HHP treatment is used as a nonthermal processing technology in the food industry to inactivate bacteria while retaining high quality of foods under suppressed chemical reactions. However, some populations of bacterial cells may survive the inactivation. Although the survivors are in a transient nongrowing state due to HHP-induced injury, they can recover from the injury and then start growing, depending on the postprocessing conditions. The recovery process in terms of cellular components after the injury remains unclear. Transcriptome analysis using vegetative cells of Bacillus subtilis revealed that the translational machinery can preferentially be reconstructed after HHP treatment. We found that both Mn2+ and Zn2+ prolonged the growth-arrested stage of HHP-injured cells by delaying ribosome reconstruction. It is likely that ribosome reconstruction is crucial for the recovery of growth ability in HHP-injured cells. This study provides further understanding of the recovery process in HHP-injured B. subtilis cells.
KEYWORDS: Bacillus subtilis, high hydrostatic pressure, injured cell, ribosome reconstruction
ABSTRACT
Vegetative cells of Bacillus subtilis can recover from injury after high-hydrostatic-pressure (HHP) treatment at 250 MPa. DNA microarray analysis revealed that substantial numbers of ribosomal genes and translation-related genes (e.g., translation initiation factors) were upregulated during the growth arrest phase after HHP treatment. The transcript levels of cold shock-responsive genes, whose products play key roles in efficient translation, and heat shock-responsive genes, whose products mediate correct protein folding or degrade misfolded proteins, were also upregulated. In contrast, the transcript level of hpf, whose product (Hpf) is involved in ribosome inactivation through the dimerization of 70S ribosomes, was downregulated during the growth arrest phase. Sucrose density gradient sedimentation analysis revealed that ribosomes were dissociated in a pressure-dependent manner and then reconstructed. We also found that cell growth after HHP-induced injury was apparently inhibited by the addition of Mn2+ or Zn2+ to the recovery medium. Ribosome reconstruction in the HHP-injured cells was also significantly delayed in the presence of Mn2+ or Zn2+. Moreover, Zn2+, but not Mn2+, promoted dimer formation of 70S ribosomes in the HHP-injured cells. Disruption of the hpf gene suppressed the Zn2+-dependent accumulation of ribosome dimers, partially relieving the inhibitory effect of Zn2+ on the growth recovery of HHP-treated cells. In contrast, it was likely that Mn2+ prevented ribosome reconstruction without stimulating ribosome dimerization. Our results suggested that both Mn2+ and Zn2+ can prevent ribosome reconstruction, thereby delaying the growth recovery of HHP-injured B. subtilis cells.
IMPORTANCE HHP treatment is used as a nonthermal processing technology in the food industry to inactivate bacteria while retaining high quality of foods under suppressed chemical reactions. However, some populations of bacterial cells may survive the inactivation. Although the survivors are in a transient nongrowing state due to HHP-induced injury, they can recover from the injury and then start growing, depending on the postprocessing conditions. The recovery process in terms of cellular components after the injury remains unclear. Transcriptome analysis using vegetative cells of Bacillus subtilis revealed that the translational machinery can preferentially be reconstructed after HHP treatment. We found that both Mn2+ and Zn2+ prolonged the growth-arrested stage of HHP-injured cells by delaying ribosome reconstruction. It is likely that ribosome reconstruction is crucial for the recovery of growth ability in HHP-injured cells. This study provides further understanding of the recovery process in HHP-injured B. subtilis cells.
INTRODUCTION
High-hydrostatic-pressure (HHP) treatment is a nonthermal food processing technology that can extend the shelf life of food via microbial inactivation while maintaining nutrients, flavors, and colors, which are often deteriorated by heat treatment (1). On the other hand, some populations of HHP-treated cells can survive the treatment, which is typically carried out at 400 to 600 MPa. Although these survivors are in a transient nongrowing state due to HHP-induced injuries, they can recover from the injury during storage or incubation (2–4).
HHP treatment inhibits various cellular processes, including motility, cell division, replication, transcription, and translation, thereby causing pleiotropic effects on cells (5–8). Previous studies have revealed that ribosomes could be a primary target for HHP-induced cell death in bacteria (9, 10). Proteomic and transcriptomic studies of Escherichia coli showed that heat shock proteins (HSPs), such as DnaK and GroESL, and cold shock proteins (CSPs), such as CspA and CspB, were induced in the cells under sublethal HHP at around 50 MPa (11, 12). In addition, sublethal heat shock was shown to induce pressure resistance in E. coli (13), while other experiments demonstrated that cold shock affected pressure resistance in Staphylococcus aureus and Listeria monocytogenes (14, 15). It is likely that HSPs and CSPs help protect cells against HHP-induced stresses. As previously demonstrated by VanBogelen and Neidhardt (16), ribosome-targeting antibiotics can induce HSPs or CSPs, leading these authors to suggest that ribosomes can act as cellular thermosensors. Based on these previous studies, ribosome damage associated with HHP treatment may trigger the inductions of HSPs and CSPs.
It has also been demonstrated that HHP treatment can induce endogenous oxidative stress in E. coli (17). Since increased levels of intracellular reactive oxygen species (ROS) can cause damage to nucleic acids, proteins, and membrane lipids, ROS scavengers, such as catalase and superoxide dismutase, are considered to play important roles in HHP resistance in E. coli (17). Thus, any comprehensive analysis of HHP-treated E. coli cells should include the effects of both HHP and oxidative stresses.
In a Gram-positive model bacterium, Bacillus subtilis, much effort has been devoted to optimizing the HHP conditions necessary for spore inactivation or germination (18, 19). In contrast, relatively few studies have examined the effect of HHP on B. subtilis vegetative cells. We have previously reported the effect of HHP on vegetative cells of a sporulation-deficient B. subtilis strain (2). The characteristics of HHP-injured B. subtilis cells are different from those of E. coli cells. Oxidative stress contributes to HHP-induced cell death in E. coli (17), while ROS scavengers had little or no effect on the viability of HHP-treated B. subtilis cells (2). Furthermore, HHP-injured B. subtilis cells are highly sensitive to salts, with cell lysis occurring in the presence of NaCl or KCl at concentrations of 100 mM or higher (2). These results suggest that HHP-induced damage and subsequent recovery may differ between E. coli and B. subtilis. Our research goal is to understand how bacteria can recover from HHP-induced injury. In this study, DNA microarray analysis was performed to investigate gene expression patterns and the ribosome reconstruction process was analyzed in HHP-injured vegetative cells of B. subtilis.
RESULTS AND DISCUSSION
Transient growth arrest of B. subtilis cells treated by HHP.
To investigate the effect of HHP on B. subtilis vegetative cells, the sporulation-deficient strain TI465, which lacks the sporulation-specific σ factor F (2), was used in this study. When stationary-phase cells (6 h of cultivation in NaCl-free L medium [10 g of tryptone and 5 g of yeast extract, both per liter]) were exposed to 250 MPa of HHP (25°C for 10 min) and subsequently diluted into fresh recovery medium (NaCl-free L medium), the cells showed a prolonged growth lag. The average growth delay (the time difference between HHP-treated and untreated culture) at an optical density at 600 nm (OD600) of 1.0 was 160 ± 21 min. Under the experimental condition, a 10-fold dilution of untreated control culture (1.0-log reduction in viable cell counts) resulted in a delay of 70 ± 13 min. Accordingly, the apparent reduction in viable cells after HHP treatment therefore can be expressed as (DTHHP/DTCONT) log CFU per milliliter, where DTHHP and DTCONT indicate the delay times for HHP-treated culture and for 10-fold-diluted control culture, respectively. Based on this calculation, the apparent reduction in viable-cell counts after 250 MPa treatment was 2.3 log CFU/ml. However, the actual reduction in viable-cell counts as determined by colony formation was approximately 0.3 log CFU/ml. The different viability values extracted from growth delay and colony counts indicate that the HHP-treated B. subtilis culture included nongrowing cells as previously demonstrated for E. coli (3, 4). Hence, we concluded that a large population of viable cells after 250 MPa of HHP treatment was temporarily in the nongrowing state (growth arrest phase).
Transcriptome analysis of HHP-injured B. subtilis cells.
To understand the recovery process in HHP-injured B. subtilis cells, gene expression patterns following HHP treatment were investigated using DNA microarray analysis. The cells treated at 250 MPa were transferred into the recovery medium and incubated with vigorous shaking. Total RNAs were extracted from the cells incubated for 0 h (immediately after transferring the HHP-treated cells into the recovery medium), 2 h (growth arrest phase), and 4 h (regrowth phase) and then subjected to DNA microarray analysis. Principal-component analysis (PCA) showed that the gene expression profile at each time point was clearly different from the others (see Fig. S1 in the supplemental material). The microarray data were analyzed using the rank products method (20) to identify differentially expressed genes (DEGs) that were either up- or downregulated after HHP treatment. With a false-discovery rate (FDR) of less than 5%, 77 genes were identified as DEGs (Tables 1 and 2). To confirm the DNA microarray results, reverse transcription-quantitative PCR (RT-qPCR) analysis was carried out, focusing on one or two genes from each transcription unit of all DEGs (Fig. 1 and Fig. S2).
TABLE 1.
