS) for AMD and diabetic retinopathy, in various retinal cell types might be involved in the disease mechanisms in question. The tightly orchestrated reaction of all retinal cell types to distinct conditions of tissue stress suggests that cell-type-specific responses must be considered for successful development of therapeutic strategies targeting retinal complement activity in the future.
Finally, it needs to be pointed out that the analysis of complement activation and its putative role during retinal development, aging, and retinal degeneration was beyond the scope of the present study that was primarily set up to generate a detailed retinal complement expression atlas. However, complement function in the aforementioned processes (irrespective of its source) has been partially addressed and demonstrated by other research groups in the field (Sohn et al., 2003, 2000; Radu et al., 2011; Scheetz et al., 2013; Sudharsan et al., 2017; Weber et al., 2014; Yang et al., 2016). We would like to point out that those local complement transcripts we were able to detect in distinct retinal cell types are sufficient to activate the classical and alternative complement pathways. Given the rather low expression of components of the terminal canonical complement pathway in cells from the neuroretina, it remains debatable whether it can be intraretinally active without input from the RPE that did produce moderate levels of respective transcripts and/or the systemic complement system. However, we also would like to stress the point that those complement components we demonstrated to be locally expressed are likely to have non-canonical functions in the retina (e.g., C3 in synapse pruning or CFH in phagocytosis or apoptosis) (Martin et al., 2016; Hawksworth et al., 2017). There is growing evidence for intracellular functions of early complement components, which could have an impact on normal cellular physiology (Liszewski et al., 2017). To follow up this intriguing line of thinking, future studies are needed to identify these intracellular functions in addition to their cell secretion and functional interactions.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Diana Pauly (diana.pauly@ukr.de). This study did not generate new unique reagents.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Single cell RNA-Seq and RNA-FISH was performed on wild-type (C57BL/6J) male mice (10 weeks old) purchased from Jackson Laboratory (Bar Harbor, ME, USA). All experimental procedures were approved by the University of Pennsylvania Animal Care and Use Committee. Mice were sacrificed with cervical dislocation under anesthesia. Experiments for immunomagnetic separation were done in accordance with the European Community Council Directive 2010/63/EU and the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research and were approved by the local Bavarian authorities (55.2 DMS-2532–2-182, Germany). All mice were housed in a 12 hour light/ dark cycle with ~400 lux. Experiments on complement expression in aging mice were conducted with 8, 16 and 24 week old male and female on BALB/c mice. Retinal ischemia was induced in one eye of 8 week old male and female C57BL/6J mice. The untreated contralateral eye served as internal control and, accordingly, an additional control group was not needed thereby sticking to the rules of the three R’s by keeping reducing the number of animals used in respective experiments.
METHOD DETAILS
Retinal ischemia/ reperfusion injury
The protocols for induction of transient retinal ischemia were approved by the local Bavarian authorities (55.2 DMS-2532–2-182, Germany). Ischemia was induced in one eye of 8 week old male and female C57BL/6J mice using the high intraocular pressure (HIOP) method (Pannicke et al., 2014; Wagner et al., 2016). The other eye remained untreated and served as an internal control. Anesthesia was induced with ketamine (100 mg/kg body weight, intraperitoneal (ip); Ratiopharm, Ulm, Germany), xylazine (5 mg/kg, ip; Bayer Vital, Leverkusen, Germany), and atropine sulfate (100 mg/kg, ip; Braun, Melsungen, Germany). The anterior chamber of the test eye was cannulated from the pars plana with a 30-gauge infusion needle, connected to a saline bottle. The intraocular pressure was increased to 160 mmHg for 90 minutes by elevating the bottle. After removing the needle, the animals survived for 24 hours and subsequently, they were sacrificed with carbon dioxide for tissue analyses.
Single Cell RNA Analysis of mouse retina
Mouse eyeballs were quickly removed and placed in cold phosphate buffered saline (PBS). The mouse retina was carefully removed under dissecting scope and tissue was dissociated immediately using the papain dissociation system (Worthington, Lakewood, NJ, USA) following the manufacturer’s instructions. Briefly, the mouse retina was incubated at 37°C for 30 minutes in Eagle’s Balanced Salt Solution (EBSS) with DNase followed by tissue trituration with a 10 mL pipette. Cell pellet was collected after centrifugation at 300 × g for 5 minutes and then resuspended in DNase albumin-inhibitor solution. The cell suspension was carefully layered on top of the albumin-inhibitor solution, then centrifuged at 70 × g for 6 minutes. The cell pellet was washed and resuspended in 1:1 DMEM/F12 + 10% FBS. All centrifugation steps were performed at room temperature. The final cell suspension was filtered with 40 μm cell strainer (Falcon, Corning, NY, USA) to remove large debris. To assess cell viability, cells were stained with 0.4% trypan blue (Mediatech, Inc., Manassas, VA, USA) and counted using a hemocytometer. Viable cells (greater than 80%) were submitted to the Center for Applied Genomics at the Children’s Hospital of Philadelphia (CHOP) for cell separation and lysis on the 10X Chromium Genomics instrument and sequencing on the Illumina Hi-Seq instrument.
