Abstract
Tryptamine is an alkaloid compound with demonstrated bioactivities and is also a precursor molecule to many important hormones and neurotransmitters. The high efficiency biosynthesis of tryptamine from inexpensive and renewable carbon substrates is of great research and application significance. In the present study, a tryptamine biosynthesis pathway was established in a metabolically engineered E. coli-E. coli co-culture. The upstream and downstream strains of the co-culture were dedicated to tryptophan provision and conversion to tryptamine, respectively. The constructed co-culture was cultivated using either glucose or glycerol as carbon source for de novo production of tryptamine. The manipulation of the co-culture strains’ inoculation ratio was adapted to balance the biosynthetic strengths of the pathway modules for bioproduction optimization. Moreover, a biosensor-assisted cell selection strategy was adapted to improve the pathway intermediate tryptophan provision by the upstream strain, which further enhanced the tryptamine biosynthesis. The resulting biosensor-assisted modular co-culture produced 194 mg/L tryptamine with a yield of 0.02 g/g glucose using shake flask cultivation. The findings of this work demonstrate that the biosensor-assisted modular co-culture engineering offers a new perspective for conducting microbial biosynthesis.
Keywords: Modular co-culture engineering, Biosensor, E. coli, Tryptamine, Pathway modularization
Highlights
-
•
De novo biosynthesis of tryptamine was achieved using E. coli-E. coli co-cultures.
-
•
Biosensor-assisted cell selection systems were integrated into the co-cultures.
-
•
Tryptamine production was significantly improved compared to the mono-cultures.
-
•
The engineered co-culture showed dynamic growth and biosynthesis behaviors.
1. Introduction
Tryptamine is a biogenic amine that belongs to the family of monoamine alkaloid. Tryptamine is not only considered as a brain neurotransmitter by itself, but a precursor for making many important bioactive molecules such as serotonin, melatonin, and substituted tryptamines (Kang et al., 2007; Mahmood et al., 2010; Mousseau, 1993). Therefore, there has been consistent interest for producing this valuable molecule with low cost and high efficiency. Tryptamine can be biosynthesized from amino acid tryptophan by a tryptophan decarboxylase. As such, the integration of tryptophan decarboxylation into the tryptophan biosynthesis pathway enables the de novo production of tryptamine, as shown in Fig. 1. It has been reported that the expression of the tryptophan decarboxylase in transgenic tobacco resulted in accumulation of tryptamine (Songstad et al., 1990). For microbial biosynthesis, previous studies have established a heterologous pathway in E. coli for biosynthesis of tryptamine and serotonin from glucose (Mora-Villalobos and Zeng, 2018; Park et al., 2011). Lee et al. further extended the pathway for producing tryptamine derivatives such as N-hydroxycinnamoyl tryptamine in E. coli (Lee et al., 2017). However, so far, no dedicated efforts have been made for de novo tryptamine biosynthesis and its optimization using simple carbon substrate such as glucose and glycerol.
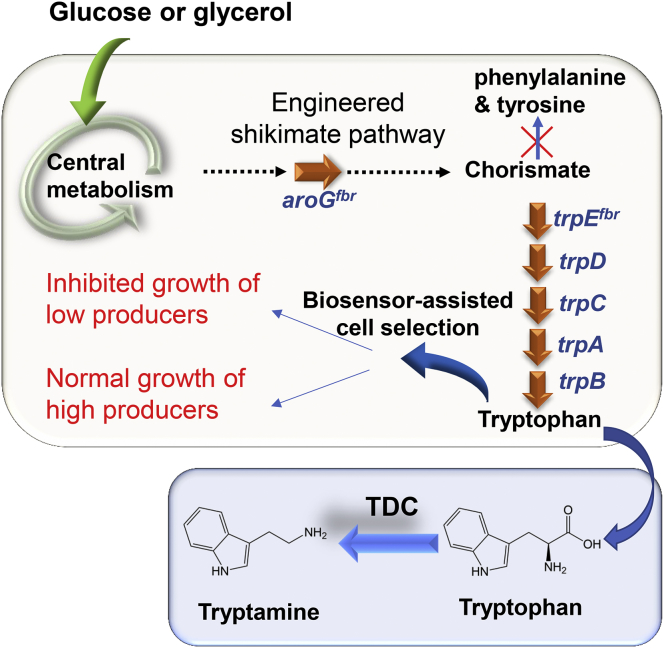
Fig. 1.
The co-culture design for biosynthesis of tryptamine. The upstream strain is responsible for producing tryptophan from simple substrate glucose or glycerol. The downstream strain is dedicated to converting tryptophan into tryptamine. The over-expressed pathway genes are indicated by solid arrows. Tryptophan decarboxylase (TDC) was expressed in the downstream strain. The biosensor-based selection systems were introduced into the upstream strain for enhancement of tryptophan provision.
It is noteworthy that previous studies on microbial biosynthesis of tryptamine and its derivatives employed the conventional mono-culture engineering strategy, i.e., utilization of the culture containing one single metabolically engineered microbial strain. In recent years, modular co-culture engineering has emerged as an innovative and effective approach for biosynthesis of a wealth of value-added biochemicals (Chen et al., 2019; Jawed et al., 2019; Jones and Wang, 2018; Roell et al., 2019; Zhang and Wang, 2016). In particular, the pathway modularization and balancing in the context of co-cultures add a new dimension for microbial biosynthesis. A variety of same-species co-cultures (e.g. E. coli-E. coli co-cultures) and multiple species co-cultures (e.g. E. coli-S. cerevisiae co-cultures), have been successfully constructed to address the challenges for biosynthesis performance enhancement. Meanwhile, new engineering strategies, such as using poly-cultures consisting of three or more microbial strains, symbiotic growth of the co-culture members, and co-culture mathematical modelling, been developed to extend the utility of the modular co-culture engineering (Bayer et al., 2009; Jones et al., 2017; Li et al., 2019; Niehaus et al., 2019). The accomplishments of the previous efforts offer strong incentive for using microbial co-cultures for achieving efficient tryptamine bioproduction. In the present study, we metabolically engineered and optimized an E. coli-E. coli co-culture for producing tryptamine from renewable carbon substrates glucose and glycerol. In this co-culture system, the upstream strain was constructed for producing the intermediate tryptophan, and the downstream strain was engineered for converting tryptophan to the final product tryptamine using tryptophan decarboxylase TDC (Fig. 1). Accordingly, the pathway balancing could be pursued by manipulating the relative biosynthetic strengths for tryptophan provision and consumption, which was conducted through adjusting the ratio between the upstream and downstream strains’ subpopulations.
In order to address the issue of insufficient pathway intermediate provision, a biosensor-assisted cell selection approach was adapted for the constructed co-cultures. To this end, there have been pioneering efforts using biosensors in metabolic engineering and synthetic biology applications. For example, genetic and metabolic heterogeneity can lead to variations of biosynthesis performance of the cells within a population. Several biosensors-assisted systems have been developed and utilized to address this issue to serve biosynthesis enhancement. (Liu et al., 2017; Rugbjerg et al., 2018; Snoek et al., 2018; Wang et al., 2019; Xiao et al., 2016). Specifically, a metabolite-responsive biosensor and a growth-regulation gene are integrated together, which provides regulation rules that promote the growth of cells producing high amounts of the target metabolite and inhibit the growth of the cells producing low amount of the target metabolite (Lv et al., 2019). In a recent study by our group, such a biosensor-assisted cell selection mechanism was integrated into microbial co-cultures to enhance the bioproduction of a commodity chemical phenol through improving the provision of pathway intermediates (Guo et al., 2019).
