Highlights
-
•
2,3-Butanediol (2,3-BD) is a propitious compound with many industrial uses.
-
•
2,3-BD production has always been hampered by low fermentation yields and high production costs.
-
•
2,3-BD production may be enhanced by optimization of culture conditions and use of high-producing strains.
-
•
TMetabolic engineering tools are currently used to generate high-yielding strains.
Abbreviations: 2,3-BD, 2,3-Butanediol; adhE, alcohol dehydrogenase; ackA, acetate kinase-phosphotransacetylase; ldhA, lactate dehydrogenase; AlsS, α-acetolactate synthase; AlsD, α-acetolactate decarboxylase; MEK, methyl ethyl ketone; PUMAs, polyurethane-melamides; gldA, glycerophosphate dehydrogenase gene
Keywords: 2, 3-Butanediol; Metabolic engineering; Butanediol dehydrogenase; Klebsiella; Species
Abstract
2,3-Butanediol (2,3-BD) is a propitious compound with many industrial uses ranging from rubber, fuels, and cosmetics to food additives. Its microbial production has especially attracted as an alternative way to the petroleum-based production. However, 2,3-BD production has always been hampered by low yields and high production costs. The enhanced production of 2,3-butanediol requires screening of the best strains and a systematic optimization of fermentation conditions. Moreover, the metabolic pathway engineering is essential to achieve the best results and minimize the production costs by rendering the strains to use efficiently low cost substrates. This review is to provide up-to-date information on the current strategies and parameters for the enhanced microbial production of 2,3-BD.
1. Introduction
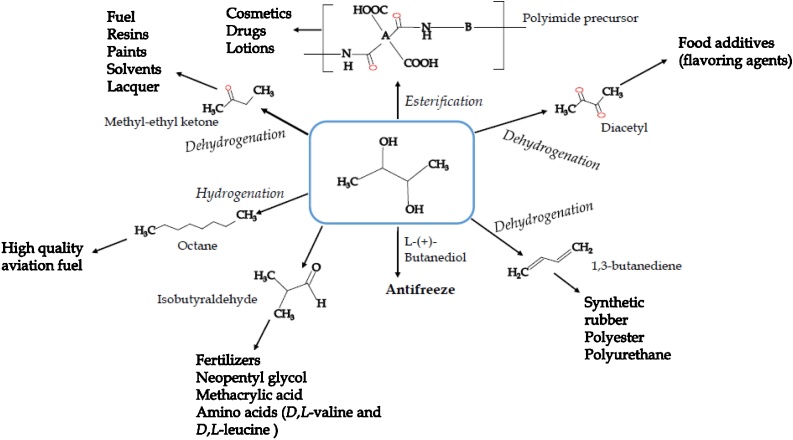
Numerous compounds which could only be produced by chemical processes in the past, can now be biologically generated from renewable resources using engineered organisms. Recently, the development of integrated biorefineries has attracted increasing attention as a means of giving sustainable alternative ways for solving several difficulties including exhaustion of fossil oil supplies, high soaring petroleum prices and environmental pollution [1]. As a solution to scarcity of nonrenewable energy resources, biofuel is the only sustainable solution to satisfy ever-growing energy demands [[2], [3], [4]]. One of the examples is microbial production of 2, 3-butanediol by different microorganisms including bacteria and yeast trains [1,5], and it is now widely used in the industry [[6], [7], [8]]. 2,3-BD is a valuable chemical feedstock due to its potential applications [9], including its use as a liquid fuel and for the production of various chemicals, foods, and pharmaceuticals (Fig. 1) [10,11].
Fig. 1.
Some important derivatives of 2,3-butanediol and their potential applications (adapted from Białkowska, 2016).
Recently, various microorganisms including Klebsiellaoxytoca, Klebsiella pneumoniae, Serratia marcescens, Enterobacter aerogenes, and Paenibacillus polymyxa and Saccharomyces cerevisiae have been increasingly used to produce 2,3-butanediol [9,12,13]. Among them, Klebsiella species are preferred for higher 2,3-BD production thanks to their capability for utilizing various substrates coupled with higher cultural adaptability [11,14]. Apart from glucose or xylose which are utilized as substrates, other sources available are starch, molasses, water hyacinth and Jerusalem artichoke tubers, or sweet potatoes [15]. Additionaly, Jatropha waste constitutes a propitious row material to manufacture both sugars and 2,3-butanediol [16].
Various microorganisms can use various substrates such as hexoses, pentoses, and disaccharides for producing 2,3-butanediol via mixed-acid butanediol pathway [12]. However, this pathway produces several liquid byproducts including lactic, succinic, acetic acid, and ethanol [17]. Microorganisms have been genetically engineered not only for improving 2,3-BD production, but also expanding the substrate spectrum of 2,3-BD producing strains, especially for low cost substrates [18]. Microorganisms can also utilize other substrates including plant biomass, agricultural and industrial wastes, and convert them into biofuel and useful co-products [19,20]. The main factors for enhancing 2,3-BD yield include the selection of appropriate high-yielding strains and systematically optimizing the culture conditions such as temperature, aeration and byproduct control. Moreover, the choice of an efficient cultivation strategy especially batch, fed-batch and continuous fermentation is one of the paramount importance for optimal yields [21]. For this reason, this work has explored and reviewed the currently most promising methods for improving microbial production of 2,3-butanediol.
2. Metabolic pathway for the production of 2,3-BD
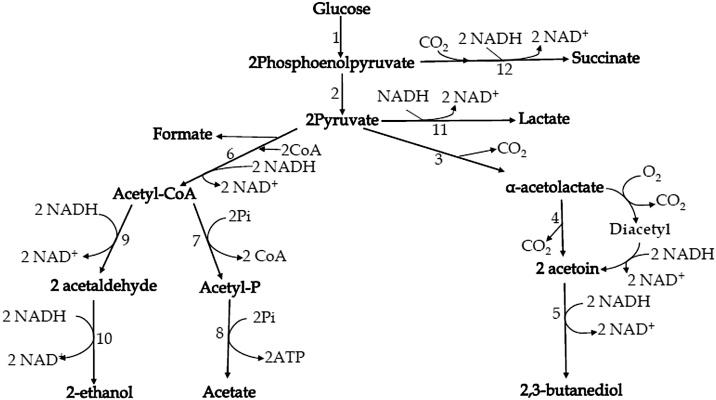
Different microorganisms capable of producing 2,3-BD include several types of bacteria, yeasts, or even algae [22]. 2,3-Butanediol is synthesized via a mixed-acid fermentation pathway, which is normally accompanied by the formation of various end-products depending on the microorganism and fermentation strategies [22,23]. Until now, little is known about the functional aspect of the metabolic regulation of 2,3-BD production. It is, however, hypothesized that 2,3-BD biosynthesis has an essential physiological importance including preventive acidification, contributing to cell regulation of the NADH/NAD+ ratio and storing carbon and energy for microbial growth [24]. The metabolic pathway for the synthesis of 2,3-BD from glucose in bacteria is shown in Fig. 2. The three main enzymes, namely α-acetolactate synthase, α-acetolactate decarboxylase and butanediol dehydrogenase involve in the production of 2,3-BD from pyruvate [5]. In Klebsiella, these enzymes are encoded by the budB, budA, and budC genes, respectively [25].
Fig. 2.
The metabolic pathways for 2,3-butanediol synthesis in bacteria (modified, based on Magee & Kosaric, 1987). 1, glycolysis and pentose phosphate pathway enzymes; 2, pyruvate kinase; 3, α-acetolactate synthase; 4,α-acetolactate decarboxylase; 5, acetoin reductase (2,3-butanediol dehydrogenase); 6, pyruvate–formate lyase; 7, phospho-transacetylase; 8, acetate kinase; 9, acetaldehyde dehydrogenase; 10, ethanol dehydrogenase; 11, actate dehydrogenase; 12, phosphoenolpyruvate decarboxylase ; 13, malate dehydrogenase; 14, fumarase; 15, succinate dehydrogenase.
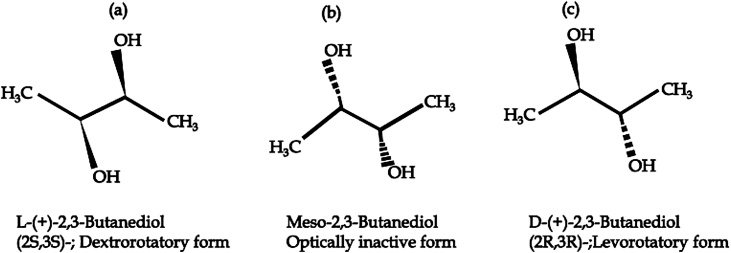
Alpha-acetolactate can spontaneously be converted to diacetyl, a reaction that requires the presence of oxygene. The generated diacetyl is subsequently transformed into acetoin at the expense of the reducing power. Depending on pH value and oxygen level, pyruvate also undegoes reduction or oxidation generating various organic acids and alcohols including lactate, formate, acetate, succinate, ethanol and acetoin. In order to minimize such unwanted byproducts, several approaches including mutant screening, genetic engineering and fermentation technology were suggested for enhancing 2,3-BD production [5,26]. In addition, pyruvate is transformed into acetolactate by two distinct enzymes. The first enzyme is catabolic α-acetolactate synthase, which has optimum pH value of 5.8 in acetate, and is part of the butanediol pathway. The second enzyme in the butanediol biosynthesis pathway is acetolactate decarboxylase, with an optimum pH value of 6.3, and it catalyzes the formation of acetoin via the decarboxylation of acetolactate. There is also a third enzyme named diacetyl (acetoin) reductase, which catalyzes a reversible reduction of acetoin to 2,3-BD and an irreversible reduction of diacetyl to acetoin. Diacetyl (acetoin) reductase is a tetrameric enzyme which uses NADH as a coenzyme. Different isomers of 2,3-BD are formed from acetoin due to activities of different acetoin reductase enzymes with different stereospecificities, or by a cyclic pathway. The three 2,3-BD stereoisomers are dextro- and levo- forms which are optically active, and an optically inactive meso-form (Fig. 3). These three 2,3-BD stereoisomers are obtained due to the activity of three enzymes, namely an acetoin racemase, L(+) 2,3-BD dehydrogenase and D(–) 2,3-BD dehydrogenase. The L(+) 2,3-BD dehydrogenase converts L(+) acetoin to L(+) 2,3-BD and meso-2,3-BD, while the D(–) 2,3-BD dehydrogenase reduces D(–) acetoin to D(–) 2,3-BD and meso-2,3-BD [27,28].
Fig. 3.
The stereoisomers of 2,3-butanediol. a: l-(+)-2,3-Butanediol (2S,3S)-;Dextrorotatory form. b: Meso-2,3-Butanediol; Optically inactive form. c: d-(+)-2,3-Butanediol (2R,3R)-;Levorotatory form.
