Abstract
Secreted, plasma membrane, and resident proteins of the secretory pathway are synthesized in the endoplasmic reticulum (ER) where they undergo post-translational modifications, oxidative folding, and subunit assembly in tightly monitored processes. An ER quality control (ERQC) system oversees protein maturation and ensures that only those reaching their native state will continue trafficking into the secretory pathway to reach their final destinations. Those that fail must be recognized and eliminated to maintain ER homeostasis. Two cellular mechanisms have been identified to rid the ER of terminally unfolded and aggregated proteins. ER-associated degradation (ERAD) was discovered nearly 30 years ago and entails the identification of improperly matured secretory pathway proteins and their retrotranslocation to the cytosol for degradation by the ubiquitin-proteasome system. ER-phagy has been more recently described and caters to larger, more complex proteins and protein aggregates that are not readily handled by ERAD. This pathway has unique upstream components and relies on the same downstream effectors of autophagy used in other cellular processes to deliver clients to lysosomes for degradation. In this review, we describe the main elements of ERQC, ERAD, and ER-phagy and focus on recent advances in these fields.
Keywords: ER Quality control (ERQC), ER chaperones, Unfolded protein response (UPR), ER-associated degradation (ERAD), ER-phagy, Ubiquitin proteasome system (UPS)
1. Introduction
The endoplasmic reticulum (ER) is a major site of protein synthesis in eukaryotic cells where it has been estimated, in the case of mammalian cells, that approximately one third of the proteins encoded in the genome will be translocated, undergo post-translational modifications, fold oxidatively, and often assemble into multimeric complexes (Braakman & Hebert, 2013). The proper maturation of these proteins is essential to inter- and intra-cellular signaling, and failures in this process can lead to disease and even death. Thus, a robust ER quality control (ERQC) machinery has coevolved with the clients that must aid and monitor the outcome of an immense array of proteins that have vastly distinct sequences and structures. Proteins that pass ERQC will be transported to their functional destination, which can be sub-regions of the ER, other organelles of the secretory pathway, the cell surface, or the extracellular space. Those that fail must be identified and targeted for proteasomal degradation in a process known as ER-associated degradation (ERAD) or trafficked to the lysosomes through autophagic processes (ER-phagy). Although the clients vary by cell type and organism, the general components of these systems are highly conserved across eukaryotic evolution. Considerable advancements have been made in the past decade in our understanding of the relationship between ERQC, ERAD, and ER-phagy that have been obtained through a multitude of studies ranging from purified proteins to genome-wide screens, biochemical assays, and cell-based studies in organisms extending from yeast to humans.
2. ER Quality Control
Nascent proteins enter the ER co- or post-translationally in yeast, and almost exclusively post-translationally in mammalian cells, where they encounter a host of molecular chaperones and folding enzymes that both aid and monitor their progress in achieving a native tertiary or quaternary state (Figure 1). This process is termed ER quality control (ERQC) and has been the focus of multiple excellent reviews Braakman & Bulleid, 2011; Araki & Nagata, 2012; Braakman & Hebert, 2013). Readers wishing more in-depth discussion of the topic are directed to them, as they are outside the focus of the current review.
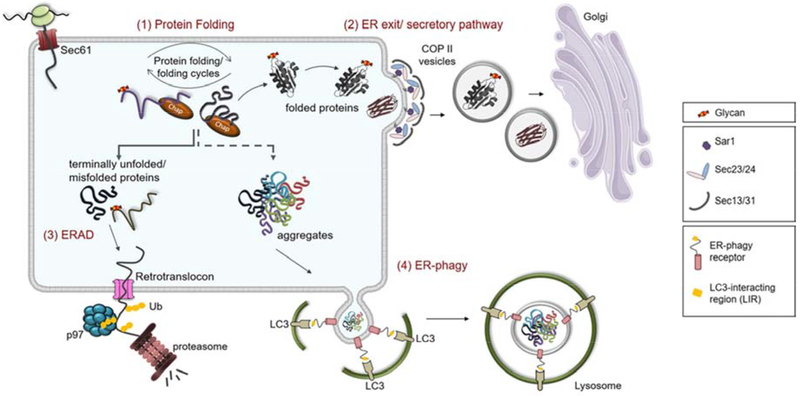
Figure 1: ER-quality control (ERQC): from folding, to secretion or disposal of ER synthesized proteins.
(1) Newly synthesized proteins enter the ER lumen via the Sec61 translocon channel and begin to fold once in the lumen where their progress is both aided and monitored by molecular chaperones (chap). (2) Correctly folded and assembled proteins that pass ERQC are incorporated into COPII vesicles and are transported further along the secretory pathway to reach their functional destination, either in the secretory pathway itself, the cell surface, or the extracellular space. To maintain ER homeostasis, proteins that fail to obtain their proper native structure must be eliminated from the ER by one of two mechanisms: ER-associated degradation (ERAD) or ER-phagy. (3) Soluble ERAD clients are targeted for retrotranslocation to the cytosol via a channel (retrotranslocon) consisting of several multi-pass membrane proteins and auxiliary factors. As these clients emerge in the cytosol, they become poly-ubiquitinated allowing them to be recognized by the p97 AAA-ATPase that provides the energy for extracting the protein from the ER for delivery to 26S proteasome for degradation. (4) Large protein aggregates that are not easily handled by ERAD, are degraded by ER-phagy via the lysosomal pathway. This process is also dependent on receptors, which possess LC3-interacting regions (LIRs) that allow interactions with LC3-decorated autophagosome membranes and engulfment of the protein aggregates and delivery to lysosomes for degradation via standard autophagic pathways.
In brief, here are two major families of molecular chaperones in the ER; the lectin chaperones calnexin (Cne1p in yeast) and calreticulin, and the Hsp70 cognate BiP (Kar2p in yeast). The lectin chaperones bind monoglucosylated N-glycans in proximity to unfolded regions and work in concert with UDP-glucose:glycoprotein glycosyltransferase (UGGT), an enzyme that binds directly to unfolded regions and ensures the adjacent N-glycan remains monoglucosylated as long as the client is incompletely folded (Parodi, 2000),(Hebert, Foellmer, & Helenius, 1995),(Ritter & Helenius, 2000). Conversely, BiP binds directly to unfolded regions on non-glycosylated nascent chains or glycoproteins that do not possess a glycan near the region that is slow to fold (Hendershot, Bole, Kohler, & Kearney, 1987; Molinari & Helenius, 2000). BiP can be aided by ER-localized DnaJ-like (ERdj) co-factors, seven of which have been identified in mammals and three in yeast (reviewed in (Kampinga & Craig, 2010; Pobre, Poet, & Hendershot, 2019)). Four of the mammalian ERdjs (ERdj3-6) bind directly to unfolded proteins and recruit BiP, whereas two of the yeast DnaJ-like proteins do. Unfolded regions on proteins do not fold while bound to either of the chaperone families, but instead are maintained in a protected state that is conducible to folding. Both the lectins (Tatu & Helenius, 1997) and BiP (Meunier, Usherwood, Chung, & Hendershot, 2002) are part of larger complexes containing protein disulfide isomerases (PDIs) and peptidyl-prolyl isomerases (PPIs) that catalyze folding reactions. Prior to completing maturation, clients are retained in the ER through their association with either BiP or calreticulin, which possess C-terminal KDEL/HDEL sequences, respectively, that dictate their ER localization (Munro & Pelham, 1987) or with calnexin, an integral membrane, resident ER protein. Proteins that complete folding and assembly, thus burying the hydrophobic sequences that are recognized by BiP (Blond-Elguindi et al., 1993; Behnke, Mann, Scruggs, Feige, & Hendershot, 2016) and UGGT (Calles-Garcia et al., 2017; Izumi et al., 2017), no longer interact with these chaperones. In most cases, proteins that are no longer recognized by chaperones have folded and passed ERQC allowing them to be targeted to ER exit sites and transported to the Golgi via COPII vesicles for delivery to their ultimate site of function (as reviewed in (Barlowe & Helenius, 2016)).
3. Failing ERQC
Secretory pathway proteins that are not ultimately able to fold must be identified and degraded, however, the half-lives of unfolded proteins can vary dramatically. It is not entirely clear how folding attempts versus decisions to degrade are controlled for various unfolded clients, although some aspects of this process are now better understood (Figure 2).
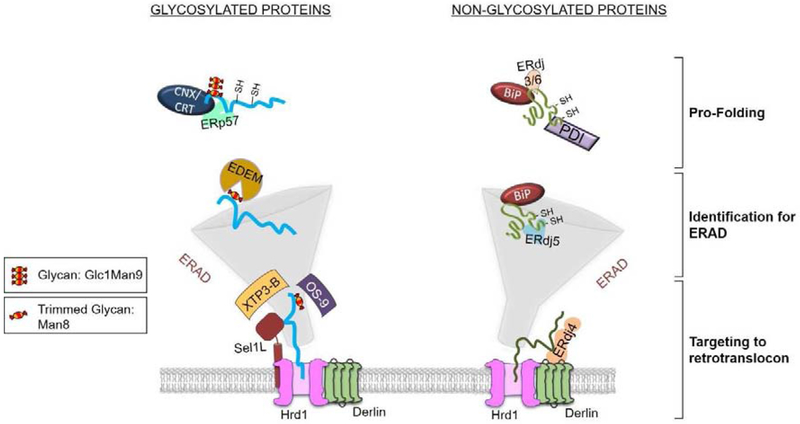
Figure 2: ER-quality control: triaging proteins between folding and ERAD.