Here, we extended the use of biosensor-assisted cell selection for facilitating the biosynthesis of tryptamine in the context of engineered E. coli-E. coli co-cultures. Specifically, two independent biosensor-assisted cell selection systems were recruited for enhancing the tryptophan bioproduction in the upstream strain of the constructed co-culture system. The biosensor of the first system employed the leader peptide gene tnaC of the tnaCAB operon which senses tryptophan abundance and control gene expression through interaction with transcription termination factor Rho. The tetracycline resistance gene tetA was used as the growth-regulating component of this cell selection system. For such a tnaC-tetA system, high tryptophan concentration switched on the tetA expression and thus enabled the growth the cell in the presence of exogenous antibiotic tetracycline. The second system contained the mtr gene promoter and the toxin gene hipA. High tryptophan concentration switched off the expression of the toxic expression of the hipA gene and thus promoted cell growth. Our results showed that using these two systems, the engineered co-cultures’ tryptamine bioproduction production was improved significantly. It is therefore demonstrated that biosensor-assisted modular co-culture engineering is an effective strategy for enhancing heterologous tryptamine biosynthesis in E. coli.
2. Materials and methods
2.1. Plasmids and strains construction
Strains and plasmids used in this work are listed in Table 1. Sequences of the primers used in this study are listed in Supplementary Information. Tryptophan decarboxylase from Catharanthus roseus TDC (GenBank accession no. J04521) was codon-optimized for E. coli expression and synthesized by Bio basic Inc. Nucleotide sequences of TDC gene is provided in the Supplementary Information. DNA purification and plasmids isolation used kits from Zymo Research (Irvine, CA, USA). All restriction enzymes, DNA ligase, Q5 Master-mix enzymes and Gibson assembly builder were purchased from New England Biolabs. Plasmid construction was conducted using E. coli DH5α (New England Biolabs) competent cells. All cloning procedures were performed according to the manufacturer’s protocols. Detail information for plasmid construction is described as follows.
Table 1.
Plasmids and strains used in this study.
| Plasmids | Description | Source |
|---|---|---|
| pET21c | T7 promoter, AmpR | Novagen |
| pBR322 | AmpR, TetR | Thermo Scientific |
| pET28a | T7 promoter, KanR | Novagen |
| pCDFDuet-1 | double T7 promoters, StrpR | Novagen |
| pACYCDuet-1 | double T7 promoters, CmR | Novagen |
| pTS | pET21c carrying tryptophan regulatory element (tnaC operon) and tetA gene, AmpR | This study |
| pTC | pET21c carrying the tetA gene, AmpR | This study |
| pTE0 | pET28a carrying the trpEfbr gene under the control of the T7 promoter, KanR | Wang et al. (2019) |
| pTE3-1 | pUC57(KanR) carrying the trpEfbr, pctV and shiA genes under the control of the constitutive Zymomonas mobilis pyruvate decarboxylase promoter (Ppdc), KanR | Wang et al. (2019) |
| pTE3 | pET28a carrying the trpEfbr and aroGfbr genes under the control of the constitutive Zymomonas mobilis pyruvate decarboxylase promoter (Ppdc), KanR | Wang et al. (2019) |
| pTY0 | pET28a carrying the trpEfbr and TDC genes under the control of the T7 promoter, KanR | This study |
| pTY1 | pET28a carrying the trpEfbr, aroGfbr and TDC genes under the control of the T7 promoter, KanR | This study |
| pTY2 | pET28a carrying the trpEfbr, aroGfbr and TDC genes under the control of the constitutive Zymomonas mobilis pyruvate decarboxylase promoter (Ppdc), KanR | This study |
| pTD2-1 | pUC57(KanR) carrying the trpD and trpC genes under the control of the constitutive Zymomonas mobilis pyruvate decarboxylase promoter (Ppdc) and trpB and trpA genes under the control of the T7 promoter, KanR | Wang et al. (2019) |
| pTY3 | pET21c carrying the trpD and trpC genes under the control of the constitutive Zymomonas mobilis pyruvate decarboxylase promoter (Ppdc), AmpR | This study |
| pTY4 | pET28a carrying the TDC gene under the control of the T7 promoter, KanR | This study |
| pTY5 | pET21c carrying the TDC gene under the control of the constitutive Zymomonas mobilis pyruvate decarboxylase promoter (Ppdc), AmpR | This study |
| pTD2 | pACYCDuet-1 carrying the trpD, trpC, trpB and trpA genes under the control of the constitutive Zymomonas mobilis pyruvate decarboxylase promoter (Ppdc), CmR | Wang et al. (2019) |
| pSE1 | pET21c carrying the hipA gene under the control of the mtr promoter (Pmtr), AmpR | Wang et al. (2019) |
| pTD3 |
pCDFDuet-1 carrying the trpD, trpC, trpB and trpA genes under the control of the constitutive Zymomonas mobilis pyruvate decarboxylase promoter (Ppdc), StrpR |
This study |
| Strains |
Description |
Source |
| BL21(DE3) | F− ompT hsdSB (rB–, mB–) gal dcm (DE3), a common expression host | Invitrogen |
| BH2 | E. coli BL21(DE3) ΔxylA ΔtyrA ΔpheA, an engineered strain with disrupted xylose catabolism, as well as tyrosine and phenylalanine pathways | Zhang et al. (2015a) |
| XL10-Gold | endA1 glnV44 recA1 thi-1 gyrA96 relA1 lac Hte Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 TetRF’[proAB lacIqZΔM15 Tn10(TetR) Amy CmR], a strain with high transformation efficiency | Stratagene |
| BTH3 | BH2 harboring pTE3 and pTD2 | Wang et al. (2019) |
| BTP1 | BTH3 harboring pET21c | Wang et al. (2019) |
| XYD | XL10-Gold harboring pET28a and pTY5 | This study |
| BMC | BH2 harboring pTY2 and pTD2 | This study |
| BMC2 | BH2 harboring pTY2, pTD2 and pTC | This study |
| BLXC | BL21(DE3) harboring pTC | This study |
| BLXS | BL21(DE3) harboring pTS | This study |
| XTH | XL10-Gold harboring pTE3 and pTD3 | This study |
| BTXC | BTH3 harboring pTC | This study |
| BTS | BTH3 harboring pTS | This study |
| BMS | BH2 harboring pTY2, pTD2 and pTS | This study |
| BTS1 | BTH3 harboring pSE1 | Wang et al. (2019) |
| BMS1 | BH2 harboring pTY2, pTD2 and pSE1 | This study |
To construct plasmid pTS, three DNA fragments, including tnaC sequence with the tnaCAB operon promoter PCR amplified from the E. coli K12 chromosome by primers Tnac-Gf and Tnac-Gr, the tetA fragment PCR amplified from pBR322 plasmid by primers Teta-Gf and Teta-Gr, and plasmid pET21c backbone digested with SphI/NotI, were assembled together using the Gibson assembly kit (New England Biolabs). To construct plasmid pTC, the tetA gene with the promoter was PCR amplified from pBR322 using primer Tet-f and Tet-r, digested with NdeI/XhoI, and then ligated with plasmid pET21c treated with the same enzymes. To construct plasmid pTY0, a DNA fragment containing the codon-optimized TDC gene was synthesized (Bio Basic Inc. New York) (Park et al., 2011), digested by HindIII and XhoI, and ligated with the plasmid pTE0 treated with the same restriction enzymes (Wang et al., 2019). To construct pTY1, the aroGfbr (aroG (Asp146Asn)) gene was PCR amplified from the E. coli P2H chromosome using primer AroG-f and AroG-r, digested with BamHI/SalI, and ligated with pTY0 treated with the same restriction enzymes (Ganesan et al., 2017). To construct pTY2, the Ppdc-trpEfbr DNA fragment was PCR amplified from a previously constructed plasmid pTE3-1 using primer CST-trpE-f and CST-trpE-r (Wang et al., 2019), digested with SphI/SpeI, and then ligated with plasmid pTY1 treated with SphI/XbaI. To construct pTY3, the Ppdc-trpD-trpC fragment was PCR amplified from a previous constructed plasmid pTD2-1 using primer CST-TrpDC-f and CST-TrpDC-r (Wang et al., 2019), digested with EcoNI/NdeI, and then ligated with pET21c plasmid treated with the same restriction enzymes. To construct pTY4, a DNA fragment containing the codon-optimized TDC gene was synthesized (Bio Basic Inc. New York), digested by NdeI/XhoI, and ligated with the plasmid pET28a treated with the same restriction enzymes. To construct pTY5, the TDC fragment was generated by NcoI/XhoI digestion of pTY4, which was then ligated with plasmid pTY3 treated with the same restriction enzymes. To construct pTD3, the Ppdc-trpDCBA fragment was generated by digestion of pTD2 by EcoNI/XhoI, which was then ligated with pCDFDuet-1 plasmid treated with the same restriction enzymes.