Various mechanisms for different 2,3-BD stereoisomer formation were elaborated in K. pneumonia. Glycerol dehydrogenase was reported to exhibit 2R,3R-butanediol dehydrogenase activity and catalyzes conversion of both R-acetoin and S-acetoin into levo-butanediol and meso-2,3-butanediol, respectively. Butanediol dehydrogenase is also a single enzyme that catalyzes the synthesis of S-acetoin from diacetyl, and further to dextro-butanediol [29]. Though different strategies were proposed to produce the three stereoisomers of 2,3-BD (Table 1), it is still difficult to produce only one and single pure stereoisomer. Therefore, there is a need of modifiying various culture conditions including pH, temperature, and shaking speed in order to optimize the production of diffent isomers [27].
Table 1.
Some genes and pathways engineered to improve optical pure 2,3-BDO production.
| Organism | 2,3-BDO biosynthesis genes | Pathway | Precursor | Isomer | Titer (g/L) | Enantiopurity | Reference |
|---|---|---|---|---|---|---|---|
| Escherichia Coli | K. pneumoniae budA, budB, budC | − | Glucose | meso- | 17.7 | 98% | [107] |
| K. pneumoniae budA, budC | Conversion of R-acetoin to meso-butanediol | Glucose | meso- | 15.3 | 99% | [108] | |
| K. pneumoniae budA, budC, ydjL from Bacillus subtilis | No isopropyl beta-D-thiogalactoside (IPTG) addition | Glucose | (R,R) | 115 | 99% | [109] | |
| Enterobacter cloacae | knock out of bdh, ldh, and frdA, and inactivation of ptsG, and overexpression of (2R,3R) bdh and galP | Blocking pathways of conversion from glucose to meso -2,3-BD, and pyrivate to lactate, succinate, and (2S, 3S)-2,3-BD | Xylose and glucose | (R,R) | 152 | 98% | [101] |
| Pichia pastoris | B. subtilis alsS, alsD genes, and S.cerevisiaeBDH1 | Conversion of pyruvate to R-acetoin, then converted to (2R, 3R)-2,3-butanediol | Glucose | (R,R) | 74.5 | 99% | [55] |
| Bacillus subtilis |
K. pneumoniae budC, deletion of bdhA, pta and ldh,and knock out of acoA |
Conversion of D-(-) acetoin to meso-2,3-BD | Glucose | meso- | 103.7 | 99% | [110] |
| Bacillus licheniformis | Deletion of gldA | − | Miscanthus floridulus Hydrolysate |
meso- | 48.5 | Not tested | [111] |
In principle, different monosaccharides including hexoses and pentoses can be fermented to produce 2,3-BD. In this case, the key intermediate is pyruvate, and three enzymes α-acetolactate synthase, α-acetolactate decarboxylase, and 2,3-BD dehydrogenase are critical for 2,3-BD generation. However, besides 2,3-BD, some by-products are generally formed including ethanol, acetate, lactate, formate, and succinate, which seriously affect the fermentation yields.
3. Optimization of different parameters for the enhanced 2,3- BD production
Several researchers have focused on improving strains and operation mode to reach higher 2,3-BD yields. The 2,3-BD production varies depending on microorganisms, culture conditions and the substrate used as shown in the Table 2. The higher 2,3-BD production requires not only the improvement of culture medium composition but also the strain selection and the improvement of biosynthetic pathway by metabolic engineering [30]. The later revealed to be very effective especially by altering ethanol, lactate and acetate production routes [[31], [32], [33]]. For example, inactivation of acetaldehyde dehydrogenase, or elimination of lactate dehydrogenase showed higher yields of 2,3-BD by klebsiella sp. [17,34]. Cultivation strategies are now in various forms including fed-batch cultivations under growth-limiting conditions, batch cultivations under resting cells conditions, semi-continuous productions with immobilized cells, continuous cultivations and production with free cells, and solid state fermentations. Therefore, the choice of the correct cultivation methods is crucial for improved 2,3-BD production.
Table 2.
Comparison of 2,3-BD production by different microorganisms.
| Host species | Carbon sources | Culture mode | T(0C) | pH | Agitation (rpm) | Aeration (vvm) | 2,3-BD Titer (g/L) | 2,3-BD Productivity (g/L.H) | 2,3-BD yield (g/g) | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| Klebsiella oxytocia | Glucose | Fed batch | 30 | 6 | 200, 300, 400 | 1 | 142.5 | 1.47 | 0.42 | [12] |
| Crude glycerol | Fed batch | 30 | 6 | 400 | 1 | 131.5 | 0.84 | 0.44 | [94] | |
| Pure glycerol | Fed batch | 30 | 6 | 400 | 1 | 115 | 1.01 | 0.39 | [95] | |
| Glucose | Fed batch | 37 | 6 | 300 | 1 | 117.4 | 1.2 | 0.49 | [42] | |
| Glucose | Fed batch | 37 | 6.5 | 200 | 1 | 130 | 1.63 | 0.48 | [1] | |
| Corncob acid hydrolysate | Fed-batch | 37 | 6.3 | 300 | 0.3 | 35.7 | 0.59 | 0.5 | [11] | |
| Molasses | Cell recycle | 35 | 5.5 | 200, 150 | 0.5, 0.3 | 118 | 2.4 | − | [64] | |
| Klebsiella pneumoniae | Glucose | Fed-batch | 37 | 6 | 500 | 1.5 | 150 | 4.21 | − | [96] |
| Glucose | Fed batch | 35 | 6.5 | 150 | 1 | 116 | − | 0.49 | [97] | |
| Corncob molasses | Fed-batch | 37 | 6 | 500,400 | 1.0,0.5 | 78.9 | 1.3 | − | [81] | |
| Glycerol | Fed-batch | 37 | 8 | 200 | 0.44 | 49.2 | − | − | [14] | |
| Starch | Fed batch; SSF | 37 | 7 | 200 | − | 53.8 | − | − | [10] | |
| Paenibacillus polymyxa | Jerusalem artichoke tuber | Fed batch; SSF | 37 | 6 | 300 | 0.15 | 91.63 | 2.29 | 0.32 | [15] |
| Glucose | Fed batch | 35 | 6.5 | 300 | 0.075 | 68.54 | 0.7 | 0.34 | [15] | |
| Jerusalem artichoke tuber | Batch | 30 | 6 | 240,120 | − | 36.92 | 0.88 | − | [98] | |
| Bacillus amyloliquefaciens | Glucose | Fed batch | 37 | 6.5 | 350 | 0.33 | 132.9 | 2.95 | − | [46] |
| Bacillus licheniformis | Glucose | Fed batch | 50 | 7 | 500 | 1 | 123.7 | 2.95 | 0.508 | [99] |
|
Enterobacter aerogenes Enterobacter cloacae |
sugarcane molasses | Fed batch | 37 | 6 | 280 | 1.5 | 140 | − | − | [100] |
| Glucose | Fed batch | 37 | 6 | 150 | 1 | 118.05 | − | − | [33] | |
| Corn stoverhydroly-sate | Fed batch | 30 | 6 | 500 | 1 | 119.4 | 2.3 | − | [101] | |
| oil palm | SSF | 37 | 6.5 | 200 | − | 25.0l | 0.13 | − | [102] | |
| Serratia marcescens | Sucrose | Fed batch | 30 | 6 | 500 | 0.5 | 152 | 2.67 | − | [103] |
| Serratia marcescens | Sucrose | Fed batch | 30 | 6 | 200 | 0.5 | 139.92 | 3.49 | − | [104] |
| Saccharo-myces cerevisiae | Glucose | Fed batch | 30 | 5.5 | 300, 500 | 1 − 2 | 154.3 | 1.98 | 0.404 | [105] |
| Xylose | Fed batch | 30 | 5.5 | 200 | 0.5 | 96.8 | 0.58 | − | [52] | |
| Methylomicrobium alcaliphilum 20Z | Methane | Fed batch | 30 | 8.9 | 650 | – | 31.8 | 0.086 | 0.03 | [106] |
The modulation of fermentation parameters has a significant impact on 2,3-BD yield. Many studies have shown a negative correlation between oxygen concentration and 2,3-BD yields. Higher oxygen concentration leads to cell mass formation, while 2,3-BD formation is negatively affected. Briefly, 2,3-BD yield increases with reduction of oxygen content [35]. In such case, however, the overall conversion rate is hampered by the lower concentration of cells [36]. Other factors including culture temperature, pH, and acetic acid supplementation also involve in metabolic regulation [37]. Besides the effects of those culture conditions, genetic engineering has been importantly utilized for high production of 2,3-BD [38].
3.1. Metabolic engineering for high-producing strains
Recently, it has been possible to use genetic tools for converting microbes into efficient biofactories. However, to optimize the metabolic pathways for improved yields of the target product, one has to understand well metabolic characteristics [39,40]. New improved mutants can be obtained by mutation or altering some genes involved in 2,3-BD pathway, especially those requiring NADH for the formation of the byproducts [33,41,42]. In addition, new studies showed that by preventing byproduct synthesis and overexpressing genes associated with 2,3-BD synthesis resulted in increased yields of 2,3-BD [43]. For example, a short-chain of acyl dehydrogenase identified and characterized from K. pneumoniae was applied, and L-2,3-butanediol was produced at high-level. The L-2,3-BD dehydrogenase participates in the reduction of both diacetyl and acetoin, and in the oxidation of L-2,3-BD [44]. K. oxytoca GSC 12206 isolated from cattle was 2,3-BD native hyper-producing and nonpathogenic bacterium. The disruption of ldhA gene in this strain led to a mutant with minimal lactic acid formation while enhancing the 2,3-BD production up to 115 g/l of 2,3-BD, with 0.41 g/g and 2.27 g/l.h as yield and productivity, respectively [17].