The N-linked glycans ( ) on nascent glycoproteins provide a recognition signal for the lectin chaperones calnexin (CNX) and calreticulin (CRT) that are associated with co-factors like the PDI family member ERp57, allowing them to undergo continued attempts to fold. Processing of the N-linked glycan by resident ER mannosidases removes the terminally unfolded glycoprotein from the folding cycle and allows it to be recognized by the EDEM proteins. The glycosylated ERAD client is transferred to two other lumenal lectins, XTP3-B and OS-9 which pass the client to Sel1L, an integral membrane protein, which is associated with the retrotranslocation complex. In the case of non-glycosylated proteins, they bind to the Hsp70 chaperone BiP as they enter the ER, which interacts with pro-folding ER-localized DnaJ co-factors like ERdj3/ERdj6. By less well understood mechanisms, the critical decision of ERQC between folding and identification for degradation involves transfer the client to the pro-degradation co-chaperones, ERdj4 and ERdj5. ERdj5 is a reductase that disrupts disulfide bonds and thus induces further unfolding of clients for ERAD, while ERdj4 is associated with Derlin, a component of the retrotranslocon.
) on nascent glycoproteins provide a recognition signal for the lectin chaperones calnexin (CNX) and calreticulin (CRT) that are associated with co-factors like the PDI family member ERp57, allowing them to undergo continued attempts to fold. Processing of the N-linked glycan by resident ER mannosidases removes the terminally unfolded glycoprotein from the folding cycle and allows it to be recognized by the EDEM proteins. The glycosylated ERAD client is transferred to two other lumenal lectins, XTP3-B and OS-9 which pass the client to Sel1L, an integral membrane protein, which is associated with the retrotranslocation complex. In the case of non-glycosylated proteins, they bind to the Hsp70 chaperone BiP as they enter the ER, which interacts with pro-folding ER-localized DnaJ co-factors like ERdj3/ERdj6. By less well understood mechanisms, the critical decision of ERQC between folding and identification for degradation involves transfer the client to the pro-degradation co-chaperones, ERdj4 and ERdj5. ERdj5 is a reductase that disrupts disulfide bonds and thus induces further unfolding of clients for ERAD, while ERdj4 is associated with Derlin, a component of the retrotranslocon.
In the case of glycoproteins, the structure of the N-linked glycan dictates continued chances to fold, recognition for transport to the Golgi, or alternatively targeting for degradation through the ubiquitin-proteasome system (UPS) (reviewed in (Aebi, Bernasconi, Clerc, & Molinari, 2010; Molinari & Hebert, 2015)). The removal of mannose residues from the N-linked glycan by the slow acting α1,2-mannosidase I causes the client to exit the calnexin/calreticulin cycle. Importantly a protein whose N-glycan is trimmed from 9 to 8 mannoses becomes a substrate for both Golgi transport and ERAD revealing a possible competition between these outcomes, which is central to ERQC. More recent studies have shown that if the protein is folded it will be incorporated into COPII vesicles through its association with cargo receptors. However, if it is not folded, its hydrophobic regions in combination with the mannose-trimmed glycan will be recognized by ER degradation-enhancing α-mannosidase I-like proteins (Htm1p/Mnl1p in yeast and EDEM1-3 in mammals) allowing it to interact with the lectins OS-9 and XTP3-B and be targeted for degradation (reviewed in (Maattanen, Gehring, Bergeron, & Thomas, 2010; Sokolowska, Pilka, Sandvig, Wegrzyn, & Slominska-Wojewodzka, 2015; Slominska-Wojewodzka & Sandvig, 2015; Shenkman et al., 2018). It also appears that compartmentalization of glycosylated ERAD clients in regions of the ER distinct from ER exit sites plays a role in the fidelity of ERQC (reviewed in (Shenkman & Lederkremer, 2019)).
In the case of non-glycosylated BiP clients the pivotal point in ERQC is less well understood, but a number of recent studies suggest that it likely involves the transfer of clients from pro-folding ERdj co-chaperones to pro-degradation ERdjs (Figure 2) (reviewed in (Pobre et al., 2019)). However, some conflicting data have been obtained. For instance, ERdj3 is associated with the translocon (Dejgaard et al., 2010; Guo & Snapp, 2013), which suggests that it can engage nascent polypeptide chains as they enter the ER lumen. It also binds stably to long-lived Ig heavy chains (Meunier et al., 2002) and has multiple interaction sites throughout two clients that overlap with BiP binding sites (Behnke et al., 2016). Furthermore, depletion of ERdj3 accelerates the turnover of the PiZ mutant of α1-anti-trypsin (Khodayari et al., 2017). All of these observations are consistent with a pro-folding function for ERdj3. Conversely, another study reported that ERdj3 depletion resulted in the stabilization of a glucocerebrosidase mutant (Tan et al., 2014). Closer examination revealed that this mutant associated with calnexin instead, which does not necessarily demonstrate that ERdj3 was performing a pro-degradation function for this protein; only that calnexin provided a longer period for folding than the BiP/ERdj3 cycle. ERdj6 selectively binds unfolded vesicular stomatitis virus G protein and is released as the protein folds (Petrova, Oyadomari, Hendershot, & Ron, 2008). Neither ERdj3 or ERdj6 are significantly up-regulated by ER stress and in fact nascent ERdj6 synthesized during stress is inefficiently targeted to the ER (Rutkowski et al., 2007). These studies in combination are most compatible with a profolding role for these ERdj proteins instead of a pro-degradation one. On the other hand, most studies to date consistently indicate that ERdj4 and ERdj5 assist BiP in targeting proteins for degradation. ERdj4 is associated with retrotranslocon components involved in targeting misfolded proteins for degradation (Lai, Otero, Hendershot, & Snapp, 2012), and reduced expression of ERdj4 prolongs the half-life of disease-associated surfactant protein C mutants (Dong, Bridges, Apsley, Xu, & Weaver, 2008), pro-insulin (Fritz et al., 2014), and epithelial sodium channels (Buck, Kolb, Boyd, Kleyman, & Brodsky, 2010). Another ER-localized DnaJ-like protein, ERdj5, possess six thioredoxin-like domains in addition to a J domain, thus it is a member of both the PDI and ERdj super-families (Hosoda, Kimata, Tsuru, & Kohno, 2003). It functions primarily as a reductase in the ER where it serves to reduce folded domains or oligomeric structures so they can more readily be degraded (Ushioda et al., 2008). Depletion of ERdj5 stabilizes surfactant protein C mutants (Dong et al., 2008), and its over-expression accelerates turnover of the null Hong Kong (NHK) variant of α1-anti-trypsin (Ushioda et al., 2008). ERdj5 also functions to reduce some toxin subunits, allowing them to enter the cytosol where their targets reside (Inoue et al., 2015; Williams, Inoue, Banks, & Tsai, 2013), and it binds lumenal degradation components thus assisting in their targeting for retrotranslocation. Both ERdj4 and ERdj5 bind were found to bind relatively fewer sequences in clients than either ERdj3 or BiP. These sequences were predicted by the TANGO algorithm to be particularly prone to β aggregate formation (Behnke et al., 2016), thus necessitating the rapid degradation of clients, if these sites are not buried by folding.
4. Responding to the problem
If unfolded proteins accumulate in the ER to a point that exceeds the available levels of molecular chaperones, in particular BiP, that prevent them from aggregating, a signal transduction program is activated termed the unfolded protein response (UPR). This response aims to restore ER homeostasis (reviewed in (Ron & Walter, 2007; Walter & Ron, 2011)) and is increasingly the target of small molecule activators and suppressors (reviewed in (Gonzalez-Teuber et al., 2019)). In fungi, Ire1 is the single transducer of the UPR, but as organisms grew in complexity so did the response (reviewed in (Hollien, 2013)). For instance, plants have two transducers, Ire1 and ATF6 and metazoan have three Ire1, PERK and ATF6. The mammalian UPR is characterized by a transcriptional up-regulation of ER chaperones and a decrease in the load of client proteins in the ER during stress conditions that are dependent on these chaperones. The latter is achieved by a combination of decreased translation, which is carried out via two distinct methods (inhibition of cap-dependent translation and cleavage of mRNAs docked at the ER membrane), and an increase in degradation. A UPR-induced expansion of the ER volume further serves to reduce the potential of unfolded proteins for aggregation. During stress conditions, the UPR triggers cell cycle arrest to prevent the propagation of cells experiencing ER stress, and if the stress is not resolved it can activate apoptotic pathways, although the tipping point between cell survival and death varies dramatically by tissue type. The basic outline and components of the UPR in several organisms are well-defined, and a major focus of research has shifted to its roles in development (reviewed in (Mitra & Ryoo, 2019)) and disease (reviewed in (Wang & Kaufman, 2012)). The demonstration that unfolded proteins represents the signal for activating the ER stress response was reported many years ago (Kozutsumi, Segal, Normington, Gething, & Sambrook, 1988), as was the demonstration that over-expression of BiP, but not of other ER chaperones, could inhibit UPR activation (Dorner, Wasley, & Kaufman, 1992). However, the precise mechanism of UPR activation continues to be debated with two theories predominating and most recent studies focusing on Ire1 (reviewed in (Kimata & Kohno, 2011; Chen & Brandizzi, 2013; Adams, Kopp, Larburu, Nowak, & Ali, 2019)). Some contend that release of BiP from the transducers by competition with unfolded proteins provides the critical activation signal (Bertolotti, Zhang, Hendershot, Harding, & Ron, 2000; Okamura, Kimata, Higashio, Tsuru, & Kohno, 2000; Kimata, Oikawa, Shimizu, Ishiwata-Kimata, & Kohno, 2004; Amin-Wetzel et al., 2017), while others argue that unfolded proteins bind directly to the transducers causing them to cluster and activate in trans (Kimata et al., 2007; Gardner & Walter, 2011; Oikawa, Kitamura, Kinjo, & Iwawaki, 2012; Carrara, Prischi, Nowak, Kopp, & Ali, 2015), relegating BiP to a more indirect role in regulating Ire1 activation. There are even disagreements between those favoring the BiP model for controlling Ire1 activation in terms of whether BiP binds to the transducer through its peptide binding domain (Amin-Wetzel et al., 2017) or its nucleotide binding domain (Carrara et al., 2015), and whether BiP co-factors or ATP play a role. Recent quantitative studies have convincingly shown that levels of an unassembled IgM heavy chain temporarily surpass levels of available BiP leading to an acute activation of the UPR (Bakunts et al., 2017; Vitale et al., 2019), which could be compatible with either model.