2.2. Medium and cultivation conditions
Tryptamine biosynthesis in this study was performed using an MY medium. One liter of MY medium contained 6.8 g of Na2HPO4, 3 g of KH2PO4, 1 g of NH4Cl, 0.5 g of NaCl, 0.24 g of MgSO4, 0.5 g of yeast extract, 40 mg of tyrosine, 40 mg of phenylalanine, 5 g glucose, and trace elements. The working concentrations of trace elements were 0.4 mg/L Na2EDTA, 0.03 mg/L H3BO3, 1 mg/L thiamine, 0.94 mg/L ZnCl2, 0.5 mg/L CoCl2, 0.38 mg/L CuCl2, 1.6 mg/L MnCl2, 3.77 mg/L CaCl2, and 3.6 mg/L FeCl2. For biosynthesis on glycerol, 5 g glycerol was used to replace glucose. The working concentrations of antibiotics were 50 mg/L kanamycin, 34 mg/L chloramphenicol, and 100 mg/L ampicillin. When needed, 10 mg/L tetracycline was used for the biosensor-assisted cell selection.
Both E. coli mono-cultures and co-cultures were cultivated in 10 mL MY medium under 250 rpm at 30 °C. For mono-culture cultivation, 10% (v/v) overnight LB culture of the desired E. coli strains was inoculated into the MY medium with proper antibiotics and incubated at 37 °C for 8 h. The cells were then harvested by centrifugation and re-suspended in the fresh MY medium with an initial OD600 of 0.5. After 48 h cultivation, the culture samples were taken for HPLC analysis. For E. coli–E. coli co-cultures, 10% (v/v) overnight LB cultures of the desired E. coli strains were cultivated in the MY medium as seed cultures, respectively. After 8 h growth at 37 °C, the upstream and downstream cell seed cultures were measured for cell density, harvested by centrifugation, and inoculated into the MY medium containing appropriate antibiotics. The needed volumes for individual seed cultures were calculated based on the desired inoculation ratios. The initial OD600 of the co-culture after inoculation was controlled at 0.5, followed by 48 h cultivation at 30 °C.
For biosynthesis time profile analysis, seed cultures were grown in shake flasks containing 30 mL MY medium and cultivated at 37 °C for 12 h. The seed cultures of the upstream strains and the downstream strains were then inoculated into 30 mL MY medium containing 10 g/L glucose. The initial OD600 of the co-culture after inoculation was controlled at 0.5 with the desired upstream: downstream ratio, followed by 72 h cultivation in 250 mL shake flasks at 30 °C.
2.3. Construction and characterization of tryptophan biosensor-assisted cell selection system
The pTS plasmid containing the tryptophan biosensor-assisted selection system was composed of the tetA gene fused downstream to the tnaC gene. The tnaC gene encodes the leading sequence of the tnaCAB operon, whose expression was induced by tryptophan (Supplementary Information) (Gong and Yanofsky, 2001; Yanofsky et al., 1991).
Strain BLXS was constructed by introducing plasmid pTS into E. coli BL21(DE3). Strain BLXC was used as a control strain for growth characterization. The E. coli strains were first cultivated in the LB medium at 37 °C. The overnight culture was then centrifuged and re-suspended in fresh MY medium containing 10 mg/L tetracycline. The initial OD600 was kept at 0.5. Desired amounts of tryptophan were added to the culture to generate different working concentrations. After cultivation under 250 rpm at 30 °C for 18 h, the cultures were subjected to cell density analysis by measuring the light absorption at 600 nm.
2.4. Quantification of tryptophan and tryptamine
To quantify tryptophan and tryptamine, E. coli culture samples were centrifuged at 10,000 rpm for 5 min, and the supernatants were filtered through 0.45 mm PTFE membrane syringe filters. 10 μL filtered sample was injected into an Acclaim™ AmG C18 HPLC column controlled by a Shimadzu HPLC system with a diode array detector set to a wavelength of 280 nm. The samples were eluted using 97% solvent A (0.5% acetic acid in HPLC water) and 3% solvent B (99.9% acetonitrile) running at a flow rate of 0.6 mL/min.
2.5. Determination of co-culture population composition
The determination of the co-culture population composition was conducted by the blue-white colony counting method described in a previous report (Zhang et al., 2015b). 8 μL of the co-culture sample was diluted 106-fold and then spread onto an LB agar plate containing IPTG and X-Gal. After 12 h of incubation at 37 °C, the upstream strains (BTP1 or BTS1) carrying the intact lacZ gene generated blue colonies, while the downstream strain XYD containing the disrupted lacZ gene generated white colonies. The numbers of the blue and white colonies on the plates were counted for calculating the strain-to-strain ratio of the co-culture population.
3. Results
3.1. Design and construction of the E. coli-E. coli co-culture for tryptamine de novo biosynthesis
In order to employ the modular co-culture engineering strategy for tryptamine biosynthesis, we designed an E. coli-E. coli co-culture harboring the pathway leading from carbon substrate glucose or glycerol to tryptamine. Specifically, there were two individually engineered E. coli strains within the co-culture system. As shown in Fig. 1, the upstream strain was constructed for tryptophan overproduction; the downstream strain was engineered for converting tryptophan to tryptamine. Notably, the pathway intermediate tryptophan is known to be capable of move across the cell membrane (Burkovski and Krämer, 2002), which enables the connection of the two pathway modules accommodated by the two separate co-culture strains.