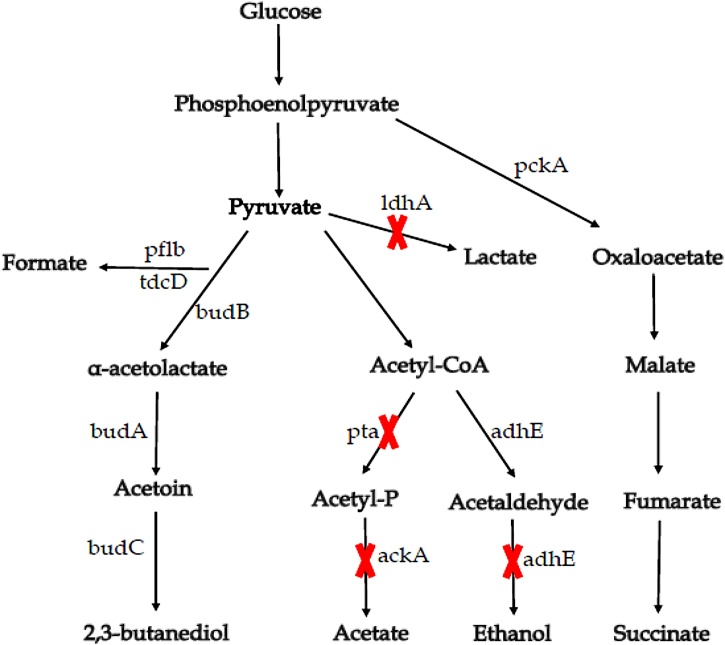
Similarly, alteration of both lactate dehydrogenase and phosphotransacetylase genes involved in lactic acid and acetic acid formation pathways in a wild type K. oxytoca ME-303 resulted in improved production of 2,3-BD. The generated mutant ME-UD-3, showed 7.8 % more yield compared to wild type, while byproducts namely lactic acid and acetic acid were also tremendously reduced up to 88 % and 92 %, respectively [45]. The same enzyme was mutated in K. pneumoniae, and the resulting lactate dehydrogenase-deficient mutant (ΔldhA) successfully reached 2,3-BD production of 148.8 g/l and productivity of 2.48 g/l.h using glucose as a substrate. By using sugarcane as a substrate, the production was 76.24 g/l, while the productivity was 2.31 g/l.h [38]. High production of 2,3-BD can also be reached by disrupting the ethanol biosynthesis pathway. In that regard, for constructing an ethanol-deficient mutant, the aldA gene coding for aldehyde dehydrogenase in K. oxytoca was replaced with a tetracycline resistance cassette. By minimizing the formation of acetoin and eliminating ethanol production, the production improved up to130 g/l with the productivity of 1.63 g/l.h and the yield of 2,3-BD relative to glucose of 0.48 g/g [1]. Moreover, the construction of Klebsiella oxytoca KMS005 mutant lacking three genes, namely alcohol dehydrogenase (adhE), acetate kinase-phosphotransacetylase (ackA) and lactate dehydrogenase (ldhA) boosted up to 2,3-BD productivity of 117.4 g/l, and 0.49 g/g yield, while acetoin, lactate and formic acid were not detected [42]. Further, that pathway was generally considered as metabolic pathway for 2,3-BD production in Klebsiella species with higher yield (Fig. 4).
Fig. 4.
Metabolic pathway for 2,3-BD biosynthesis in Klebsiella sp. (modified, based on Jantama et al., 2015). The crosses represent gene deletions for improving 2,3-BD production. Genes and enzymes: budB, acetolactate synthase; budA, acetolactate decarboxylase; budC, alcohol dehydrogenase; ldhA, lactate dehydrogenase; pflB, pyruvate formate-lyase; pta, phosphate acetyltransferase; ackA, acetate kinase; adhE, alcohol dehydrogenase; aldA, aldehyde dehydrogenase and tdcD, propionate kinase.
Besides Klebsiella species, several other microorganism have been proven to produce 2,3-BD efficiently using low-cost substrate. Bacillus amyloliquefaciens has been reported to produce 2,3-BD high titer of 102.3 g/l, yield of 0.44 g/g and 1.16 g/l.h of productivity due to the overexpression of NADH/NAD+ regeneration system, which increased 2,3-BD production and prevention of byproducts accumulation. The introduction of the transcriptional regulator ALsR under the control of a moderate promoter PbdhA facilitates carbon flux increasing to 2,3-BD branch [26]. Co-overexpression of bdH and gapA genes was also used to enhance 2,3-BD production and inhibit accumulation of unwanted byproducts. As a result, the production increased up to 132.9 g/l in B. amyloliquefaciens B10-127 in fed-batch fermentations [46]. In B. subtilis, the exchange of (2R, 3R)-butanediol dehydrogenase gene with butanediol dehydrogenase (encoded by K. pneumoniae budC) led to a new strain BSF9 producing 103.7 g/l pure meso-2-BD with 0.487 g/g yield when glucose is applied as the substrate [47]. In addition, B. licheniformis was identified as a safe microorganism for the production of 2, 3-BD, which can utilize hexose and pentose as carbon source. Moreover, it can ferment lignocellulose hydrolysate for production of 2,3-BD. For instance, using feed-batch fermentation two strains, WX-02ΔbudC and WX-02ΔgldA produced optically pure compounds with 32.2 g/l D-2,3-BD and 48.5 g/l meso-2,3-BD, respectively. Furthermore, these two strains could also utilize xylose as a carbon source to produce D-2,3-BD (10.9 g/l) and meso-2,3-BD (20.2 g/l), respectively [48].
To enhance 2,3-BD productivity in Serratia marcescens, meso-2,3-butanediol and (2S,3S)-2,3-butanediol decreased due to the inactivation of slaC gene, while an increased accumulation of (3R)-acetoin was observed. Introduction of bdhA gene from B. subtilis 168 encoding (2R,3R)-2,3-butanediol dehydrogenase into the slaC strain of S. marcescens MG1 led to excess (2R,3R)-2,3-butanediol dehydrogenase through acceleration of the reaction from (3R)-acetoin to (2R,3R)-2,3-butanediol, and caused amassing of (2R,3R)-2,3-BD. The highest production achieved was 89.81 g/l (2R,3R)-2,3-BD in fed-batch fermentation due to high expression of butanediol dehydrogenase [49]. Similarly, the overexpression of genes involved in 2,3- BD production in Raoultella ornithinolytica resulted in 68.27 g/l of 2,3- BD with pH control at 5.5, and agitation speed at 400 rpm. However, accumulation of acetic acid reached up to 23.32 g/l. In addition, the overexpression of homologous budABC genes improved the 2,3-BD production up to 112.19 g/l and 1.35 g/l.h productivity, whereas accumulation of acetic acid decreased to 9.71 g/l [50].
Recently, yeast was target in research as one of a promising host cell for 2,3-BD production, and various strategies have been crafted to enhance its production. In S. cerevisiae, pyruvate is a crucial precursor in 2,3- BD synthesis as it favors ethanol production at the expense of 2,3-BD. In recent study, S. cerevisiae was shown to produce 81.0 g/l 2,3-BD in a fed-batch fermentation strategy using glucose as a carbon source. For reaching such high concentration, pyruvate carbon flux tugging strategy was used to reduce dominant ethanol production, thus increased 2,3-BD production [51]. In another study, a high 2,3-BD production was achieved by modulating the xylose metabolic pathway in engineered S. cerevisiae. In fed-batch fermentation, engineered strain produced 2,3-BD titer of 96.8 g/l with 0.58 g/Lh of productivity using xylose as substrate [52]. On the other hand, it was reported that deletion of ADH genes, ADH1, ADH3 and ADH5 encoding major alcohol dehydrogenases in S. cerevisiae increased 2,3-butanediol production under aerobic condition and batch fermentation strategy. Nevertheless, to reach highest production, 2,3-BD biosynthetic pathway genes from both B. subtilis and E. aerogenes were introduced in the engineered strain, which lead to a high 2,3-BD titer of 2.29 g/l and yield of 0.113 g/g [53].
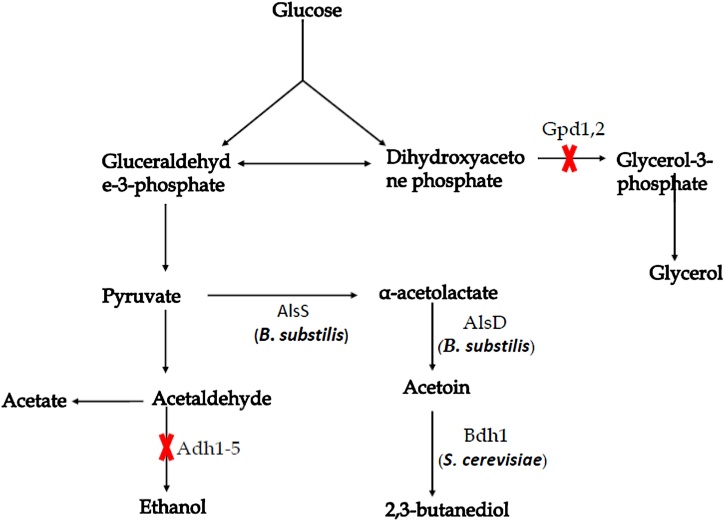
Furthemore, alsS and alsD genes from B. subtilis were introduced in S. cerevisiae and E. coli for higher 2,3-BD production. In this case, pyruvate is converted to α-acetolactate by alsS, and then α-acetolactate to acetoin by alsD, and finally acetoin is converted into 2,3-BD by butanediol dehydrogenase (Fig. 5). For effectively utilize the desired carbon source, glycerol accumulation was eliminated by deleting GPD1 and GPD2 genes. The production increased up to to 72.9 g/l with 84 % theoretical yield [54]. Generally, designed synthesis pathway (Fig. 5) was shown as successful 2,3-BD production pathway in yeast with higher productivity.
Fig. 5.
Metabolic pathway for 2,3-butanediol production in yeast (modified, based on S. Kim & Hahn, 2015). The genes and enzymes: α-acetolactate synthase (AlsS), α-acetolactate decarboxylase (AlsD), and 2,3-butanedioldehydrogenase(Bdh1). Five alcohol dehydrogenases (ADH1 to ADH5) and two glycerol-3-phosphate dehydrogenases (GPD1 and GPD2) were deleted for improving 2,3-BD production.
Beside S. cerevisiae, Pichia expression system was also attempted for 2,3-BD production by introducing into the genome alsS and alsD genes from B. subtilis and the BDH1 gene from S. cerevisiae. For this reason, and through statistical medium optimization, the resulted strain yielded 74.5 g/l [55].
Similarly, the 2,3-BD biosynthesis pathway was also imported in some bacterial strains. For instance, three genes from B. lincheniformis and B. subtilis coding for enzymes acetolactate synthase (Als), acetolactate decarboxylase (AldC), and butanediol dehydrogenase (Bdh) were introduced into Z. mobilis in order to redirect the carbon flux from ethanol to 2,3-BD production [56]. Much higher yields of 10 g/l 2,3-BD were produced not only from glucose and xylose, but also from mixed C6/C5 sugar streams derived from the deacetylation and mechanical refining process [56]. On the other hand, N. concharum can produce bulk chemicals using acetate, which is an abundant and renewable material. For that reason, the integration of acetoin and meso-2,3-buanediol biosynthesis pathway into N. concharum has resulted in high titer of 0.38 g/l for meso-2,3-buanediol [57]. Similarly, natural Clostridium acetobutylicum species are able to produced D- and L-stereoisomers of acetoin but they can not convert it into 2,3-butanediol due to the absence of acetoin reductase (ACR). ACR thus was introduced and overexpressed in Clostridium acetobutylicum ATCC 824. The engineered strains were able to convert nearly 90 % of the natively produced acetoin into 22 mM D-2,3-butanediol [58].