All analyses were carried out in the statistical software R v 3.5.1. The R package Seurat was used for data analysis, dataset merging and cell clustering analysis. For clustering, we used 2000 or more genes that had detectable expression with high variability in the ~92,000 mouse retinal cells. Six genetically identical C57BL/6J mice were sequenced, our data were consistent across different batches (Figure S1). We filtered out low-quality cells in which < 90% of the reads did not map to the genome using the Cell Ranger pipeline from 10× genomics, and ultimately obtained 92,343 cells used in our subsequent analyses. Given the consistent number of genes (nGene), UMIs (nUMI), and the percentage of mitochondrial genes (percent.mito) detected in each batch, we merged the sequencing runs and used 30 principal components as an input to t-distributed stochastic neighbor embedding (t-SNE) method for dimension reduction and data visualization. We found 25 cell clusters within the retina using an unsupervised analysis that did not rely on known markers of retinal cells. Marker genes were identified for all clusters with the function markers.all in the R package Seurat; all marker genes with power less than 0.4 were discarded. Moreover, only cells with mitochondrial gene percentages < 50%, and those with unique gene counts between 200 and 3,500 were used, leaving us with 91,798 retinal cells. After filtering, we sought to consolidate the 25 clusters into a total of 11 (for N = 91,798 cells), each of which represents a major, functionally important cell class in the retina. For this, we used the known, established marker genes for common retinal cell types summarized in Table S1. Although previous studies of scRNA-seq on the mouse retina have identified more than 30 different cell types, this difference is largely explained by their subdivision of bipolar cells (BCs) into numerous sub-categories. We decided to classify cell types based on general categories because we wanted to study complement expression in retinal cells more broadly. Using this approach, the cell type proportions in the retina proper are comparable between our study and past studies. Based on mean complement gene expression, we categorized positive expression for a gene within a cell type only if either ≥ 5% of cells or at least 50 cells within that class had nonzero expression for that gene.
RNA-FISH
Mouse protocol was approved by the University of Pennsylvania IACUC committee. A 10 week old C57BL/6J male was euthanized with 5 mg pentobarbital sodium. The eye was dissected free from the orbit, washed in PBS, embedded in OCT (Tissue Tek, Sakura Finetek USA, Torrance, CA, USA) and immediately snap-frozen in liquid nitrogen. The frozen tissue was sliced at 10 μm on a cryostat and stored at −80° C on glass slides.
Tissue fixation and RNA in situ hybridization (RNA-FISH) was carried out using Stellaris® RNA FISH (LGC Biosearch Technologies, Petaluma, CA, USA) following the manufacturer’s instructions. Briefly, the tissue section was fixed in 3.7% formaldehyde in 1X PBS for 10 minutes at room temperature. After washing twice with 1X PBS, the tissue section was permeabilized in 70% ethanol for at least 1 hour at room temperature. Oligo probes were designed against mRNA coding sequence for each gene using the Stellaris Custom probe sets and labeled with Quasar® 570 dye or Quasar® 670 dye. For a positive control, mouse GAPDH probe (Stellaris ShipReady probe sets) was purchased. To secure tissues on the slide during hybridization, the HybriWell® sealing system (Grace Bio-Labs, Bend, OR, USA) was used. Each tissue section was incubated in hybridization buffer containing probe (final concertation was between 62.5 nM to 250 nM) overnight at 37°C and then washed in washing buffer A for 30 minutes at 37°C. Cell nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI, 0.1 μg/ml) followed by washing in Buffer B. All buffers were purchased from LGC Bioresearch Technologies. Finally, mounting medium (ProLong Gold antifade reagent, Invitrogen, Life Technologies, Eugene, OR, USA) was added and a coverglass was mounted on the slide. Confocal microscopy to image the RNA-FISH samples was performed at the Bioimaging Core Facility of the Biomedical Center of the LMU Munich. RNA FISH images were obtained with an inverted Leica SP8X WLL microscope with a 63×/1.3 Glyc objective. We sequentially recorded Quasar 670 (excitation 647 nm; emission 670 nm – 760 nm) and Quasar 570 (excitation 556 nm; emission 566 nm – 630 nm) with hybrid photo detectors (HyDs) and DAPI (excitation 405 nm; emission 415 nm – 450 nm) with a conventional photomultiplier tube. The same illumination and acquisition settings were used for all sections. Brightness and contrast of the images were adjusted with the open source software FIJI (ImageJ; National Institutes of Health, Bethesda, MD, USA).