Genes induced during the recovery phase of HHP-injured B. subtilis
| Gene | Gene product (function) | Expression ratioa
|
||
|---|---|---|---|---|
| 2 h/0 h | 4 h/0 h | 4 h/2 h | ||
| citB | Aconitase | 2.89 | ||
| infA | Translation initiation factor IF-1 | 3.46 | ||
| infC | Translation initiation factor IF-3 | 2.88 | ||
| map | Methionine aminopeptidase | 2.67 | ||
| mtlD | Mannitol-1-phosphate 5-dehydrogenase | 3.01 | ||
| pdhA | Pyruvate dehydrogenase (E1 alpha subunit) | 2.32 | ||
| pstA | Phosphate ABC transporter (permease) | 4.03 | ||
| pstS | Phosphate ABC transporter (binding protein) | 3.94 | ||
| rplE | Ribosomal protein L5 | 3.04 | ||
| rplJ | Ribosomal protein L10 | 2.68 | ||
| rplN | Ribosomal protein L14 | 3.33 | ||
| rplP | Ribosomal protein L16 | 3.47 | ||
| rplR | Ribosomal protein L18 | 3.02 | ||
| rpmC | Ribosomal protein L29 | 3.48 | ||
| rpmE | Ribosomal protein L31 | 2.16 | ||
| rpsN | Ribosomal protein S14 | 2.94 | ||
| rpsQ | Ribosomal protein S17 | 3.15 | ||
| tuaG | Biosynthesis of teichuronic acid | 2.89 | ||
| ylbN | Unknown | 3.16 | ||
| yubF | Unknown | 2.81 | ||
| yugI | General stress protein found in the ribosome fraction | 3.14 | ||
| clpE | Clp ATPase | 4.98 | 1.52 | |
| cspB | Cold shock protein | 2.77 | 3.52 | |
| rplA | Ribosomal protein L1 | 2.78 | 2.95 | |
| rplS | Ribosomal protein L19 | 3.63 | 2.93 | |
| rpmJ | Ribosomal protein L36 | 3.38 | 2.56 | |
| tig | Trigger factor | 2.89 | 3.72 | |
| yneF | Unknown | 5.68 | 5.24 | |
| yozC | Unknown | 2.42 | 3.06 | |
| cspD | Cold shock protein | 2.71 | 4.58 | 1.87 |
| groES | 10-kDa chaperonin | 3.74 | 5.27 | 1.53 |
| abh | Transcriptional regulator | 3.61 | 2.30 | |
| acpP | Acyl carrier protein | 4.33 | 2.61 | |
| feuA | Siderophore binding protein | 3.59 | 4.38 | |
| ydbN | Unknown | 5.40 | 3.94 | |
| yfiY | Siderophore uptake? | 4.15 | 3.67 | |
| ykuN | Flavodoxin | 3.30 | 4.07 | |
| yolA | Unknown; SP-β protein | 4.04 | 4.47 | |
| yqgA | Unknown; cell wall binding protein | 4.37 | 3.22 | |
| ysbA | Murein hydrolase regulator LrgA; essential for pyruvate utilization | 5.92 | 6.58 | |
| ysbB | Antiholin-like protein LrgB | 4.93 | 5.67 | |
| hag | Flagellin protein | 3.53 | ||
| hbs | Nonspecific DNA-binding protein HBsu | 2.39 | ||
| rbsA | Ribose ABC transporter (ATP-binding protein) | 3.88 | ||
| spoVG | Stage V sporulation protein G (spore cortex synthesis) | 2.79 | ||
| ykuP | Flavodoxin | 4.18 | ||
| yolB | Unknown; SP-β protein | 3.34 | ||
| ywsB | Unknown; cell wall binding protein | 1.92 | ||
Expression ratio indicates the average value of the log2 ratio between two independent microarray results.
TABLE 2.
Genes repressed during the recovery phase of HHP-injured B. subtilis
| Gene | Gene product (function) | Expression ratioa
|
||
|---|---|---|---|---|
| 2 h/0 h | 4 h/0 h | 4 h/2 h | ||
| ahpC | Alkyl hydroperoxide reductase (small subunit) | −3.94 | ||
| ahpF | Alkyl hydroperoxide reductase (large subunit) | −3.64 | ||
| bfmBAA (bkdA1) | Branched-chain alpha-keto acid dehydrogenase E1 subunit | −2.58 | ||
| csbDb | General stress protein | −3.39 | ||
| gapB | Glyceraldehyde-3-phosphate dehydrogenase | −2.47 | ||
| hag | Flagellin protein | −4.46 | ||
| mntA | Manganese ABC transporter (membrane protein) | −2.89 | ||
| mntB | Manganese ABC transporter (ATP-binding protein) | −3.33 | ||
| pckA | Phosphoenolpyruvate carboxykinase | −2.92 | ||
| rbsA | Ribose ABC transporter (ATP-binding protein) | −3.16 | ||
| spoVG | Stage V sporulation protein G (spore cortex synthesis) | −3.32 | ||
| yflTb | General stress protein | −3.16 | ||
| yqgZ (mgsR) | Spx paralog controls a subregulon within general stress response | −3.71 | ||
| ytxH | General stress protein | −2.55 | ||
| hpf (yvyD) | Dimerization of 70S ribosome | −4.45 | ||
| ywsB | Unknown; putative cell wall binding protein | −3.27 | ||
| citZ | Citrate synthase II | −2.69 | −2.14 | |
| gsiB | General stress protein | −2.73 | −1.74 | |
| katA | Vegetative catalase 1 | −4.06 | −3.52 | |
| mntC | Manganese ABC transporter (membrane protein) | −3.40 | −1.94 | |
| mrgA | Metalloregulation DNA-binding stress protein; E. coli Dps homolog | −3.34 | −3.02 | |
| srfAA | Surfactin synthetase | −2.45 | −2.60 | |
| ybyB | Unknown | −4.72 | −3.16 | |
| yhzC | General stress protein | −2.86 | −2.55 | |
| ykzI | General stress protein | −3.94 | −3.57 | |
| appC | Oligopeptide ABC transporter (permease) | −1.97 | −3.91 | −1.94 |
| phrA | Phosphatase RapA inhibitor | −1.84 | −3.76 | −1.92 |
| rapA | Response regulator aspartate phosphatase | −2.08 | −3.75 | −1.67 |
| ybcO (skfA) | Sporulation killing factor | −2.22 | −3.62 | −1.40 |
| yydF | Inducer of LiaRS | −2.65 | −3.83 | −1.19 |
| mntD | Manganese ABC transporter | −2.76 | ||
| nhaX | Regulator of nhaC | −2.67 | ||
| srfAB | Surfactin synthetase | −2.59 | ||
| clpE | Clp ATPase | −3.45 | ||
| pstA | Phosphate ABC transporter (permease) | −3.09 | ||
| mtlD | Mannitol-1-phosphate 5-dehydrogenase | −3.40 | ||
| citB | Aconitase | −3.26 | ||
| tuaG | Biosynthesis of teichuronic acid | −2.39 | ||
Expression ratio indicates the average value of the log2 ratio between two independent microarray results.
Not reproducible by RT-qPCR analysis.
FIG 1.
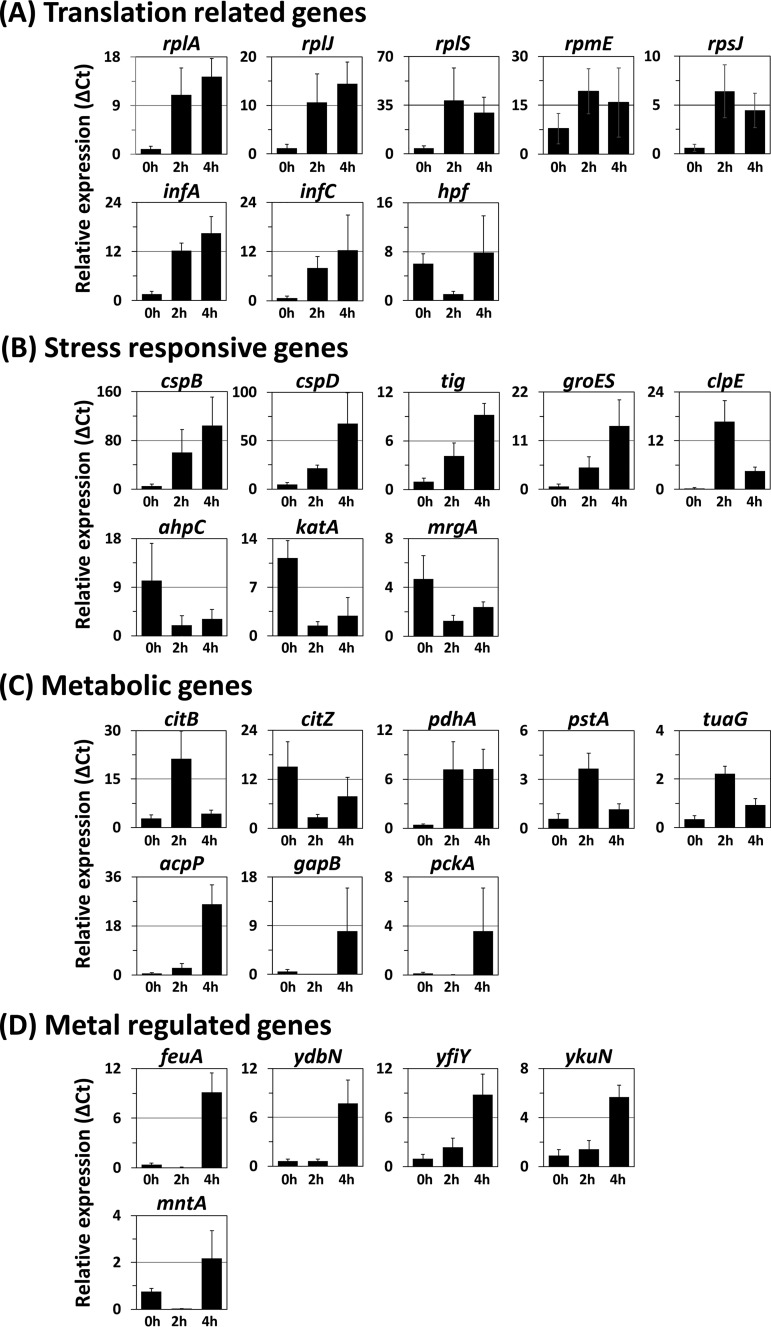
Transcriptional analysis of genes affected by HHP treatment in B. subtilis. B. subtilis strain TI465 was subjected to HHP treatment at 250 MPa at 25°C for 10 min. After HHP treatment, the culture was diluted 5-fold in fresh NaCl-free L medium and incubated at 37°C with vigorous shaking. After incubation for 0 h (immediately after dilution), 2 h, or 4 h, cells were collected by centrifugation. Then total RNAs were extracted and used for RT-qPCR analysis. The relative expression was expressed as the relative amount of each transcript per 1,000 units of 16S rRNA (ΔCt). Genes are classified into four groups according to their functions: translation-related genes (A), stress-responsive genes (B), metabolic genes (C), and metal-regulated genes (D). The average values of at least four independent experiments are shown.