Overall, much progress in genetic engineering was made for improving the microbial production of 2,3-BD. Efforts were mostly focused on overexpression of 2,3-BD pathway genes, deletion of competing pathway genes or introducing the whole biosynthesis pathway in new organisms. Additionally, in silico simulation based on genomic scale metabolic model was also tried for better 2,3-BD yields. Despites all the efforts, there is still a need for establishing excellent strains with improved growth and productivity while using low-cost materials for fermentation.
3.2. pH optimization
pH of culture medium is a crucial factor that influences microbial production of 2,3-BD as it affects the distribution of metabolites in fermentation for various microorganisms and substrates [59]. It has been reported that there is a negative correlation between 2,3-BD productivity and pH of the cultivation broth. 2,3-BD is generally the main product at pH range 5.0–6.5, whilet it is lactic acid at pH 7.1 – 8.0. This means that the metabolic route can be changed due to the pH level, and hence controlling the cultivation broth at appropriate pH range may increase the efficiency of 2,3-BD production.
Many studies have pointed out that the production of 2,3-BD occurs in the pH range of 4.5–9.0 with optimal pH value of 6.0 [21,59]. However, the optimal pH for the production of 2,3-BD depends on strains and substrates used. In Klebsiella oxytoca, a number of studies have shown that the production of 2,3-BD and other organic acids is affected at pH 5.5. The activities of enzymes for 2,3-BD producing flux (Als) increased at pH 5.5 than at pH 7.0. However, at pH 7.0, the enzymes involved in formation of other organic acids pathway (lactate dehydrogenease, acetate kinase) showed high activities compared to pH 5.5 [59]. For Klebsiella sp. Zmd30 strain, the optimum pH was 6.0, which could be associated with cell growth. In batch fermentation, the production of 2,3-BD was 57.17 g/l, while fed-batch operation produced 110 g/l with a yield of 94 % [21]. Fed-batch operation might be the most suitable for the profit-oriented 2,3-BD production as showed by the several studies. However, another study using indigenous Klebsiella sp. Ana-WS5 in batch culture with pH controlled at 7.0 yielded the highest productivity of 0.86 g/l.h (PD + BD) and the highest PD/BD ratio of 7.67 [60].
Using glycerol and forced pH fluctuations method, the highest yield was 70 g/l, while in a previous study only 52.5 g/l 2,3-BD had been produced without pH control. Therefore, the forced pH fluctuations highlighted pH to be a factor which determines microbial conversion processes, and can enhance 2,3-BD production [61,62]. Similarily, the pH and acetic acid were reported to affect both the growth of Enterobacter aerogenes and 2,3-butanediol production from glucose.
In general, from many reports, the pH ranging from 5.0–7.0 may be tolerated by many cells and may lead to the maximum or nearly constant concentration of 2,3-BD [63].
3.3. Choice of cultivation strategies
Designing a good cultivation strategy is an important key aspect for improving 2,3BD production. It involves the control of substrate concentration and assessment of feeding strategies, regardless of batch or fed-batch [11]. Batch processes usually lead to low productivity, while substrate inhibition can easily occur. To solve this problem, constant addition of substrate such as glucose or molasses at appropriate rates during the cultivation via fed-batch operation is preferred. This results in high titers of 2,3-BD and greatly reduces the effects of initial substrate inhibition [21]. A double-fed batch approach under aerobic conditions was also developed for K. pneumonia. In addition, fed-batch strategies utilized by K. pneumoniae includes pulse, constant feed rate, constant residual glucose concentration, and exponential fed-batch. Constant residual glucose concentration feeding strategy showed a high titer of 150 g/l 2,3-BD. The continuous culture can significantly improve the productivity of 2,3-BD, nonetherless, it was reported that the productivity is ordinarily lower as considerable amounts of sugars still remains in the product stream, and consistently hampering the 2,3-BD formation [5].
In fed-batch, when the feeding is not timely, bacteria may reuse 2,3-BD as a carbon source for cell growth. To counteract this situation, there was a development of an improved strategy of pH-stat fed-batch culture by which glucose and sodium hydrate fed in glucose level controlled through an automatic pH adjustment. This pH-stat fed-batch strategy was used for enhancing 2,3-BD productivity by K. oxytoca that resulted in highly improved productivity up to 127.9 g/l, 1.78 g/l.h, and 0.48 g/g (2,3-BD/glucose) for 2,3 BD productivity and yield, respectively [62]. On the other hand, using test molasses, repeated batch culture with cell recovery resulted 2,3-BD of 118 g/l [64]. Using batch fermentation, E. aerogenes SUMI014 strain produced 93.75 g/l, with 0.49 g/g of yield and 1.74 g/l.h of productivity. Fed-batch fermentation with acetate addition strategy increased the 2,3-BD production up to 126.10 g/l, 0.38 g/g yield and 2.10 g/l.h of productivity [65]. Paenibacillus polymyxa achieved maximum 2,3-BD production through optimization of fermentation medium and conditions in a fed-batch fermentation. The toxic threshold of 2,3-BD production was found to be 50 g/l in a non-optimized medium, while in optimum conditions, it reached 68.54 g/l. The fed-batch fermentation showed higher productivity because excess glucose are used, thus helping both cell maintenance and growth. 2,3-BD productivity provided by optimum condition in fed-batch fermentations was 68.54 g/l of titer, 0.34 g/g of yield and 0.70 g/l.h of productivity [66]. Notably, the substrate concentration is also important, and must be measured and monitored.
From the experiments, it has proven that glucose generates much more 2,3 BD. In exponential fed-batch fermentation, the nutrient feeding rate was determined by the equation:
Where F is the feeding rate; μ is the specific growth rate; (VX)0 is the biomass at start of feed; t is culture time; YX/S is the theoretical cell yield on substrate; Si and S are substrate concentrations in the feeding solution and in the reactor, respectively [67]. Despite the usual low productivity, for evaluating new processes, batch reactors are still used for small-scale operations.
3.4. Aeration and agitation
3.4.1. Aeration
The regulation of oxygen supply is a crucial variable in the 2,3-BD fermentation. Despite that Klebsiella sp. are facultative anaerobes, 2,3-BD production occurs under lower oxygen condition which maintains an internal redox balance with respect to the pyridine nucleotide pool during glycolysis and biosynthesis [68]. The regeneration of NADH from glycolysis occurs by butanediol dehydrogenase in a reversible reaction: acetoin ⇌ 2,3-BD. As a result, the relative formation of acetoin to 2,3-BD could maintain NAD+/NADH balance [69]. It was reported that under an aerobic conditions, the α-acetolactate synthase is rapidly and irreversibly inactivated. 2,3-BD production is therefore prevented under a higher O2 supply [70], while aeration especially microaeration increases its production [71]. Particularly, Klebsiella species can obtain energy by respiration and fermentation processes, both pathways are simultaneously activated when a limited O2 is supplied, therefore, 2,3-BD production is affected by the relative activities of each pathway. By lowering O2 supply, however, cell mass is also reduced, and subsquently decreased 2,3-BD yield [37]. The aerobic culture of K. oxytoca showed the highest cell concentrations and greatest 2,3-butanediol manufacturing in comparison to the anaerobic culture conditions. The adequate aeration-agitation was utilized for partial suppression of the concentration of butanediol inhibitors such as ethanol and lactic acid. However, excessive aeration-agitation may generate acetoin and acetic acid at the expense of butanediol [72].
The supply of very low oxygen was shown as the cause of suppression of cell growth and decrease in diol/acetoin formation rates [73]. In fed-batch fermentation, Enterobacter species produced 69.5 g/l of 2,3-BD, while E. aerogenes produced 118.05 g/l by optimizing conditions for both medium and aeration [74]. In addition, B. licheniformis ATCC9789 was identified for high 2,3-BD production due to optimized aeration conditions. The optimal oxygen transfer rate ranging from 7 to 15 mmol/l.h proved to increase 2,3-BD yield of 0.44 g/g and productivity of 0.91 g/l.h [75]. Generally, 2,3-BD is produced under low O2 supply, and thus, it is very important to establish a proper oxygen supply control method to ensure optimum 2,3-BD production.
3.4.2. Agitation of the fermentation medium
The agitation speed is essential for 2,3-BD fermentation as it promotes the fermentation yield. For instance, a higher production of 2,3-BD was 89.9 g/l using the optimum speed in the range of 200 rpm and 300, while agitation speeds out of the the optimum range resulted in reduced 2,3-BD concentration yielding only 78.5 g/l at 400 rpm, and 79.4 g /l at 100 rpm. Such change may be caused by high accumulation of ethanol and acetoin as the main byproducts. Hence, the control of a constant agitation speed throughout the whole culture process could not attain high concentration, high yield and high productivity of 2,3-BD concurrently [76]. On the other hand, two-stage agitation speed control strategy for 2,3-BD fermentation may improve 2,3-BD production. For instance, the technique led to higher titers of 95.5 g/ l 2,3-BD, with 22.14 % increase in productivity. Therefore, two-stage agitation speed control strategy may be applied for enough oxygen supply and improved 2,3-BD production [35,76].
The investigation carried out on agitation speed optimization for high production of 2,3-BD by utilizing K. oxytoca M1 showed that the increasing of agitation speeds caused higher glucose consumption as well as higher dry weight and 2,3-BD concentration. However, 2,3-BD yield reduced with the lower agitation speed. Furthermore, proper agitation speeds led to 78.8 g/l, 102.1 g/l, 105.1 g/l at 200 rpm, 300 rpm, 400 rpm, respectively [12]. In an other study where K. oxytoca was used, the increased agitation speed from 150 to 450 rpm showed a significant productivity change from 0.43 to 2.7 g/l.h at 450 rpm, while favoring cell growth [77]. In addition, the agitation speeds examined from 200 rpm to 500 rpm in E. coli showed that both biomass and 2,3-BD production increased with agitation speed up to 400 rpm and decreased thereafter. At 200 rpm and 400 rpm, the productivity of 2,3 BD increased from 0.51 g/l.h to 1.48 g/l.h, while yield increased from 0.38 g/g to 0.46 g/g, respectively [78]. Three agitation speeds (200 rpm, 300 rpm and 400 rpm) examined for Pichia pastoris showed that the highest productivity of 8.34 g/l was achieved at 300 rpm [55]. Considering the above results, optimum agitation speed is essential for efficient 2,3-BD production, and it is mostly dependent on the strain used for fermentation.