MACS enrichment of retinal cell types
Retinal cell types were enriched as described previously using magnetic-activated cell sorting (MACS) (Grosche et al., 2016). Briefly, retinae were treated with papain (0.2 mg/ml; Roche Molecular Biochemicals) for 30 minutes at 37°C in the dark in Ca2+- and Mg2+-free extracellular solution (140 mM NaCl, 3 mM KCl, 10 mM HEPES, 11 mM glucose, pH 7.4). After several washes and 4 minutes of incubation with DNase I (200 U/ml), retinae were triturated in extracellular solution (now with 1 mM MgCl2 and 2 mM CaCl2). To purify microglial and vascular cells, the retinal cell suspension was subsequently incubated with CD11b- and CD31 microbeads according to the manufacturer’s protocol (Miltenyi Biotec, Bergisch Gladbach, Germany). The respective binding cells were depleted from the retinal suspension using LS-columns, prior to Müller cell enrichment. To purify Müller glia, the cell suspension was incubated in extracellular solution containing biotinylated anti-CD29 (0.1 mg/ml, Miltenyi Biotec) for 15 minutes at 4°C. Cells were washed in an extracellular solution, spun down, resuspended in the presence of anti-biotin MicroBeads (1:5; Miltenyi Biotec,) and incubated for 10 minutes at 4°C. After washing, CD29+ Müller cells were separated using large cell (LS) columns according to the manufacturer’s instructions (Miltenyi Biotec). Cells in the flow through of the last sorting step- depleted of microglia, vascular cells and Müller glia- were considered as the neuronal population. RPE was collected by scratching the eye cup after the retina had been removed and thus, scratch samples also contained cells from the underlying choroid. Samples were digested, and in subsequent steps, macrophages were depleted using anti-CD11b-microbeads and vascular cells using CD31-microbeads (Miltenyi Biotec).
qRT-PCR
Total RNA was isolated from the enriched cell populations using the PureLink® RNA Micro Scale Kit (Thermo Fisher Scientific, Schwerte, Germany). A DNase digestion step was included to remove genomic DNA (Roche). We performed RNA integrity validation and quantification using the Agilent RNA 6000 Pico chip analysis according to the manufactures instructions (Agilent Technologies, Waldbronn, Germany). First-strand cDNAs from the total RNA purified from each cell population were synthesized using the RevertAid H Minus First-Strand cDNA Synthesis Kit (Fermentas by Thermo Fisher Scientific, Schwerte, Germany). We designed primers using the Universal ProbeLibrary Assay Design Center (Roche, Table S6) and measured transcript levels of candidate genes by qRT-PCR using the TaqMan hPSC Scorecard Panel (384 well, ViiA7, Life Technologies, Darmstadt, Germany) according to the company’s guidelines.
LC-MS/MS mass spectrometry analysis
LC-MS/MS analysis was performed as described previously (Frik et al., 2018; Lepper et al., 2018) on a Q-Exactive HF mass spectrometer (Thermo Fisher Scientific Inc., Waltham, MA, U.S.A.) coupled to an Ultimate 3000 RSLC nano-HPLC (Dionex, Sunnyvale, CA). Briefly, 0.5 μg sample was automatically loaded onto a nano trap column (300 μm inner diameter × 5 mm, packed with Acclaim PepMap100 C18.5 μm, 100 Å; LC Packings, Sunnyvale, CA) before separation by reversed phase chromatography (HSS-T3 M-class column, 25 cm, Waters) in an 80 minutes non-linear gradient from 3 to 40% acetonitrile (ACN) in 0.1% formic acid (FA) at a flow rate of 250 nl/min. Eluted peptides were analyzed by the Q-Exactive HF mass spectrometer equipped with a nano-flex ionization source. Full scan MS spectra (from m/z 300 to 1500) and MS/MS fragment spectra were acquired in the Orbitrap with a resolution of 60,000 or 15000 respectively, with maximum injection times of 50 ms each. Up to ten most intense ions were selected for HCD fragmentation depending on signal intensity (TOP10 method). Target peptides already selected for MS/MS were dynamically excluded for 30 s. Spectra were analyzed using the Progenesis QI software for proteomics (Version 3.0, Nonlinear Dynamics, Waters, Newcastle upon Tyne, UK) for label-free quantification, as previously described (Grosche et al., 2016). All features were exported as a Mascot generic file (mgf) and used for peptide identification with Mascot (version 2.4) in the UniProtKB/Swiss-Prot taxonomy mouse database (Release 2017.02, 16871 sequences). Search parameters used were: 10 ppm peptide mass tolerance, 20 mmu fragment mass tolerance, one missed cleavage allowed, carbamidomethylation set as fixed modification, and methionine oxidation, asparagine or glutamine deamidation were allowed as variable modifications. A Mascot-integrated decoy database search calculated an average false discovery rate (FDR) of < 1%.