5. Cellular pathways for disposing misfolded ER proteins
Proteins that fail to pass ER quality control must be disposed of to protect ER homeostasis. Two mechanisms have been discovered that serve to identify and remove unfolded/misfolded proteins or orphan subunits of multimeric complexes from the ER; ERAD and ER-phagy. Many components of these two pathways are conserved in organisms ranging from yeast to mammals. These pathways feed into distinct cellular hubs (Figure 1) widely employed for the degradation of proteins from all organelles, the proteasome and the lysosome (vacuole in yeast). The delivery of ER clients to the proteasome was identified ~30 years ago and is currently better understood, although cutting-edge research continues on ERAD providing a more detailed and mechanistic understanding of this pathway. ER-phagy on the other hand has been more recently discovered, and as such, our understanding of this pathway is currently more limited. We will focus on the newest findings on ERAD, offer references for a few of the many excellent reviews on earlier delineations of the pathway, and describe the available data on ER-phagy.
5.1. ER-associated degradation (ERAD)
The identification of resident ER molecular chaperones in the mid-1980s and improved techniques to study the biosynthesis of secretory pathway cargo led a number of labs to discover that failure rates for achieving native protein structures could be quite high, particularly in the case of disease-associated mutant proteins (Le, Graham, & Sifers, 1990),(Ward, Omura, & Kopito, 1995), and that complete assembly of multimeric proteins could be inefficient (Sitia, Neuberger, & Milstein, 1987; Lippincott Schwartz, Bonifacino, Yuan, & Klausner, 1988); Amara, Lederkremer, & Lodish, 1989). These unsuccessfully folded proteins, as well as unassembled, orphan subunits turned over rapidly. However, unexpectedly lysosomal inhibitors and agents that disrupted transport to the Golgi did not stabilize them, arguing they were not degraded in the lysosome. A subsequent effort by multiple groups revealed that these proteins were extracted from the ER and degraded by the ubiquitin-proteasome pathway via a process termed ER-associated degradation (ERAD) (Werner, Brodsky, & McCracken, 1996; Wiertz et al., 1996); a pathway that was found to be conserved from yeast to mammals (reviewed in (Needham & Brodsky, 2013). In the ensuing years, the number of proteins classified as ERAD substrates has grown and many ERAD components have been identified through a combination of genetic and biochemical approaches (reviewed in (Olzmann, Kopito, & Christianson, 2013; Christianson & Ye, 2014; Berner, Reutter, & Wolf, 2018). Together these have provided a general understanding of the ERAD process, although mechanistic details a several points remain incompletely understand and are a focus of intense research. A number of excellent reviews expound upon the role of ERAD in protein folding disorders, toxin entry, and a variety of viral diseases (reviewed in (Guerriero & Brodsky, 2012; He, Ravindran, & Tsai, 2015; Qi, Tsai, & Arvan, 2017; Needham, Guerriero, & Brodsky, 2019)). In this review we will briefly describe the general steps in ERAD and the components that execute them and focus instead on significant advances that have occurred more recently.
5.1.1. The retrotranslocon and ERAD substrate retrotranslocation
Once folding and/or assembly of nascent ER proteins has failed and clients for ERAD have been identified, they must be transported to the cytosol where the UPS resides. This requires a protein conducting channel, a force for pulling the client from the ER, and ubiquitin conjugation for proteasomal degradation (Figure 3). The retrotranslocon channel that accommodates the extraction of ERAD clients to the cytosol has been the focus of intensive genetic and biochemical research and has resulted in the identification of numerous retrotranslocon components (reviewed in (Hampton & Sommer, 2012; Romisch, 2017; Berner et al., 2018; Wu & Rapoport, 2018)). In an attempt to identify mammalian channel components, a transmembrane ERAD client was affinity purified and subjected to MS/MS analyses of co-purified proteins, which identified several proteins known to be involved in ERAD, as well as the multi-pass membrane proteins Derlins 1-3 (Lilley & Ploegh, 2004; Lilley & Ploegh, 2005). The Derlins are homologues of Der1 p, which is essential for the degradation of some yeast ERAD clients (Knop, Finger, Braun, Hellmuth, & Wolf, 1996). Subsequently, photocrosslinking experiments conducted in yeast with a lumenal ERAD client identified interactions with Hrd1p (Hrd1 in mammals) at early stages of retrotranslocation (Carvalho, Stanley, & Rapoport, 2010), and revealed major interactions with Hrd3p, a homologue of mammalian SEL1 (Mueller, Lilley, & Ploegh, 2006), throughout the length of the polypeptide chain (Stanley, Carvalho, & Rapoport, 2011). Hrd1p is a multi-pass membrane protein with E3 ligase activity that oligomerizes, both features of which are required for its ERAD function (Carvalho et al., 2010), and over-expression of Hrd1p complements the genetic disruption of other essential ERAD components including Hrd3p (Carvalho et al., 2010). Reconstitution of proteoliposomes with Hrd1p was sufficient to retrotranslocate a membrane-anchored ERAD client when p97 was added (Baldridge & Rapoport, 2016). Hrd1p was found to undergo auto-ubiquitination, resulting in a conformational change in the protein that allowed initiation of retrotranslocation of the substrate (Figure 3). A cryo-electron microscopy (cryo-EM) structure of Hrd1p bound to Hrd3p has been solved revealing multiple interactions between the lumenal domains of these proteins, and also found that the transmembrane domains of Hrd1p assembled into a funnel-like structure (Schoebel et al., 2017). This is reminiscent of the nascent polypeptide-conducting channels of Sec61 and prokaryotic SecYp (Park & Rapoport, 2012), which serves to translocate nascent chains into the ER lumen. In combination these data are consistent with Hrd1 being the central part of the retrotranslocon.
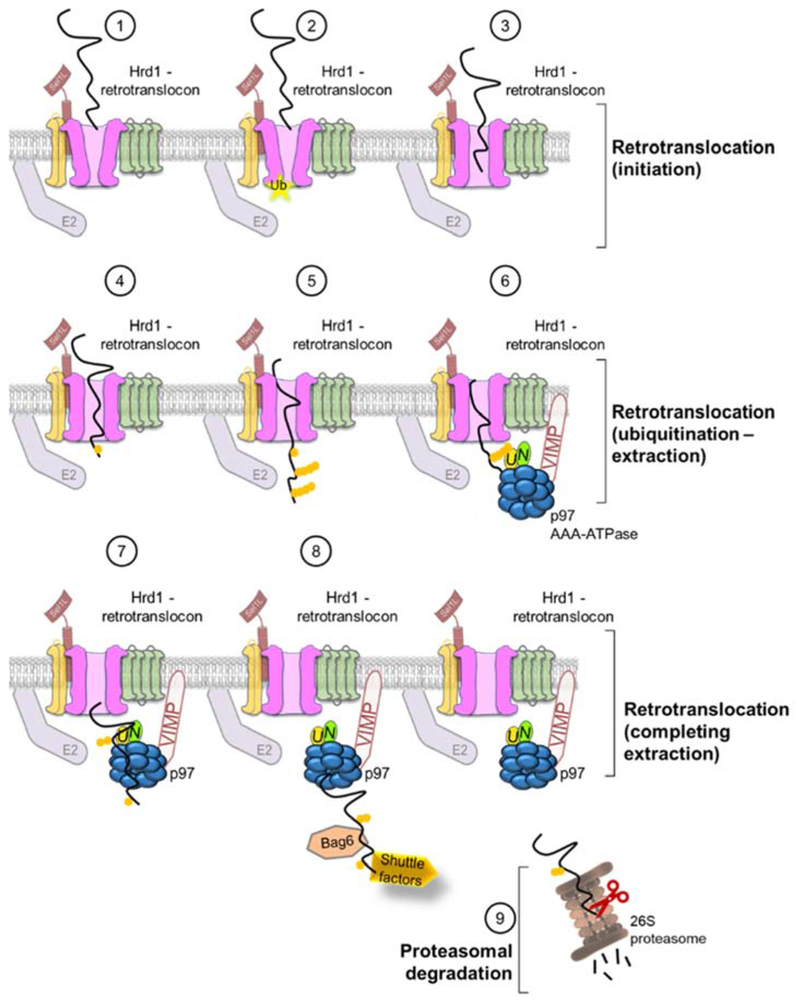
Figure 3: General model for retrotranslocation of ERAD clients through a Hrd1 retrotranslocon channel.
(1) An ERAD client is targeted to a closed retrotranslocon via its association with Sel1L/Hrd3p, which interacts directly with the multi-pass E3 ligase Hrd1. (2) Auto-ubiquitination of Hrd1 results in conformational changes that “open” the channel. (3) The polypeptide will be inserted into the Hrd1 channel and begin crossing the ER membrane via hydrophobic interactions with the TM regions of Hrd1 and sequential rounds of binding and release. (4) Once the substrate begins to emerge into the cytosol, it will be ubiquitinated by Hrd1 and associated E2 ubiquitin-conjugating proteins. (5) As the client continues to move through the channel, additional Ub chains are added and extended resulting in client poly-ubiquitination. (6) The AAA-ATPase, p97, is recruited to the ER membrane via its association with VIMP. Poly-ubiquitin chains on the ERAD client are recognized by the Ufd1/Npl4 (U/N) co-factors, which are located at the N-terminus of the p97. (7) Through conformational changes due to cycles of ATP binding and hydrolysis, p97 provides the necessary energy to extract ERAD clients from the ER membrane. (8) In many cases the ERAD client is then recognized by cytosolic chaperones and shuttling factors that deliver it to the proteasome. (9) Finally, the 26S proteasome receives ERAD substrates and again via cycles of ATP-hydrolysis translocates the polypeptide into its proteolytic core for degradation.