The upstream co-culture strain BTP1 was constructed in a previous study (Wang et al., 2019). This strain contained two plasmids pTD2 and pTE3 for over-expression of the key tryptophan pathway genes, including feedback control resistant DAHP synthase aroGfbr, feedback control resistant anthranilate synthase subunit trpEfbr, anthranilate synthase subunit trpD, tryptophan synthase subunit genes trpA and trpB, and indole-3-glycerol phosphate synthase/phosphoribosylanthranilate isomerase trpC (Table 1). E. coli XL10-Gold was used as the host strain for downstream conversion from tryptophan to tryptamine, as this strain showed outstanding performance for biosynthesis of another aromatic molecule 3-amino-benzoic acid using an engineered co-culture (Zhang and Stephanopoulos, 2016). The gene TDC encoding tryptophan decarboxylase derived from Catharanthus roseus was codon-optimized and introduced into E. coli XL10-Gold to yield the downstream co-culture strain XYD.
3.2. Tryptamine bioproduction by an E. coli-E. coli co-culture without cell selection systems
The engineered tryptophan over-producing strain BTP1 and the tryptophan-to-tryptamine conversion strain XYD were co-cultured in one consolidated culture for de novo tryptamine bioproduction on 5 g/L glucose or glycerol. The inoculum ratio between the upstream and downstream co-culture strains was optimized to balance the biosynthesis capabilities of the corresponding pathway modules. The mono-culture control strain BMC was constructed by introducing the whole tryptamine biosynthesis pathway into the upstream strain (Table 1). Another potential mono-culture control strain would be the downstream strain containing the whole tryptamine biosynthesis pathway. However, this strain was not constructed and used in this study, because the downstream strain was a poor host for producing tryptophan (Supplementary Information Fig. S1).
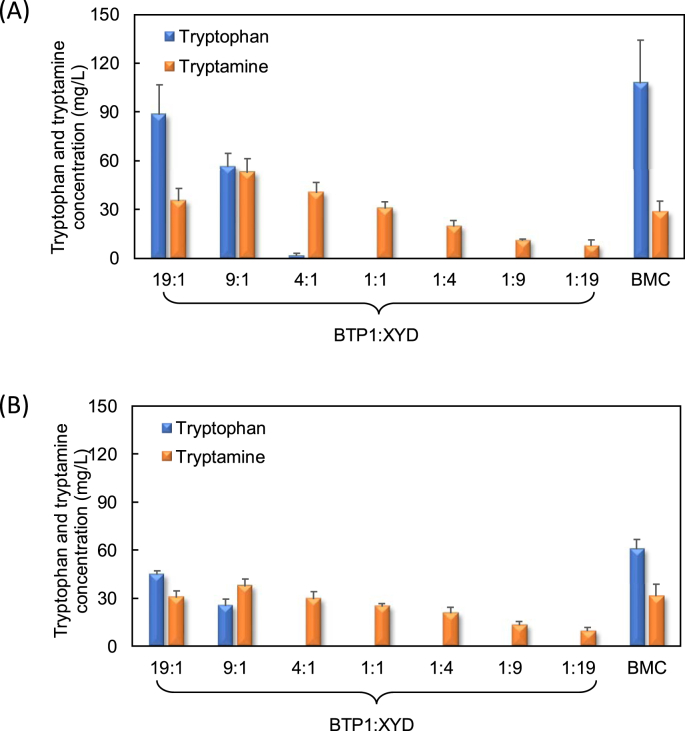
For the tryptamine bioproduction on glucose, the constructed mono-culture and co-culture systems were cultivated under the same condition for comparison. As shown in Fig. 2A, the tryptamine biosynthesis by the engineered co-culture varied significantly with the inoculum ratio. Excessive inoculation of the upstream strain BTP1 resulted in overly high tryptophan supply, which led to relatively small sub-population of the downstream strain and insufficient tryptophan-to-tryptamine bioconversion. On the contrary, when the downstream strain was inoculated with high ratio, the supply of tryptophan from the upstream strain was diminished. Due to these two effects, the optimal tryptamine production was achieved at the ratio of 9:1, under which the tryptamine bioproduction reached 53 mg/L, although there was also accumulation of 56 mg/L tryptophan. In comparison, the mono-culture control strains BMC produced 30 mg/L tryptamine under the same conditions.
Fig. 2.
Tryptamine bioproduction by utilization of the BMC mono-culture and BTP1:XYD co-culture inoculated at different ratios using (A) glucose and (B) glycerol as the carbon substrate.
The constructed co-culture also showed desired tryptamine production when 5 g/L glycerol was used as the carbon source. As shown in Fig. 2B, the final tryptamine concentration varied over a wide range of the co-culture strains’ inoculation ratio. The highest tryptamine production of 37 mg/L was achieved at the inoculation ratio of 9:1, which is the same with the optimal inoculation rate for the biosynthesis on glucose. In comparison, the mono-culture control strains BMC produced 32 mg/L tryptamine. Overall, the co-culture’s tryptamine production on glucose was higher than on glycerol, suggesting that glucose is a better carbon source for the tryptamine bioproduction by the co-culture.
It should be also noted that there was significant tryptophan accumulation in the mono-culture control grown on both glucose and glycerol. In contrast, almost no tryptophan accumulation was observed in the constructed co-culture at most of the inoculum ratios, indicating that the constructed co-cultures had robust tryptophan-to-tryptamine conversion capability largely due to the reduced metabolic burden on the downstream strain. In the meantime, this situation suggested that the upstream supply of tryptophan was the limiting factor of the overall tryptamine production in the co-cultures.
3.3. Construction and characterization of the cell selection systems
In order to improve the tryptophan supply and tryptamine production, two different biosensor-assisted cell selection systems were introduced in the upstream strain, respectively. Each cell selection system comprised a tryptophan-responsive biosensor and a growth-regulating gene, as reported by previous studies (Wang et al., 2019; Xiao et al., 2016). Depending on the tryptophan concentration, expression of the growth-regulating gene was up- or down-regulated. As a result, the growth of the high tryptophan-producing cells was promoted, while the growth of low tryptophan-producing cells was inhibited, as shown in Fig. 1. Therefore, the upstream strain’s population was dominated by the high tryptophan-producing cell and the overall tryptophan production performance was enhanced.
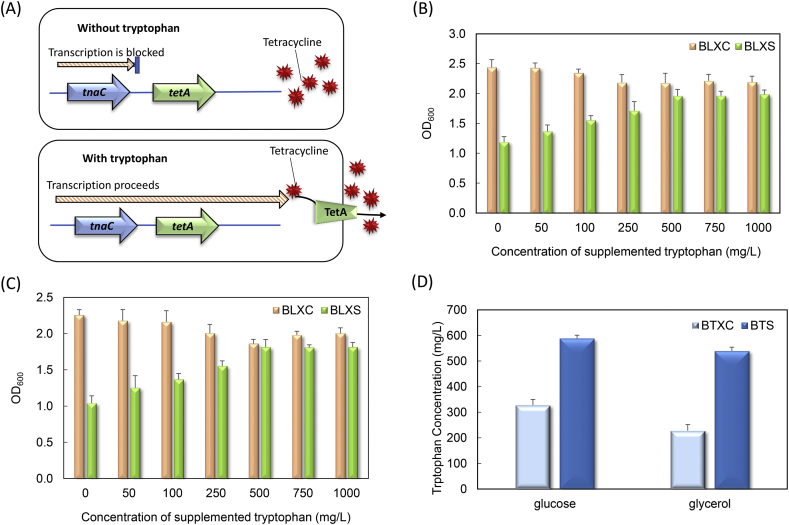
For the first cell selection system, we used E. coli tnaCAB operon’s leader peptide gene tnaC to control the tetracycline resistance gene tetA expression (Fig. 3A). The strain harboring this cell selection system was first characterized for growth regulation in response to varying tryptophan concentrations. Specifically, E. coli BLXC and BLXS, two strains without and with the tnaC-tetA cell selection system, were cultivated in the MY medium with 10 mg/L tetracycline. As shown in Fig. 3B, BLXC’s growth showed slight decrease with the increase of the exogenous tryptophan concentration, possibly due to the cell toxicity associated with high tryptophan concentration. The growth of BLXS, however, showed clear dependence on the tryptophan concentration as expected: low tryptophan concentration inhibited the growth, whereas high tryptophan concentration promoted the growth. In addition, BLXC and BLXS growth comparison on glycerol demonstrated similar trend (Fig. 3C). These results indicate that the tnaC-tetA cell selection system has the desired tryptophan-sensing and cell growth regulation functions.