Western blot
Cell pellets of enriched cell populations from pooled pair of mouse eyes were dissolved in reducing Laemmli sample buffer, denatured and sonicated. Neuronal protein extraction reagent (Thermo Fisher Scientific, Braunschweig, Germany) was added to the neuron populations. Samples were separated on a 12% SDS-PAGE. The immunoblot was performed as previously described (Schäfer et al., 2017). Detection was performed with primary and secondary antibodies diluted in blocking solution (Table S7). Blots were developed with WesternSure PREMIUM Chemiluminescent Substrate (LI-COR, Bad Homburg, Germany). To validate specificity of the antibodies, all of them were tested on mouse serum as positive control (Figure S3).
Immunofluorescent labeling of retina and RPE
To quantify cell nuclei and perform stainings for C1s, C3 and glutamine synthetase (GLUL) in retinal sections of 4% paraformaldehyde (PFA)-fixated and paraffin-embedded murine eyes, the sections were deparaffinised and incubated with Hoechst33342/DAPI (1:1000; #H1399, Thermo Fisher Scientific, Braunschweig, Germany) or detection antibodies as previously described (Schäfer et al., 2017) (Table S7). Images were acquired using confocal microscopy (VisiScope, Visitron Systems, Puchheim, Germany).
Retinal microglia quantification was performed in the retinal flat mounts. Anterior segments of mouse eyes were removed, and the retina carefully separated. Flat mounts were fixated in 4% PFA (retina 1 h room temperature), permeabilized (1% Triton X-100) and blocked (1% BSA, 5% goat serum, 0.1 M NaPO4, pH 7). Retinal flat mounts were stained with anti-Iba1 antibody (3% Triton X-100, 1% DMSO, 5% normal goat serum, overnight at 4°C) and secondary antibody (1% BSA in PBS, overnight at 4°C) (Table S7). Retinal flat mounts were embedded with photoreceptors facing down, and the GCL facing up. Images were taken with a confocal microscope (VisiScope, Visitron Systems).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses were performed using Prism (Graphpad Software, San Diego, CA, USA). In most of the experiments in the present study results from 4 biological replicates were collected to keep to the rules of the three Rs for the sake of animal welfare. Since this low number of input values does not allow an appropriate estimation about a normal Gaussian distribution, significance levels were determined by the non-parametric Mann-Whitney U test unless stated otherwise. All data are expressed as mean ± standard error (SEM) unless stated otherwise. Detailed information about specific n-values, implemented statistical tests and coding of significance levels are provided in the respective figure legends.
DATA AND CODE AVAILABILITY
The accesssion number for the single cell RNA-Seq data reported in this paper is GSE116426 (Gene Expression Omnibus (GEO)). Other data supporting the findings of this study are available from the corresponding author upon request.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| mouse anti-CD29-Biotin | Milteny Biotec (Bergisch-Gladbach, Germany) | 130–101–943; RRID:AB_2660700 |
| rabbit anti-PDHB | Abcam (Cambridge, UK) | ab155996; RRID:AB_2814826 |
| rabbit anti-C1s | Proteintech (Rosemont, IL, USA) | #14554–1-AP / (Schäfer et al., 2017); RRID:AB_2814827 |
| goat anti-C4 | Complement Technologies (Tyler, TX, USA) | #A205; RRID:AB_2814828 |
| goat anti-C3-HRP | MP Biomedicals (Santa Ana, CA, USA) | #55557 / (Schäfer et al., 2017) |
| rabbit anti-C3 | Abcam (Cambridge, UK) | ab11887; RRID:AB_298669 |
| goat anti-CFB | Merck (Darmstadt, Germany) | #341272 / (Schäfer et al., 2017); RRID:AB_2082392 |
| sheep anti-CFD | R&D Systems (Minneapolis, MN, USA) | #AF5430; RRID:AB_1655868 |
| mouse anti-C5 | Quidel (San Diego, CA, USA) | #A217; RRID:AB_452484 |
| goat anti-C6 | Complement Technologies (Tyler, TX, USA) | #A223; RRID:AB_2814831 |
| goat anti-C7 | Tecomedical (Sissach, CH) | #A308; RRID:AB_2814832 |
| rabbit anti-C9 | Antibodies online (Aachen, Germany) | #ABIN1714714; RRID:AB_2814833 |
| rat anti-CFP | in-house | (Schäfer et al., 2017) |
| goat anti-CFH | Merck (Darmstadt, Germany) | #341276; RRID:AB_2080303 |
| goat anti-CFI | Quidel (San Diego, CA, USA) | A313 / (Rose et al., 2008): RRID:AB_452514 |
| rabbit anti-IBA1 | Wako Chemicals (Neuss, Germany) | #019–19741 / (Schäfer et al., 2016); RRID:AB_839504 |
| mouse anti-glutamine sythetase | Merck (Darmstadt, Germany) | MAB302 /(Mages et al., 2019); RRID:AB_2110656 |
| goat anti-rat Ig-HRP | Dianova (Hamburg, Germany) | #112–035–003; RRID:AB_2338128 |
| goat anti-rabbit Ig-HRP | Dianova (Hamburg, Germany) | #111–035–003; RRID:AB2313567 |