CRISPR-mediated disruption of Hrd1 in mammalian cells dramatically diminished the degradation of both lumenal and membrane clients (Zhang, Xu, Liu, & Ye, 2015), although direct evidence for it being the major channel component have not yet been obtained in mammalian systems. A large scale interaction map of the mammalian ERAD network was obtained by isolating 15 individual proteins that had previously been implicated in ERAD and performing MS/MS analysis on interacting proteins for each. A multilayer approach was employed that integraded proteomics, functional genomics, and gene expression data to delineate organization of the ERAD pathway in mammals (Christianson et al., 2012). This tour-de-force approach established Hrd1 as a central hub and linked lumenal component of the response to cytosolic, downstream effectors of ERAD, and identifed new components of ERAD. Together the data obtained provided pathways for the identification and targeting of clients, as well as organization of cytosolic elements of the pathway (reviewed in (Olzmann et al., 2013)).
5.1.2. ERAD substrate ubiquitination occurs on multiple types of amino acids
As the ERAD substrate emerges into the cytosol, ubiquitin (Ub) is added by ER-localized E3 ubiquitin ligases (Figure 3) of which nearly 40 have been identified in mammals thus far (Kaneko et al., 2016) and three in yeast (Hirsch, Gauss, Horn, Neuber, & Sommer, 2009). However, most model mammalian ERAD clients queried to- date rely on the Hrd1 E3 ligase. In addition to the canonical modification of lysines and the N-terminus of proteins, a variety of linkages used in constructing poly-Ub chains have been identified and mutational analyses have argued that Ub can be added to several other amino acids for both cytosolic and ERAD clients (Reviewed in (Kravtsova-Ivantsiv & Ciechanover, 2012; McDowell & Philpott, 2013)). For instance, mK3, a mouse γ-herpesvirus E3 modifies the cytosolic tail of the major histocompatibility chain to escape immune detection even when all lysines on the cytosolic tail are mutated (Wang et al., 2007). The attachment of Ub chains occurred as long as serines, threonines, or cysteines were present, and susceptibility of these chains to high pH or reducing agents was compatible to their attachment to these residues. Similar data were obtained for Hrd1-dependent ubiquitination of two ERAD clients, a non-secreted immunoglobulin light chain (Shimizu, Okuda-Shimizu, & Hendershot, 2010) and the T cell receptor α chain (Ishikura, Weissman, & Bonifacino, 2010) in mammalian cells, and on Doa10-dependent modification of a lysine-less version of the inner nuclear membrane protein Asi2 in yeast (Boban, Ljungdahl, & Foisner, 2015). The E2s responsible for serine/threonine modification in mammals are Ube2J2 (Wang et al., 2009) and Ube2J1 (Cuellar, Perales-Calvo, Muga, Valpuesta, & Moro, 2013), whereas Ubc6 and Ubc7 are necessary for non-lysine modification in yeast (Boban et al., 2015). In vitro ubiquitination studies followed by MS/MS analyses unequivocally demonstrated that Ubc6 attached Ub to hydroxylated amino acids, whereas Ubc7 was responsible for the elongation of these chains by more typical lysine 48 linkages (Weber et al., 2016). This is consistent with K48 linkages detected on poly-Ub chains attached to serine or threonine in mammals (Shimizu et al., 2010). The diversity in amino acids that can be modified by ERAD-specific E2/E3 pairs likely provides the flexibility to tag clients soon after they emerge into the cytosol and to ubiquitinate any of the ~6500 different proteins that enter the ER and can fail to fold.
5.1.3. Providing the force for retrotranslocation
In addition to playing a role in recognition by the proteasome, ubiquitination allows the ERAD client to be engaged by the p97/VCP (valosin-containing protein) AAA-ATPase complex (Cdc48 in yeast). p97 is an essential, highly conserved, homo-hexameric AAA+ ATPase with a vast array of cellular functions and is the only known energy source for the complete dislocation of ERAD clients (reviewed in (Hanzelmann & Schindelin, 2017; Ye, Tang, Zhang, & Xia, 2017; van den Boom & Meyer, 2018)). p97 consists of an N-terminal domain (N domain) and two ATPase domains (D1 and D2 domains), which form a hexameric, double-ring structure that creates a central pore with 12 ATP binding sites. The N domains provide binding sites for multiple, distinct cofactors that regulate p97’s diverse functions throughout the cell. Two of these, Ufd1 and Npl4, bind to the N domain of p97 and recognize ubiquitinated substrates (Figure 3). The N domain also associates with VIMP, an ER integral membrane protein important for ERAD (Lilley & Ploegh, 2005; Ye et al., 2005), thus orienting p97 to the ER membrane so it can capture ubiquitinated clients as they emerge from the ER (reviewed in (Jarosch, Geiss-Friedlander, Meusser, Walter, & Sommer, 2002)). A recent crystal structure of the N domain bound to a minimal, essential fragment of VIMP revealed that their interaction, and thus recruitment of p97 to the ER membrane, is modulated through nucleotide-dependent conformational changes in p97 (Tang, Zhang, Ye, & Xia, 2017), which could serve to enhance or diminish the levels of p97 associated with the ER during cellular stresses. This structure also explains why certain p97 mutations are pathogenic (Weihl, Dalal, Pestronk, & Hanson, 2006).
A number of studies have focused on how clients interact with p97 during the ATP-hydrolysis-dependent extraction process. Using normal mode analyses it was found that the largest movements in the p97 structure occur between the D1:D2 rings, supporting a model in which ERAD clients can be threaded between the two D rings, pass through the central cavity of the D2 ring, and exit from the distal side of D2 (Na & Song, 2016). Data giving rise to a second model for p97-dependent extraction have been obtained from several other studies. For instance, a cryo-EM structure of full-length human p97 revealed multiple conformational states in the complex, some of which indicated the central cavity of D1 could be large enough to accommodate an unfolded polypeptide chain (Banerjee et al., 2016). Additionally, a photo-crosslinking study demonstrated interactions between an ERAD client and several points in the central cavities of the D1 and D2 rings, and found that it exited from the D2 ring, arguing the client passed through both rings (Bodnar & Rapoport, 2017). In further support of a model in which clients passed through both D rings of p97, a cryo-EM structure of the D1:D2 core of VAT, an archaeon p97 homologue, was obtained in which an unfolded VAT subunit was present throughout the central cavity formed by both NBD domains (Ripstein, Huang, Augustyniak, Kay, & Rubinstein, 2017). Lastly, a recent structure of yeast p97/Ufd1/Npl4 complex bound to a trapped client captured images of a single internal Ub of the polyUb chain bound to Npl4 in an unfolded state and inserted through the D1 ring to the periphery of the D2 ring (Twomey et al., 2019). The Ub moieties on either side of the inserted Ub remained folded and bound to the co-factors on the outside of the p97 cavity. This indicates that the D1 channel is large enough to accommodate a peptide loop. Further studies will be required to determine if there is more than one route through the p97 complex during the extraction of proteins from the ER.
5.1.4. Delivery to the proteasome and degradation
The next step is the delivery of the ERAD substrate to the 26S proteasome for degradation. One study using a p97 mutant to isolate stalled retrotranslocating chains reported that immunoprecipitation of a proteasome subunit resulted in co-immunoprecipitation of Hrd1p, Hrd3p, lumenal ERAD components, and p97 only when yeast with mutant p97 was used (Nakatsukasa, Brodsky, & Kamura, 2013). Additionally, proteins like hHR2 (Rad23 in yeast) which have both ubiquitin- and proteasome-binding domains might serve as shuttling factors to transfer the substrate from p97 to the proteasome (reviewed in (Christianson & Ye, 2014). Cytosolic chaperones such as Hsc70 (Matsumura, David, & Skach, 2011) and Bag6 (Claessen, Sanyal, & Ploegh, 2014) can also interact with ERAD clients in the cytosol after p97-mediated extraction from the ER via hydrophobic regions exposed on these unfolded clients. Such interactions possibly serve to maintain substrate solubility and prevent aggregation in the aqueous cytosolic environment, but they also assist in substrate channeling to the degradation machinery. Once delivered to the 26S proteasome, degradation of the ERAD clients proceeds in the same way as described for all proteins (Figure 3) (reviewed in (Dikic, 2017; Bard et al., 2018)).
5.1.5. Regulation of ERAD components by the UPR
One aspect of the UPR that aids in both limiting the accumulation of unfolded proteins during ER stress and restoring homeostasis when the stress has been mitigated is the up-regulation of a number of ERAD components, which is the subject of several excellent reviews (Kincaid & Cooper, 2007; Tsai & Weissman, 2010; Hwang & Qi, 2018). The interplay between these two pathways that work to prevent ER stress and promote ER health was first revealed in a genome-wide analyses to identify transcriptional targets of the UPR in yeast (Travers et al., 2000), which found that in addition to increasing ER chaperones and folding enzymes, a several proteins that play a role in ERAD including Der1p, Hrd1p, Ubc7, and components of the ubiquitin proteasome system were transcriptionally up-regulated. This finding was extended to the XBP-1 transcription factor in mammalian cells, which targets ERAD components ERdj4 and EDEM (Lee, Iwakoshi, & Glimcher, 2003), and the ATF6 transcription factor that up-regulates Sel1L, Herp, Derlin-1-3, and Hrd1, either alone or through combined action with XBP-1 (Yamamoto et al., 2007).