Fig. 3.
Construction and characterization of the tnaC-tetA cell selection system. (A) Scheme of the tnaC-tetA cell selectin mechanism. The presence of tryptophan decides whether the tetA gene can be expressed to resist the antibiotic tetracycline. (B) and (C) Growth behaviors of E. coli BLXS harboring the tnaC-tetA system on glucose and glycerol, respectively. 10 mg/L tetracycline and varying concentrations of tryptophan was added to the medium in both cases. E. coli BLXC was a control strain without the cell selection system. (D) Comparison of tryptophan biosynthesis between BTXC and BTS using glucose and glycerol as the carbon substrates.
Next, the tryptophan production capability of the strain BTS harboring the tnaC-tetA system was investigated. As shown in Fig. 3D, when 5 g/L glucose was used as the carbon source, tryptophan production by BTS reached 588 mg/L, 80% higher than the control strain BTXC. The tryptophan production on glycerol was also improved from 228 to 539 mg/L. The significant production improvement on both glucose and glycerol demonstrate effectiveness of the tnaC-tetA cell selection system.
For the second cell selection system, we recruited a tryptophan-responsive promoter derived from the E. coli mtr gene to control the expression of the toxin hipA gene via the tryptophan-binding protein TrpR. Notably, different from the first cell selection system, the biosensor of this system is an off-switch that downregulates gene expression with high tryptophan production. The mtr-hipA system had been previously characterized and showed strong effect for improving tryptophan over-production in E. coli (Wang et al., 2019).
3.4. Enhancing the tryptamine bioproduction using thetnaC-tetAsystem
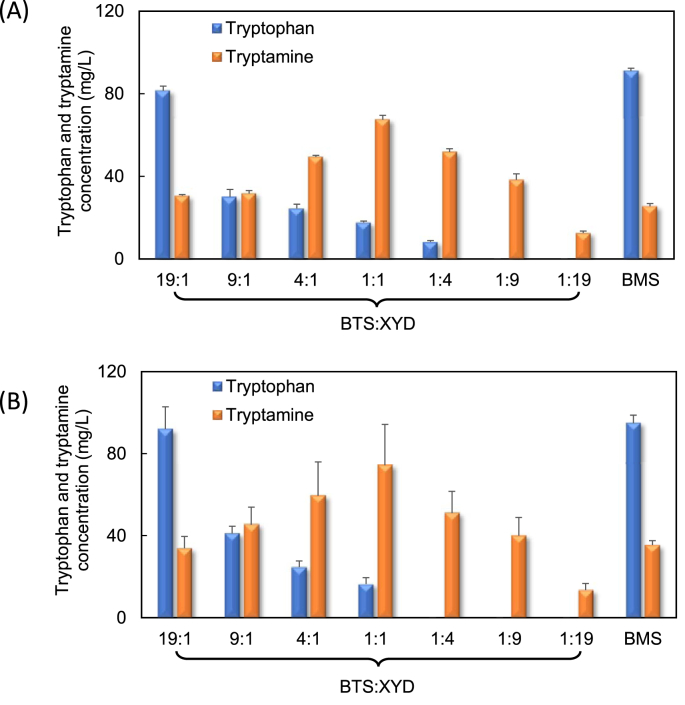
Based on the constructed tnaC-tetA cell selection system, a new upstream strain BTS was developed and co-cultivated with the downstream strain XYD for the tryptamine biosynthesis. Fig. 4A shows the profile of tryptamine bioproduction on glucose. The highest tryptamine concentration reached 67 mg/L, 26% higher than the co-culture without the cell selection system. The optimal inoculation ratio also shifted from 9:1 to 1:1, indicating that the application of the cell selection strategy improved the tryptophan supply and reduced the need of high upstream strain inoculation ratio. This was further confirmed by the appearance of tryptophan accumulation at the inoculation ratio of 4:1, 1:1 and 1:4. A mono-culture control strain BMS was also constructed to harbor the whole tryptamine pathway as well as the cell selection system. It was found that the cell selection strategy did not improve the tryptamine production in this strain, compared with strain BMC without the tnaC-tetA system. This finding is in good agreement with our previous study (Guo et al., 2019) and suggests that the use of cell selection mechanism in mono-culture resulted in selection of cells with high tryptophan accumulation but not high tryptamine production. This situation highlights the unique advantage of using biosensor-assisted cell selection in co-cultures over mono-cultures.
Fig. 4.
Tryptamine bioproduction using the tnaC-tetA biosensor-assisted cell selection system. (A) glucose and (B) glycerol were used as the carbon substrates for tryptamine bioproduction by the BMS mono-culture and BTS:XYD co-culture inoculated at different ratios.
The BTS:XYD co-culture was also used for tryptamine production on glycerol. As shown in Fig. 4B, 74 mg/L tryptamine was produced from 5 g/L glycerol at the optimal inoculation ratio of 1:1. Such production was 2-fold higher than the co-culture without the cell selection system. Moreover, the tryptophan supply was found to be enhanced in this production system, as evidenced by the improved tryptophan accumulation and shifted optimal inoculation ratio. In comparison, the mono-culture control strain BMS only produced 35 mg/L tryptamine under the same condition.
3.5. Enhancing the tryptamine bioproduction using themtr-hipAcell selection system
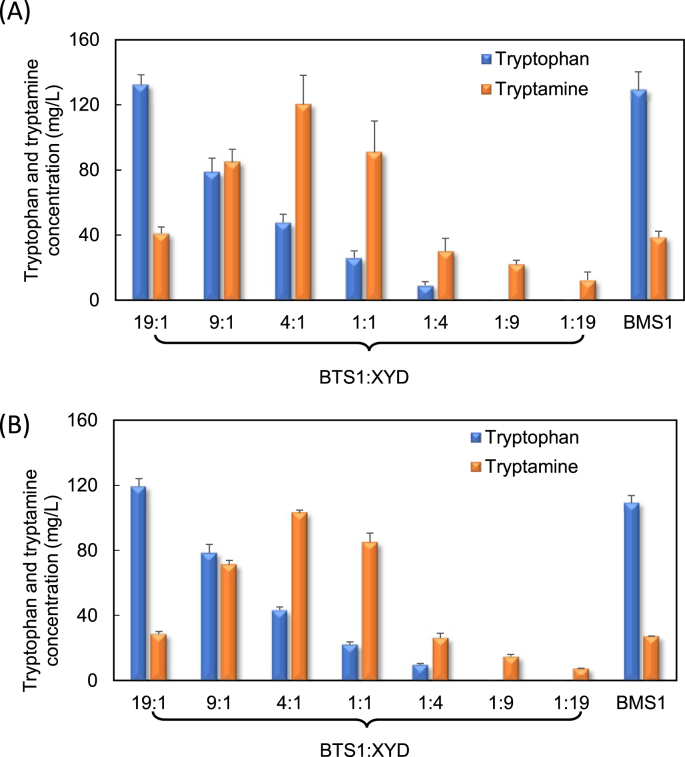
In addition to the tnaC-tetA cell selection system, this study also explored the use of the previously constructed mtr-hipA cell selection system (Wang et al., 2019). The corresponding co-culture BTS1:XYD was used for de novo tryptamine bioproduction on glucose and glycerol. As shown in Fig. 5A, the tryptamine production on glucose was significantly improved to 120 mg/L at the optimal ratio of 4:1. This production is 79% higher than that of the co-cultures using the tnaC-tetA cell selection system, suggesting that the mtr-hipA system is more robust for enhancing the overall tryptamine biosynthesis performance. In fact, our previous study has shown that the recruitment of the E. coli toxin system as the growth regulator resulted in higher bioproduction performance than using the tetA gene (Wang et al., 2019). In the present study, it was further confirmed that using toxin as the regulator is a robust method to improve the biosensor-targeted metabolite biosynthesis. It should be noted, however, that the difference of binding affinity of tryptophan with TrpR sensor protein of the toxin system and tnaC control element of the tetA system could also contribute to the selection efficiency difference and bioproduction performance variation. On the other hand, the BMS1 mono-culture control strains only produced 39 mg/L tryptamine. This is consistent with the previous finding that the introduction of the cell selection system into a mono-culture did not significantly improve the tryptamine production.