(i) Translation-related genes. DNA microarray analysis showed that 12 ribosomal protein genes (rplA, rplE, rplJ, rplN, rplP, rplR, rplS, rpmC, rpmE, rpmJ, rpsN, and rpsQ), 2 genes for translation initiation factors (infA and infC), and a gene for amino-terminal processing of proteins (map) were upregulated during the growth arrest phase (Table 1). Among them, 10 genes (rplE, rplN, rplP, rplR, rpmC, rpmJ, rpsN, rpsQ, map, and infA) belong to an identical ribosomal operon, S10-spc-α, which includes 25 ribosomal genes (21). The entire S10-spc-α operon is transcribed from the S10 promoter located immediately upstream of rpsJ, which is the first gene of this operon. Although three additional weak promoters are found in the internal region of the S10-spc-α operon (21), the S10 promoter is a sole promoter for seven genes that were listed as upregulated genes (rplE, rplN, rplP, rplR, rpmC, rpsN, and rpsQ). Since no significant difference in the rpsJ transcript was detected by DNA microarray analysis, we further validated the transcript level of rpsJ after HHP treatment by RT-qPCR. The result showed that the rpsJ gene was also upregulated in the growth arrest phase (Fig. 1A). The map, infA, and rpmJ genes can be transcribed from not only the S10 promoter but also additional promoters existing in the internal region of this operon. Hence, the infA gene was also employed for RT-qPCR analysis. In addition, the transcript levels of the remaining five genes (rplA, rplJ, rplS, rpmE, and infC), which are transcribed from a different transcription unit, were further analyzed. RT-qPCR analyses confirmed that the transcript levels of these six genes as well as rpsJ were enhanced within 2 h after HHP treatment (Fig. 1A). The hpf (yvyD) gene, whose gene product is required for ribosome inactivation by dimerizing 70S ribosomes (22), was downregulated in the growth arrest phase.
(ii) Stress-responsive genes. As observed in previous studies on E. coli (11, 12), the transcript levels of the CSP genes (cspB, cspD, and tig) as well as those of the HSP genes (groES and clpE) were increased after HHP treatment, as shown in Table 1 and Fig. 1B. CspB and CspD act as RNA chaperones that are essential for efficient translation at both low and optimal growth temperatures (23). The ribosome-associated trigger factor, which is encoded by tig, is known to initiate folding of newly synthesized proteins (24). The chaperonin GroES also supports translation through mediating correct protein folding under both normal and heat shock conditions (25). A heat-inducible Clp ATPase, ClpE, degrades misfolded proteins and a class III heat shock regulator CtsR, thereby inducing CtsR regulon genes (i.e., clpP, clpC, and clpE) (26, 27). These stress-responsive proteins might contribute to efficient translation (CspB, CspD, Tig, and GroES) and protein quality control (ClpE) in HHP-injured cells.
Oxidative stress has been reported to contribute to HHP-induced cell death in E. coli (17). In B. subtilis, however, disruption of the genes for vegetative catalase (katA and katE) had no significant effect on the viability of HHP-treated cells (2). In agreement with this previous observation, several PerR regulon genes (ahpC, katA, and mrgA), which play important roles in cellular protection against oxidative stress (28–30), were downregulated after HHP treatment (Table 2 and Fig. 1B).
The general stress regulon, which is responsible for the development of nonspecific stress resistance, is induced by nutrient limitation (31). The transcript levels of general stress genes (gsiB, yhzC, ykzI, and ytxH) were reduced in the growth arrest phase (Fig. S2). Although the DNA microarray analysis data showed that two general stress genes, csbD and yflT, were reduced after HHP treatment, this was not reproducible by RT-qPCR analysis (Fig. S2).
(iii) Metabolic genes. Several metabolic genes (pdhA, citB, tuaG, and pstA) were upregulated in the growth arrest phase (Fig. 1C). The pdhA gene is the first gene of the pdhABCD operon, which encodes the pyruvate dehydrogenase multienzyme complex. This enzyme complex catalyzes the irreversible oxidative decarboxylation of pyruvate to acetyl coenzyme A (acetyl-CoA), linking glycolysis with the tricarboxylic acid (TCA) cycle (32). Although the transcript level of the citB gene (which encodes aconitase, the enzyme which catalyzes the interconversion of citrate and isocitrate in the second step of the TCA cycle) was increased during the growth arrest phase, the transcript level of the citZ gene (which encodes the major citrate synthase in the first step of the TCA cycle) was decreased (Fig. 1C). These results suggest that HHP treatment may result in the reduction in the intracellular level of TCA cycle metabolites and the accumulation of acetyl-CoA. Our preliminary metabolome analysis using 250-MPa-treated B. subtilis cells also supported this prediction (unpublished results). Recently, it has been reported that compromising TCA cycle enzymes can affect HHP resistance in E. coli (33). Changes in the intracellular level of TCA cycle metabolites might affect the recovery of HHP-injured B. subtilis cells. Membrane damage is thought to be one of the major causes of HHP-induced death (34). The accumulation of acetyl-CoA, which is important for fatty acid biosynthesis (35), may be required for membrane repair. The transcript level of acpP, encoding an acyl carrier protein which plays an essential role in fatty acid biosynthesis (36), was increased in the regrowth phase (Fig. 1C). It seems that membrane repair would begin after ribosome reconstruction. As previously demonstrated, HHP-treated E. coli cells can be stained with propidium iodide (PI), which enters cells with a compromised membrane (3, 34). In B. subtilis, no obvious incorporation of PI into 250-MPa-treated cells was observed (data not shown). HHP treatment at 250 MPa might not cause severe damage on membrane. The tuaG gene belongs to the tuaABCDEFGH operon, which encodes enzymes required for the polymerization of teichuronic acid, and the pstA gene belongs to the pst operon, which encodes a phosphate transporter. Both operons are induced under phosphate-limited conditions (37, 38). The transcript levels of the tuaG and pstA genes were both increased in the growth arrest phase and then decreased in the regrowth phase (Fig. 1C). HHP treatment may cause phosphate limitation in B. subtilis vegetative cells. Transcription of gluconeogenic genes (e.g., gapB and pckA) is known to be repressed under glycolytic conditions and derepressed under gluconeogenic conditions (39). In HHP-treated cells, gapB and pckA genes were downregulated in the growth arrest phase and upregulated in the regrowth phase (Table 2 and Fig. 1C).
(iv) Metal-regulated genes. Because metal ions, such as iron and manganese, are required as cofactors for a large number of enzymes, these metal ions are also important for cell growth. DNA microarray and RT-qPCR analyses showed that five genes (feuA, ydbN, yfiY, ykuN, and ykuP) belonging to the Fur regulon were upregulated in the regrowth phase (Table 1 and Fig. 1D). Although DNA microarray analysis showed that the transcript level of the mntABCD operon, belonging to the MntR regulon, was reduced throughout the recovery period, RT-qPCR analysis indicated that the transcript level of this operon was also upregulated in the regrowth phase (Table 2 and Fig. 1D). The Fur regulon is induced under iron-limited conditions, while the MntR regulon is induced under manganese limitation (40, 41). Metal ions may be required for the activation of newly synthesized metalloproteins during the regrowth phase of HHP-treated cells.
Ribosomes are reconstructed during the growth arrest phase.
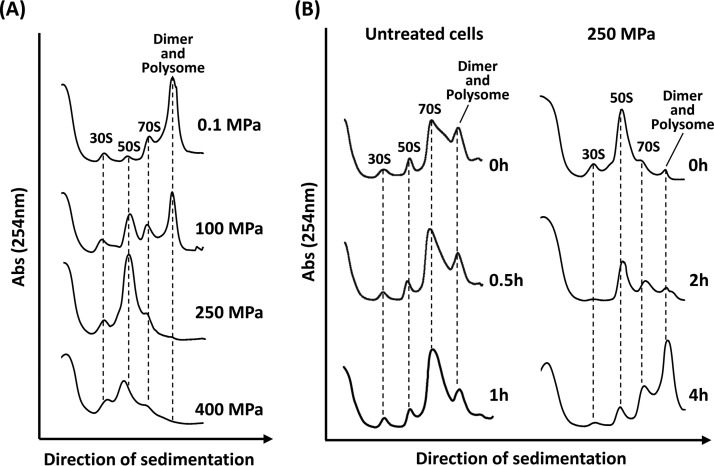
DNA microarray analysis indicated that many translation-related factors were induced during the growth arrest phase. To examine the effect of HHP on ribosomes, we analyzed ribosome profiles using sucrose density gradient sedimentation (Fig. 2A). In the stationary-phase untreated cells grown under atmospheric pressure (0.1 MPa), ribosomes largely formed multimers (dimers or polysomes), as described previously (22). Although 100-MPa-treated cells grew normally (data not shown), accumulations of 30S and 50S ribosomal subunits were observed, indicating that ribosomes partly dissociated (Fig. 2A). In the cells treated with 250 MPa, the peaks corresponding to intact ribosomes (70S, dimers, and polysomes) almost completely disappeared, with a concomitant increase in the 50S peak. In the 400-MPa-treated cells, however, even the amount of 50S decreased compared with that of the 250-MPa-treated cells, suggesting further dissociation or degradation of the ribosomes. Thus, ribosomes were dissociated or degraded after HHP treatment in a pressure-dependent manner.
FIG 2.