5.1.6. ERAD in homeostasis and disease
Apart from the clearance of folding-defective proteins in the ER, ERAD plays an essential role in managing the levels of various proteins in a process that is highly regulated and occurs as a response to specific signals. One of more fully characterized examples involves the ERAD-regulated levels of 3-hydroxy-3-methylglutaryl-coenzymeA reductase (HMGR), an ER-localized, multi-pass membrane protein that catalyzes the rate-limiting step of sterol biosynthesis (reviewed in (Jo & DeBose-Boyd, 2010; Wangeline, Vashistha, & Hampton, 2017)). In a very simplified overview, sterol accumulation in the ER membrane alters transmembrane regions of HMGR and enhances its binding to membrane-embedded Insig-1/2 proteins, which in turn associate with ER membrane ubiquitin ligases (gp78 and Trc8 in humans and Hrd1p in yeast), thereby promoting ubiquitination of HMGR and proteasomal degradation (Song, Sever, & DeBose-Boyd, 2005). Conversely, lower membrane levels of sterol prevent these associations and targeting to the ubiquitin proteasome system, resulting in the stabilization of HMGR. Another intermediate in the pathway of sterol synthesis, squalene, is subjected to ubiquitination (by Teb4 in mammals and Doa10a in yeast) in response to increased levels of cholesterol (Foresti, Ruggiano, Hannibal-Bach, Ejsing, & Carvalho, 2013). In another example of feedback regulation, levels of apolipoprotein B, which plays a critical role in forming and trafficking liposomes, is regulated by ERAD in response to reduced lipid availability or synthesis (reviewed in (Brodsky & Fisher, 2008)). Defects in the degradation pathways of these proteins lead to a variety of diseases linked to lipid homeostasis and atherosclerosis. Similarly, ER stress and protein misfolding have been linked to a number of disorders of the liver, pancreas, and muscles (reviewed in (Lukas et al., 2019)), as well as to systemic metabolism (Ichhaporia et al., 2018; Reddy, Shruthi, Prabhakar, Sailaja, & Reddy, 2018; Bhattacharya et al., 2018).
Several studies have focused in understanding the effect of various ERAD components in health and disease. Global deficiencies in ERAD proteins are embryonic lethal, and thus cell type-specific deficiencies in ERAD components have been used to provide insights into physiological ERAD functions and endogenous ERAD substrates (reviewed in (Qi et al., 2017)). Adipocytes-specific ablation of Sel1L, a critical ERAD component for glycoproteins, resulted in mice on a Western-type diet with postprandial hypertriglyceridemia. This was found to be due to a resulting ER accumulation and aggregation of lipoprotein lipase (LPL) (Sha et al., 2014). Other studies reported that Sel1L knock-out mice developed pancreatic insufficiency (Sun et al., 2014), whereas epithelial Sel1L was shown to be required for intestinal homeostasis (Sun et al., 2016). Maybe less immediately obvious, loss of ERAD components can also affect proteins residing outside the ER. For instance, during ER stress associated with cirrhosis, Hrd1 targets NRF2, a transcription factor that that protect against oxidative stress. Hepatocyte-specific deletion of Hrd1, led to a dramatic increase in NRF2 levels and its targets in an experimental model for cirrhosis (Wu et al., 2014). This finding highlights the pathological importance of cross-talk between ER stress and ERAD pathways.
5.2. ER-phagy
In addition to the disposal of misfolded, unfolded, and incompletely assembled proteins by ERAD, there is growing awareness that more selective autophagic pathways (ER-phagy) can remove large protein aggregates that are not readily handled by ERAD and transport them to the lysosome for degradation (reviewed in (Grumati, Dikic, & Stolz, 2018; Loi, Fregno, Guerra, & Molinari, 2018; Wilkinson, 2019)). These processes can also extract damaged ER membranes, help shrink the ER after resolution of an ER stress response (RecovER-phagy), and maintain homeostasis of different types of ER membranes. While this area of research is fairly new, a number of the components and their functions are emerging. As the name implies, ER-phagy relies on many downstream components of traditional autophagic pathways including Atg/LC3 and a membrane source, for delivery of cargo to the lysosomes (Figure 1). It is also dependent on receptors, proteins capable of bending or distorting the ER membrane, and proteins capable of membrane scission. In the case of mammals, the most upstream components vary by the type of ER-phagy.
The first component to be identified was FAM134B/RETREG1, which localizes to the ends of ER sheets and is important for the turnover of this type of ER membrane (Khaminets et al., 2015). FAM132B inserts into the ER membrane post-translationally through a Reticulon Homology Domain (RHD), which does not fully enter the ER lumen and serves to bend the membrane. Some evidence suggests that atlastin 2, a member of the dynamin GTPase super-family, may provide the scission step in fragmentation of tsheet ER (Liang, Lingeman, Ahmed, & Corn, 2018). FAM134B possesses an LC3-interacting region (LIR) at its C-terminus that allows it to interact with LC3-decorated autophagosome membranes. It can play a role in the degradation of some lumenal protein aggregates, including the PiZ mutant of α1-anti-trypsin (Fregno et al., 2018) and procollagen (Forrester et al., 2019). FAM134B does not enter the ER and thus must have interactors that traverse the ER membrane to recognize lumenal clients. The latter study suggests it can interact with calnexin, a type I transmembrane protein that has chaperone activity. Reticulon 3 (RTN3) has numerous splice variants and the longest of these, RTN3L, serves to turnover or remodel ER tubules (Grumati et al., 2017). Similar to FAM134B, it inserts into the ER membrane, but doesn’t cross it, and provides the membrane bending function, but there are no data currently to implicate an atlastin in the scission process. RTN3 has 6 LIRs to promote interaction with LC3 on autophagosome membranes, although it is unclear if or how it interacts with lumenal clients to assist in their degradation.
UPR activation leads to up-regulation of CCPG1, an integral membrane, non-canonical autophagy cargo receptor (Smith & Wilkinson, 2018). It possesses a single LIR in the cytosolic portion of the protein and two FIP200 interacting regions that together allow it to cluster and interact with the autophagic machinery. Its lumenal region is poised to interact with misfolded or aggregated proteins, and studies in acinar pancreatic cells reveal a role for CCPG1 in proteostasis. As discussed above, one aspect of the UPR is to enlarge the ER to accommodate unfolded proteins. In mammalian cells, once the stress is resolved a process referred to as RecovER-phagy is enlisted to re-establish ER homeostasis. Data argue that the mammalian transmembrane protein Sec62, a component of the Sec61 channel through which nascent proteins enter the ER (Meyer et al., 2000), is repurposed to degrade excess ER membranes during recovery (Fumagalli et al., 2016). Data argue that Sec62 dissociates from the translocon and recruits the autophagic apparatus through the single LIR near its C-terminus. It is noteworthy that ER chaperones and folding enzymes that were up-regulated by the UPR are also taken up in the RecovER-phagy process, but ERAD components are excluded (Fumagalli et al., 2016), allowing these two mechanisms of ER degradation to act simultaneously.
6. Conclusions
Over the past decade, significant advances have been made toward understanding the checkpoints in ERQC, the interplay between components of the ERAD system, and the molecular mechanisms that execute retrotranslocation of clients for degradation by the UPS. In addition, there has been an increased appreciation for the role of autophagic processes in clearing the ER of aggregated proteins and novel elements of this pathway have been identified. This was made possible through a combination of sophisticated imaging and interaction technologies and increasingly high-resolution structures of multi-component complexes, but also through the clever use of more traditional genetic and biochemical approaches.