Fig. 5.
Tryptamine bioproduction using the mtr-hipA biosensor-assisted cell selection system. (A) glucose and (B) glycerol were used as the carbon substrates for tryptamine bioproduction by the BMS1 mono-culture and BTS1:XYD co-culture inoculated at different ratios.
As shown in Fig. 5B, the tryptamine production on glycerol was also considerably improved using the BTS1:XYD co-culture. At the optimal inoculation ratio of 4:1, the co-culture produced 103 mg/L tryptamine from 5 g/L glycerol, which is 39% higher than the BTS:XYD co-culture using the tnaC-tetA system and 2.8-fold higher than the BTP1:XYD co-culture without the biosensor. The BMS1 mono-culture control strain only produced 27 mg/L tryptamine. The experimental results above clearly demonstrate that the recruitment of the mtr-hipA cell selection system is an effective strategy for enhancing the co-culture’s capability for producing tryptamine on both glucose and glycerol.
3.6. Characterization of the co-culture biosynthesis behavior
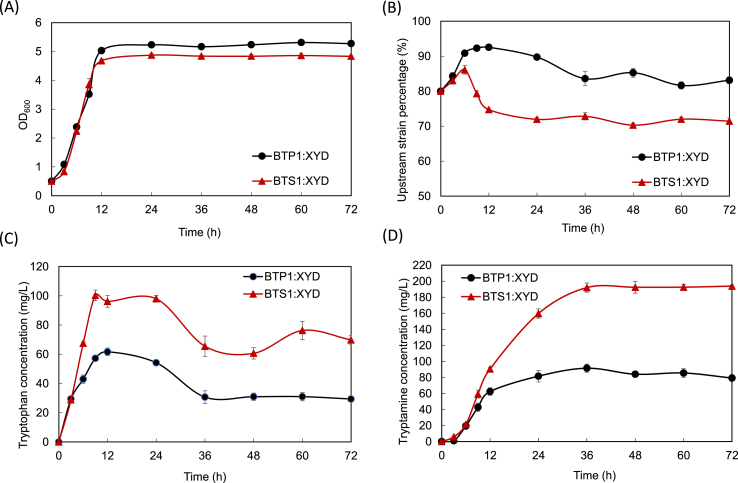
In an effort to characterize the dynamic tryptamine biosynthesis behavior, the engineered BTS1:XYD co-culture (with the biosensor) and BTP1:XYD control co-culture (without the biosensor) were grown in shake flasks containing 10 g/L glucose at the inoculum ratio of 4:1. The overall culture cell density, strain-to-strain ratio, and pathway metabolite concentrations were analyzed throughout the cultivation. As shown in Fig. 6A, the growth of the co-culture systems followed the typical pattern of a batch culture. After the exponential phase between 3 and 12 h, the highest cell density was stabilized at OD600 ˜ 5.2 for the control co-culture and OD600 ˜ 4.8 for the BTS1:XYD co-culture with the biosensor. Notably, the BTS1:XYD co-culture’s cell density was lower than the control co-culture, indicating that the hipA-based cell selection system inhibited the growth of the low-performing cells of the upstream strain and thus reduced the overall co-culture growth.
Fig. 6.
Shake flask cultivation of the BTP1:XYD and BTS1:XYD co-cultures for the tryptamine bioproduction. Time profiles of (A) overall co-culture cell density, (B) upstream strain percentage, (C) accumulation of the pathway intermediate tryptophan and (D) concentration of tryptamine were derived from triplicate shake flask runs. Error bars represent standard errors of the experimental results.
The time profiles of the upstream strains to downstream strain subpopulation ratio were analyzed using a blue-white colony counting method (Zhang et al., 2015b). For the BTP1:XYD co-culture, the percentage of BTP1 changed from 80% to 93% within 12 h and decreased to 83% towards the end of the cultivation. This trend shows that the co-growth relationship between the co-culture strains is a dynamic and fluctuating process. Interestingly, for the BTS1:XYD co-culture, the percentage of BTS1 increased from 80% (0 h) to 86% (6 h), then decreased to 75% (12 h) and lastly plateaued at 72% for the rest of the cultivation period. This sharp decline starting from 6 h is considered to be due to the growth inhibition of the upstream strain’s low-performing cells resulted from the mtr-hipA cell selection system.
As shown in Fig. 6D, the co-culture BTS1:XYD was found to produce a considerably higher amount of tryptamine than the control co-culture BTP1:XYD. The concentration of the tryptamine in BTS1:XYD started to increase 3 h after the inoculation and plateaued at around 192 mg/L after 36 h; whereas the control group’s tryptamine biosynthesis peaked at around 91 mg/L at 36 h. In the meantime, it was found that the pathway intermediate tryptophan was accumulated along with the co-culture growth (Fig. 6C). For BTP1:XYD, it was observed that tryptophan was accumulated to around 62 mg/L at 12 h, decreased to 31 mg/L at 36 h and stabilized at this level toward the end of cultivation. The tryptophan concentration was kept at a relatively low level at most of the cultivation period, indicating that the insufficient intermediate supply limited the production of tryptamine. In comparison, for BTS1:XYD, the tryptophan accumulation reached 100 mg/L at 9 h, decreased to 65 mg/L at 36 h and fluctuated around this level for the remaining period of the cultivation. The accumulation of the pathway intermediate tryptophan was due to the low subpopulation percentage of the downstream strain (less than 20%) shown in Fig. 6B. Interestingly, the tryptamine production and tryptophan accumulation difference between the two co-cultures both started to occur at around 6–12 h (Fig. 6C and D), suggesting that the enhancement of tryptophan provision by the mtr-hipA based selection system took effect in the mid exponential growth phase. This finding is consistent with the upstream strain percentage change at 6 h discussed above.