| rabbit anti-goat Ig-HRP | Dianova (Hamburg, Germany) | #305–035–003; RRID:AB2339400 |
| goat anti-rabbit-IG-Cy3 | ThermoFisher (Braunschweig, Germany) | #A10520; RRID:AB2534029 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| CD11b (Microglia) MicroBeads, human and mouse | Milteny Biotec | 130–093–634 |
| CD31 MicroBeads, mouse | Milteny Biotec | 130–097–418 |
| Anti-Biotin MicroBeads UltraPure | Milteny Biotec | 130–105–637 |
| Critical Commercial Assays | ||
| RevertAid H Minus First-Strand cDNA Synthesis Kit | Thermo Fisher Scientific | K1632 |
| PureLink® RNA Micro Scale Kit | Thermo Fisher Scientific | 12183016 |
| Deposited Data | ||
| scRNA sequencing data | Gene Expression Omnibus (GEO) | GSE116426 |
| Experimental Models: Organisms/Strains | ||
| BALB/cJRj mice | Janvier Labs | SC-BALBJ-M |
| C57BL/6J | Jackson Laboratories | 000664 |
| Oligonucleotides | ||
| see Table S6 | This paper | N/A |
| Software and Algorithms | ||
| Progenesis QI software for proteomics (Version 3.0) | Nonlinear Dynamics, Waters, Newcastle upon Tyne, U.K. | N/A |
| R package Seurat | Stuart et al., 2019 | N/A |
| FIJI (ImageJ) | National Institutes of Health, Bethesda, MD, USA | N/A |
| R v 3.5.1 | https://www.R-project.org | N/A |
Highlights.
Each retinal cell type expresses a specific signature of complement components
Müller and RPE cells are the main source of retinal complement transcripts
Components of the alternative and classical activating pathways were detected
The cell-type-specific complement signature changes with aging and degeneration
ACKNOWLEDGMENTS
We thank Gabriele Jäger, Dirkje Felder, Renate Foeckler, Andrea Dannullis, and Elfriede Eckert for excellent technical support for cell preparation, immunodetection, and molecular biology. This project was supported by the Deutsche Forschungsgemeinschaft (grant DFG-GR 4403/5-1 to A.G. and grant DFG-PA 1844/3-1 to D.P.) and the Macula Vision Research Foundation (D.S., M.L., and C.A.C.), with institutional support from the EyeSight Foundation of Alabama and Research to Prevent Blindness (C.A.C.); the NIH (grant R01EY030192 to D.S. and M.L.; grants R01GM108600 and R01GM125301 to M.L.; and grant 5R01-HG006137 to N.R.Z.); and by a Blavatnik Family Fellowship (D.A.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.10.084.
SUPPORTING CITATIONS
The following references appear in the Supplemental Information: Cheng et al., 2013a
REFERENCES
- Anderson DH, Radeke MJ, Gallo NB, Chapin EA, Johnson PT, Curletti CR, Hancox LS, Hu J, Ebright JN, Malek G, et al. (2010). The pivotal role of the complement system in aging and age-related macular degeneration: hypothesis re-visited. Prog. Retin. Eye Res 29, 95–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva K, Zhang M, Fan W, Li X, Chen Y, Rebolledo-Mendez JD, and Cooper NG (2014). Time-dependent gene profiling indicates the presence of different phases for ischemia/reperfusion injury in retina. Ophthalmol. Eye Dis 6, 43–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astafurov K, Dong CQ, Panagis L, Kamthan G, Ren L, Rozenboym A, Perera TD, Coplan JD, and Danias J (2014). Complement expression in the retina is not influenced by short-term pressure elevation. Mol. Vis 20, 140–152. [PMC free article] [PubMed] [Google Scholar]
- Chen H, Liu B, Lukas TJ, and Neufeld AH (2008). The aged retinal pigment epithelium/choroid: a potential substratum for the pathogenesis of age-related macular degeneration. PLoS ONE 3, e2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Muckersie E, Forrester JV, and Xu H (2010). Immune activation in retinal aging: a gene expression study. Invest. Ophthalmol. Vis. Sci 51, 5888–5896. [DOI] [PubMed] [Google Scholar]
- Cheng L, Bu H, Portillo J-AC, Li Y, Subauste CS, Huang SS, Kern TS, and Lin F (2013a). Modulation of retinal Müller cells by complement receptor C5aR. Invest. Ophthalmol. Vis. Sci 54, 8191–8198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CL, Djajadi H, and Molday RS (2013b). Cell-specific markers for the identification of retinal cells by immunofluorescence microscopy. Methods Mol. Biol. 935, 185–199. [DOI] [PubMed] [Google Scholar]
- Crabb JW (2014). The proteomics of drusen. Cold Spring Harb. Perspect. Med. 4, a017194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribbs DH, Berchtold NC, Perreau V, Coleman PD, Rogers J, Tenner AJ, and Cotman CW (2012). Extensive innate immune gene activation accompanies brain aging, increasing vulnerability to cognitive decline and neurodegeneration: a microarray study. J. Neuroinflammation 9, 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damani MR, Zhao L, Fontainhas AM, Amaral J, Fariss RN, and Wong WT (2011). Age-related alterations in the dynamic behavior of microglia. Aging Cell 10, 263–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AE 3rd, Harrison RA, and Lachmann PJ (1984). Physiologic inactivation of fluid phase C3b: isolation and structural analysis of C3c, C3d,g (alpha 2D), and C3g. J. Immunol 132, 1960–1966. [PubMed] [Google Scholar]