3.5. Optimization of the temperature
During fermentation, the regulation of temperature is very important since enzymatic activity and cellular maintenance depend on temperature. The optimum temperature range for bacterial fermentation is controlled between 30−37 °C, which is the range for maximum biomass production [70]. At this temperature range, desired fermentation product can be produced alternatively to biomass due to changing from aerobic to microaerobic condition. Above the optimum temperature, cells and enzymes are altered, and consequently rendering the metabolism and 2,3-butanediol minimal. Suboptimal temperature may also fail the regulation and metabolic rate. It was found that in K. pneumoniae cultures, a temperature decrease from 35 °C to 30 °C not only provoked a considerable reduction in ethanol production in favor of 2,3-BD synthesis, but also prevented byproducts formation. The optimum temperature for K. pneumoniae is thought to be 33 °C under applied condition, but temperature fractuations slightly affect 2,3-BD synthesis [37]. On the other side, a temperature of 39 °C was found to be optimal for E. aerogenes [68], whereas for Paenibacillus polymyxa, 30 °C was reported as the optimum temperature in batch and fed-batch cultivation [79]. Nevertheless, the optimum temperature greatly depends on the strain and substrate used for cultivation. Thus, the optimal value should be determined individually for each strain and substrate utilized [37].
3.6. Substrates
The production of 2,3-BD using glucose and sucrose as substrates in several studies may not economically be viable. To minimize the production cost, inexpensive and renewable substrates are used [80], notably from inexpensive resources including lignocellulosic biomass, corncob molasses,corncob acid hydrolysate [11,81] and Jerusalem artichoke tuber hydrolysate [15]. K. oxytoca is one of the strains having large substate spectrum for fermentation including molasses, enzymatically hydrolyzed cereal mashes, acid hydrolyzed starch and wheat, wood hydrolysates and sulfite waste liquor.
The pretreatment method of raw material before fermentation is used in order to obtain 90 % of cellulose, which is further hydrolyzed and utilized as a substrate in 2,3-BD production [37]. K. oxytoca can utilize both pentoses and hexoses, and that is practically considered as essential since hydrolysate from biomass materials can have pentose/glucose ratios of 1:1.5. Interestingly, 2,3-BD can be obtained by conversion of nearly all of the sugars present in hemicellulose and cellulose hydrolysates [82]. The productivity of 2,3-BD was improved by increasing succinic acid from 0 g/l to 30 g/l, while 10 g/l led to the best results [83]. Enterobacter cloacae subsp. dissolvens SDM could also utilize inexpensive materials such as crop-biomass cassava powder to produce 93.9 g/l 2,3-BD using simultaneous saccharification and fermentation strategies [84]. Briefly, a variety of substrates have been attempted as carbon source (Table 1 and Table 2). However, there is still a pressing need to find better strains that are both capable of utilizing low cost substrates and producing higher 2,3-BD yields.
4. 2, 3-Butanediol recovery strategies
The methods used for separation of 2,3-BD include steam stripping, solvent extraction, reverse osmosis and pervaporation [85]. A countercurrent steam stripping has been formerly developed for recovery of 2,3-BD from whole fermentation broths at pilot plant. However, the excess energy and work are required to accomplish such process comes with extra production costs [7,86]. In particular, processing cost can slightly be reduced by integrated methods of reverse osmosis and distillation. The integrated method is inexpensive compared to combination of distillation and extraction which utilizes tributyl phosphate as extractant. Liquid–liquid extraction has been greatly attractive, and it involves for instance in a solvent extraction and aqueous two-phase extraction of 2,3-BD in PEG/dextran system [7]. Liquid -liquid equilibrium data was also used to separate 2,3-butanediol from aqueous streams in the presence of tetraoctyl ammonium 2-methyl-1-naphthoate. Organic solvents including alcohols and esters, especially (ethyl acetate, tributylphosphate, diethyl ether, n-butanol, dodecanol, and oleyl alcohol served for the extraction of 2,3-BD [87]. The investigation was carried out for recovery of 2,3-BD by repulsive extraction or salting out by potassium chloride (KCl) or dehydrated K2CO3. In fact, K2CO is affected by salting-out on butanol extraction in acetone-butanol-ethanol fermentation.
It is also necessary to remove water from fermentation broth before salting out because 2,3-BD showing lowest concentration in the broth can be salted out even at a saturated KCl or K2CO3 solution. Formal produced under acidic catalysis is a reactive extraction of 1,3-propanediol, and 2,3-butanediol which can make reaction with formaldehyde. 2,3-BD and methylal were formed from the reaction of acid methanol and 2,3-butanediol formal collected in the top oil phase. The hydrolysis of methylal can produce methanol and formaldehyde [7]. Some methods like anionic extraction method were shown as convenient for 2,3-BD recovery based on a reversible complexation with phenylboronates into an anionic complex. In addition, 2,3-BD can be recovered by back-extraction into an acidic solution. Extraction and back-extraction conditions were optimized utilizing commercial 2,3-butanediol and finally applied to different fermentation broths. Under optimum conditions, up to 72–93% 2,3-butanediol can be extracted, while 80–90% may be recovered using the same strategy. However, other constituents including glycerol or glucose can be co-extracted with 2,3-BD from the broth during the process [88].
Different parameters to enhance the production of 2,3-BD have been optimized and various purification technologies have been developed [7]. However, it is still challenging to develop an efficient and economically sustainable downstream process in 2,3-BD purification from its fermentation broth. This is mainly due to its low concentration, great affinity to water molecules, and the presence of other dissolved components of the fermentation mash [89].
5. Techno-economic analysis for 2,3-butanediol production
Techno-economic evaluation of diverse technologies for biofuel production is a crucial step for decision making in the development of bio-economy. Techno economic assessment (TEA) should be carried out to define the feasibility of the biorefineries at large scale. Many researchers have pointed out that the feedstock cost and the technology used are critical factors influencing the conversion of biomass into biofuels [90]. However, only few reports are available on TEA of 2,3-BD production. Koutinas and others (2016) showed that it is possible to establish an industrial production of 2,3-BD, but also highlighted its elevated minimum selling price (MSP) associated with higher costs of raw materials and insufficient productivity [91]. To improve this process, three carbon sources namely glycerol, sucrose and sugarcane molasses were used in fermentation while using reported experimental data and downstream separation [92,93]. The TEA on this experimental test showed that estimated MPS was higher than 1 $/kg [91]. Maina and others (2019) have also tried to optimize bioreactor conditions for higher BD production in a fed-batch culture using Enterobacter ludwigii to ferment very high polarity (VHP) cane sugar. By maintaining the culture at 33.9 °C and pH 6.3, higher yield (0.37 g/g) and productivity (3.95 g/ L. h) of 2,3-BD values were obtained. However, in a such fermentation system, the estimated prices rise as high as $2.67/kg for a projected annual production of 50,000 tones. In both TEA cases, MPS was higher than $ 1.00/kg. The main factors include high costs of fermentation media, separation and purification process, fermentation efficiency, and the specific growth conditions required by the strain used in the bioprocess. For this reason, there is still a need for high producing strains capable of using inexpensive and easily accessible substrates so that 2,3-BD becomes an affordable platform chemical.
6. Conclusion
Microbial production of 2,3-BD is very promising because of the recent advances in genetic engineering and fermentation strategies. Efficient biological production systems could certainly solve the scarcity of synthetic chemicals and fossil fuels. However, 2,3-BD production is still hampered by low yields and high production costs associated with the use of expensive materials such as glucose. Therefore, there is a need for developing and screening safe and high producing microorganisms, which are capable of using low-cost substrates for improved yields. Alternatively, 2,3BD production could promoted by eliminating byproducts synthesis, especially ethanol, lactate and acetate. Most importantly, more efforts should focus on the optimization of fermentation parameters including culture medium, pH, pressure, temperature, and oxygen supply, as well as the design of good bioreactors. In summary, the enhanced production of 2,3-BD relies on rigorous works, especially genetic and metabolic engineering as well as improved fermentation conditions.
Author contributions
O.H., E.M., and B. H. L. wrote the manuscript and approved the final manuscript.
Declaration of Competing Interest
The authors declare no conflict of interest.
Contributor Information
Olivier Hakizimana, Email: olha.7478@yahoo.com.
Emmanuel Matabaro, Email: ematabaro@ethz.ch.
Byong H. Lee, Email: byong.lee@mail.mcgill.ca.