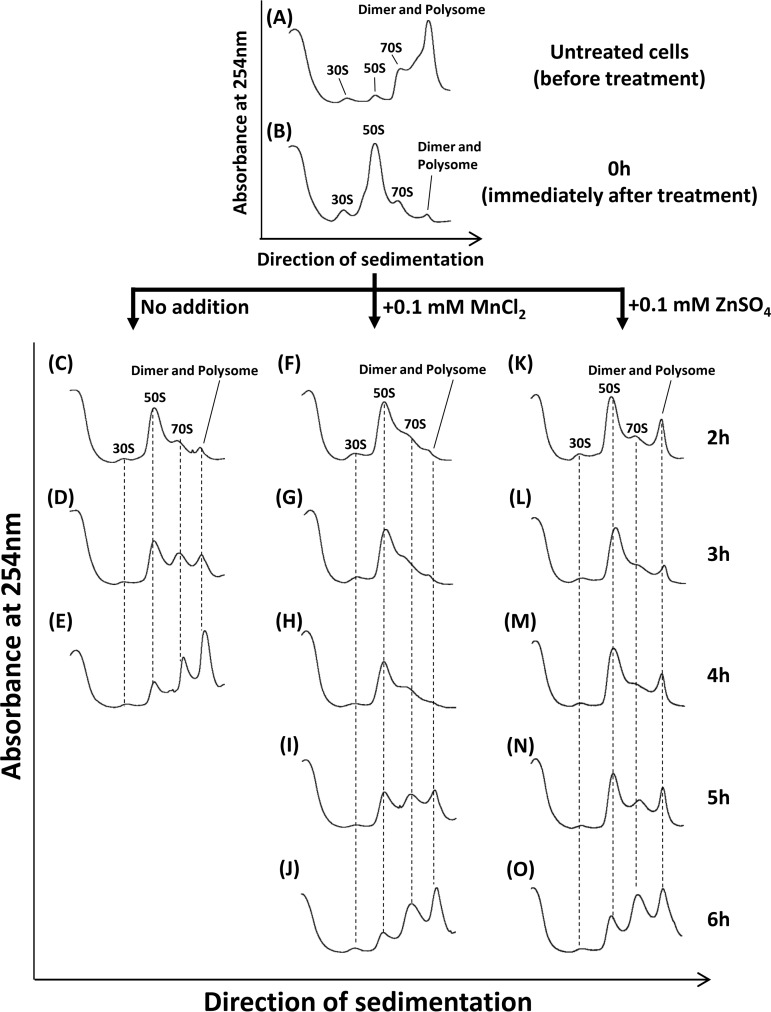
Effect of HHP on ribosomes in B. subtilis cells. (A) Pressure-dependent dissociation of ribosomes. B. subtilis strain TI465 was exposed to HHP at 100, 250, or 400 MPa at 25°C for 10 min. After HHP treatment, cells were collected by centrifugation. Then each cell extract was prepared and analyzed using 10% to 40% sucrose gradient sedimentation. Cell extract from an untreated culture (0.1 MPa) was also examined as a control. (B) Change in the ribosome sedimentation profile during recovery after HHP treatment. B. subtilis strain TI465 was subjected to 250 MPa at 25°C for 10 min. After the HHP treatment, the culture was diluted 5-fold in fresh NaCl-free L medium and incubated at 37°C with vigorous shaking. To compare with the ribosome profile in untreated cells, the untreated culture was also prepared by the same method except for HHP treatment. After incubation for the indicated times, cells were collected by centrifugation. Cell extracts were then analyzed using 10% to 40% sucrose gradient sedimentation.
We next monitored ribosome profiles during the recovery of HHP-injured cells (Fig. 2B). In untreated cells, inactive ribosome dimers were rapidly dissociated into active 70S within 30 min after cells were transferred into fresh medium (Fig. 2B, left). In contrast, ribosomes dissociated into 30S and 50S subunits immediately after 250-MPa treatment (0 h) (Fig. 2B, right). In the growth arrest phase (2 h of incubation), the amounts of ribosomal particles 30S and 50S apparently decreased, while the amounts of 70S ribosomes remained low. It is likely that free ribosomal particles (30S and 50S) were degraded. Zundel et al. previously reported that free ribosomal particles (30S and 50S) are more susceptible to degradation by RNase(s) than 70S ribosomes (42). Hence, the formation of free ribosomal particles (30S and 50S) can initiate ribosome breakdown. This is a well-known response to generate nutrients from degraded ribosomes under nutrient-limiting conditions (42–44). In HHP-treated cells, the dissociated ribosomal particles also can be degraded and recycled as a nutrient source. In the regrowth phase (after 4 h of incubation), ribosomes formed multimers (dimers or polysomes) similar to that in stationary-phase cells (Fig. 2A, 0.1 MPa), whereas it formed 70S in growing untreated cells (Fig. 2B, untreated cells, 0.5 and 1 h). Since our preliminary metabolome analysis suggested a remarkable reduction in the intracellular pools of nucleotides and amino acids in 250-MPa-treated B. subtilis cells (unpublished results), the nutrient availability might be restricted due to HHP-induced impairments.
Growth recovery of HHP-injured B. subtilis cells is delayed in the presence of Mn2+ or Zn2+.
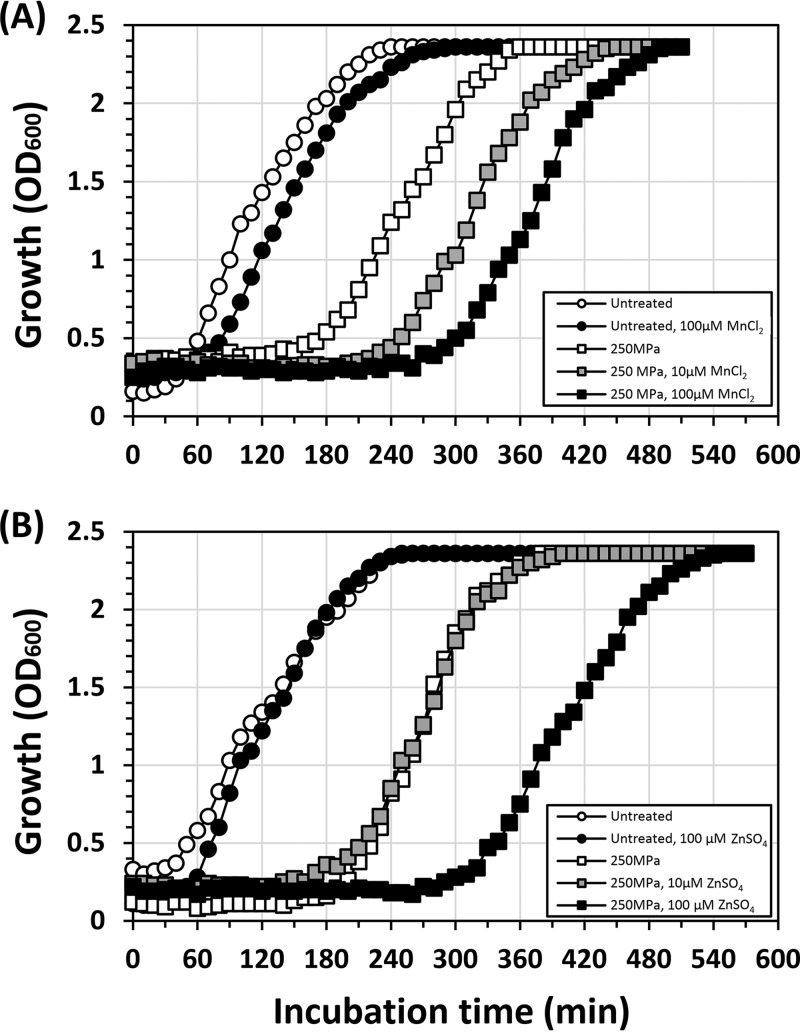
Our transcriptional analyses indicated that both the Fur and MntR regulons were upregulated in the regrowth phase (Fig. 1). Since metal ions are required for a large number of metalloproteins, we hypothesized that HHP-injured cells might require several metal ions for their growth. Accordingly, we examined the effects of several metal ions on the growth recovery of HHP-injured cells. Because ribosomes contain Mg2+ and Zn2+, we tested not only FeSO4 and MnCl2 but also MgSO4 and ZnSO4. Contrary to our hypothesis, MnCl2 and ZnSO4 had inhibitory effects on the growth recovery of HHP-treated cells (Fig. 3), while FeSO4 (100 μM) and MgSO4 (10 mM) had no significant effect (data not shown). The addition of MnCl2 to the recovery medium prolonged the growth arrest phase (Fig. 3A). The average growth delay in Mn2+-supplemented medium compared to that in nonsupplemented medium (160 ± 21 min) was extended to 210 ± 5.8 min at 10 μM and 260 ± 5.8 min at 100 μM, respectively. ZnSO4 also inhibited growth recovery at a concentration of 100 μM but not at 10 μM (Fig. 3B). The average growth delay in the presence of 100 μM ZnSO4 was 250 ± 29 min. Since neither MnCl2 nor ZnSO4 had any significant effect on the cell viability as determined by colony formation (data not shown), it seems that these metal ions inhibited the recovery process. When E. coli W3110 cells were treated under the same condition (250 MPa at 25°C for 10 min) and diluted in fresh L medium supplemented with MnCl2 (100 μM) or ZnSO4 (100 μM), no significant effects of these metal ions on the growth recovery were observed compared to growth in nonsupplemented L medium (data not shown).
FIG 3.
Effects of Mn2+ and Zn2+ on the growth recovery of HHP-treated B. subtilis cells. B. subtilis strain TI465 was treated with 250 MPa of HHP at 25°C for 10 min and diluted 10-fold in a fresh NaCl-free L medium supplemented with MnCl2 (A) or ZnSO4 (B). Cell growth was monitored by measuring the OD600.