Overall, the characterization of the BTS1:XYD co-culture biosynthesis showed that the upstream strain occupied the majority of the co-culture population throughout the cultivation and produce a sufficient amount of tryptophan for the conversion to tryptamine. On the other hand, the downstream strain possessed strong biosynthetic capability that allowed it to complete effective tryptophan to tryptamine conversion at a relatively low sub-population. The overall yield of the tryptamine production of BTS1:XYD on glucose in shake flash was 0.02 g/g glucose. The tryptamine production was mostly achieved within 36 h under the adapted cultivation condition.
4. Discussion
Tryptamine heterologous biosynthesis is of great research interest for improving this compound’s availability and facilitating the production of tryptamine derivatives with recognized values. This study utilized the emerging modular engineering approach for reconstituting the tryptamine biosynthesis pathway in the context of an E. coli-E. coli co-culture. Several engineering strategies were applied for co-culture biosynthesis optimization. First, the inoculum ratios between the upstream and downstream co-culture strains were varied to balance the biosynthetic capabilities of the corresponding pathway modules. In fact, this strategy has been widely used in previous studies for pathway balancing and bioproduction optimization in engineered co-cultures (Chen et al., 2019; Jawed et al., 2019; Jones and Wang, 2018; Roell et al., 2019; Zhang and Wang, 2016). It should be noted that, even for the co-cultures composed of same-species strains, the constituent strains often had quite different growth and biosynthesis behaviors. As a result, the relative strain-to-strain ratio needs to be fine-tuned by varying the inoculum ratio. For this study, it was found that the inoculum ratio indeed played a critical role in determining the bioproduction performance. The optimal production was achieved at the inoculation ratio of 9:1, suggesting that the biosynthesis capability of the upstream strain was at disadvantage and needed to be compensated by high inoculum at the beginning of cultivation.
To further increase the supply of tryptophan for tryptamine production, we recruited two cell selection mechanisms to select for tryptophan high-producing cells of the co-culture’s upstream strain. These cell selection mechanisms removed low performing cells from the microbial population and have been verified to be an effective approach for target product biosynthesis enhancement in the context of mono-culture (Lv et al., 2019; Rugbjerg et al., 2018; Wang et al., 2019; Xiao et al., 2016). In this study, it was shown that the E. coli toxin-based selection system was more effective than antibiotic-based selection system for tryptamine production improvement. In addition, the toxin-based selection system is straightforward to implement in the co-culture, as it does not require any additional modification of the downstream strain, such as introduction of new antibiotic resistance for compatible growth with the upstream strain carrying the antibiotic-based cell selection system, which minimizes the impact on the downstream strain’s growth. On the other hand, such a cell selection strategy is challenging to make positive impact in the mono-culture. In fact, for the mono-culture, the employment of the cell selection mechanism using a tryptophan biosensor resulted in selection of the cells with high tryptophan accumulation, whereas the conversion from tryptophan to tryptamine was not promoted by the cell selection mechanism. This is supported by comparison of the tryptophan and tryptamine production by strains BMC2 ad BMS without and with the tnaC-tetA system. As shown in Fig. S2, BMS showed higher tryptophan accumulation capability but the same tryptamine production level with BMC2 on both glucose and glycerol, confirming that when used in the context of mono-culture, the cell selection system was mainly devoted to select for the cells with better tryptophan accumulation, but not better tryptamine production. In contrast, the application of the cell selection mechanism in the context of a rationally designed co-culture overcomes this issue, as the modular nature of the co-culture allows for physical segregation and independent engineering of the co-culture strains such that establishment of the cell selection mechanism in the upstream strain does not reduce downstream strain’s biosynthetic capability. As such, our results highlight the outstanding advantages of integration of biosensor into co-cultures over into mono-cultures.
Glycerol is an important byproduct of the biodiesel industry and a renewable carbon material with high abundance. Glycerol’s utilization as carbon source for microbial biosynthesis has attracting increasing research interest (Clomburg and Gonzalez, 2013; Mattam et al., 2013; Pradima and Kulkarni, 2017). There have been efforts using modular co-culture engineering to convert glycerol to important biochemical products, such as cis,cis-muconic acid and 3-hydroxybenzoic acid (Zhang et al., 2015a; Zhou et al., 2019). In this study, we confirmed that glycerol can be used to produce tryptamine using the engineered co-culture, although the production performance is lower than using glucose as the substrate. Also, it was found that the integration of the designed cell selection system in the co-culture also improved the tryptamine biosynthesis on glycerol. This result, together with the bioproduction improvement on glucose, suggested that biosensor-assisted co-cultures can be more broadly used for biosynthesis using other renewable carbon substrates, such as xylose and arabinose.
The shake flask cultivation achieved production of 194 mg/L tryptamine and 0.02 g/g glucose production yield, which provides a robust platform for future biosynthesis of more advanced end products, such as serotonin and melatonin. To this end, the required pathway enzymes for making the target products can be conveniently expressed in the downstream strains of the co-cultures without the undesired interference with the upstream pathway module which is segregated in another strain. Therefore, the findings of this study and its potential utility for future research demonstrate the potential and broad applicability of the modular co-culture engineering in the future.
Declaration of competing interest
The authors declare no financial or commercial conflict of interest.
Acknowledgement
This material is based upon work supported in part by the National Science Foundation under Grant No. 1706058. The authors are also grateful for the support by the startup research fund provided by Rutgers, The State University of New Jersey. Zhenghong Li is a recipient of the CSC Ph.D. fellowship.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mec.2019.e00110.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Bayer T.S., Widmaier D.M., Temme K., Mirsky E.A., Santi D.V., Voigt C.A. Synthesis of methyl halides from biomass using engineered microbes. J. Am. Chem. Soc. 2009;131:6508–6515. doi: 10.1021/ja809461u. [DOI] [PubMed] [Google Scholar]
- Burkovski A., Krämer R. Bacterial amino acid transport proteins: occurrence, functions, and significance for biotechnological applications. Appl. Microbiol. Biotechnol. 2002;58:265–274. doi: 10.1007/s00253-001-0869-4. [DOI] [PubMed] [Google Scholar]
- Chen T., Zhou Y., Lu Y., Zhang H. Advances in heterologous biosynthesis of plant and fungal natural products by modular co-culture engineering. Biotechnol. Lett. 2019;41:27–34. doi: 10.1007/s10529-018-2619-z. [DOI] [PubMed] [Google Scholar]