Highlights:
Updates on checkpoints in protein quality control in the ER
New insights on mechanisms of ERAD and ER-phagy
Connectivity between ERQC, UPR, ERAD, and ER-phagy systems
Acknowledgements
Funding was provided by the American Lebanese Syrian Associated Charities of St. Jude Children’s Research Hospital, as well as a grant to LMH from the National Institute of General Medicine Sciences (5R01GM054068-20).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Adams CJ, Kopp MC, Larburu N, Nowak PR, & Ali MMU (2019). Structure and Molecular Mechanism of ER Stress Signaling by the Unfolded Protein Response Signal Activator IRE1. Front Mol.Biosci, 6, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebi M, Bernasconi R, Clerc S, & Molinari M (2010). N-glycan structures: recognition and processing in the ER. Trends Biochem.Sci, 35, 74–82. [DOI] [PubMed] [Google Scholar]
- Amara JF, Lederkremer F, & Lodish HF (1989). Intracellular degradation of unassembled asialoglycoprotein receptor subunits: a pre-Golgi, nonlysosomal endoproteolytic cleavage. J.Cell Biol, 109, 3315–3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin-Wetzel N, Saunders RA, Kamphuis MJ, Rato C, Preissler S, Harding HP et al. (2017). A J-Protein Co-chaperone Recruits BiP to Monomerize IRE1 and Repress the Unfolded Protein Response. Cell, 171, 1625–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki K & Nagata K (2012). Protein folding and quality control in the ER. Cold Spring Harb.Perspect.Biol, 4, a015438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakunts A, Orsi A, Vitale M, Cattaneo A, Lari F, Tade L et al. (2017). Ratiometric sensing of BiP-client versus BiP levels by the unfolded protein response determines its signaling amplitude. Elife., 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldridge RD & Rapoport TA (2016). Autoubiquitination of the Hrd1 Ligase Triggers Protein Retrotranslocation in ERAD. Cell, 166, 394–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Bartesaghi A, Merk A, Rao P, Bulfer SL, Yan Y et al. (2016). 2.3 A resolution cryo-EM structure of human p97 and mechanism of allosteric inhibition. Science, 351, 871–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard JAM, Goodall EA, Greene ER, Jonsson E, Dong KC, & Martin A (2018). Structure and Function of the 26S Proteasome. Annu.Rev.Biochem, 87, 697–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe C & Helenius A (2016). Cargo Capture and Bulk Flow in the Early Secretory Pathway. Annu.Rev.Cell Dev.Biol, 32, 197–222. [DOI] [PubMed] [Google Scholar]
- Behnke J, Mann MJ, Scruggs FL, Feige MJ, & Hendershot LM (2016). Members of the Hsp70 Family Recognize Distinct Types of Sequences to Execute ER Quality Control. Mol.Cell, 63, 739–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berner N, Reutter KR, & Wolf DH (2018). Protein Quality Control of the Endoplasmic Reticulum and Ubiquitin-Proteasome-Triggered Degradation of Aberrant Proteins: Yeast Pioneers the Path. Annu.Rev.Biochem, 87, 751–782. [DOI] [PubMed] [Google Scholar]
- Bertolotti A, Zhang Y, Hendershot LM, Harding HP, & Ron D (2000). Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat.Cell Biol, 2, 326–332. [DOI] [PubMed] [Google Scholar]
- Bhattacharya A, Sun S, Wang H, Liu M, Long Q, Yin L et al. (2018). Hepatic Sel1L-Hrd1 ER-associated degradation (ERAD) manages FGF21 levels and systemic metabolism via CREBH. EMBO J., 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blond-Elguindi S, Cwirla SE, Dower WJ, Lipshutz RJ, Sprang SR, Sambrook JF et al. (1993). Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell, 75, 717–728. [DOI] [PubMed] [Google Scholar]
- Boban M, Ljungdahl PO, & Foisner R (2015). Atypical ubiquitylation in yeast targets lysine-less Asi2 for proteasomal degradation. J.Biol.Chem, 290, 2489–2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodnar NO & Rapoport TA (2017). Molecular Mechanism of Substrate Processing by the Cdc48 ATPase Complex. Cell, 169, 722–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakman I & Bulleid NJ (2011). Protein folding and modification in the mammalian endoplasmic reticulum. Annu.Rev.Biochem, 80, 71–99. [DOI] [PubMed] [Google Scholar]
- Braakman I & Hebert DN (2013). Protein folding in the endoplasmic reticulum. Cold Spring Harb.Perspect.Biol, 5, a013201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL & Fisher EA (2008). The many intersecting pathways underlying apolipoprotein B secretion and degradation. Trends Endocrinol.Metab, 19, 254–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck TM, Kolb AR, Boyd CR, Kleyman TR, & Brodsky JL (2010). The endoplasmic reticulum-associated degradation of the epithelial sodium channel requires a unique complement of molecular chaperones. Mol.Biol.Cell, 21, 1047–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calles-Garcia D, Yang M, Soya N, Melero R, Menade M, Ito Y et al. (2017). Single-particle electron microscopy structure of UDP-glucose:glycoprotein glucosyltransferase suggests a selectivity mechanism for misfolded proteins. J.Biol.Chem, 292, 11499–11507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrara M, Prischi F, Nowak PR, Kopp MC, & Ali MM (2015). Noncanonical binding of BiP ATPase domain to Ire1 and Perk is dissociated by unfolded protein CH1 to initiate ER stress signaling. Elife., 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P, Stanley AM, & Rapoport TA (2010). Retrotranslocation of a misfolded luminal ER protein by the ubiquitin-ligase Hrd1p. Cell, 143, 579–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y & Brandizzi F (2013). IRE1: ER stress sensor and cell fate executor. Trends Cell Biol, 23, 547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson JC, Olzmann JA, Shaler TA, Sowa ME, Bennett EJ, Richter CM et al. (2012). Defining human ERAD networks through an integrative mapping strategy. Nat.Cell Biol, 14, 93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson JC & Ye Y (2014). Cleaning up in the endoplasmic reticulum: ubiquitin in charge. Nat.Struct.Mol.Biol, 21, 325–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claessen JH, Sanyal S, & Ploegh HL (2014). The chaperone BAG6 captures dislocated glycoproteins in the cytosol. PLoS.ONE, 9, e90204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuellar J, Perales-Calvo J, Muga A, Valpuesta JM, & Moro F (2013). Structural insights into the chaperone activity of the 40-kDa heat shock protein DnaJ: binding and remodeling of a native substrate. J.Biol.Chem, 288, 15065–15074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejgaard K, Theberge JF, Heath-Engel H, Chevet E, Tremblay ML, & Thomas DY (2010). Organization of the Sec61 translocon, studied by high resolution native electrophoresis. J.Proteome.Res, 9, 1763–1771. [DOI] [PubMed] [Google Scholar]
- Dikic I (2017). Proteasomal and Autophagic Degradation Systems. Annu.Rev.Biochem, 86, 193–224. [DOI] [PubMed] [Google Scholar]
- Dong M, Bridges JP, Apsley K, Xu Y, & Weaver TE (2008). ERdj4 and ERdj5 are required for endoplasmic reticulum-associated protein degradation of misfolded surfactant protein C. Mol.Biol.Cell, 19, 2620–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner AJ, Wasley LC, & Kaufman RJ (1992). Overexpression of GRP78 mitigates stress induction of glucose regulated proteins and blocks secretion of selective proteins in Chinese hamster ovary cells. EMBO.J, 11, 1563–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foresti O, Ruggiano A, Hannibal-Bach HK, Ejsing CS, & Carvalho P (2013). Sterol homeostasis requires regulated degradation of squalene monooxygenase by the ubiquitin ligase Doa10/Teb4. Elife., 2, e00953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester A, De LC, Grumati P, Fasana E, Piemontese M, Staiano L et al. (2019). A selective ER-phagy exerts procollagen quality control via a Calnexin-FAM134B complex. EMBO J, 38, e99847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fregno I, Fasana E, Bergmann TJ, Raimondi A, Loi M, Solda T et al. (2018). ER-to-lysosome-associated degradation of proteasome-resistant ATZ polymers occurs via receptor-mediated vesicular transport. EMBO J., 37, e99259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz JM, Dong M, Apsley KS, Martin EP, Na CL, Sitaraman S et al. (2014). Deficiency of the BiP cochaperone ERdj4 causes constitutive endoplasmic reticulum stress and metabolic defects. Mol.Biol.Cell, 25, 431–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli F, Noack J, Bergmann TJ, Cebollero E, Pisoni GB, Fasana E et al. (2016). Translocon component Sec62 acts in endoplasmic reticulum turnover during stress recovery. Nat.Cell Biol, 18, 1173–1184. [DOI] [PubMed] [Google Scholar]
- Gardner BM & Walter P (2011). Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response. Science, 333, 1891–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Teuber V, Albert-Gasco H, Auyeung VC, Papa FR, Mallucci GR, & Hetz C (2019). Small Molecules to Improve ER Proteostasis in Disease. Trends Pharmacol.Sci. [DOI] [PubMed] [Google Scholar]
- Grumati P, Dikic I, & Stolz A (2018). ER-phagy at a glance. J.Cell Sci, 131. [DOI] [PubMed] [Google Scholar]
- Grumati P, Morozzi G, Holper S, Mari M, Harwardt MI, Yan R et al. (2017). Full length RTN3 regulates turnover of tubular endoplasmic reticulum via selective autophagy. Elife., 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerriero CJ & Brodsky JL (2012). The delicate balance between secreted protein folding and endoplasmic reticulum-associated degradation in human physiology. Physiol Rev., 92, 537–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F & Snapp EL (2013). ERdj3 regulates BiP occupancy in living cells. J Cell Sci., 126, 1429–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton RY & Sommer T (2012). Finding the will and the way of ERAD substrate retrotranslocation. Curr.Opin.Cell Biol, 24, 460–466. [DOI] [PubMed] [Google Scholar]
- Hanzelmann P & Schindelin H (2017). The Interplay of Cofactor Interactions and Post-translational Modifications in the Regulation of the AAA+ ATPase p97. Front Mol.Biosci, 4, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K, Ravindran MS, & Tsai B (2015). A bacterial toxin and a nonenveloped virus hijack ER-to-cytosol membrane translocation pathways to cause disease. Crit Rev.Biochem.Mol.Biol, 50, 477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert DN, Foellmer B, & Helenius A (1995). Glucose trimming and reglucosylation determine glycoprotein association with calnexin in the endoplasmic reticulum. Cell, 81, 425–433. [DOI] [PubMed] [Google Scholar]