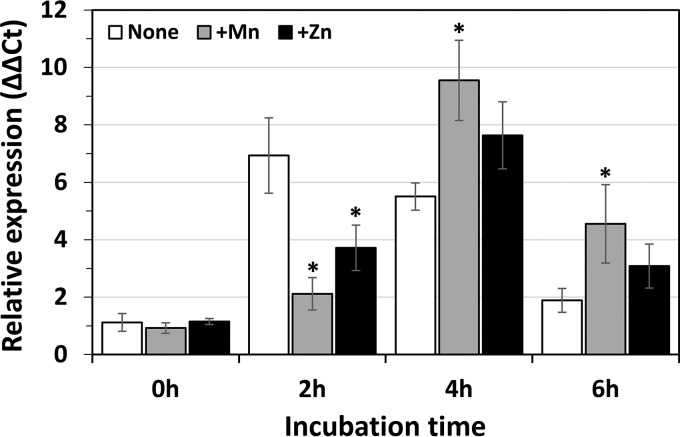
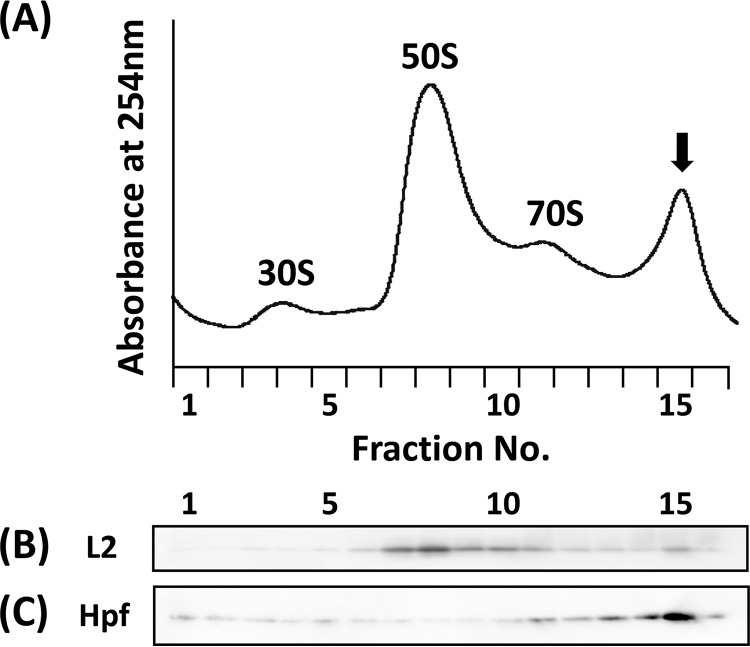
To examine the effects of MnCl2 and ZnSO4 on the transcription of ribosomal genes during the recovery phase, we monitored the transcript level of the rpsJ gene by performing RT-qPCR. The results showed a significant delay in the transcript level of rpsJ when HHP-treated cells were incubated in the recovery medium supplemented with MnCl2 or ZnSO4 (Fig. 4). Sucrose density gradient sedimentation analysis also indicated that ribosome reconstruction was delayed under these conditions (Fig. 5). In the presence of MnCl2 or ZnSO4, the 50S peak was still high even after incubation for 4 h, suggesting that both Mn2+ and Zn2+ ions might prevent the degradation of dissociated 50S particle, either directly or indirectly. Interestingly, an unusual accumulation of ribosome multimers (dimers or polysomes) was found after incubation for 2 h in the presence of Zn2+, but not with Mn2+ (Fig. 5K and F). Western blot analysis using antibodies against ribosomal protein L2 and ribosome dimerization factor Hpf revealed that both were detected in this peak (Fig. 6B and C). No such accumulation of Hpf was observed in cells incubated in the Mn2+-supplemented recovery medium (Fig. S3). These results suggest that Zn2+ can stimulate dimer formation of ribosomes in HHP-treated cells. In contrast, it seems that Mn2+ prevented ribosome reconstruction without stimulating ribosome dimerization (Fig. 5).
FIG 4.
Effects of Mn and Zn on transcript level of rpsJ following HHP treatment. B. subtilis strain TI465 was subjected to HHP treatment at 250 MPa at 25°C for 10 min. After HHP treatment, the culture was diluted 5-fold in fresh NaCl-free L medium (white) or the same medium supplemented with 0.1 mM MnCl2 (gray) or 0.1 mM ZnSO4 (black). After the incubation for 0 h (immediately after dilution), 2 h, 4 h, or 6 h, cells were collected by centrifugation. Then total RNAs were extracted and used for RT-qPCR analysis. Total RNA in untreated culture immediately before HHP treatment was also prepared. The transcript levels of rpsJ were normalized by adjusting the amount of 16S rRNA in each RNA sample. The relative expression of rpsJ was calculated by dividing the amount of rpsJ mRNA in HHP-treated cells at each time point by that in cells immediately before HHP treatment (ΔΔCt). The average values of three independent experiments are shown. Statistical analysis was performed by using Dunnett’s test. Asterisks indicate significant differences compared with the nonsupplemented control at each time point (P < 0.05).
FIG 5.
Effects of Mn2+ and Zn2+ on ribosome reconstruction in HHP-treated B. subtilis cells. B. subtilis strain TI465 was subjected to 250 MPa at 25°C for 10 min. After the HHP treatment, the culture was diluted 5-fold in fresh NaCl-free L medium (C to E) or the same medium supplemented with 0.1 mM MnCl2 (F to J) or 0.1 mM ZnSO4 (K to O). After each indicated time, the cells were collected by centrifugation. Cell extracts were then analyzed using 10% to 40% sucrose gradient sedimentation. The cell extract just before the HHP treatment is also shown (A). The cell extract at 0 h (B) was prepared from the HHP-treated culture before dilution with the fresh medium.
FIG 6.
Western blot analysis of a Zn2+-dependent unusual peak. A B. subtilis strain TI465 culture was treated with 250 MPa at 25°C for 10 min. After the HHP treatment, the culture was diluted 5-fold in fresh NaCl-free L medium supplemented with 0.1 mM ZnSO4 and incubated for 2 h at 37°C with vigorous shaking. Cell extract was then fractionated using a sucrose density gradient (A). Each fractionated sample was analyzed by Western blotting using anti-L2 (B) or anti-Hpf (C) rabbit antisera. The arrow indicates the peak that appeared in the presence of ZnSO4.
Hpf is involved in Zn2+-induced dimerization of ribosomes in HHP-treated cells.
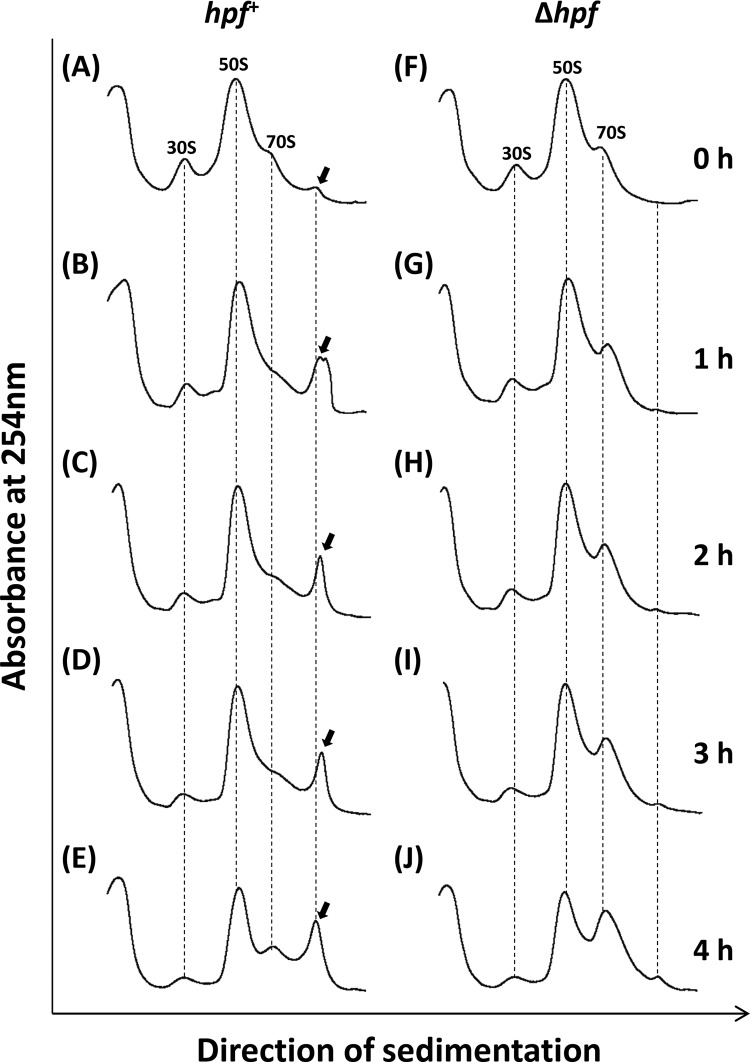
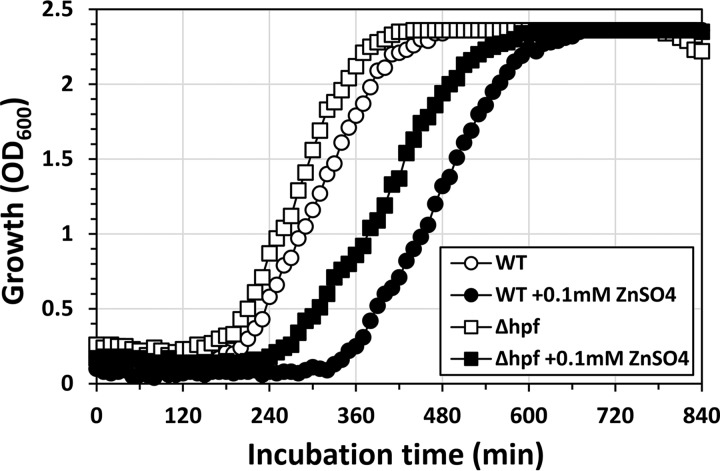
We analyzed whether Zn2+-induced dimerization of ribosomes can occur even in a Δhpf mutant (Fig. 7). In the hpf+ strain, ribosomes formed the dimers within 1 h after the HHP-treated cells were transferred into a Zn2+-supplemented medium (Fig. 7B). In contrast, the ribosome dimers were no longer observed in the Δhpf mutant, indicating that Hpf was required for the Zn2+-induced dimerization in the HHP-treated cells (Fig. 7F to J). In addition, the accumulation of 70S in the Δhpf mutant was observed after 4 h of incubation, whereas no accumulation was observed with the hpf+ strain (Fig. 7E and J). Moreover, the Δhpf mutant showed an approximately 70-min shorter growth delay than that of the hpf+ strain (Fig. 8). These results suggest that Zn2+ can prevent the growth recovery of HHP-injured cells by stimulating Hpf-dependent dimerization. In this study, we used stationary-phase cells that contain Hpf protein (22). Since the addition of ZnSO4 to the recovery medium had no effect on the hpf expression (data not shown), preexisting Hpf protein can be involved in Zn2+-induced dimer formation. We could not observe the Zn2+-induced dimer formation in untreated cells (data not shown). Conformational changes caused by HHP might be required for Zn2+-induced dimer formation. B. subtilis has several ribosomal proteins with the Zn-binding motif CXXC (45, 46). These Zn-binding motifs might be involved in the Zn2+-induced dimer formation. It should be noted that the inhibitory effect of Zn2+ ions on the growth was still observed even in the Δhpf mutant (Fig. 8). This indicates that Zn2+ also has a different inhibitory effect on the growth recovery of HHP-injured B. subtilis cells. Zn-sensing transcriptional regulator Zur represses at least three Zn-containing ribosomal protein paralogs (L31 paralog YtiA, L33 paralog RpmGC, and S14 paralog YhzA) under Zn-rich conditions (45–48). Moreover, an elevated level of Mn2+ results in repression of not only the MntR regulon but also the Fur and PerR regulons (47, 48). We therefore cannot exclude the possibility that Mn2+ and Zn2+ ions affect the growth recovery of HHP-injured B. subtilis cells through the function of these metal-sensing regulators.