- Friedman E, and Ts’o MO (1968). The retinal pigment epithelium. II. Histologic changes associated with age. Arch. Ophthalmol 79, 315–320. [DOI] [PubMed] [Google Scholar]
- Frik J, Merl-Pham J, Plesnila N, Mattugini N, Kjell J, Kraska J, Gómez RM, Hauck SM, Sirko S, and Götz M (2018). Cross-talk between monocyte invasion and astrocyte proliferation regulates scarring in brain injury. EMBO Rep. 19, e45294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsche LG, Igl W, Bailey JNC, Grassmann F, Sengupta S, Bragg-Gresham JL, Burdon KP, Hebbring SJ, Wen C, Gorski M, et al. (2016). A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat. Genet 48, 134–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassmann F, Cantsilieris S, Schulz-Kuhnt A-S, White SJ, Richardson AJ, Hewitt AW, Vote BJ, Schmied D, Guymer RH, Weber BHF, and Baird PN (2016). Multiallelic copy number variation in the complement component 4A (C4A) gene is associated with late-stage age-related macular degeneration (AMD). J. Neuroinflammation 13, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosche A, Hauser A, Lepper MF, Mayo R, von Toerne C, Merl-Pham J, and Hauck SM (2016). The proteome of native adult Müller glial cells from murine retina. Mol. Cell. Proteomics 15, 462–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus HE, Nickerson JM, Edelhauser HF, Bergman LAMK, and Berglin L (2013). Anatomic alterations in aging and age-related diseases of the eye. Invest. Ophthalmol. Vis. Sci 54, ORSF23–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawksworth OA, Coulthard LG, and Woodruff TM (2017). Complement in the fundamental processes of the cell. Mol. Immunol 84, 17–25. [DOI] [PubMed] [Google Scholar]
- Jeon CJ, Strettoi E, and Masland RH (1998). The major cell populations of the mouse retina. J. Neurosci 18, 8936–8946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempe H, Schwabe A, Crémazy F, Verschure PJ, and Bruggeman FJ (2015). The volumes and transcript counts of single cells reveal concentration homeostasis and capture biological noise. Mol. Biol. Cell 26, 797–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DS, Ross SE, Trimarchi JM, Aach J, Greenberg ME, and Cepko CL (2008). Identification of molecular markers of bipolar cells in the murine retina. J. Comp. Neurol 507, 1795–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B-J, Braun TA, Wordinger RJ, and Clark AF (2013). Progressive morphological changes and impaired retinal function associated with temporal regulation of gene expression after retinal ischemia/reperfusion injury in mice. Mol. Neurodegener. 8, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn MH, Kim CY, Jiang B, Dumitrescu AV, and Kwon YH (2008). Disruption of the complement cascade delays retinal ganglion cell death following retinal ischemia-reperfusion. Exp. Eye Res 87, 89–95. [DOI] [PubMed] [Google Scholar]
- Lepper MF, Ohmayer U, von Toerne C, Maison N, Ziegler A-G, and Hauck SM (2018). Proteomic landscape of patient-derived CD4+ T cells in recent-onset type 1 diabetes. J. Proteome Res. 17, 618–634. [DOI] [PubMed] [Google Scholar]
- Levin AM, Rusu I, Orlin A, Gupta MP, Coombs P, D’Amico DJ, and Kiss S (2017). Retinal reperfusion in diabetic retinopathy following treatment with anti-VEGF intravitreal injections. Clin. Ophthalmol 11, 193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Jia C, Kazmierkiewicz KL, Bowman AS, Tian L, Liu Y, Gupta NA, Gudiseva HV, Yee SS, Kim M, et al. (2014). Comprehensive analysis of gene expression in human retina and supporting tissues. Hum. Mol. Genet 23, 4001–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liszewski MK, Elvington M, Kulkarni HS, and Atkinson JP (2017). Complement’s hidden arsenal: New insights and novel functions inside the cell. Mol. Immunol. 84, 2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C, Chen M, and Xu H (2011). Complement gene expression and regulation in mouse retina and retinal pigment epithelium/choroid. Mol. Vis 17, 1588–1597. [PMC free article] [PubMed] [Google Scholar]
- Luo C, Zhao J, Madden A, Chen M, and Xu H (2013). Complement expression in retinal pigment epithelial cells is modulated by activated macrophages. Exp. Eye Res 112, 93–101. [DOI] [PubMed] [Google Scholar]