- Hendershot L, Bole D, Kohler G, & Kearney JF (1987). Assembly and secretion of heavy chains that do not associate posttranslationally with immunoglobulin heavy chain-binding protein. J.Cell.Biol, 104, 761–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch C, Gauss R, Horn SC, Neuber O, & Sommer T (2009). The ubiquitylation machinery of the endoplasmic reticulum. Nature, 458, 453–460. [DOI] [PubMed] [Google Scholar]
- Hollien J (2013). Evolution of the unfolded protein response. Biochim.Biophys.Acta, 1833, 2458–2463. [DOI] [PubMed] [Google Scholar]
- Hosoda A, Kimata Y, Tsuru A, & Kohno K (2003). JPDI, a novel endoplasmic reticulum-resident protein containing both a BiP-interacting J-domain and thioredoxin-like motifs. J.Biol.Chem, 278, 2669–2676. [DOI] [PubMed] [Google Scholar]
- Hwang J & Qi L (2018). Quality Control in the Endoplasmic Reticulum: Crosstalk between ERAD and UPR pathways. Trends Biochem.Sci, 43, 593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichhaporia VP, Kim J, Kavdia K, Vogel P, Horner L, Frase S et al. (2018). SIL1, the endoplasmic-reticulum-localized BiP co-chaperone, plays a crucial role in maintaining skeletal muscle proteostasis and physiology. Dis.Model.Mech, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Dosey A, Herbstman JF, Ravindran MS, Skiniotis G, & Tsai B (2015). ERdj5 Reductase Cooperates with Protein Disulfide Isomerase To Promote Simian Virus 40 Endoplasmic Reticulum Membrane Translocation. J.Virol, 89, 8897–8908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikura S, Weissman AM, & Bonifacino JS (2010). Serine residues in the cytosolic tail of the T-cell antigen receptor alpha-chain mediate ubiquitination and endoplasmic reticulum-associated degradation of the unassembled protein. J Biol.Chem, 285, 23916–23924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi M, Kuruma R, Okamoto R, Seko A, Ito Y, & Kajihara Y (2017). Substrate Recognition of Glycoprotein Folding Sensor UGGT Analyzed by Site-Specifically (15)N-Labeled Glycopeptide and Small Glycopeptide Library Prepared by Parallel Native Chemical Ligation. J.Am.Chem.Soc, 139, 11421–11426. [DOI] [PubMed] [Google Scholar]
- Jarosch E, Geiss-Friedlander R, Meusser B, Walter J, & Sommer T (2002). Protein dislocation from the endoplasmic reticulum--pulling out the suspect. Traffic., 3, 530–536. [DOI] [PubMed] [Google Scholar]
- Jo Y & DeBose-Boyd RA (2010). Control of cholesterol synthesis through regulated ER-associated degradation of HMG CoA reductase. Crit Rev.Biochem.Mol.Biol, 45, 185–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga HH & Craig EA (2010). The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat.Rev.Mol.Cell Biol, 11, 579–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M, Iwase I, Yamasaki Y, Takai T, Wu Y, Kanemoto S et al. (2016). Genome-wide identification and gene expression profiling of ubiquitin ligases for endoplasmic reticulum protein degradation. Sci.Rep, 6, 30955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaminets A, Heinrich T, Mari M, Grumati P, Huebner AK, Akutsu M et al. (2015). Regulation of endoplasmic reticulum turnover by selective autophagy. Nature, 522, 354–358. [DOI] [PubMed] [Google Scholar]
- Khodayari N, Marek G, Lu Y, Krotova K, Wang RL, & Brantly M (2017). Erdj3 Has an Essential Role for Z Variant Alpha-1-Antitrypsin Degradation. J.Cell Biochem, 118, 3090–3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata Y, Ishiwata-Kimata Y, Ito T, Hirata A, Suzuki T, Oikawa D et al. (2007). Two regulatory steps of ER-stress sensor Ire1 involving its cluster formation and interaction with unfolded proteins. J.Cell Biol, 179, 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata Y & Kohno K (2011). Endoplasmic reticulum stress-sensing mechanisms in yeast and mammalian cells. Curr.Opin.Cell Biol, 23, 135–142. [DOI] [PubMed] [Google Scholar]
- Kimata Y, Oikawa D, Shimizu Y, Ishiwata-Kimata Y, & Kohno K (2004). A role for BiP as an adjustor for the endoplasmic reticulum stress-sensing protein Ire1. J.Cell Biol, 167, 445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kincaid MM & Cooper AA (2007). ERADicate ER stress or die trying. Antioxid.Redox.Signal, 9, 2373–2387. [DOI] [PubMed] [Google Scholar]
- Knop M, Finger A, Braun T, Hellmuth K, & Wolf DH (1996). Der1, a novel protein specifically required for endoplasmic reticulum degradation in yeast. EMBO J., 15, 753–763. [PMC free article] [PubMed] [Google Scholar]
- Kozutsumi Y, Segal M, Normington K, Gething MJ, & Sambrook J (1988). The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature, 332, 462–464. [DOI] [PubMed] [Google Scholar]
- Kravtsova-Ivantsiv Y & Ciechanover A (2012). Non-canonical ubiquitin-based signals for proteasomal degradation. J.Cell Sci, 125, 539–548. [DOI] [PubMed] [Google Scholar]
- Lai CW, Otero JH, Hendershot LM, & Snapp E (2012). ERdj4 protein is a soluble endoplasmic reticulum (ER) DnaJ family protein that interacts with ER-associated degradation machinery. J Biol.Chem, 287, 7969–7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le A, Graham KS, & Sifers RN (1990). Intracellular degradation of the transport-impaired human PiZ alpha 1-antitrypsin variant. Biochemical mapping of the degradative event among compartments of the secretory pathway. J.Biol.Chem, 265, 14001–14007. [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, & Glimcher LH (2003). XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell Biol, 23, 7448–7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang JR, Lingeman E, Ahmed S, & Corn JE (2018). Atlastins remodel the endoplasmic reticulum for selective autophagy. J.Cell Biol, 217, 3354–3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley BN & Ploegh HL (2004). A membrane protein required for dislocation of misfolded proteins from the ER. Nature, 429, 834–840. [DOI] [PubMed] [Google Scholar]
- Lilley BN & Ploegh HL (2005). Multiprotein complexes that link dislocation, ubiquitination, and extraction of misfolded proteins from the endoplasmic reticulum membrane. Proc.Natl.Acad.Sci.U.S.A, 102, 14296–14301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott Schwartz J, Bonifacino JS, Yuan LC, & Klausner RD (1988). Degradation from the endoplasmic reticulum: disposing of newly synthesized proteins. Cell, 54, 209–220. [DOI] [PubMed] [Google Scholar]
- Loi M, Fregno I, Guerra C, & Molinari M (2018). Eat it right: ER-phagy and recovER-phagy. Biochem.Soc.Trans, 46, 699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas J, Pospech J, Oppermann C, Hund C, Iwanov K, Pantoom S et al. (2019). Role of endoplasmic reticulum stress and protein misfolding in disorders of the liver and pancreas. Adv.Med.Sci, 64, 315–323. [DOI] [PubMed] [Google Scholar]
- Maattanen P, Gehring K, Bergeron JJ, & Thomas DY (2010). Protein quality control in the ER: the recognition of misfolded proteins. Semin.Cell Dev.Biol, 21, 500–511. [DOI] [PubMed] [Google Scholar]
- Matsumura Y, David LL, & Skach WR (2011). Role of Hsc70 binding cycle in CFTR folding and endoplasmic reticulum-associated degradation. Mol.Biol.Cell, 22, 2797–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell GS & Philpott A (2013). Non-canonical ubiquitylation: mechanisms and consequences. Int.J.Biochem.Cell Biol, 45, 1833–1842. [DOI] [PubMed] [Google Scholar]
- Meunier L, Usherwood YK, Chung KT, & Hendershot LM (2002). A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol.Biol.Cell, 13, 4456–4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer HA, Grau H, Kraft R, Kostka S, Prehn S, Kalies KU et al. (2000). Mammalian Sec61 is associated with Sec62 and Sec63. J.Biol.Chem, 275, 14550–14557. [DOI] [PubMed] [Google Scholar]
- Mitra S & Ryoo HD (2019). The unfolded protein response in metazoan development. J.Cell Sci, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari M & Hebert DN (2015). Glycoprotein maturation and quality control. Semin.Cell Dev.Biol, 41, 70. [DOI] [PubMed] [Google Scholar]
- Molinari M & Helenius A (2000). Chaperone selection during glycoprotein translocation into the endoplasmic reticulum. Science, 288, 331–333. [DOI] [PubMed] [Google Scholar]
- Mueller B, Lilley BN, & Ploegh HL (2006). SEL1L, the homologue of yeast Hrd3p, is involved in protein dislocation from the mammalian ER. J.Cell Biol, 175, 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S & Pelham HR (1987). A C-terminal signal prevents secretion of luminal ER proteins. Cell, 48, 899–907. [DOI] [PubMed] [Google Scholar]
- Na H & Song G (2016). Predicting the functional motions of p97 using symmetric normal modes. Proteins, 84, 1823–1835. [DOI] [PubMed] [Google Scholar]
- Nakatsukasa K, Brodsky JL, & Kamura T (2013). A stalled retrotranslocation complex reveals physical linkage between substrate recognition and proteasomal degradation during ER-associated degradation. Mol.Biol.Cell, 24, 1765–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needham PG & Brodsky JL (2013). How early studies on secreted and membrane protein quality control gave rise to the ER associated degradation (ERAD) pathway: the early history of ERAD. Biochim.Biophys.Acta, 1833, 2447–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needham PG, Guerriero CJ, & Brodsky JL (2019). Chaperoning Endoplasmic Reticulum-Associated Degradation (ERAD) and Protein Conformational Diseases. Cold Spring Harb.Perspect.Biol, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa D, Kitamura A, Kinjo M, & Iwawaki T (2012). Direct association of unfolded proteins with mammalian ER stress sensor, IRE1beta. PLoS.ONE, 7, e51290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Kimata Y, Higashio H, Tsuru A, & Kohno K (2000). Dissociation of Kar2p/BiP from an ER sensory molecule, Ire1p, triggers the unfolded protein response in yeast. Biochem.Biophys.Res.Commun, 279, 445–450. [DOI] [PubMed] [Google Scholar]
- Olzmann JA, Kopito RR, & Christianson JC (2013). The mammalian endoplasmic reticulum-associated degradation system. Cold Spring Harb.Perspect.Biol, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E & Rapoport TA (2012). Mechanisms of Sec61/SecY-mediated protein translocation across membranes. Annu.Rev.Biophys, 41, 21–40. [DOI] [PubMed] [Google Scholar]