References
- 1.Ji X.J., Huang H., Zhu J.G., Ren L.J., Nie Z.K., Du J., Li S. Engineering Klebsiella oxytoca for efficient 2, 3-butanediol production through insertional inactivation of acetaldehyde dehydrogenase gene. Appl. Microbiol. Biotechnol. 2010;85:1751–1758. doi: 10.1007/s00253-009-2222-2. [DOI] [PubMed] [Google Scholar]
- 2.Clomburg J.M., Gonzalez R. Biofuel production in Escherichia coli: the role of metabolic engineering and synthetic biology. Appl. Microbiol. Biotechnol. 2010;86:419–434. doi: 10.1007/s00253-010-2446-1. [DOI] [PubMed] [Google Scholar]
- 3.Kiran B., Kumar R., Deshmukh D. Perspectives of microalgal biofuels as a renewable source of energy. Energy Convers. Manage. 2014;88:1228–1244. [Google Scholar]
- 4.Nigam P.S., Singh A. Production of liquid biofuels from renewable resources. Prog. Energy Combust. Sci. 2011;37:52–68. [Google Scholar]
- 5.Ji X.J., Huang H., Ouyang P.K. Microbial 2,3-butanediol production: a state-of-the-art review. Biotechnol. Adv. 2011;29:351–364. doi: 10.1016/j.biotechadv.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 6.Kim T., Cho S., Lee S.M., Woo H.M., Lee J., Um Y., Seo J.H. High production of 2,3-Butanediol (2,3-BD) by Raoultella ornithinolytica B6 via optimizing fermentation conditions and overexpressing 2,3-BD synthesis genes. PLoS One. 2016;11 doi: 10.1371/journal.pone.0165076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiu Z.L., Zeng A.P. Present state and perspective of downstream processing of biologically produced 1,3-propanediol and 2,3-butanediol. Appl. Microbiol. Biotechnol. 2008;78:917–926. doi: 10.1007/s00253-008-1387-4. [DOI] [PubMed] [Google Scholar]
- 8.Qin J., Xiao Z., Ma C., Xie N., Liu P., Xu P. Production of 2,3-Butanediol by Klebsiella pneumoniae using glucose and ammonium phosphate. Chin. J. Chem. Eng. 2006;14:132–136. [Google Scholar]
- 9.Anvari M., Safari Motlagh M.R. Enhancement of 2,3-butanediol production by Klebsiella oxytoca PTCC 1402. J. Biomed. Biotechnol. 2011;2011 doi: 10.1155/2011/636170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsvetanova F., Petrova P., Petrov K. 2,3-butanediol production from starch by engineered Klebsiella pneumoniae G31-A. Appl. Microbiol. Biotechnol. 2014;98:2441–2451. doi: 10.1007/s00253-013-5418-4. [DOI] [PubMed] [Google Scholar]
- 11.Cheng K.K., Liu Q., Zhang J.A., Li J.P., Xu J.M., Wang G.H. Improved 2,3-butanediol production from corncob acid hydrolysate by fed-batch fermentation using Klebsiella oxytoca. Process. Biochem. 2010;45:613–616. [Google Scholar]
- 12.Cho S., Kim T., Woo H.M., Lee J., Kim Y., Um Y. Enhanced 2,3-butanediol production by optimizing fermentation conditions and engineering Klebsiella oxytoca M1 through overexpression of acetoin reductase. PLoS One. 2015;10 doi: 10.1371/journal.pone.0138109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moon S.K., Kim D.K., Park J.M., Min J., Song H. Development of a semi-continuous two-stage simultaneous saccharification and fermentation process for enhanced 2,3-butanediol production by Klebsiella oxytoca. Lett. Appl. Microbiol. 2018;66:300–305. doi: 10.1111/lam.12845. [DOI] [PubMed] [Google Scholar]
- 14.Petrov K., Petrova P. High production of 2,3-butanediol from glycerol by Klebsiella pneumoniae G31. Appl. Microbiol. Biotechnol. 2009;84:659–665. doi: 10.1007/s00253-009-2004-x. [DOI] [PubMed] [Google Scholar]
- 15.Sun L.H., Wang X.D., Dai J.Y., Xiu Z.L. Microbial production of 2,3-butanediol from Jerusalem artichoke tubers by Klebsiella pneumoniae. Appl. Microbiol. Biotechnol. 2009;82:847–852. doi: 10.1007/s00253-008-1823-5. [DOI] [PubMed] [Google Scholar]
- 16.Jiang Lq., Fang Z., Li X.K., Luo J. Production of 2,3-butanediol from cellulose and Jatropha hulls after ionic liquid pretreatment and dilute-acid hydrolysis. AMB Express. 2013;3:48. doi: 10.1186/2191-0855-3-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim D.K., Rathnasingh C., Song H., Lee H.J., Seung D., Chang Y.K. Metabolic engineering of a novel Klebsiella oxytoca strain for enhanced 2,3-butanediol production. J. Biosci. Bioeng. 2013;116:186–192. doi: 10.1016/j.jbiosc.2013.02.021. [DOI] [PubMed] [Google Scholar]
- 18.Yang T., Rao Z., Zhang X., Xu M., Xu Z., Yang S.T. Metabolic engineering strategies for acetoin and 2,3-butanediol production: advances and prospects. Crit. Rev. Biotechnol. 2017;37:990–1005. doi: 10.1080/07388551.2017.1299680. [DOI] [PubMed] [Google Scholar]
- 19.Savaliya M.L., Dhorajiya B.D., Dholakiya B.Z. RETRACTED ARTICLE: recent advancement in production of liquid biofuels from renewable resources: a review. Res. Chem. Intermed. 2015;41:475–509. [Google Scholar]
- 20.Brennan L., Owende P. Biofuels from microalgae—a review of technologies for production, processing, and extractions of biofuels and co-products. Renew. Sustain. Energy Rev. 2010;14:557–577. [Google Scholar]
- 21.Wong C.L., Yen H.W., Lin C.L., Chang J.S. Effects of pH and fermentation strategies on 2,3-butanediol production with an isolated Klebsiella sp. Zmd30 strain. Bioresour. Technol. 2014;152:169–176. doi: 10.1016/j.biortech.2013.10.101. [DOI] [PubMed] [Google Scholar]
- 22.Sabra W., Groeger C., Zeng A.P. Microbial cell factories for diol production. Adv. Biochem. Eng. Biotechnol. 2016;155:165–197. doi: 10.1007/10_2015_330. [DOI] [PubMed] [Google Scholar]
- 23.Shi L., Gao S., Yu Y., Yang H. Microbial production of 2,3-butanediol by a newly-isolated strain of Serratia marcescens. Biotechnol. Lett. 2014;36:969–973. doi: 10.1007/s10529-013-1433-x. [DOI] [PubMed] [Google Scholar]
- 24.Van Houdt R., Aertsen A., Michiels C.W. Quorum-sensing-dependent switch to butanediol fermentation prevents lethal medium acidification in Aeromonas hydrophila AH-1N. Res. Microbiol. 2007;158:379–385. doi: 10.1016/j.resmic.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 25.Bialkowska A.M. Strategies for efficient and economical 2,3-butanediol production: new trends in this field. World J. Microbiol. Biotechnol. 2016;32:200. doi: 10.1007/s11274-016-2161-x. [DOI] [PubMed] [Google Scholar]
- 26.Yang T., Rao Z., Zhang X., Xu M., Xu Z., Yang S.-T. Enhanced 2,3-butanediol production from biodiesel-derived glycerol by engineering of cofactor regeneration and manipulating carbon flux in Bacillus amyloliquefaciens. Microb. Cell Fact. 2015;14:122. doi: 10.1186/s12934-015-0317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen J.S., Zidwick M.J., Rogers P. Organic acid And solvent production: butanol, acetone, And isopropanol; 1,3- And 1,2-propanediol production; And 2,3-butanediol production. In: Rosenberg E., editor. The Prokaryotes: Applied Bacteriology and Biotechnology. Springer; Berlin Heidelberg, Berlin, Heidelberg: 2013. pp. 77–134. [Google Scholar]
- 28.Zhang L., Guo Z., Chen J., Xu Q., Lin H., Hu K., Guan X., Shen Y. Mechanism of 2,3-butanediol stereoisomers formation in a newly isolated Serratia sp. T241, Sci Rep. 2016;6:19257. doi: 10.1038/srep19257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen C., Wei D., Shi J., Wang M., Hao J. Mechanism of 2,3-butanediol stereoisomer formation in Klebsiella pneumoniae. Appl. Microbiol. Biotechnol. 2014;98:4603–4613. doi: 10.1007/s00253-014-5526-9. [DOI] [PubMed] [Google Scholar]
- 30.Li L., Zhang L., Li K., Wang Y., Gao C., Han B., Ma C., Xu P. A newly isolated Bacillus licheniformis strain thermophilically produces 2,3-butanediol, a platform and fuel bio-chemical. Biotechnol. Biofuels. 2013;6 doi: 10.1186/1754-6834-6-123. 123-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho C., Choi S.Y., Luo Z.W., Lee S.Y. Recent advances in microbial production of fuels and chemicals using tools and strategies of systems metabolic engineering. Biotechnol. Adv. 2015;33:1455–1466. doi: 10.1016/j.biotechadv.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 32.Shen X., Lin Y., Jain R., Yuan Q., Yan Y. Inhibition of acetate accumulation leads to enhanced production of (R,R)-2,3-butanediol from glycerol in Escherichia coli. J. Ind. Microbiol. Biotechnol. 2012;39:1725–1729. doi: 10.1007/s10295-012-1171-4. [DOI] [PubMed] [Google Scholar]
- 33.Jung M.Y., Ng C.Y., Song H., Lee J., Oh M.K. Deletion of lactate dehydrogenase in Enterobacter aerogenes to enhance 2,3-butanediol production. Appl. Microbiol. Biotechnol. 2012;95:461–469. doi: 10.1007/s00253-012-3883-9. [DOI] [PubMed] [Google Scholar]
- 34.Guo X., Cao C., Wang Y., Li C., Wu M., Chen Y., Zhang C., Pei H., Xiao D. Effect of the inactivation of lactate dehydrogenase, ethanol dehydrogenase, and phosphotransacetylase on 2,3-butanediol production in Klebsiella pneumoniae strain. Biotechnol. Biofuels. 2014;7:44. doi: 10.1186/1754-6834-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan S., Jantama S.S., Kanchanatawee S., Jantama K. Process optimization on micro-aeration supply for high production yield of 2,3-Butanediol from maltodextrin by metabolically-engineered Klebsiella oxytoca. PLoS One. 2016;11 doi: 10.1371/journal.pone.0161503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ji X.J., Huang H., Du J., Zhu J.G., Ren L.J., Hu N., Li S. Enhanced 2,3-butanediol production by Klebsiella oxytoca using a two-stage agitation speed control strategy. Bioresour. Technol. 2009;100:3410–3414. doi: 10.1016/j.biortech.2009.02.031. [DOI] [PubMed] [Google Scholar]
- 37.Celińska E., Grajek W. Biotechnological production of 2,3-butanediol—current state and prospects. Biotechnol. Adv. 2009;27:715–725. doi: 10.1016/j.biotechadv.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 38.Lee S.M., Oh B.R., Park J.M., Yu A., Heo S.Y., Hong W.K., Seo J.W., Kim C.H. Optimized production of 2,3-butanediol by a lactate dehydrogenase-deficient mutant of Klebsiella pneumoniae. Biotechnol. Bioprocess Eng. 2013;18:1210–1215. [Google Scholar]
- 39.Lee J.W., Na D., Park J.M., Lee J., Choi S., Lee S.Y. Systems metabolic engineering of microorganisms for natural and non-natural chemicals. Nat. Chem. Biol. 2012;8:536–546. doi: 10.1038/nchembio.970. [DOI] [PubMed] [Google Scholar]