FIG 7.
Effect of Hpf on Zn2+-dependent dimerization of ribosomes in HHP-treated B. subtilis cells. B. subtilis strain TI477 (hpf+ [A to E]) and TI478 (Δhpf [F to J]) cultures were treated with 250 MPa at 25°C for 10 min. After the HHP treatment, each culture was diluted 5-fold in fresh NaCl-free L medium supplemented with 0.1 mM ZnSO4. After incubation for 0 h (immediately after dilution), 1 h, 2 h, 3 h, and 4 h, the cells were collected by centrifugation. Cell extracts were then analyzed using 10% to 40% sucrose gradient sedimentation. The arrows indicate peaks corresponding to ribosome dimers.
FIG 8.
Effects of Hpf and Zn supplementation on the growth of HHP-treated B. subtilis. B. subtilis strain TI477 (wild type [WT]; hpf+) and TI478 (Δhpf) cultures were treated with 250 MPa for 10 min at 25°C. After the HHP treatment, the cultures were diluted 10-fold in fresh NaCl-free L medium (open symbols) or the same medium supplemented with 0.1 mM ZnSO4 (closed symbols). Cell growth was monitored by measuring the OD600.
Concluding remarks.
In this study, we investigated the ribosome profile during the recovery of HHP-injured B. subtilis cells. When B. subtilis cells were treated with HHP at 250 MPa or higher, intact ribosomes (70S, dimer, and polysome) were largely dissociated or degraded (Fig. 2A). It is therefore considered that >250-MPa-treated cells cannot grow until active ribosomes are reconstructed.
We found that the addition of Mn2+ or Zn2+ to the recovery medium apparently delayed ribosome reconstruction, preventing the growth recovery of HHP-injured cells (Fig. 3 and 5). In the recovery medium supplemented with Mn2+ or Zn2+, the dissociated 50S particle was accumulated for a longer time than in nonsupplemented medium (Fig. 5). Moreover, Zn2+, but not Mn2+, stimulated Hpf-dependent dimer formation of ribosomes (Fig. 6 and 7). Since the dimerized ribosome is considered more resistant to cleavage by ribonucleases than free ribosomal particles 30S and 50S, Zn2+ can prevent ribosome breakdown and the subsequent release of nutrients such as nucleotides and amino acids, thereby delaying ribosome reconstruction. In fact, the inhibitory effect of Zn2+ on the growth recovery of HHP-treated cells was partially relieved by disruption of the hpf gene (Fig. 8). In contrast, it seems that Mn2+ prevented ribosome reconstruction without stimulating ribosome dimerization (Fig. 5). Although the underlying mechanism is still unclear, we propose that the degradation and recycling of dissociated ribosomes are important for ribosome reconstruction in the early stage of the recovery process in HHP-injured B. subtilis cells.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All B. subtilis strains used in this study were derived from strain 168. Strain TI465 (trpC2 spoIIAC::kan) was used because it lacks the sporulation-specific σ factor F (2). The strain RIK2508 (trpC2 Δhpf) was previously constructed from strain 168 (22). Strains TI478 (trpC2 spoIIAC::kan Δhpf) and TI477 (trpC2 spoIIAC::kan) were obtained by transforming strains RIK2508 and its parent strain with genomic DNA of TI465. To confirm the effect of MnCl2 or ZnSO4 in HHP-treated E. coli cells, strain W3110 was purchased from the National Bio-Resource Project (NBRP) of the National Institute of Genetics (strain ID; ME9780) and used.
The growth media used were L medium (10 g of tryptone, 5 g of yeast extract, and 5 g of NaCl, all per liter) and NaCl-free L medium (10 g of tryptone and 5 g of yeast extract, both per liter). Kanamycin (3 μg/ml) was used to select the transformants described above.
HHP treatment.
Cells were grown in NaCl-free L medium for 6 h at 37°C with vigorous shaking. For sucrose density gradient sedimentation analysis, the culture was concentrated by centrifugation followed by removal of approximately 95% of the supernatant. Then the culture was packed into a sterile plastic bag that was heat sealed following manual exclusion of visible air bubbles and then subjected to HHP treatment using an HHP food processor (Dr. CHEF; Kobe Steel, Kobe, Japan) at 25°C for 10 min as previously described (2). The rate of increasing and decreasing pressure was set at 200 MPa/min.
Growth delay assay.
Cells were subjected to HHP treatment at an appropriate pressure as described above. Following HHP treatment, 2 ml of the treated culture was inoculated into 18 ml of the recovery medium (NaCl-free L medium) and incubated at 37°C with vigorous shaking (200 rpm). To investigate the effect of metal ions, the recovery medium was supplemented with MnCl2 or ZnSO4 at concentrations of 10 or 100 μM. Cell growth was monitored by measuring OD600 using an OD-Monitor A&S (TAITEC, Japan). The measurable range of this system was 0.01 to 2.55. The growth delay time at an OD600 of 1.0 was expressed as the time difference between test culture and control culture. Under this experimental condition, a 10-fold dilution of untreated culture resulted in a growth delay of 70 ± 13 min (data not shown). At least three independent experiments were performed to assess the reproducibility.
DNA microarray analysis.
Cells were subjected to HHP treatment at 250 MPa as described above. Following HHP treatment, the culture was diluted 5-fold in fresh recovery medium and incubated at 37°C with vigorous shaking. After incubation for 0 h (immediately after dilution), 2 h, or 4 h, cells were collected by centrifugation (5,000 × g, 5 min, and 25°C). Then cells were treated with killing buffer (20 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 20 mM NaN3), collected (again using centrifugation), and stored at −80°C until RNA extraction was performed. Total cellular RNA was obtained using Isogen reagent (Nippon Gene, Japan) as previously described (49). cDNA synthesis, labeling, hybridization to the Affymetrix Bacillus subtilis genome array GeneChip, and scanning were carried out by TaKaRa Bio (Japan). All microarray data were MIAME (Minimum Information About a Microarray Experiment) compliant.
An outline of the DNA microarray data analysis was composed of the three steps as follows: data quantification, overall gene expression profile evaluation, and identification of differentially expressed genes (DEGs). The raw microarray data (CEL files) were quantified and processed using R (https://www.r-project.org/) (50), which is a statistical programming language, with relevant R packages provided by Bioconductor (http://www.bioconductor.org/) (51). Quantification was implemented using the distribution-free weighted method (DFW) (52). Global gene expression profiles of all samples were evaluated by principal-component analysis (PCA) (53) using the “prcomp()” function in R. To identify DEGs, the rank products method (20) was applied to the three pairs of groups (0 versus 2 h, 0 versus 4 h, and 2 versus 4 h) selected from the DFW-quantified data, with the number of permutations set at 1,000. Probe sets with a false-discovery rate (FDR) of <0.05 were regarded as having different transcript levels between the two groups (i.e., they were differently expressed). In order to show the changes in expression values intuitively, the expression ratios of the DEGs between the selected pairs of groups were calculated using conventional Mas5-quantified microarray data. The annotation file for the B. subtilis genome array was downloaded from the NetAffx website (http://www.affymetrix.com/analysis/index.affx; as of 23 October 2015). Microarray analysis was performed two times independently for each sample, and the expression ratio was indicated with average values in the log2 ratio between duplicated independent experiments.
RT-qPCR.
For reverse transcription-quantitative PCR (RT-qPCR), cells were subjected to HHP treatment at 250 MPa as described above. Following HHP treatment, the culture was diluted 5-fold in fresh recovery medium and incubated at 37°C with vigorous shaking. After incubation for the desired time, cells were collected by centrifugation (5,000 × g, 5 min, and 25°C). Then total cellular RNA was prepared as previously described (49). First-strand cDNA was synthesized using PrimeScript RT reagent kit with genomic DNA (gDNA) eraser (TaKaRa Bio, Japan) with random-hexamer primers. Real-time PCR was performed using Thunder Bird SYBR qPCR mix (Toyobo, Japan) with an ABI 7300 real-time PCR system (Applied Biosystems, USA). The oligonucleotides used for qPCR are listed in Table 3. 16S rRNA was used for normalization of RNA levels.
TABLE 3.