- Parodi AJ (2000). Protein glucosylation and its role in protein folding. Annu.Rev.Biochem, 69, 69–93. [DOI] [PubMed] [Google Scholar]
- Petrova K, Oyadomari S, Hendershot L, & Ron D (2008). Regulated association of misfolded endoplamic reticulum lumenal proteins with P58/DNAJc3. EMBO J., 27, 2862–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pobre KFR, Poet GJ, & Hendershot LM (2019). The endoplasmic reticulum (ER) chaperone BiP is a master regulator of ER functions: Getting by with a little help from ERdj friends. J.Biol.Chem, 294, 2098–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L, Tsai B, & Arvan P (2017). New Insights into the Physiological Role of Endoplasmic Reticulum-Associated Degradation. Trends Cell Biol., 27, 430–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy SS, Shruthi K, Prabhakar YK, Sailaja G, & Reddy GB (2018). Implication of altered ubiquitin-proteasome system and ER stress in the muscle atrophy of diabetic rats. Arch.Biochem.Biophys, 639, 16–25. [DOI] [PubMed] [Google Scholar]
- Ripstein ZA, Huang R, Augustyniak R, Kay LE, & Rubinstein JL (2017). Structure of a AAA+ unfoldase in the process of unfolding substrate. Elife., 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter C & Helenius A (2000). Recognition of local glycoprotein misfolding by the ER folding sensor UDP-glucose:glycoprotein glucosyltransferase. Nat.Struct.Biol, 7, 278–280. [DOI] [PubMed] [Google Scholar]
- Romisch K (2017). A Case for Sec61 Channel Involvement in ERAD. Trends Biochem.Sci, 42, 171–179. [DOI] [PubMed] [Google Scholar]
- Ron D & Walter P (2007). Signal integration in the endoplasmic reticulum unfolded protein response. Nat.Rev.Mol.Cell Biol, 8, 519–529. [DOI] [PubMed] [Google Scholar]
- Rutkowski DT, Kang SW, Goodman AG, Garrison JL, Taunton J, Katze MG et al. (2007). The role of p58IPK in protecting the stressed endoplasmic reticulum. Mol. Biol. Cell, 18, 3681–3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoebel S, Mi W, Stein A, Ovchinnikov S, Pavlovicz R, DiMaio F et al. (2017). Cryo-EM structure of the protein-conducting ERAD channel Hrd1 in complex with Hrd3. Nature, 548, 352–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha H, Sun S, Francisco AB, Ehrhardt N, Xue Z, Liu L et al. (2014). The ER-associated degradation adaptor protein Sel1L regulates LPL secretion and lipid metabolism. Cell Metab, 20, 458–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenkman M & Lederkremer GZ (2019). Compartmentalization and Selective Tagging for Disposal of Misfolded Glycoproteins. Trends Biochem.Sci. [DOI] [PubMed] [Google Scholar]
- Shenkman M, Ron E, Yehuda R, Benyair R, Khalaila I, & Lederkremer GZ (2018). Mannosidase activity of EDEM1 and EDEM2 depends on an unfolded state of their glycoprotein substrates. Commun.Biol, 1, 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y, Okuda-Shimizu Y, & Hendershot LM (2010). Ubiquitylation of an ERAD substrate occurs on multiple types of amino acids. Mol.Cell, 40, 917–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitia R, Neuberger MS, & Milstein C (1987). Regulation of membrane IgM expression in secretory B cells: translational and post-translational events. EMBO.J, 6, 3969–3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slominska-Wojewodzka M & Sandvig K (2015). The Role of Lectin-Carbohydrate Interactions in the Regulation of ER-Associated Protein Degradation. Molecules., 20, 9816–9846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MD & Wilkinson S (2018). CCPG1, an unconventional cargo receptor for ER-phagy, maintains pancreatic acinar cell health. Mol.Cell Oncol, 5, e1441631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolowska I, Pilka ES, Sandvig K, Wegrzyn G, & Slominska-Wojewodzka M (2015). Hydrophobicity of protein determinants influences the recognition of substrates by EDEM1 and EDEM2 in human cells. BMC.Cell Biol, 16, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song BL, Sever N, & DeBose-Boyd RA (2005). Gp78, a membrane-anchored ubiquitin ligase, associates with Insig-1 and couples sterol-regulated ubiquitination to degradation of HMG CoA reductase. Mol.Cell, 19, 829–840. [DOI] [PubMed] [Google Scholar]
- Stanley AM, Carvalho P, & Rapoport T (2011). Recognition of an ERAD-L substrate analyzed by site-specific in vivo photocrosslinking. FEBS Lett., 585, 1281–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Lourie R, Cohen SB, Ji Y, Goodrich JK, Poole AC et al. (2016). Epithelial Sel1L is required for the maintenance of intestinal homeostasis. Mol.Biol.Cell, 27, 483–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Shi G, Han X, Francisco AB, Ji Y, Mendonca N et al. (2014). Sel1L is indispensable for mammalian endoplasmic reticulum-associated degradation, endoplasmic reticulum homeostasis, and survival. Proc.Natl.Acad.Sci.U.S.A, 111, E582–E591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan YL, Genereux JC, Pankow S, Aerts JM, Yates JR III, & Kelly JW (2014). ERdj3 is an endoplasmic reticulum degradation factor for mutant glucocerebrosidase variants linked to Gaucher’s disease. Chem.Biol, 21, 967–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WK, Zhang T, Ye Y, & Xia D (2017). Structural basis for nucleotide-modulated p97 association with the ER membrane. Cell Discov., 3, 17045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatu U & Helenius A (1997). Interactions between newly synthesized glycoproteins, calnexin and a network of resident chaperones in the endoplasmic reticulum. J.Cell Biol, 136, 555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, & Walter P (2000). Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell, 101, 249–258. [DOI] [PubMed] [Google Scholar]
- Tsai YC & Weissman AM (2010). The Unfolded Protein Response, Degradation from Endoplasmic Reticulum and Cancer. Genes Cancer, 1, 764–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twomey EC, Ji Z, Wales TE, Bodnar NO, Ficarro SB, Marto JA et al. (2019). Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding. Science, 365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushioda R, Hoseki J, Araki K, Jansen G, Thomas DY, & Nagata K (2008). ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science, 321, 569–572. [DOI] [PubMed] [Google Scholar]
- van den Boom J & Meyer H (2018). VCP/p97-Mediated Unfolding as a Principle in Protein Homeostasis and Signaling. Mol.Cell, 69, 182–194. [DOI] [PubMed] [Google Scholar]
- Vitale M, Bakunts A, Orsi A, Lari F, Tade L, Danieli A et al. (2019). Inadequate BiP availability defines endoplasmic reticulum stress. Elife., 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P & Ron D (2011). The unfolded protein response: from stress pathway to homeostatic regulation. Science, 334, 1081–1086. [DOI] [PubMed] [Google Scholar]
- Wang S & Kaufman RJ (2012). The impact of the unfolded protein response on human disease. J.Cell Biol, 197, 857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Herr RA, Chua WJ, Lybarger L, Wiertz EJ, & Hansen TH (2007). Ubiquitination of serine, threonine, or lysine residues on the cytoplasmic tail can induce ERAD of MHC-I by viral E3 ligase mK3. J.Cell Biol, 177, 613–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Herr RA, Rabelink M, Hoeben RC, Wiertz EJ, & Hansen TH (2009). Ube2j2 ubiquitinates hydroxylated amino acids on ER-associated degradation substrates. J.Cell Biol, 187, 655–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wangeline MA, Vashistha N, & Hampton RY (2017). Proteostatic Tactics in the Strategy of Sterol Regulation. Annu.Rev.CellDev.Biol, 33, 467–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward CL, Omura S, & Kopito RR (1995). Degradation of CFTR by the ubiquitin-proteasome pathway. Cell, 83, 121–127. [DOI] [PubMed] [Google Scholar]
- Weber A, Cohen I, Popp O, Dittmar G, Reiss Y, Sommer T et al. (2016). Sequential Poly-ubiquitylation by Specialized Conjugating Enzymes Expands the Versatility of a Quality Control Ubiquitin Ligase. Mol.Cell, 63, 827–839. [DOI] [PubMed] [Google Scholar]
- Weihl CC, Dalal S, Pestronk A, & Hanson PI (2006). Inclusion body myopathy-associated mutations in p97/VCP impair endoplasmic reticulum-associated degradation. Hum.Mol.Genet, 15, 189–199. [DOI] [PubMed] [Google Scholar]
- Werner ED, Brodsky JL, & McCracken AA (1996). Proteasome-dependent endoplasmic reticulum-associated protein degradation: an unconventional route to a familiar fate. Proc.Natl.Acad.Sci.U.S.A, 93, 13797–13801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiertz EJ, Jones TR, Sun L, Bogyo M, Geuze HJ, & Ploegh HL (1996). The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell, 84, 769–779. [DOI] [PubMed] [Google Scholar]
- Wilkinson S (2019). ER-phagy: shaping up and destressing the endoplasmic reticulum. FEBS J., 286, 2645–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JM, Inoue T, Banks L, & Tsai B (2013). The ERdj5-Sel1L complex facilitates cholera toxin retrotranslocation. Mol.Biol.Cell, 24, 785–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Zhao F, Gao B, Tan C, Yagishita N, Nakajima T et al. (2014). Hrd1 suppresses Nrf2-mediated cellular protection during liver cirrhosis. Genes Dev., 28, 708–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X & Rapoport TA (2018). Mechanistic insights into ER-associated protein degradation. Curr.Opin.Cell Biol, 53, 22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Sato T, Matsui T, Sato M, Okada T, Yoshida H et al. (2007). Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev.Cell, 13, 365–376. [DOI] [PubMed] [Google Scholar]
- Ye Y, Shibata Y, Kikkert M, van VS, Wiertz E, & Rapoport TA (2005). Recruitment of the p97 ATPase and ubiquitin ligases to the site of retrotranslocation at the endoplasmic reticulum membrane. Proc.Natl.Acad.Sci.U.S.A, 102, 14132–14138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Tang WK, Zhang T, & Xia D (2017). A Mighty “Protein Extractor” of the Cell: Structure and Function of the p97/CDC48 ATPase. Front Mol.Biosci, 4, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Xu Y, Liu Y, & Ye Y (2015). gp78 functions downstream of Hrd1 to promote degradation of misfolded proteins of the endoplasmic reticulum. Mol.Biol.Cell, 26, 4438–4450. [DOI] [PMC free article] [PubMed] [Google Scholar]