- 40.Park J.M., Song H., Lee H.J., Seung D. Genome-scale reconstruction and in silico analysis of Klebsiella oxytoca for 2,3-butanediol production. Microb. Cell Fact. 2013;12:20. doi: 10.1186/1475-2859-12-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park J.M., Song H., Lee H.J., Seung D. In silico aided metabolic engineering of Klebsiella oxytoca and fermentation optimization for enhanced 2,3-butanediol production. J. Ind. Microbiol. Biotechnol. 2013;40:1057–1066. doi: 10.1007/s10295-013-1298-y. [DOI] [PubMed] [Google Scholar]
- 42.Jantama K., Polyiam P., Khunnonkwao P., Chan S., Sangproo M., Khor K., Jantama S.S., Kanchanatawee S. Efficient reduction of the formation of by-products and improvement of production yield of 2,3-butanediol by a combined deletion of alcohol dehydrogenase, acetate kinase-phosphotransacetylase, and lactate dehydrogenase genes in metabolically engineered Klebsiella oxytoca in mineral salts medium. Metab. Eng. 2015;30:16–26. doi: 10.1016/j.ymben.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 43.Kim B., Lee S., Park J., Lu M., Oh M., Kim Y., Lee J. Enhanced 2,3-butanediol production in recombinant Klebsiella pneumoniae via overexpression of synthesis-related genes. J. Microbiol. Biotechnol. 2012;22:1258–1263. doi: 10.4014/jmb.1201.01044. [DOI] [PubMed] [Google Scholar]
- 44.Park J.M., Hong W.K., Lee S.M., Heo S.Y., Jung Y.R., Kang I.Y., Oh B.R., Seo J.W., Kim C.H. Identification and characterization of a short-chain acyl dehydrogenase from Klebsiella pneumoniae and its application for high-level production of L-2,3-butanediol. J. Ind. Microbiol. Biotechnol. 2014;41:1425–1433. doi: 10.1007/s10295-014-1483-7. [DOI] [PubMed] [Google Scholar]
- 45.Ji X.J., Huang H., Li S., Du J., Lian M. Enhanced 2,3-butanediol production by altering the mixed acid fermentation pathway in Klebsiella oxytoca. Biotechnol. Lett. 2008;30:731–734. doi: 10.1007/s10529-007-9599-8. [DOI] [PubMed] [Google Scholar]
- 46.Yang T., Rao Z., Zhang X., Xu M., Xu Z., Yang S.T. Improved production of 2,3-butanediol in Bacillus amyloliquefaciens by over-expression of glyceraldehyde-3-phosphate dehydrogenase and 2,3-butanediol dehydrogenase. PLoS One. 2013;8 doi: 10.1371/journal.pone.0076149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fu J., Huo G., Feng L., Mao Y., Wang Z., Ma H., Chen T., Zhao X. Metabolic engineering of Bacillus subtilis for chiral pure meso-2,3-butanediol production. Biotechnol. Biofuels. 2016;9:90. doi: 10.1186/s13068-016-0502-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao Y., Huang H., Chen S., Qi G. Production of optically pure 2,3-butanediol from Miscanthus floridulus hydrolysate using engineered Bacillus licheniformis strains. World J. Microbiol. Biotechnol. 2018;34:66. doi: 10.1007/s11274-018-2450-7. [DOI] [PubMed] [Google Scholar]
- 49.Bai F., Dai L., Fan J., Truong N., Rao B., Zhang L., Shen Y. Engineered Serratia marcescens for efficient (3R)-acetoin and (2R,3R)-2,3-butanediol production. J. Ind. Microbiol. Biotechnol. 2015;42:779–786. doi: 10.1007/s10295-015-1598-5. [DOI] [PubMed] [Google Scholar]
- 50.Kim T., Cho S., Lee S.-M., Woo H.M., Lee J., Um Y., Seo J.-H. High production of 2,3-Butanediol (2,3-BD) by Raoultella ornithinolytica B6 via optimizing fermentation conditions and overexpressing 2,3-BD synthesis genes. PLoS One. 2016;11 doi: 10.1371/journal.pone.0165076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ishii J., Morita K., Ida K., Kato H., Kinoshita S., Hataya S., Shimizu H., Kondo A., Matsuda F. A pyruvate carbon flux tugging strategy for increasing 2,3-butanediol production and reducing ethanol subgeneration in the yeast Saccharomyces cerevisiae. Biotechnol. Biofuels. 2018;11:180. doi: 10.1186/s13068-018-1176-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim S.J., Sim H.J., Kim J.W., Lee Y.G., Park Y.C., Seo J.H. Enhanced production of 2,3-butanediol from xylose by combinatorial engineering of xylose metabolic pathway and cofactor regeneration in pyruvate decarboxylase-deficient Saccharomyces cerevisiae. Bioresour. Technol. 2017;245:1551–1557. doi: 10.1016/j.biortech.2017.06.034. [DOI] [PubMed] [Google Scholar]
- 53.Ng C.Y., Jung M.-Y., Lee J., Oh M.-K. Production of 2,3-butanediol in Saccharomyces cerevisiae by in silico aided metabolic engineering. Microb. Cell Fact. 2012;11 doi: 10.1186/1475-2859-11-68. 68-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim S., Hahn J.-S. Efficient production of 2,3-butanediol in Saccharomyces cerevisiae by eliminating ethanol and glycerol production and redox rebalancing. Metab. Eng. 2015;31:94–101. doi: 10.1016/j.ymben.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 55.Yang Z., Zhang Z. Production of (2R, 3R)-2,3-butanediol using engineered Pichia pastoris: strain construction, characterization and fermentation. Biotechnol. Biofuels. 2018;11:35. doi: 10.1186/s13068-018-1031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang S., Mohagheghi A., Franden M.A., Chou Y.C., Chen X., Dowe N., Himmel M.E., Zhang M. Metabolic engineering of Zymomonas mobilis for 2,3-butanediol production from lignocellulosic biomass sugars. Biotechnol. Biofuels. 2016;9:189. doi: 10.1186/s13068-016-0606-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li W., Pu N., Liu C.-X., Yuan Q.-P., Li Z.-J. Metabolic engineering of the marine bacteria Neptunomonas concharum for the production of acetoin and meso-2,3-butanediol from acetate. Biochem. Eng. J. 2019 [Google Scholar]
- 58.Siemerink M.A., Kuit W., Lopez Contreras A.M., Eggink G., van der Oost J., Kengen S.W. D-2,3-butanediol production due to heterologous expression of an acetoin reductase in Clostridium acetobutylicum. Appl. Environ. Microbiol. 2011;77:2582–2588. doi: 10.1128/AEM.01616-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park C., Lu M., Yun S., Park K., Lee J. Effect of pH on the metabolic flux of Klebsiella oxytoca producing 2,3-butanediol in continuous cultures at different dilution rates. Bioprocess Biosyst. Eng. 2013;36:845–855. doi: 10.1007/s00449-013-0932-4. [DOI] [PubMed] [Google Scholar]
- 60.Yen H.W., Li F.T., Wong C.L., Chang J.S. The pH effects on the distribution of 1,3-propanediol and 2,3-butanediol produced simultaneously by using an isolated indigenous Klebsiella sp. Ana-WS5. Bioprocess Biosyst. Eng. 2014;37:425–431. doi: 10.1007/s00449-013-1008-1. [DOI] [PubMed] [Google Scholar]
- 61.Petrov K., Petrova P. Enhanced production of 2,3-butanediol from glycerol by forced pH fluctuations. Appl. Microbiol. Biotechnol. 2010;87:943–949. doi: 10.1007/s00253-010-2545-z. [DOI] [PubMed] [Google Scholar]
- 62.Nie Z.K., Ji X.J., Huang H., Du J., Li Z.Y., Qu L., Zhang Q., Ouyang P.K. An effective and simplified fed-batch strategy for improved 2,3-butanediol production by Klebsiella oxytoca. Appl. Biochem. Biotechnol. 2011;163:946–953. doi: 10.1007/s12010-010-9098-6. [DOI] [PubMed] [Google Scholar]
- 63.Zeng A.P., Biebl H., Deckwer W.D. Effect of pH and acetic acid on growth and 2,3-butanediol production of Enterobacter aerogenes in continuous culture. Appl. Microbiol. Biotechnol. 1990;33:485–489. [Google Scholar]
- 64.Afschar A.S., Bellgardt K.H., Vaz Rossell C.E., Czok A., Schaller K. The production of 2,3-butanediol by fermentation of high test molasses. Appl. Microbiol. Biotechnol. 1991;34:582–585. [Google Scholar]
- 65.Lee S.J., Choi H.S., Kim C.K., Thapa L.P., Park C., Kim S.W. Process strategy for 2,3-butanediol production in fed-batch culture by acetate addition. J. Ind. Eng. Chem. 2017;56:157–162. doi. [Google Scholar]
- 66.Okonkwo C.C., Ujor C.V., Mishra K.P., Ezeji C.T. Process development for enhanced 2,3-Butanediol production by Paenibacillus polymyxa DSM 365. Fermentation. 2017;3 [Google Scholar]
- 67.Lee J., Lee S.Y., Park S. Fed-batch culture of Escherichia coli W by exponential feeding of sucrose as a carbon source. Biotechnol. Tech. 1997;11:59. [Google Scholar]
- 68.Converti A., Perego P., Del Borghi M. Effect of specific oxygen uptake rate on Enterobacter aerogenes energetics: carbon and reduction degree balances in batch cultivations. Biotechnol. Bioeng. 2003;82:370–377. doi: 10.1002/bit.10570. [DOI] [PubMed] [Google Scholar]
- 69.Blomqvist K., Nikkola M., Lehtovaara P., Suihko M.L., Airaksinen U., Stråby K.B., Knowles J.K., Penttilä M.E. Characterization of the genes of the 2,3-butanediol operons from Klebsiella terrigena and Enterobacter aerogenes. J. Bacteriol. 1993;175:1392–1404. doi: 10.1128/jb.175.5.1392-1404.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Perego P., Converti A., Del Borghi M. Effects of temperature, inoculum size and starch hydrolyzate concentration on butanediol production by Bacillus licheniformis. Bioresour. Technol. 2003;89:125–131. doi: 10.1016/s0960-8524(03)00063-4. [DOI] [PubMed] [Google Scholar]
- 71.Barrett E.L., Collins E.B., Hall B.J., Matoi S.H. Production of 2,3-Butylene Glycol from Whey by Klebsiella pneumoniae and Enterobacter aerogenes. J. Dairy Sci. 1983;66:2507–2514. doi: 10.3168/jds.S0022-0302(83)82119-5. [DOI] [PubMed] [Google Scholar]
- 72.Qureshi N., Cheryan M. Effects of aeration on 2,3-butanediol production from glucose by Klebsiella oxytoca. J. Ferment. Bioeng. 1989;67:415–418. [Google Scholar]
- 73.Silveira M.M., Schmidell W., Berbert M.A. Effect of the air supply on the production of 2,3-butanediol by Klebsiella pneumoniae NRRL B199. J. Biotechnol. 1993;31:93–102. [Google Scholar]
- 74.Jung M.-Y., Ng C.Y., Song H., Lee J., Oh M.-K. Deletion of lactate dehydrogenase in Enterobacter aerogenes to enhance 2,3-butanediol production. Appl. Microbiol. Biotechnol. 2012;95:461–469. doi: 10.1007/s00253-012-3883-9. [DOI] [PubMed] [Google Scholar]