Primers used in this study for RT-qPCR
| Forward primer | Sequence (5′→3′) | Reverse primer | Sequence (5′→3′) |
|---|---|---|---|
| abhF | CCGATTGAGTTGAGACGGG | abhR | CCCTCAGGGCTTAGCGTAA |
| bfmBAAF | TCCGGGTTTGCAAAAGCA | bfmBAAR | TCTCCATACGTCCCGCAA |
| csbDF | ACAAAATGAAGGGCGGCT | csbDR | TGGATTTCGCCTTTTGCC |
| groESF | GCCATTAGGTGATCGCGT | groESR | GAACCAGCTGCCACGATT |
| gsiBF | TGAGCAGAGAAGAAGCAGG | gsiBR | TCGTTGTTGCGGGCGTTT |
| infCF | TCGGACAAAATGGCGACC | infCR | CAGGCGGTTTTGCATTCG |
| mrgAF | ATCACAGACGGCGGAAAC | mrgAR | TCCGACAAACAAGTCCGC |
| mtlDF | GAACCGAATGCACTGGCT | mtlDR | GAATGCCGCACACCTCTT |
| pdhAF | CCGCTTGCTGTATACGCA | pdhAR | AGGATCGTCACCAGCCAT |
| pstAF | CTGACAACACCGCGTCTT | pstAR | AGAAATCACCAGCACGGC |
| tigF | CAAAGGCCTGGGCATTGA | tigR | CAGCTTTTCCGCCTTCGA |
| tuaGF | ACAGCTTAATGGCGCAGG | tuaGR | TCATCACCGTCAAGCAGC |
| ydbNF | AAACATGTCAGGAGGGCG | ydbNR | TGCGGTCGATCCTGTCAT |
| yfiYF | TGCTGAATCTTTGGCCGG | yfiYR | TTGTTTGTCTGGTCGCCC |
| yflTF | GCAGAAAATGGGTGTTGCG | yflTR | TTGGCTCCGATCGTGTTG |
| ykuNF | TGTCATGCTGCAAGAGGC | ykuNR | AACCTCTCGCAAACGCTC |
| ykzIF | TGCTGTCTGGAGACTAGAGG | ykzIR | TCCATTCTTTTCTAAGTCGC |
| yqgZF | TCCAAGCTGCACGTCATG | yqgZR | AGTCTGGCTTCTTGTCGC |
| ysbAF | TCGCTTGGTGTGATGCAG | ysbAR | CGCTTCGGTTTTGCGTTT |
| yugIF | CAAGCTGCGCCTGAGAAA | yugIR | GGTCTTTGCGGTTGGACA |
| ywsBF | ACTTGCGGCTGGAATGAC | ywsBR | ATGCGGAAGGCTTCGTTC |
| acpPF | AGATCGCCTTGGCGTTGA | acpPR | TGTAGTTCACAGCGTCGC |
| citBF | GCGCGAAACTTGTCGGCAA | citBR | TGCAAGCGGCAGTTCAGCA |
| clpEF | ACGAACTGGCGGCAAAAC | clpER | TGTTCGTCTGCCTGCAGT |
| cspBF | TCTCTGCTATTCAAGGCG | cspBR | ACGTTAGCAGCTTGTGGTC |
| cspDF | TGAAGTTGAAGGCGGAGAC | cspDR | AGAAGCTTGAGGTCCACGA |
| feuAF | TCTCCGGCAAATTCCCTGA | feuAR | GTGCCTGCTGTGCTGATT |
| hagF | ATGGGTGCTGACGCTCTT | hagR | ACCGCACCAAGCTTAGCA |
| hbsF | TGCGTGAACGTTCTGCAC | hbsR | TTTTCCGGCAACTGCGTC |
| infAF | CACTGCCAAACGCGATGT | infAR | ACGTAATCCTGCCACGAGT |
| rplAF | ACGACGTCTCTGAAGCAGT | rplAR | CGAAAACGAGAACGCGCT |
| rplJF | AACGCGATCGCATTCAGC | rplJR | AGCCTTCGCGTGATGGAA |
| rplSF | GTTCCGTCCTGGTGACACT | rplSR | TCGCTGATTCCACCACCA |
| yqgAF | GGACATTGGCCCGCAATA | yqgAR | ACCTACTGCCCAAGGTGT |
| ahpCF | ATCGATCCAGACGGCGTT | ahpCR | TCCCATTTAGCCGGGCAA |
| appCF | TCTGCTGTTTTCGCCCCT | appCR | AAAGAGACACGCGCTCCA |
| citZF | ACCATGAGCTGAACGCGT | citZR | ACGGCTCAGCGTTTTCCA |
| gapBF | AAAACAGCTGCCTTGGCG | gapBR | TTCCCGGAGCGGTCAAAA |
| katAF | CGCGCACGCAACAAAGTA | katAR | TTTTGGACCGCCGAAGCT |
| mntAF | GCCGGAAAAAGCGCTGAT | mntAR | TGCAACCCGCTGTACAGA |
| pckAF | AGAAGCGTTTGAGCGGCT | pckAR | CAGCTGCCGCGCAAATAA |
| rapAF | AGGATTGGAAATCGCCCGT | rapAR | TCATGCGCCAGCTCTTCA |
| rbsAF | AACGCACGCACGCCATTT | rbsAR | CGTTTGGCGCCAACATCCA |
| srfAAF | TCATTGGCACAGCAGCGA | srfAAR | TGTCTCGGCGTGCTGTTT |
| yhzCF | GTCCCGCAACACGCAAAA | yhzCR | TGGCAAGCCGACGGTTAT |
| ytxHF | TCATCGGTGCCACTACAGC | ytxHR | TTCCTTCGCATCTGCCGT |
| yvyDF | CGATGACAGATCTGGCGCT | yvyDR | AGCCAAGACCGTTTGCCA |
| yydFF | CACTAACAATGAGACTGTG | yydFR | ACTTCCAAGAATCCAGCGA |
| rpsJF | GTATCTGGTCCGATTCCG | rpsJR | GTGGTGTTGGGTTCACAAT |
Sucrose density gradient centrifugation.
Cells (>100 AU [milliliter of culture × OD600]) were collected by centrifugation (5,000 × g, 25°C, and 10 min) and washed with 20 mM Tris-HCl buffer solution (pH 7.0). Sucrose density gradient centrifugation analysis was performed as previously described (22). At least two independent experiments were performed to assess the reproducibility.
Accession number(s).
All microarray data have been deposited in a MIAME (Minimum Information About a Microarray Experiment)-compliant database, the National Center for Biotechnology Information Gene Expression Omnibus (GEO Series accession number GSE126203), as detailed on the FGED Society website (http://www.fged.org/projects/miame/).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Ministry of Agriculture, Forestry and Fishery of Japan (ID-13406381), JICA-Kirin fellowship program 2017-2018 (to H.T.M.N.) and 2018-2019 (to T.T.M.H.), and the Japan Society for the Promotion of Science (JSPS) Grant-in-Aid for Young Scientists (B) (17K15253 to G.A.).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01640-19.
REFERENCES
- 1.Yamamoto K. 2017. Food processing by high hydrostatic pressure. Biosci Biotechnol Biochem 81:672–679. doi: 10.1080/09168451.2017.1281723. [DOI] [PubMed] [Google Scholar]
- 2.Inaoka T, Kimura K, Morimatsu K, Yamamoto K. 2017. Characterization of high hydrostatic pressure-injured Bacillus subtilis cells. Biosci Biotechnol Biochem 81:1235–1240. doi: 10.1080/09168451.2017.1292835. [DOI] [PubMed] [Google Scholar]
- 3.Kimura K, Morimatsu K, Inaoka T, Yamamoto K. 2017. Injury and recovery of Escherichia coli ATCC25922 cells treated by high hydrostatic pressure at 400-600 MPa. J Biosci Bioeng 123:698–706. doi: 10.1016/j.jbiosc.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 4.Koseki S, Yamamoto K. 2006. Recovery of Escherichia coli ATCC 25922 in phosphate buffered saline after treatment with high hydrostatic pressure. Int J Food Microbiol 110:108–111. doi: 10.1016/j.ijfoodmicro.2006.01.039. [DOI] [PubMed] [Google Scholar]
- 5.Meganathan R, Marquis RE. 1973. Loss of bacterial motility under pressure. Nature 246:525–527. doi: 10.1038/246525a0. [DOI] [PubMed] [Google Scholar]
- 6.Ishii A, Sato T, Wachi M, Nagai K, Kato C. 2004. Effect of high hydrostatic pressure on bacterial cytoskeleton FtsZ polymers in vivo and in vitro. Microbiology 150:1965–1972. doi: 10.1099/mic.0.26962-0. [DOI] [PubMed] [Google Scholar]
- 7.Yayanos AA, Pollard BC. 1969. A study of the effects of hydrostatic pressure on macromolecular synthesis in Escherichia coli. Biophys J 9:1464–1482. doi: 10.1016/S0006-3495(69)86466-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zobell CE, Cobet AB. 1964. Filament formation by Escherichia coli at increased hydrostatic pressures. J Bacteriol 87:710–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niven GW, Miles CA, Mackey BM. 1999. The effects of hydrostatic pressure on ribosome conformation in Escherichia coli: an in vivo study using differential scanning calorimetry. Microbiology 145:419–425. doi: 10.1099/13500872-145-2-419. [DOI] [PubMed] [Google Scholar]
- 10.Pavlovic M, Hörmann S, Vogel RF, Ehrmann MA. 2005. Transcriptional response reveals translation machinery as target for high pressure in Lactobacillus sanfranciscensis. Arch Microbiol 184:11–17. doi: 10.1007/s00203-005-0021-4. [DOI] [PubMed] [Google Scholar]
- 11.Welch TJ, Farewell A, Neidhardt FC, Bartlett DH. 1993. Stress response of Escherichia coli to elevated hydrostatic pressure. J Bacteriol 175:7170–7177. doi: 10.1128/jb.175.22.7170-7177.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishii A, Oshima T, Sato T, Nakasone K, Mori H, Kato C. 2005. Analysis of hydrostatic pressure effects on transcription in Escherichia coli DNA microarray procedure. Extremophiles 9:65–73. doi: 10.1007/s00792-004-0414-3. [DOI] [PubMed] [Google Scholar]
- 13.Aertsen A, Vanoirbeek K, De Spiegeleer P, Sermon J, Hauben K, Farewell A, Nyström T, Michiels CW. 2004. Heat shock protein-mediated resistance to high hydrostatic pressure in Escherichia coli. Appl Environ Microbiol 70:2660–2666. doi: 10.1128/aem.70.5.2660-2666.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noma S, Hayakawa I. 2003. Barotolerance of Staphylococcus aureus is increased by incubation at below 0°C prior to hydrostatic pressure treatment. Int J Food Microbiol 80:261–264. doi: 10.1016/s0168-1605(02)00142-3. [DOI] [PubMed] [Google Scholar]
- 15.Wemekamp-Kamphuis HH, Karatzas AK, Wouters JA, Abee T. 2002. Enhanced levels of cold shock proteins in Listeria monocytogenes LO28 upon exposure to low temperature and high hydrostatic pressure. Appl Environ Microbiol 68:456–463. doi: 10.1128/aem.68.2.456-463.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.VanBogelen RA, Neidhardt FC. 1990. Ribosomes as sensors of heat and cold shock in Escherichia coli. Proc Natl Acad Sci U S A 87:5589–5593. doi: 10.1073/pnas.87.15.5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aertsen A, De Spiegeleer P, Vanoirbeek K, Lavilla M, Michiels CW. 2005. Induction of oxidative stress by high hydrostatic pressure in Escherichia coli. Appl Environ Microbiol 71:2226–2231. doi: 10.1128/AEM.71.5.2226-2231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gould GW, Sale AJ. 1970. Initiation of germination of bacterial spores by hydrostatic pressure. J Gen Microbiol 60:335–346. doi: 10.1099/00221287-60-3-335. [DOI] [PubMed] [Google Scholar]