- Clomburg J.M., Gonzalez R. Anaerobic fermentation of glycerol: a platform for renewable fuels and chemicals. Trends Biotechnol. 2013;31:20–28. doi: 10.1016/j.tibtech.2012.10.006. [DOI] [PubMed] [Google Scholar]
- Ganesan V., Li Z., Wang X., Zhang H. Heterologous biosynthesis of natural product naringenin by co-culture engineering. Synthetic and systems biotechnology. 2017;2:236–242. doi: 10.1016/j.synbio.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong F., Yanofsky C. Reproducing tna operon regulation in vitro in an S-30 system tryptophan induction inhibits cleavage of TnaC peptidyl-tRNA. J. Biol. Chem. 2001;276:1974–1983. doi: 10.1074/jbc.M008892200. [DOI] [PubMed] [Google Scholar]
- Guo X., Li Z., Wang X., Wang J., Chala J., Lu Y., Zhang H. De novo phenol bioproduction from glucose using biosensor-assisted microbial co-culture engineering. Biotechnol. Bioeng. 2019;116(12):3349–3359. doi: 10.1002/bit.27168. [DOI] [PubMed] [Google Scholar]
- Jawed K., Yazdani S.S., Koffas M.A. Advances in the development and application of microbial consortia for metabolic engineering. Metabolic Engineering Communications. 2019 doi: 10.1016/j.mec.2019.e00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.A., Vernacchio V.R., Collins S.M., Shirke A.N., Xiu Y., Englaender J.A., Cress B.F., McCutcheon C.C., Linhardt R.J., Gross R.A. Complete biosynthesis of anthocyanins using E. coli polycultures. mBio. 2017;8 doi: 10.1128/mBio.00621-17. e00621-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.A., Wang X. Use of bacterial co-cultures for the efficient production of chemicals. Curr. Opin. Biotechnol. 2018;53:33–38. doi: 10.1016/j.copbio.2017.11.012. [DOI] [PubMed] [Google Scholar]
- Kang S., Kang K., Lee K., Back K. Characterization of tryptamine 5-hydroxylase and serotonin synthesis in rice plants. Plant Cell Rep. 2007;26:2009–2015. doi: 10.1007/s00299-007-0405-9. [DOI] [PubMed] [Google Scholar]
- Lee S.J., Sim G.-Y., Lee Y., Kim B.-G., Ahn J.-H. Engineering of Escherichia coli for the synthesis of N-hydroxycinnamoyl tryptamine and serotonin. J. Ind. Microbiol. Biotechnol. 2017;44:1551–1560. doi: 10.1007/s10295-017-1975-3. [DOI] [PubMed] [Google Scholar]
- Li Z., Wang X., Zhang H. Balancing the non-linear rosmarinic acid biosynthetic pathway by modular co-culture engineering. Metab. Eng. 2019;54:1–11. doi: 10.1016/j.ymben.2019.03.002. [DOI] [PubMed] [Google Scholar]
- Liu S.-D., Wu Y.-N., Wang T.-M., Zhang C., Xing X.-H. Maltose utilization as a novel selection strategy for continuous evolution of microbes with enhanced metabolite production. ACS Synth. Biol. 2017;6:2326–2338. doi: 10.1021/acssynbio.7b00247. [DOI] [PubMed] [Google Scholar]
- Lv Y., Qian S., Du G., Chen J., Zhou J., Xu P. UMBC Faculty Collection; 2019. Coupling Feedback Genetic Circuits with Growth Phenotype for Dynamic Population Control and Intelligent Bioproduction. [DOI] [PubMed] [Google Scholar]
- Mahmood Z.A., Ahmed S.W., Azhar I., Sualeh M., Baig M.T., Zoha S. Bioactive alkaloids produced BY fungi I. Updates ON alkaloids from the species OF the genera boletus, FUSARIUM and psilocybe. Pak. J. Pharm. Sci. 2010;23 [PubMed] [Google Scholar]
- Mattam A.J., Clomburg J.M., Gonzalez R., Yazdani S.S. Fermentation of glycerol and production of valuable chemical and biofuel molecules. Biotechnol. Lett. 2013;35:831–842. doi: 10.1007/s10529-013-1240-4. [DOI] [PubMed] [Google Scholar]
- Mora-Villalobos J.-A., Zeng A.-P. Synthetic pathways and processes for effective production of 5-hydroxytryptophan and serotonin from glucose in Escherichia coli. J. Biol. Eng. 2018;12:3. doi: 10.1186/s13036-018-0094-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousseau D.D. Tryptamine: a metabolite of tryptophan implicated in various neuropsychiatric disorders. Metab. Brain Dis. 1993;8:1–44. doi: 10.1007/BF01000528. [DOI] [PubMed] [Google Scholar]
- Niehaus L., Boland I., Liu M., Chen K., Fu D., Henckel C., Chaung K., Miranda S.E., Dyckman S., Crum M. Microbial coexistence through chemical-mediated interactions. Nat. Commun. 2019;10:2052. doi: 10.1038/s41467-019-10062-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Kang K., Lee S.W., Ahn M.-J., Bae J.-M., Back K. Production of serotonin by dual expression of tryptophan decarboxylase and tryptamine 5-hydroxylase in Escherichia coli. Appl. Microbiol. Biotechnol. 2011;89:1387–1394. doi: 10.1007/s00253-010-2994-4. [DOI] [PubMed] [Google Scholar]
- Pradima J., Kulkarni M.R. Review on enzymatic synthesis of value added products of glycerol, a by-product derived from biodiesel production. Resource-Efficient Technologies. 2017;3:394–405. [Google Scholar]
- Roell G.W., Zha J., Carr R.R., Koffas M.A., Fong S.S., Tang Y.J. Engineering microbial consortia by division of labor. Microb. Cell Factories. 2019;18:35. doi: 10.1186/s12934-019-1083-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rugbjerg P., Sarup-Lytzen K., Nagy M., Sommer M.O.A. Synthetic addiction extends the productive life time of engineered Escherichia coli populations. Proc. Natl. Acad. Sci. 2018;115:2347–2352. doi: 10.1073/pnas.1718622115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoek T., Romero-Suarez D., Zhang J., Ambri F., Skjoedt M.L., Sudarsan S., Jensen M.K., Keasling J.D. An orthogonal and pH-tunable sensor-selector for muconic acid biosynthesis in yeast. ACS Synth. Biol. 2018;7:995–1003. doi: 10.1021/acssynbio.7b00439. [DOI] [PubMed] [Google Scholar]
- Songstad D.D., De Luca V., Brisson N., Kurz W.G., Nessler C.L. High levels of tryptamine accumulation in transgenic tobacco expressing tryptophan decarboxylase. Plant Physiol. 1990;94:1410–1413. doi: 10.1104/pp.94.3.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Cabales A., Li Z., Zhang H. Biosensor-assisted high performing cell selection using an E. coli toxin/antitoxin system. Biochem. Eng. J. 2019;144:110–118. [Google Scholar]
- Xiao Y., Bowen C.H., Liu D., Zhang F. Exploiting nongenetic cell-to-cell variation for enhanced biosynthesis. Nat. Chem. Biol. 2016;12:339. doi: 10.1038/nchembio.2046. [DOI] [PubMed] [Google Scholar]
- Yanofsky C., Horn V., Gollnick P. Physiological studies of tryptophan transport and tryptophanase operon induction in Escherichia coli. J. Bacteriol. 1991;173:6009–6017. doi: 10.1128/jb.173.19.6009-6017.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Li Z., Pereira B., Stephanopoulos G. Engineering E. coli–E. coli cocultures for production of muconic acid from glycerol. Microb. Cell Factories. 2015;14:134. doi: 10.1186/s12934-015-0319-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Pereira B., Li Z., Stephanopoulos G. Engineering Escherichia coli coculture systems for the production of biochemical products. Proc. Natl. Acad. Sci. 2015;112:8266–8271. doi: 10.1073/pnas.1506781112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Stephanopoulos G. Co-culture engineering for microbial biosynthesis of 3-amino-benzoic acid in Escherichia coli. Biotechnol. J. 2016;11:981–987. doi: 10.1002/biot.201600013. [DOI] [PubMed] [Google Scholar]
- Zhang H., Wang X. Modular co-culture engineering, a new approach for metabolic engineering. Metab. Eng. 2016;37:114–121. doi: 10.1016/j.ymben.2016.05.007. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Li Z., Wang X., Zhang H. Establishing microbial co-cultures for 3-hydroxybenzoic acid biosynthesis on glycerol. Eng. Life Sci. 2019;19:389–395. doi: 10.1002/elsc.201800195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.