- Lyzogubov VV, Bora NS, Tytarenko RG, and Bora PS (2014). Polyethylene glycol induced mouse model of retinal degeneration. Exp. Eye Res 127, 143–152. [DOI] [PubMed] [Google Scholar]
- Lyzogubov VV, Bora PS, Wu X, Horn LE, de Roque R, Rudolf XV, Atkinson JP, and Bora NS (2016). The complement regulatory protein CD46 deficient mouse spontaneously develops dry-type age-related macular degeneration-like phenotype. Am. J. Pathol 186, 2088–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Cojocaru R, Gotoh N, Gieser L, Villasmil R, Cogliati T, Swaroop A, and Wong WT (2013). Gene expression changes in aging retinal microglia: relationship to microglial support functions and regulation of activation. Neurobiol. Aging 34, 2310–2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. (2015). Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mages K, Grassmann F, Jägle H, Rupprecht R, Weber BHF, Hauck SM, and Grosche A (2019). The agonistic TSPO ligand XBD173 attenuates the glial response thereby protecting inner retinal neurons in a murine model of retinal ischemia. J. Neuroinflammation 16, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marguerat S, and Bähler J (2012). Coordinating genome expression with cell size. Trends Genet. 28, 560–565. [DOI] [PubMed] [Google Scholar]
- Martin M, Leffler J, Smoląg KI, Mytych J, Björk A, Chaves LD, Alexander JJ, Quigg RJ, and Blom AM (2016). Factor H uptake regulates intracellular C3 activation during apoptosis and decreases the inflammatory potential of nucleosomes. Cell Death Differ. 23, 903–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merle NS, Church SE, Fremeaux-Bacchi V, and Roumenina LT (2015). Complement system part I - molecular mechanisms of activation and regulation. Front. Immunol 6, 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micklisch S, Lin Y, Jacob S, Karlstetter M, Dannhausen K, Dasari P, von der Heide M, Dahse H-M, Schmölz L, Grassmann F, et al. (2017). Age-related macular degeneration associated polymorphism rs10490924 in ARMS2 results in deficiency of a complement activator. J. Neuroinflammation 14, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai R, Okunuki Y, Husain D, Kim CB, Lambris JD, and Connor KM (2018). The complement system is critical in maintaining retinal integrity during aging. Front. Aging Neurosci 10, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannicke T, Frommherz I, Biedermann B, Wagner L, Sauer K, Ulbricht E, Härtig W, Krügel U, Ueberham U, Arendt T, et al. (2014). Differential effects of P2Y1 deletion on glial activation and survival of photoreceptors and amacrine cells in the ischemic mouse retina. Cell Death Dis. 5, e1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinelli M, Carissimo A, Cutillo L, Lai C-H, Mutarelli M, Moretti MN, Singh MV, Karali M, Carrella D, Pizzo M, et al. (2016). An atlas of gene expression and gene co-regulation in the human retina. Nucleic Acids Res. 44, 5773–5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu RA, Hu J, Yuan Q, Welch DL, Makshanoff J, Lloyd M, McMullen S, Travis GH, and Bok D (2011). Complement system dysregulation and inflammation in the retinal pigment epithelium of a mouse model for Stargardt macular degeneration. J. Biol. Chem 286, 18593–18601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheaume BA, Jereen A, Bolisetty M, Sajid MS, Yang Y, Renna K, Sun L, Robson P, and Trakhtenberg EF (2018). Single cell transcriptome profiling of retinal ganglion cells identifies cellular subtypes. Nat. Commun 9, 2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch K, Jadhav AP, Trimarchi JM, Stadler MB, Roska B, Sun BB, and Cepko CL (2008). The transcriptome of retinal Müller glial cells. J. Comp. Neurol 509, 225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose KL, Paixao-Cavalcante D, Fish J, Manderson AP, Malik TH, Bygrave AE, Lin T, Sacks SH, Walport MJ, Cook HT, et al. (2008). Factor I is required for the development of membranoproliferative glomerulonephritis in factor H-deficient mice. J. Clin. Invest 118, 608–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutar M, Natoli R, Albarracin R, Valter K, and Provis J (2012). 