- 19.Paidhungat M, Setlow B, Daniels WB, Hoover D, Papafragkou E, Setlow P. 2002. Mechanisms of induction of germination of Bacillus subtilis spores by high pressure. Appl Environ Microbiol 68:3172–3175. doi: 10.1128/aem.68.6.3172-3175.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Breitling R, Armengaud P, Amtmann A, Herzyk P. 2004. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett 573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 21.Li X, Lindahl L, Sha Y, Zengel JM. 1997. Analysis of the Bacillus subtilis S10 ribosomal protein gene cluster identifies two promoters that may be responsible for transcription of the entire 15-kilobase S10-spc-α cluster. J Bacteriol 179:7046–7054. doi: 10.1128/jb.179.22.7046-7054.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akanuma G, Kazo Y, Tagami K, Hiraoka H, Yano K, Suzuki S, Hanai R, Nanamiya H, Kato-Yamada Y, Kawamura F. 2016. Ribosome dimerization is essential for the efficient regrowth of Bacillus subtilis. Microbiology 162:448–458. doi: 10.1099/mic.0.000234. [DOI] [PubMed] [Google Scholar]
- 23.Graumann P, Wendrich TM, Weber MHW, Schröder K, Marahiel MA. 1997. A family of cold shock proteins in Bacillus subtilis is essential for cellular growth and for efficient protein synthesis at optimal and low temperatures. Mol Microbiol 25:741–756. doi: 10.1046/j.1365-2958.1997.5121878.x. [DOI] [PubMed] [Google Scholar]
- 24.Teter SA, Houry WA, Ang D, Tradler T, Rockabrand D, Fischer G, Blum P, Georgopoulos C, Hartl FU. 1999. Polypeptide flux through bacterial Hsp70: DnaK cooperates with trigger factor in chaperoning nascent chains. Cell 97:755–765. doi: 10.1016/S0092-8674(00)80787-4. [DOI] [PubMed] [Google Scholar]
- 25.Li M, Wong SL. 1992. Cloning and characterization of the groESL operon from Bacillus subtilis. J Bacteriol 174:3981–3992. doi: 10.1128/jb.174.12.3981-3992.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Derre I, Rapoport G, Devine K, Rose M, Msadek T. 1999. ClpE, a novel type of HSP100 ATPase, is part of the CtsR heat shock regulon of Bacillus subtilis. Mol Microbiol 32:581–593. doi: 10.1046/j.1365-2958.1999.01374.x. [DOI] [PubMed] [Google Scholar]
- 27.Miethke M, Hecker M, Gerth U. 2006. Involvement of Bacillus subtilis ClpE in CtsR degradation and protein quality control. J Bacteriol 188:4610–4619. doi: 10.1128/JB.00287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Antelmann H, Engelmann S, Schmid R, Hecker M. 1996. General and oxidative stress response in Bacillus subtilis: cloning, expression, and mutation of the alkyl hydroperoxide reductase operon. J Bacteriol 178:6571–6578. doi: 10.1128/jb.178.22.6571-6578.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen L, Keramati L, Helmann JD. 1995. Coordinate regulation of Bacillus subtilis peroxide stress genes by hydrogen peroxide and metal ions. Proc Natl Acad Sci U S A 92:8190–8194. doi: 10.1073/pnas.92.18.8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen L, Helmann JD. 1995. Bacillus subtilis MrgA is a Dps (PexB) homologue: evidence for metalloregulation of an oxidative-stress gene. Mol Microbiol 18:295–300. doi: 10.1111/j.1365-2958.1995.mmi_18020295.x. [DOI] [PubMed] [Google Scholar]
- 31.Hecker M, Völker U. 1998. Non-specific, general and multiple stress resistance of growth-restricted Bacillus subtilis cells by the expression of the σB regulon. Mol Microbiol 29:1129–1136. doi: 10.1046/j.1365-2958.1998.00977.x. [DOI] [PubMed] [Google Scholar]
- 32.Sonenshein AL. 2002. The Krebs citric acid cycle, p 151–162. In Sonenshein AL, Hoch JA, Losick R (ed), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, DC. [Google Scholar]
- 33.Gayán E, Rutten N, Van Impe J, Michiels CW, Aertsen A. 2019. Identification of novel genes involved in high hydrostatic pressure resistance of Escherichia coli. Food Microbiol 78:171–178. doi: 10.1016/j.fm.2018.10.007. [DOI] [PubMed] [Google Scholar]
- 34.Prieto-Calvo M, Prieto M, López M, Alvarez-Ordóñez A. 2014. Effect of high hydrostatic pressure on Escherichia coli ultrastructure, membrane integrity and molecular composition as assessed by FTIR spectroscopy and microscopic imaging techniques. Molecules 19:21310–21323. doi: 10.3390/molecules191221310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perez C, Marini P, de Mendoza D. 1998. Effects on Bacillus subtilis of conditional expression of the accBC operon encoding subunits of acetyl coenzyme A carboxylase, the first enzyme of fatty acid synthesis. Microbiol 144:895–903. doi: 10.1099/00221287-144-4-895. [DOI] [PubMed] [Google Scholar]
- 36.Martinez MA, de Mendoza D, Schujman GE. 2010. Transcriptional and functional characterization of the gene encoding acyl carrier protein in Bacillus subtilis. Microbiology 156:484–495. doi: 10.1099/mic.0.033316-0. [DOI] [PubMed] [Google Scholar]
- 37.Soldo B, Lazarevic V, Pagni M, Karamata D. 1999. Teichuronic acid operon of Bacillus subtilis 168. Mol Microbiol 31:795–805. doi: 10.1046/j.1365-2958.1999.01218.x. [DOI] [PubMed] [Google Scholar]
- 38.Qi Y, Kobayashi Y, Hulett FM. 1997. The pst operon of Bacillus subtilis has a phosphate-regulated promoter and is involved in phosphate transport but not in regulation of the pho regulon. J Bacteriol 179:2534–2539. doi: 10.1128/jb.179.8.2534-2539.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tännler S, Fischer E, Coq DL, Doan T, Jamet E, Sauer U, Aymerich S. 2008. CcpN controls central carbon fluxes in Bacillus subtilis. J Bacteriol 190:6178–6187. doi: 10.1128/JB.00552-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pi H, Helmann JD. 2017. Sequential induction of Fur-regulated genes in response to iron limitation in Bacillus subtilis. Proc Natl Acad Sci U S A 114:12785–12790. doi: 10.1073/pnas.1713008114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Que Q, Helmann JD. 2002. Manganese homeostasis in Bacillus subtilis is regulated by MntR, a functional regulator related to the diphtheria toxin repressor family of proteins. Mol Microbiol 35:1454–1468. doi: 10.1046/j.1365-2958.2000.01811.x. [DOI] [PubMed] [Google Scholar]
- 42.Zundel MA, Basturea GN, Deutscher MP. 2009. Initiation of ribosome degradation during starvation in Escherichia coli. RNA 15:977–983. doi: 10.1261/rna.1381309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaplan R, Apirion D. 1975. The fate of ribosomes in Escherichia coli cells starved for a carbon source. J Biol Chem 250:1854–1863. [PubMed] [Google Scholar]
- 44.Davis BD, Luger SM, Tai PC. 1986. Role of ribosome degradation in the death of starved Escherichia coli cells. J Bacteriol 166:439–445. doi: 10.1128/jb.166.2.439-445.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Akanuma G, Nanamiya H, Natori Y, Nomura N, Kawamura F. 2006. Liberation of Zn-containing L31(RpmE) from ribosomes by its paralogous gene product, YtiA, in Bacillus subtilis. J Bacteriol 188:2715–2720. doi: 10.1128/JB.188.7.2715-2720.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nanamiya H, Kawamura F, Kosono S. 2006. Proteomic study of the Bacillus subtilis ribosome: finding of zinc-dependent replacement for ribosomal protein L31 paralogues. J Gen Appl Microbiol 52:249–258. doi: 10.2323/jgam.52.249. [DOI] [PubMed] [Google Scholar]
- 47.Moore CM, Helmann JD. 2005. Metal ion homeostasis in Bacillus subtilis. Curr Opin Microbiol 8:188–195. doi: 10.1016/j.mib.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 48.Helmann JD. 2014. Specificity of metal sensing: iron and manganese homeostasis in Bacillus subtilis. J Biol Chem 289:28112–28120. doi: 10.1074/jbc.R114.587071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Inaoka T, Satomura T, Fujita Y, Ochi K. 2009. Novel gene regulation mediated by overproduction of secondary metabolite neotrehalosadiamine in Bacillus subtilis. FEMS Microbiol Lett 291:151–156. doi: 10.1111/j.1574-6968.2008.01450.x. [DOI] [PubMed] [Google Scholar]
- 50.R Development Core Team. 2008. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 51.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JYH, Zhang J. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Z, McGee M, Liu Q, Scheuermann RH. 2007. A distribution free summarization method for Affymetrix GeneChip arrays. Bioinformatics 23:321–327. doi: 10.1093/bioinformatics/btl609. [DOI] [PubMed] [Google Scholar]
- 53.Kachigan SK. 1986. Statistical analysis: an interdisciplinary introduction to univariate and multivariate methods. Radius Press, New York, NY. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.