- 75.Rebecchi S., Pinelli D., Zanaroli G., Fava F., Frascari D. Effect of oxygen mass transfer rate on the production of 2,3-butanediol from glucose and agro-industrial byproducts by Bacillus licheniformis ATCC9789. Biotechnol. Biofuels. 2018;11:145. doi: 10.1186/s13068-018-1138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ji X.J., Huang H., Du J., Zhu J.G., Ren L.J., Hu N., Li S. Enhanced 2,3-butanediol production by Klebsiella oxytoca using a two-stage agitation speed control strategy. Bioresour. Technol. 2009;100:3410–3414. doi: 10.1016/j.biortech.2009.02.031. [DOI] [PubMed] [Google Scholar]
- 77.Park J.M., Song H., Lee H.J., Seung D. In silico aided metabolic engineering of Klebsiella oxytoca and fermentation optimization for enhanced 2,3-butanediol production. J. Ind. Microbiol. Biotechnol. 2013;40:1057–1066. doi: 10.1007/s10295-013-1298-y. [DOI] [PubMed] [Google Scholar]
- 78.Xu Y., Chu H., Gao C., Tao F., Zhou Z., Li K., Li L., Ma C., Xu P. Systematic metabolic engineering of Escherichia coli for high-yield production of fuel bio-chemical 2,3-butanediol. Metab. Eng. 2014;23:22–33. doi: 10.1016/j.ymben.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 79.Nakashimada Y., Marwoto B., Kashiwamura T., Kakizono T., Nishio N. Enhanced 2,3-butanediol production by addition of acetic acid in Paenibacillus polymyxa. J. Biosci. Bioeng. 2000;90:661–664. doi: 10.1263/jbb.90.661. [DOI] [PubMed] [Google Scholar]
- 80.Zeng A.P., Sabra W. Microbial production of diols as platform chemicals: recent progresses. Curr. Opin. Biotechnol. 2011;22:749–757. doi: 10.1016/j.copbio.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 81.Wang A., Wang Y., Jiang T., Li L., Ma C., Xu P. Production of 2,3-butanediol from corncob molasses, a waste by-product in xylitol production. Appl. Microbiol. Biotechnol. 2010;87:965–970. doi: 10.1007/s00253-010-2557-8. [DOI] [PubMed] [Google Scholar]
- 82.Jansen N.B., Tsao G.T. Bioconversion of pentoses to 2,3-butanediol by Klebsiella pneumoniae. Adv. Biochem. Eng. Biotechnol. 1983;27:85–99. doi: 10.1007/BFb0009105. [DOI] [PubMed] [Google Scholar]
- 83.Eiteman M.A., Miller J.H. Effect of succinic acid on 2,3-butanediol production by Klebsiella oxytoca. Biotechnol. Lett. 1995;17:1057–1062. [Google Scholar]
- 84.Wang A., Xu Y., Ma C., Gao C., Li L., Wang Y., Tao F., Xu P. Efficient 2,3-Butanediol Production from Cassava Powder by a Crop-Biomass-Utilizer,Enterobacter cloacae subsp. dissolvens SDM. PLoS One. 2012;7 doi: 10.1371/journal.pone.0040442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jiang B., Li Z.-G., Dai J.-Y., Zhang D.-J., Xiu Z.-L. Aqueous two-phase extraction of 2,3-butanediol from fermentation broths using an ethanol/phosphate system. Process. Biochem. 2009;44:112–117. [Google Scholar]
- 86.Wheat J.A., Leslie J.D., Tomkins R.V., Mitton H.E., Scott D.S., Ledingham G.A. Production and properties of 2,3-butanediol: XXVIII. Pilot plant recovery of levo-2,3-butanediol from whole wheat mashes fermented by aerobacillus polymyxa. Can. J. Res. 1948;26f:469–496. [Google Scholar]
- 87.Garcia-Chavez L.Y., Shazad M., Schuur B., de Haan A.B. (Liquid+liquid) equilibrium data for the separation of 2,3-butanediol from aqueous streams using tetraoctyl ammonium 2-methyl-1-naphthoate. J. Chem. Thermodyn. 2012;55:85–91. [Google Scholar]
- 88.Drabo P., Tiso T., Heyman B., Sarikaya E., Gaspar P., Forster J., Buchs J., Blank L.M., Delidovich I. Anionic extraction for efficient recovery of biobased 2,3-Butanediol-A platform for bulk and fine chemicals. ChemSusChem. 2017;10:3252–3259. doi: 10.1002/cssc.201700899. [DOI] [PubMed] [Google Scholar]
- 89.Harvianto G.R., Haider J., Hong J., Van Duc Long N., Shim J.J., Cho M.H., Kim W.K., Lee M. Purification of 2,3-butanediol from fermentation broth: process development and techno-economic analysis. Biotechnol. Biofuels. 2018;11:18. doi: 10.1186/s13068-018-1013-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lange J.P. Lignocellulose conversion: an introduction to chemistry, process and economics, Biofuels. Bioprod. Biorefin. 2007;1:39–48. [Google Scholar]
- 91.Koutinas A.A., Yepez B., Kopsahelis N., Freire D.M.G., de Castro A.M., Papanikolaou S., Kookos I.K. Techno-economic evaluation of a complete bioprocess for 2,3-butanediol production from renewable resources. Bioresour. Technol. 2016;204:55–64. doi: 10.1016/j.biortech.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 92.Li Y., Zhu J., Wu Y., Liu J. Reactive-extraction of 2,3-butanediol from fermentation broth by propionaldehyde: equilibrium and kinetic study. Korean J. Chem. Eng. 2013;30:73–81. [Google Scholar]
- 93.Hao J., Xu F., Liu H.J., Liu D. Downstream processing of 1,3‐propanediol fermentation broth. J. Chem. Technol. Biotechnol. 2006;81:102–108. [Google Scholar]
- 94.Maina S., Stylianou E., Vogiatzi E., Vlysidis A., Mallouchos A., Nychas G.E., de Castro A.M., Dheskali E., Kookos I.K., Koutinas A. Improvement on bioprocess economics for 2,3-butanediol production from very high polarity cane sugar via optimisation of bioreactor operation. Bioresour. Technol. 2019;274:343–352. doi: 10.1016/j.biortech.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 95.Cho S., Kim T., Woo H.M., Kim Y., Lee J., Um Y. High production of 2,3-butanediol from biodiesel-derived crude glycerol by metabolically engineered Klebsiella oxytoca M1. Biotechnol. Biofuels. 2015;8:146. doi: 10.1186/s13068-015-0336-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ma C., Wang A., Qin J., Li L., Ai X., Jiang T., Tang H., Xu P. Enhanced 2,3-butanediol production by Klebsiella pneumoniae SDM. Appl. Microbiol. Biotechnol. 2009;82:49–57. doi: 10.1007/s00253-008-1732-7. [DOI] [PubMed] [Google Scholar]
- 97.Guo X., Cao C., Wang Y., Li C., Wu M., Chen Y., Zhang C., Pei H., Xiao D. Effect of the inactivation of lactate dehydrogenase, ethanol dehydrogenase, and phosphotransacetylase on 2,3-butanediol production in Klebsiella pneumoniae strain. Biotechnol. Biofuels. 2014;7:44. doi: 10.1186/1754-6834-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gao J., Xu H., Li Q.J., Feng X.H., Li S. Optimization of medium for one-step fermentation of inulin extract from Jerusalem artichoke tubers using Paenibacillus polymyxa ZJ-9 to produce R,R-2,3-butanediol. Bioresour. Technol. 2010;101:7087–7093. doi: 10.1016/j.biortech.2010.03.143. [DOI] [PubMed] [Google Scholar]
- 99.Ge Y., Li K., Li L., Gao C., Zhang L., Ma C., Xu P. Contracted but effective: production of enantiopure 2,3-butanediol by thermophilic and GRAS: bacillus licheniformis. Green Chem. 2016;18:4693–4703. [Google Scholar]
- 100.Jung M.Y., Jung H.M., Lee J., Oh M.K. Alleviation of carbon catabolite repression in Enterobacter aerogenes for efficient utilization of sugarcane molasses for 2,3-butanediol production. Biotechnol. Biofuels. 2015;8:106. doi: 10.1186/s13068-015-0290-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li L., Li K., Wang Y., Chen C., Xu Y., Zhang L., Han B., Gao C., Tao F., Ma C., Xu P. Metabolic engineering of Enterobacter cloacae for high-yield production of enantiopure (2R,3R)-2,3-butanediol from lignocellulose-derived sugars. Metab. Eng. 2015;28:19–27. doi: 10.1016/j.ymben.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 102.Hazeena S.H., Nair Salini C., Sindhu R., Pandey A., Binod P. Simultaneous saccharification and fermentation of oil palm front for the production of 2,3-butanediol. Bioresour. Technol. 2019;278:145–149. doi: 10.1016/j.biortech.2019.01.042. [DOI] [PubMed] [Google Scholar]
- 103.Zhang L., Sun J., Hao Y., Zhu J., Chu J., Wei D., Shen Y. Microbial production of 2,3-butanediol by a surfactant (serrawettin)-deficient mutant of Serratia marcescens H30. J. Ind. Microbiol. Biotechnol. 2010;37:857–862. doi: 10.1007/s10295-010-0733-6. [DOI] [PubMed] [Google Scholar]
- 104.Zhang L., Yang Y., Sun J., Shen Y., Wei D., Zhu J., Chu J. Microbial production of 2,3-butanediol by a mutagenized strain of Serratia marcescens H30. Bioresour. Technol. 2010;101:1961–1967. doi: 10.1016/j.biortech.2009.10.052. [DOI] [PubMed] [Google Scholar]
- 105.Kim J.W., Kim J., Seo S.O., Kim K.H., Jin Y.S., Seo J.H. Enhanced production of 2,3-butanediol by engineered Saccharomyces cerevisiae through fine-tuning of pyruvate decarboxylase and NADH oxidase activities. Biotechnol. Biofuels. 2016;9:265. doi: 10.1186/s13068-016-0677-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nguyen A.D., Hwang I.Y., Lee O.K., Kim D., Kalyuzhnaya M.G., Mariyana R., Hadiyati S., Kim M.S., Lee E.Y. Systematic metabolic engineering of Methylomicrobium alcaliphilum 20Z for 2,3-butanediol production from methane. Metab. Eng. 2018;47:323–333. doi: 10.1016/j.ymben.2018.04.010. [DOI] [PubMed] [Google Scholar]
- 107.Ui S., Okajima Y., Mimura A., Kanai H., Kobayashi T., Kudo T. Sequence analysis of the gene for and characterization of d-acetoin forming meso-2,3-butanediol dehydrogenase of Klebsiella pneumoniae expressed in Escherichia coli. J. Ferment. Bioeng. 1997;83:32–37. [Google Scholar]
- 108.Lee S., Kim B., Park K., Um Y., Lee J. Synthesis of pure meso-2,3-butanediol from crude glycerol using an engineered metabolic pathway in Escherichia coli. Appl. Biochem. Biotechnol. 2012;166:1801–1813. doi: 10.1007/s12010-012-9593-z. [DOI] [PubMed] [Google Scholar]
- 109.Ji X.J., Liu L.G., Shen M.Q., Nie Z.K., Tong Y.J., Huang H. Constructing a synthetic metabolic pathway in Escherichia coli to produce the enantiomerically pure (R, R)-2,3-butanediol. Biotechnol. Bioeng. 2015;112:1056–1059. doi: 10.1002/bit.25512. [DOI] [PubMed] [Google Scholar]
- 110.Fu J., Huo G., Feng L., Mao Y., Wang Z., Ma H., Chen T., Zhao X. Metabolic engineering of Bacillus subtilis for chiral pure meso-2,3-butanediol production. Biotechnol. Biofuels. 2016;9 doi: 10.1186/s13068-016-0502-5. 90-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gao Y., Huang H., Chen S., Qi G. Production of optically pure 2,3-butanediol from Miscanthus floridulus hydrolysate using engineered Bacillus licheniformis strains. World J. Microbiol. Biotechnol. 2018;34:66. doi: 10.1007/s11274-018-2450-7. [DOI] [PubMed] [Google Scholar]