670-nm light treatment reduces complement propagation following retinal degeneration. J. Neuroinflammation 9, 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer N, Grosche A, Reinders J, Hauck SM, Pouw RB, Kuijpers TW, Wouters D, Ehrenstein B, Enzmann V, Zipfel PF, et al. (2016). Complement regulator FHR-3 is elevated either locally or systemically in a selection of autoimmune diseases. Front. Immunol 7, 542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer N, Grosche A, Schmitt SI, Braunger BM, and Pauly D (2017). Complement components showed a time-dependent local expression pattern in constant and acute white light-induced photoreceptor damage. Front. Mol. Neurosci 10, 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheetz TE, Fingert JH, Wang K, Kuehn MH, Knudtson KL, Alward WLM, Boldt HC, Russell SR, Folk JC, Casavant TL, et al. (2013). A genome-wide association study for primary open angle glaucoma and macular degeneration reveals novel loci. PLoS ONE 8, e58657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhar K, Lapan SW, Whitney IE, Tran NM, Macosko EZ, Kowalczyk M, Adiconis X, Levin JZ, Nemesh J, Goldman M, et al. (2016). Comprehensive classification of retinal bipolar neurons by single-cell transcriptomics. Cell 166, 1308–1323.e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva PS, Dela Cruz AJ, Ledesma MG, van Hemert J, Radwan A, Cavallerano JD, Aiello LM, Sun JK, and Aiello LP (2015). Diabetic retinopathy severity and peripheral lesions are associated with nonperfusion on ultrawide field angiography. Ophthalmology 122, 2465–2472. [DOI] [PubMed] [Google Scholar]
- Sim RB, Day AJ, Moffatt BE, and Fontaine M (1993). Complement factor I and cofactors in control of complement system convertase enzymes. Methods Enzymol. 223, 13–35. [DOI] [PubMed] [Google Scholar]
- Sohn JH, Kaplan HJ, Suk HJ, Bora PS, and Bora NS (2000). Chronic low level complement activation within the eye is controlled by intraocular complement regulatory proteins. Invest. Ophthalmol. Vis. Sci 41, 3492–3502. [PMC free article] [PubMed] [Google Scholar]
- Sohn J-H, Bora PS, Suk H-J, Molina H, Kaplan HJ, and Bora NS (2003). Tolerance is dependent on complement C3 fragment iC3b binding to antigen-presenting cells. Nat. Med 9, 206–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudharsan R, Beiting DP, Aguirre GD, and Beltran WA (2017). Involvement of innate immune system in late stages of inherited photoreceptor degeneration. Sci. Rep 7, 17897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenner AJ, Stevens B, and Woodruff TM (2018). New tricks for an ancient system: Physiological and pathological roles of complement in the CNS. Mol. Immunol. 102, 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Kazmierkiewicz KL, Bowman AS, Li M, Curcio CA, and Stambolian DE (2015). Transcriptome of the human retina, retinal pigmented epithelium and choroid. Genomics 105, 253–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traveset A, Rubinat E, Ortega E, Alcubierre N, Vazquez B, Hernández M, Jurjo C, Espinet R, Ezpeleta JA, and Mauricio D (2016). Lower hemoglobin concentration is associated with retinal ischemia and the severity of diabetic retinopathy in type 2 diabetes. J. Diabetes Res 2016, 3674946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner L, Pannicke T, Frommherz I, Sauer K, Chen J, and Grosche A (2016). Effects of IP3R2 receptor deletion in the ischemic mouse retina. Neuro- chem. Res. 41, 677–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner L, Pannicke T, Rupprecht V, Frommherz I, Volz C, Illes P, Hirrlinger J, Jägle H, Egger V, Haydon PG, et al. (2017). Suppression of SNARE-dependent exocytosis in retinal glial cells and its effect on ischemia-induced neurodegeneration. Glia 65, 1059–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DG, and McGeer PL (1992). Complement gene expression in human brain: comparison between normal and Alzheimer disease cases. Brain Res. Mol. Brain Res 14, 109–116. [DOI] [PubMed] [Google Scholar]
- Wang J, Yang MM, Li YB, Liu GD, Teng Y, and Liu XM (2013). Association of CFH and CFB gene polymorphisms with retinopathy in type 2 diabetic patients. Mediators Inflamm. 2013, 748435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber BHF, Charbel Issa P, Pauly D, Herrmann P, Grassmann F, and Holz FG (2014). The role of the complement system in age-related macular degeneration. Dtsch. Arztebl. Int. 111, 133–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MM, Wang J, Ren H, Sun YD, Fan JJ, Teng Y, and Li YB (2016). genetic investigation of complement pathway genes in type 2 diabetic retinopathy: an inflammatory perspective. Mediators Inflamm 2016, 1313027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Gerhardinger C, and Lorenzi M (2002). Early complement activation and decreased levels of glycosylphosphatidylinositol-anchored complement inhibitors in human and experimental diabetic retinopathy. Diabetes 51, 3499–3504. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accesssion number for the single cell RNA-Seq data reported in this paper is GSE116426 (Gene Expression Omnibus (GEO)). Other data supporting the findings of this study are available from the corresponding author upon request.